Abstract
Oleaginous microorganisms have considerable potential for biofuel and commodity chemical production. Under nitrogen-limitation, Rhodococcus jostii RHA1 grown on benzoate, an analog of lignin depolymerization products, accumulated triacylglycerols (TAGs) to 55% of its dry weight during transition to stationary phase, with the predominant fatty acids being C16:0 and C17:0. Transcriptomic analyses of RHA1 grown under conditions of N-limitation and N-excess revealed 1,826 dysregulated genes. Genes whose transcripts were more abundant under N-limitation included those involved in ammonium assimilation, benzoate catabolism, fatty acid biosynthesis and the methylmalonyl-CoA pathway. Of the 16 atf genes potentially encoding diacylglycerol O-acyltransferases, atf8 transcripts were the most abundant during N-limitation (~50-fold more abundant than during N-excess). Consistent with Atf8 being a physiological determinant of TAG accumulation, a Δatf8 mutant accumulated 70% less TAG than wild-type RHA1 while atf8 overexpression increased TAG accumulation 20%. Genes encoding type-2 phosphatidic acid phosphatases were not significantly expressed. By contrast, three genes potentially encoding phosphatases of the haloacid dehalogenase superfamily and that cluster with, or are fused with other Kennedy pathway genes were dysregulated. Overall, these findings advance our understanding of TAG metabolism in mycolic acid-containing bacteria and provide a framework to engineer strains for increased TAG production.
Rhodococci are mycolic acid-producing Actinobacteria that are of burgeoning importance in environmental and biotechnological applications due in part to their ability to catabolize a remarkably wide range of organic compounds1. Many strains of Rhodococcus genus are oleaginous, producing and accumulating large quantities of triacylglycerols (TAGs). TAG accumulation occurs under conditions of carbon excess when the bacterium is subject to non-carbon nutritional stress, such as nitrogen limitation2. TAGs can constitute up to 85% of the cellular dry weight (CDW)3,4 and are stored as lipid droplets (LD), organelle-like structures with defined protein composition5. The oleaginous nature of rhodococci has taken on new importance given the potential of these strains to degrade lignocellulosic biomass6,7,8 and the potential of TAGs as a feedstock for biotechnological applications such as feed additives, cosmetics, oleochemicals, lubricants and biofuels9,10.
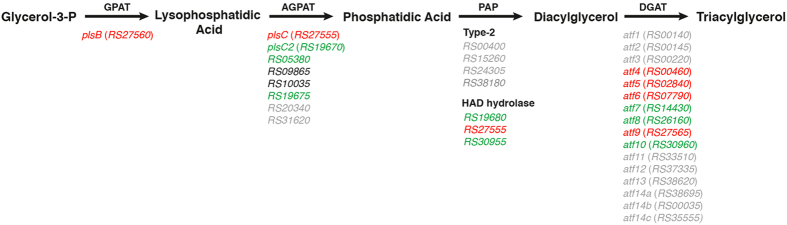
In rhodococci and other Actinobacteria, TAG biosynthesis occurs via the Kennedy pathway (Fig. 1)11. In this pathway, sn-glycerol–3-phosphate is sequentially acylated with fatty acyl-CoAs by glycerol-3-phosphate O-acyltransferase (GPAT, encoded by plsB) and 1-acylglycerol–3-phosphate O-acyltransferase (AGPAT, encoded by plsC) to yield phosphatidic acid. Phosphatidic acid is then dephosphorylated to diacylglycerol (DAG), in a reaction proposed to be catalyzed by a type 2 phosphatidic acid phosphatase (PAP2)12. In the final step, sn-3 acylation of the DAG is catalyzed by a wax ester synthase/DAG O-acyltransferase (WS/DGAT) encoded by an atf gene. Despite this knowledge, efforts to understand TAG biosynthesis in rhodococci have been complicated by the occurrence of multiple homologs of Kennedy pathway enzymes in these bacteria and the lack of biochemical or molecular genetic characterization.
Figure 1. The Kennedy pathway of TAG biosynthesis.
GPAT, glycerol-3-phosphate acyl transferase; AGPAT, acylglycerol-3-phosphate acyl transferase; PAP, phosphatidic acid phosphatase; DGAT, diacylglycerol acyl transferase. Homologous genes potentially encoding each step in RHA1 are marked as follows: green, transcripts more abundant during transition phase under N-limitation; red, transcripts less abundant during transition phase under N-limitation; grey, expressed at low levels under both conditions as defined by RPKM values that were less than 2.5% of total the RPKM value from all homologs predicted to catalyze that step. Type-2 PAPs and HAD-type hydrolases listed separately.
The multiplicity of TAG biosynthetic enzymes is exemplified by the number of predicted WS/DGATs in Rhodococcus opacus PD630 and Rhodococcus jostii RHA1: PD630 contains 17 atf homologs13 and RHA1 contains 162 including three homologs of atf14 whose gene products share 98% amino acid sequence identity14. The homologs are numbered differently in the two strains, but are numbered here according to RHA1 unless otherwise indicated (Fig. 1). Deletion of either atf1PD630 or atf2PD630 of PD630, corresponding respectively to atf3 and atf6 in RHA1, resulted in a 30–50% decreased fatty acid (FA) content during stationary phase compared to the wild-type using gluconate as a growth substrate13,15. Nevertheless, Atf1PD630 appears to be a WS based on its activity in Escherichia coli extracts while Atf2PD630 had a higher DGAT activity15. Transcriptomic studies in PD630 further indicated that the homologs of Atf6, Atf8 and Atf10 are highly expressed during TAG accumulation16. The Atf6 and Atf8 homologs were further implicated in TAG accumulation by virtue of their association with LDs. Interestingly, the homolog of Atf9 was highly repressed during TAG accumulation16. These results are consistent with recent proteomic studies of RHA1 which revealed that Atf6, Atf8 and Atf10 are more abundant in RHA1 under lipid storage conditions17. Nevertheless, the precise roles of the different WS/DGATs in wax ester (WE) and TAG biosynthesis, respectively, remain largely unknown.
Proteomics and transcriptomics studies have identified a number of other genes involved in TAG biosynthesis and have begun to provide a more integrated view of this biosynthesis with respect to cellular metabolism. Whole cell proteomic studies in gluconate-grown RHA1 have indicated that a number of metabolic pathways are up-regulated during TAG accumulation including the Entner-Doudoroff pathway, the pentose-phosphate shunt, branched-chain amino acid catabolism and the methylmalonyl-CoA pathway17. Upwards of 261 genes have been implicated in the metabolism of TAGs in PD630 based on metabolic reconstruction and bioinformatic analyses14. Proteomic studies have also identified 228 LD-associated proteins in RHA1, the two most abundant of which were RHA1_RS10270 (formerly Ro02104) and PspA5. The former includes a predicted apolipoprotein domain and was annotated as “microorganism lipid droplet small” (MLDS) because its deletion yielded larger LDs. Similarly, 177 LD-associated proteins were found to be differentially produced under lipid-accumulating conditions in PD630, including the MLDS homolog16, previously identified as TadA18. In these studies, the LD-associated proteins were predicted to be involved in a remarkably wide variety of cellular processes. Finally, ltp1, encoding a novel lipid transporter and clustered with several TAG biosynthetic genes, was identified to play a role in lipid homeostasis of RHA119.
Herein we describe whole transcriptomic analyses to elucidate the basis of N-mediated accumulation of TAGs in RHA1. Benzoate was used as a growth substrate because RHA1 grows well on this aromatic acid, similar to lignin depolymerization products, and catabolizes it via the β-ketoadipate pathway to yield acetyl-CoA and succinate20. The transcriptomic data were analyzed to further our understanding of how TAG biosynthesis is coordinated with metabolism in rhodococci. In addition, the role of a key atf gene was validated using targeted gene deletion. Phenotypic characterization included the determination of TAG content of whole cells and LDs. Finally, the studies identified a previously overlooked superfamily of enzymes that appear to be involved in TAG biosynthesis. The findings are discussed with respect to the engineering of rhodococcal strains for biotechnological applications.
Results
Accumulation of lipids in RHA1 under conditions of nitrogen limitation
To define N-limited conditions under which RHA1 accumulates TAGs, the strain was grown on 20 mM benzoate as sole growth substrate in M9 minimal medium, which contains 1 g l−1 ammonium chloride, or an otherwise chemically identical medium containing 0.05 g l−1 ammonium chloride. The cells had shorter doubling times on N-rich vs. N-limiting media, and grew to ~3-fold higher density in the N-rich medium (Fig. S1). The residual ammonium in the media at the time of harvesting the cells at early stationary phase was 0.73 g l−1 for N-rich and 0.001 g l−1 for N-limiting media. At these time points, the residual benzoate was undetectable (<80 μM) in N-rich media and 9.45 mM in N-limiting media. Based on these results, the two growth conditions were identified as N-excess and N-limited, respectively. The two cultures had similar pH values of 7.0 at early stationary phase. Finally, the growth yield (CDW) at the transition phase sampling time was 0.29 ± 0.03 mg ml−1 under conditions of N-limitation (72 h) and 0.78 ± 0.06 mg ml−1 under conditions of N-excess (91 h).
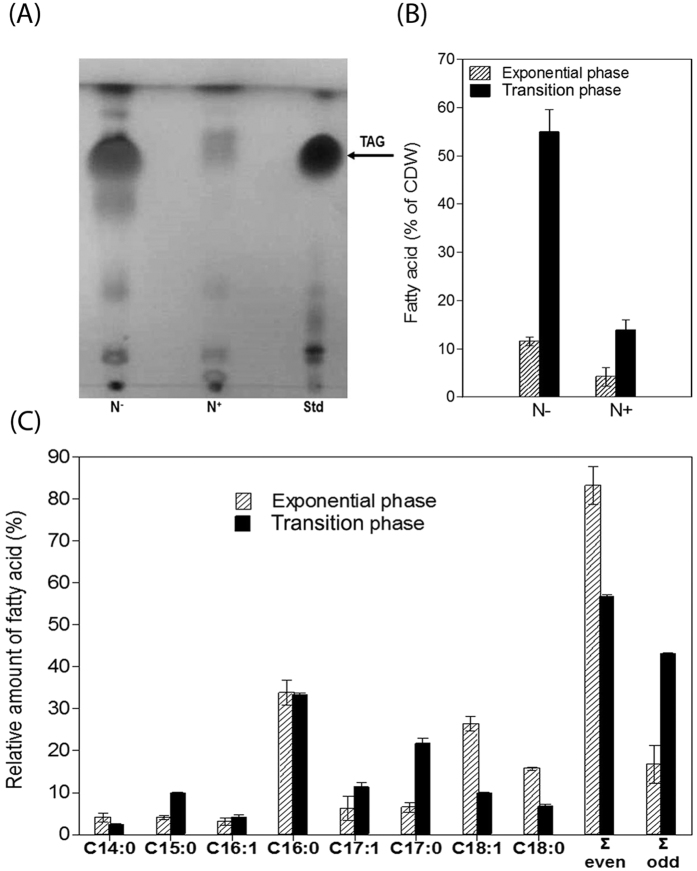
The neutral lipid content of whole cells was analyzed under N-excess and N-limited conditions at each of two time points: exponential growth and during the transition phase (i.e., from exponential growth to stationary phase). TLC analyses of total lipid extracts revealed that RHA1 accumulated significantly more TAGs in the N-limited condition during the transition phase (Fig. 2A). GC-MS analyses were performed to quantify the differences in TAG accumulation. During exponential growth under conditions of N-excess, FAs comprised <5% of the cells’ dry weight (Fig. 2B). This increased ~3-fold during the transition phase. By contrast, FAs comprised 12 ± 1% of the dry weight of exponentially growing cells under N-limited conditions, and this increased ~4.6-fold during the transition phase. This result was confirmed by Nile red staining (Fig. S3).
Figure 2. Lipid analysis of whole-cell extracts of benzoate-grown RHA1 under different nitrogen conditions.
(A) TLC. Lanes: Std, TAG standard; N−, N-limiting condition; N+, N-excess condition during transition phase. (B) Total amounts of FAs (% of cellular dry weight) during exponential growth and transition phase under N-limiting (N−) or N-excess (N+) conditions. (C) FA composition of lipid accumulated by RHA1 under N-limitation during exponential growth and transition phase. Total FAs and relative amounts of each were measured by GC-MS. The data represent the means of three biological replicates.
The GC-MS analyses further revealed that the FA profile depended on the growth phase of the cells. Thus, during exponential growth under N-limited conditions, over 80% of the FAs were of even-numbered chain length, with the major species being C16:0, C18:0 and C18:1 (Fig. 2C). By contrast, less than 60% of the FAs were of even-numbered chain length during the transition phase, with a significant decrease in the proportions of C18:0 and C18:1 species and an increase in the proportions of C15:0, C17:1, and especially C17:0 (~3-fold increase). The proportion of the most abundant FA, C16:0, was unchanged.
Transcriptomic studies of RHA1 under different nitrogen conditions
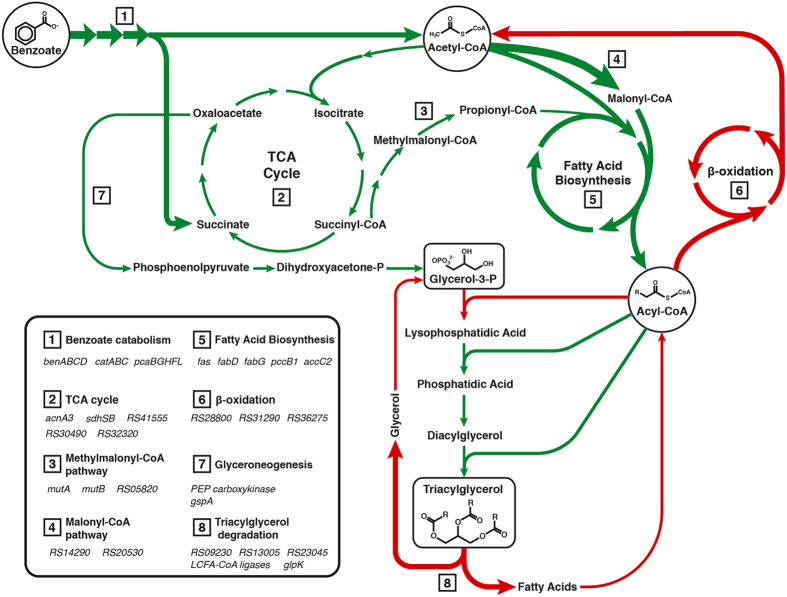
To investigate the basis of TAG accumulation under N-limitation as well as how this biosynthesis is coordinated with cellular metabolism, we performed whole transcriptome analyses using RNA-seq. RHA1 was grown under N-excess and N-limited conditions, respectively, and the transcriptomes were analyzed at the same growth phases as the lipid analyses: exponential and transitional. To maximize rRNA depletion prior to cDNA library construction, a custom-designed depletion protocol using RHA1-specific 23S and 16S rRNA probes was developed. As summarized in Table S2, the Illumina sequencing provided between 283,000 and 1,263,000 reads that were used for RPKM calculations for each of the sampled conditions. A total of 2,527 genes were validated as being dysregulated in at least one of the comparisons between each of four experimental conditions (>2-fold change in abundance and p-value < 0.05; Table S3). In comparing the data sets, the greatest differences were observed between the two sets of data from the transition phase, with 1,826 genes dysregulated. Between the two data sets from exponentially growing cells, 779 genes were dysregulated. In cells growing under N-limited conditions, 482 genes had transcripts of different abundances during the transition phase versus exponential growth. In cells growing under conditions of N-excess, 1,324 genes were dysregulated between the two growth phases. In subsequent analyses, we focused on differences between cells in transition because (a) lipids accumulated to higher levels under these conditions and (b) the greatest differences in the transcriptomes were observed. The 1,826 genes that were dysregulated under these conditions include many involved in a variety of processes as summarized in Fig. 3, Table S4, and below.
Figure 3. Overview of pathways related to TAG biosynthesis and transcript levels during the transition to stationary phase under N-limitation.
. Relevant genes are listed in the lower left chart. Arrows are colored as follows: green, transcripts of genes more abundant during N-limited transition; red, transcripts of genes less abundant during N-limited transition. Arrow thickness represents fold change: narrow, 2–10-times; regular, 10–100-times; wide, >100-times. Pyruvate was not explicitly included as a central metabolite for clarity. For the Kennedy pathway, the average RPKM values of the homologs listed in Fig. 1 were summed. For the TAG lipase step, the values of the three annotated homologs (listed as RS numbers under TAG degradation) were summed.
Nitrogen metabolism – Genes known or predicted to be involved in N metabolism were among the most highly dysregulated (Table S4). Indeed, the transcripts of genes encoding two nitrite reductases (nasDE and RHA1_RS31125-RHA1_RS31145) were ~100- and 500-fold more abundant, respectively, during the transition phase of N-limited cells. Dysregulated genes potentially involved in assimilating ammonium included three encoding homologs of glutamate dehydrogenase (RHA1_RS02795, RHA1_RS06755 and RHA1_RS14810) whose transcripts were less abundant and of two glnA homologs (RHA1_RS05590 and RHA1_RS13730) whose transcripts were more abundant.
Growth substrate catabolism – Transcripts of the benzoate catabolic genes20 were also highly abundant during the transition phase due to N-limitation (Table S4). These include the genes encoding the uptake of benzoate and its transformation to catechol (benABCDK), the cleavage and transformation of the catechol (catA1BC) and the β-ketoadipate pathway (pcaJ1I and pcaHBGLRF) that yields acetyl-CoA and succinate. The RNA-seq data further revealed that these genes are organized into the 4 operons indicated in parentheses. The transcripts of several pathways that catabolize other growth substrates were less abundant under these conditions, such as phenylacetate (paa;)21. However, these had very low average RPKM values under conditions of N-limitation (Table S2).
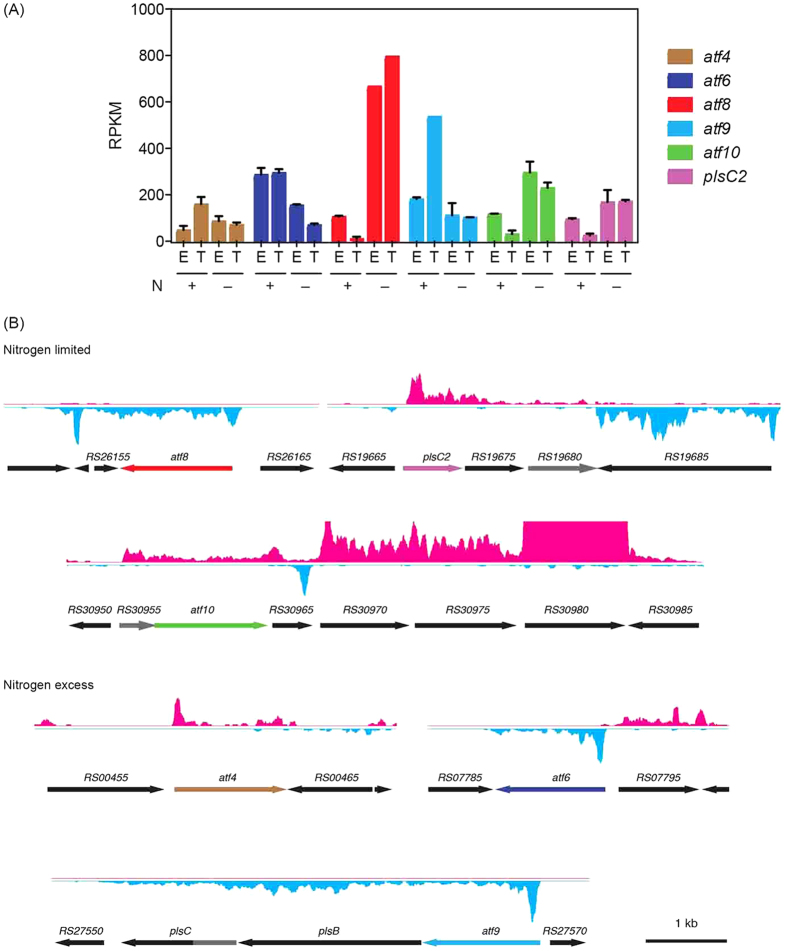
Kennedy pathway – As summarized in Fig. 1 and Table S4, RHA1 contains multiple homologs of the Kennedy pathway enzymes, some of which have not been previously identified. Many of these genes are not clustered in the genome and their physiological roles have not been experimentally validated. A notable exception is atf9, plsB and plsC, predicted to encode each of the three acyltransferases of the Kennedy pathway. These genes were transcribed as an operon and atf9 was the most abundant atf transcript under conditions of N-excess (Fig. 4). However, this transcript was 5.4-fold less abundant during transition phase under N-limitation (Table S4).
Figure 4. The expression and operonic organization of key TAG biosynthetic genes.
(A) Transcript levels of atf4, atf6, atf8, atf9, atf10 and plsC under each of the four experimental conditions. (B) Operon structure of the genes listed in panel (A).
Of RHA1’s 16 atf homologs predicted to encode WS/DGATs, eight were dysregulated during the transition phase under conditions of different N availability. Those with the most abundant transcripts during N-limitation were atf8 and atf10 (Fig. 4A), which were respectively 49- and 6-fold more abundant under this condition (Table S4). By contrast, the most abundant atf transcripts during N-excess were atf9, atf6 and atf4, which were 5.4-, 4.3- and 2.3-fold more abundant, respectively, under this condition. During exponential growth under conditions of N-excess, atf6 and atf9 were the most abundant atf transcripts. The respective operonic structures of these genes could be deduced from the RNA-seq data, except for that of atf10 (Fig. 4B). The atf10 operon has been provisionally identified to include six genes, including two that encode predicted FA desaturases (Table S4), and may be subject to regulation by an antisense RNA at the 3′ end of RHA1_RS30965 (Fig. 4B). The atf5, atf7 and atf13 genes were also dysregulated during the transition phases, but their transcript levels were significantly lower overall.
The data provide interesting insight into RHA1’s other potential Kennedy pathway genes. Thus, of the eight genes predicted to encode AGPATs17,19 sharing 20–60% amino acid sequence identity, the transcripts of three were more abundant under N-limitation: RHA1_RS05380, plsC2 and RHA1_RS19675. Of these, RHA1_RS05380 was the most highly transcribed under N-limitation based on RPKM average values (Table S3), accounting for 50% of the AGPAT transcripts under these conditions. The RNA-seq data further indicate that plsC2 and RHA1_RS19675 occur in an operon with RHA1_RS19680 (Fig. 4B), predicted to encode a haloacid dehalogenase (HAD)-type hydrolase. By contrast, no transcripts of genes encoding PlsB or PAP2 were more abundant under these conditions: transcripts of the lone plsB homolog were less abundant, as noted above, while transcripts encoding the four PAP2 homologs were present at very low levels.
RHA1 genome contains at least 34 genes predicted to encode TAG lipases2, of which three are annotated as TAG lipases. The transcript of one of these, RHA1_RS09230 was 16-fold less abundant during transition phase under N-limitation (Table S4) while those of the other two were at similar levels under the two conditions. Overall, the transcripts of the three TAG lipases were 10-fold less abundant (Fig. 3). Interestingly, the most strongly down-regulated gene in this data set, RHA1_RS16675, is annotated as an esterase/lipase (Table S4). However, this gene occurs in an operon encoding a predicted monooxygenase (RHA1_RS16680), suggesting that it is involved in a process other than TAG hydrolysis.
LD-associated proteins – Ding et al.5 identified 228 proteins associated with LDs in RHA1 while Chen et al.16 identified 177 in PD630. Transcripts associated with 103 of these were more abundant during the transition phase under N-limitation (Table S3). The dysregulated genes include RHA1_RS10270, corresponding to the LD-associated structure-like protein characterized by Ding et al.5 and TadA in PD63018.
Fatty acid biosynthesis and β-oxidation pathways – The RNA-seq data are consistent with increased synthesis of FAs and decreased FA β-oxidation during the transition phase of N-limitation. First, the transcripts of genes encoding two predicted acetyl-CoA carboxylases, RHA1_RS20530 and RHA1_RS14290, were ~100-fold more abundant. These enzymes catalyze the first committed step in fatty acid biosynthesis by transforming acetyl-CoA to malonyl-CoA, an FA building block. Interestingly, RHA1_RS14290 is co-transcribed with RHA1_RS14295 which encodes a formyl-CoA transferase. Secondly, RHA1 harbors two FA synthases (FASs): FAS-I, encoded by fas, synthesizes FAs from C14 to C24 chain length in Actinobacteria22,23; and FAS-II, encoded by acpS, fabD, acpP-fabF, fabG, pccB and accC2, produces mycolic acids from the FAs generated by FAS-I24. Transcripts of fas were 115-fold more abundant while those of genes encoding FAS-II were up to 100-fold more abundant. Consistent with the synthesized FAs being incorporated into TAGs, the transcript of RHA1_RS42105, encoding a predicted long chain fatty acid CoA ligase, was 6-fold more abundant. Finally, with respect to β-oxidation, the transcripts of genes encoding acyl-CoA dehydrogenases (RHA1_RS28800, RHA1_RS31290 and RHA1_RS36275) were less abundant.
Central metabolic pathways – Transcripts of genes encoding a variety of central metabolic pathways and enzymes were more abundant during the transition phase at N-limitation. These included those encoding the TCA cycle (14 transcriptional units, up to 39-fold more abundant), the methylmalonyl-CoA pathway (three transcriptional units, up to 10-fold), and the Entner-Doudoroff pathway (two transcriptional units, up to 14-fold). According to the KEGG database, RHA1 encodes 24 pyruvate dehydrogenases encoded by 14 different gene clusters (http://www.genome.jp/kegg/). Of these, five are operons whose transcripts were up to 39-fold more abundant under TAG accumulating conditions. Among central metabolic enzymes, TadD, an NAD(P)+-dependent glyceraldehyde 3-phosphate dehydrogenase, has been proposed to be a hallmark of TAG accumulation in PD630, effectively replacing glyceraldehyde 3-phosphate dehydrogenase (gapA) and phosphoglycerate kinase (pgk) which are present during vegetative growth18,25. This enables the cell to use NADPH under conditions of TAG accumulation instead of NADH and ATP. In RHA1 during TAG accumulation, the transcripts of gap1 (RHA1_RS16630; formerly ro03427), gap2 (RHA1_RS35030) and pgk (RHA1_RS35035) were 3-, 16- and 8-fold more abundant, respectively. Although there appears to be no direct homology of tadD in RHA1, three candidates were identified: RHA1_RS29865, RHA1_RS27320 and RHA1_RS3068017. The RNA-seq data indicate that none of these genes were dysregulated under any of the studied conditions (Table S3).
Glyceroneogenesis – Glyceroneogenesis ensures the production of glycerol 3-phosphate, the starting point of the Kennedy pathway17,26. Two enzymes of glyceroneogenesis are pyruvate carboxylase, which catalyzes the formation of oxaloacetate from pyruvate, and phosphoenolpyruvate carboxykinase, which catalyzes the decarboxylation of oxaloacetate26. In RHA1, these are encoded by pycA (RHA1_RS31870) and ppc (RHA1_RS35050), respectively. Under N-limiting conditions, transcripts of pycA were almost 2-fold less abundant whereas ppc transcripts were 6-fold more abundant. The last step of glyceroneogenesis is catalyzed by NAD(P)H-dependent glycerol 3-phosphate dehydrogenase. Of the two genes predicted to encode this enzyme in RHA1, transcripts of gpsA (RHA1_RS11710) were 6-fold less abundant and those of gpsA2 (RHA1_RS31815) were 3.4-fold more abundant.
RT-qPCR analysis of RHA1 under nitrogen-limitation
The RNA-seq data of selected TAG biosynthesis genes were verified using RT-qPCR and the primers and probes listed in Table S1. Four atf genes in addition to atf8 and atf10 were included in this analysis: atf3 and atf6, whose homologs in PD630 are physiologically important13,15; atf9, which is part of an operon with plsC and plsB; and atf4, whose product shares 47% amino acid sequence identity with Atf6. The RT-qPCR data largely validated the RNA-seq data although the exact values differed slightly (Table 1). Thus, only the transcripts atf8 and atf10 were more abundant during the transition phase under N-limitation (Table S4). By contrast, the transcripts of all the other tested atf genes were more abundant under conditions of N-excess, as were the plsC and RHA1_RS16675 (putative esterase/lipase) transcripts. Finally, this experiment confirmed that expression of the gene encoding PAP-2 was low (data not shown) and did not appear to depend on N concentrations.
Table 1. RT-qPCR analysis of selected TAG metabolic genes in RHA1 during transition phase under different N conditions.
| Gene IDa | Gene Name | Product | Fold change |
|---|---|---|---|
| RS00220 | atf3 | WS/DGAT | −3 |
| RS00400 | PAP-2 | 1 | |
| RS00460 | atf4 | WS/DGAT | −5 |
| RS07790 | atf6 | WS/DGAT | −4 |
| RS16675 | putative lipase | −1596 | |
| RS26160 | atf8 | WS/DGAT | 28 |
| RS27555 | plsC | AGPAT | −50 |
| RS27565 | atf9 | WS/DGAT | −35 |
| RS30960 | atf10 | WS/DGAT | 3 |
aRHA1_ prefix has been omitted for simplicity.
Role of atf8 on TAG accumulation in RHA1
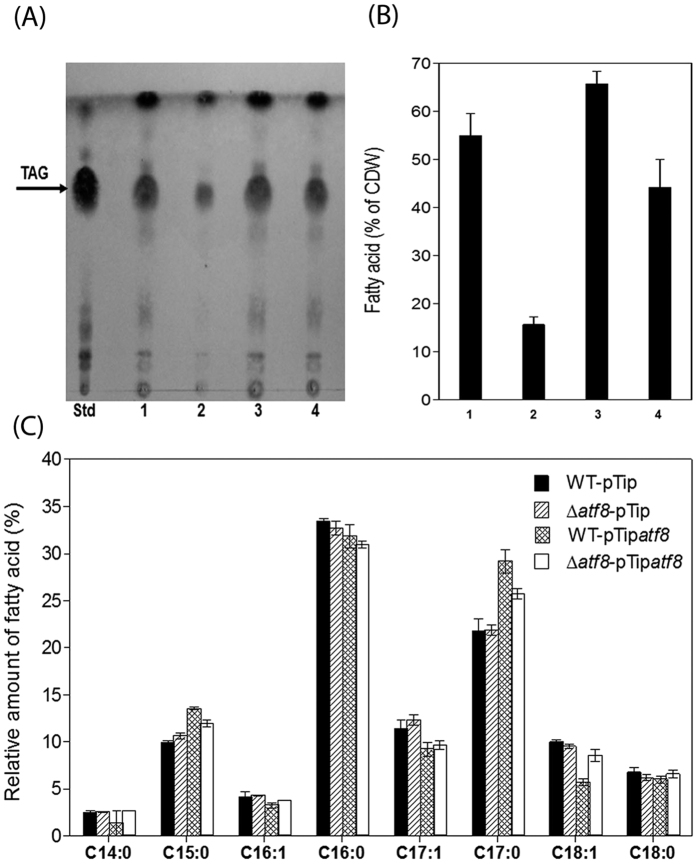
To investigate the role of Atf8 in TAG biosynthesis, an atf8 knockout mutant was constructed as summarized in Material and Methods. Growth of the mutant was comparable to that of the wild type under N-limited conditions (Fig. S1). TLC analysis of total lipids (Fig. 5A) showed a decrease in TAG content of the Δatf8 mutant compared to the wild type strain (WT) during N-limited transition phase. GC-MS analyses revealed that the mutant accumulated approximately one-third the amount of TAGs (w/w of the CDW) as the WT (Fig. 5B). The Δatf8 mutant was complemented using a replicative vector, pTipatf8, in which atf8 is under control of a thiostrepton-inducible promoter. The production of Atf8 in the complemented mutant was verified using SDS-PAGE (Fig. S2). This revealed a protein of ~50 kDa, corresponding to the predicted molecular weight of Atf8, that was absent in the WT. Total lipid analysis showed that the complemented strain accumulated higher amounts of TAG in comparison with the Δatf8 mutant, but less than the WT cells (Fig. 5A,B). The partial complementation of Δatf8 may be due to the low solubility of Atf8 when over-produced in RHA1 (data not shown).
Figure 5. Lipid analysis of whole-cell extracts of benzoate-grown atf8-manipulated strains during transition phase under N-limitation.
(A) TLC. Lanes: Std, TAG standard; 1, WT:pTip; 2, Δatf8:pTip; 3, WT:pTipatf8; and 4, Δatf8:pTipatf8. (B) Total amounts of fatty acids measured by GC-MS. Lanes numbered as in (A). (C) FA composition of RHA1 strains. Total amount of FA and the relative proportions of each type are presented as in Fig. 2. The data represent the means of three biological replicates.
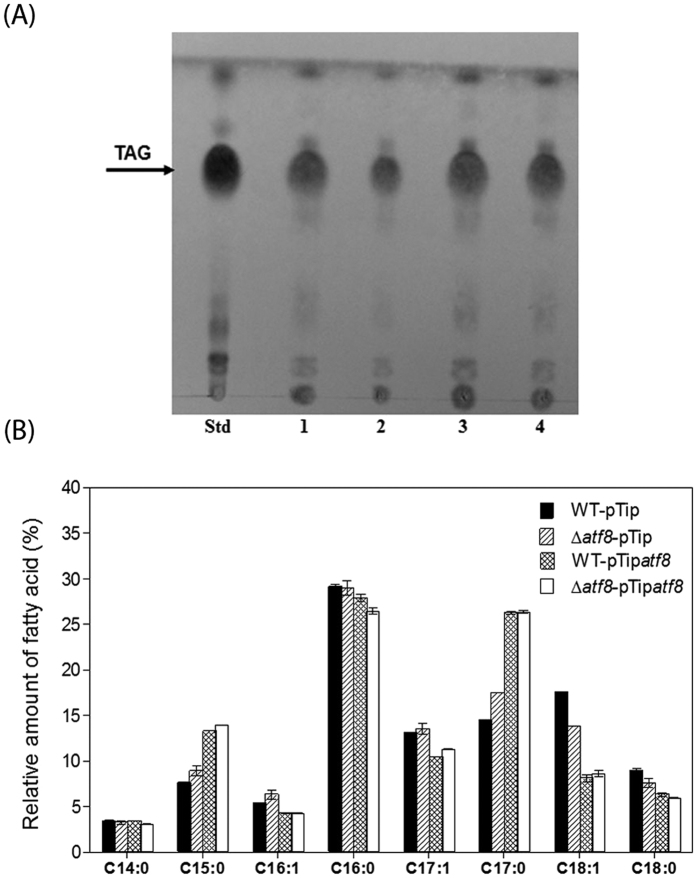
The pTipatf8 construct was also used to investigate the effect of overproducing atf8 in the WT strain. As in the complemented mutant, production of Atf8 was confirmed by SDS-PAGE (Fig. S2). Growth of the overproducing strain was comparable to that of WT under N-limitation (Fig. S1). TLC analyses of lipid content of the recombinant strain WT-pTipatf8 revealed a slight increase in TAG content after the induction of atf8 expression (Fig. 5A). Quantitative analyses revealed an increase of the total fatty acids of the WT-pTipatf8 of ~20% (w/w of the CDW) as compared to the wild type (Fig. 5B). LDs were also purified from all RHA1 strains and lipid contents were analyzed by TLC (Fig. 6A). The qualitative differences in TAG recovered in the LDs of the RHA1 strains was consistent with what was observed in the total lipid extract, confirming the importance of atf8 in TAG accumulation. Analysis of fatty acid composition revealed only slight differences between the WT and Δatf8 mutant strains (Fig. 5C). However, in strains overproducing Atf8, an increase of saturated fatty acids C15:0 and C17:0 and a decrease of unsaturated fatty acids C16:1, C17:1 and C18:1 was observed (Fig. 5C). The same pattern of fatty acid distribution was obtained in purified LDs (Fig. 6B).
Figure 6. Analysis of lipid droplets from benzoate-grown RHA1 strains during transition phase under N-limitation.
(A) TLC. Lanes: Std, TAG standard; 1, WT:pTip; 2, WT:pTipatf8; 3, Δatf8:pTip; 4, Δatf8:pTipatf8. (B) FA composition of RHA1 strains. The data represent the means of three biological replicates.
Finally, to verify the activity of Atf8, DGAT activity was measured in the lysates of four strains of RHA1. In these experiments, lysates were prepared from cells at the transition phase under N-limitation, and diolein (DiC18:1) and palmitoyl-CoA (C16-CoA) were used as substrates. Lysate from WT RHA1 with the empty pTip-QC2 vector had a specific activity of 43 ± 3 nmol mg protein−1 min−1. By contrast, lysate of the strain containing pTipatf8 contained 68 ± 9 nmol mg protein−1 min−1. Activities of the Δatf8 mutant carrying the pTip-QC2 and the pTipatf8 vectors were 32 ± 5 and 47 ± 8 nmol mg protein−1 min−1, respectively. These data support the conclusion that atf8 encodes a DGAT.
An alternative PAP for the Kennedy pathway
The low levels of the transcripts encoding the four PAP2 homologs prompted us to consider whether the third reaction of the Kennedy pathway might be catalyzed by another class of enzymes. Inspection of the operons encoding Kennedy pathway enzymes (Fig. 4B) revealed three genes predicted to encode HAD-type hydrolases according to NCBI: RHA1_RS19680, which occurs in the operon with plsC2; RHA1_RS30955, which is one of the genes of the potential atf10 operon; and plsC, which appears to encode an AGPAT-hydrolase fusion enzyme. The encoded enzymes/domains share less than 20% sequence amino acid identity. Despite the name of the superfamily, many HAD-type hydrolases are phosphatases27.
Discussion
Bacteria are of burgeoning interest in the development of consolidated bioprocesses for lignin6. RHA1 in particular has been identified as a promising starting point for metabolic engineering based on its ability to grow on a lignin-enriched stream derived from corn stover. In this study of TAG accumulation in RHA1, we used benzoate as a model of lignin-depolymerization products degraded via the β-ketoadipate pathway to succinate and acetyl-CoA. RHA1 accumulated significant amounts of TAG using this aromatic substrate under N-limitation. The transcriptomic and gene deletion experiments establish Atf8 as a key determinant of TAG accumulation under these conditions. More generally, this study advances our understanding of TAG metabolism in rhodococci, providing a framework for understanding how different homologs of the Kennedy pathway are utilized under different metabolic states of the cell. The data further indicate how metabolism directs carbon flow to TAG biosynthesis when growing on an aromatic compound, identify gaps in our knowledge of TAG biosynthesis, and provide some insight into other aspects of rhodococcal physiology.
TAG accumulation in benzoate-grown RHA1 was not as dramatic as what has been reported in rhodococci growing on other substrates and the identity of the FA species differed significantly. Previous studies of TAG biosynthesis in rhodococci have used glucose, gluconate or rich media as growth substrates3,17,28. FA contents as high as 75% and 85% CDW have been reported in PD630 grown on gluconate and olive oil, respectively28, higher than the maximum of 55% observed here for benzoate-grown RHA1 cells under conditions of N-limitation. It is unclear whether this reflects differences in the metabolism of these substrates. Consistent with previous reports of rhodococci grown on glucose and gluconate28, TAGs were the dominant neutral lipid. In contrast to the predominance of C16:0 and C18:1 FAs observed when glucose or gluconate was the growth substrate under N-limitation2,17, benzoate-grown RHA1 contained predominantly C16:0 (33%) and C17:0 (22%) FAs. Indeed, odd-numbered chain length FAs comprised more than 40% of the FAs under these conditions, indicating that propionyl-CoA was a significant building block for FAs. This building block could be generated from benzoate-derived succinate by the methylmalonyl-CoA pathway, which converts succinyl-CoA to propionyl-CoA in rhodococci29,30. Consistent with this explanation, this pathway was upregulated during TAG accumulation. The methylmalonyl-CoA pathway was also up-regulated during TAG accumulation in PD63016 and in gluconate-grown RHA117. However, odd-numbered FAs comprised less than 30% of total FA in gluconate-grown RHA1, consistent with less propionyl-CoA being produced. Finally, the FA composition was also growth phase-dependent in the current studies, with exponentially growing cells containing ~2-fold greater proportion of C18:0 and C18:1 FAs and ~2-fold lesser proportion of C15:0 and C17:0 FAs. Overall, these results indicate that growth substrate and nutritional status influence the species of FA accumulated by Rhodococcus28,31,32,33.
The presented data identify Atf8 as the major DGAT responsible for the accumulation of TAGs under N-limitation. First, the atf8 transcript was the most highly upregulated atf under these conditions and its transcript was 3.5-fold more abundant than any other. Second, deletion of atf8 led to a 70% drop in TAG levels under N-limitation. By contrast, deletion of the atf3 and atf6 homologs in PD630 (i.e., atf1PD630 and atf2PD630) had a less dramatic effect, reducing TAG levels by 20% and 50%, respectively13,15. Importantly, heterologously produced Atf1PD630 had WS activity but not DGAT13. This is consistent with the low transcript levels of atf3. Consistent with our primary conclusion, Atf8 was also more abundant in RHA1 under lipid storage conditions in proteomic studies and was associated with LDs17. Moreover, the homolog of atf8 in PD630, coincidentally atf8PD630, was also highly expressed during TAG accumulation16. Finally, the ability of the Δatf8 mutant to accumulate some TAG together with the transcriptomics data suggest that other DGATs such as Atf10 contribute to TAG biosynthesis during N-limitation. Again, this is consistent with previous reports of Atf10 being more abundant in RHA1 under lipid storage conditions17 and the corresponding gene being more highly expressed in PD63016. Nevertheless, Atf8 appears to be rate-limiting in TAG accumulation since its overexpression resulted in a 20% increase in accumulated TAG.
The data further implicate Atf6 and Atf9 as the primary DGATs responsible for TAG biosynthesis during exponential growth and that atf6 and atf9 are expressed during C-limitation. The regulation of atf9 is particularly interesting since it occurs in the only operon predicted to encode all four activities of the Kennedy pathway as discussed below. Moreover, this operon is widely conserved in rhodococci and mycobacteria. Overall, the content, regulation and distribution of this operon suggest that it encodes “house-keeping” homologs of the Kennedy pathway. Interestingly, ltp1 encoding a lipid transporter19 does not appear to be part of this operon. The lower levels of plsB transcript are somewhat unexpected since this is the only identified homolog in RHA1. Nevertheless, the transcriptomics results are largely consistent with published data obtained under different growth conditions. Thus, the homologs of Atf9 and PlsB in PD630 were repressed during TAG accumulation16, although direct comparison is difficult since the authors did not provide specific details on their growth conditions. Interestingly, Atf6 was more abundant in RHA1 under lipid storage conditions in proteomic studies17. Moreover, the corresponding enzyme in PD630, Atf2PD630, had significant DGAT activity in E. coli extracts13. It is possible that Atf6 is regulated at the transcriptional and translational levels.
The available evidence suggests the possibility that the three HAD-type hydrolases identified herein may be the major PAPs involved in TAG biosynthesis. First, all three are arranged in operons with genes encoding other enzymes of the Kennedy pathway; none of the genes encoding PAP2 homologs are. Second, transcripts of two of the HAD-type hydrolases were more abundant under TAG-accumulating conditions. By contrast, RHA1’s four genes encoding PAP2 homologs were present at very low levels in this study’s experimental conditions. Interestingly, the HAD-type hydrolases are conserved in the TAG gene clusters of mycolic acid-containing Actinobacteria such as rhodococci and mycobacteria (results not shown). This includes the atf9-plsB-plsC operon in which PlsC is a predicted fusion of AGPAT and HAD domains. The corresponding gene in M. tuberculosis, Rv2483c, was predicted to be required for growth in mice based on transposon mutagenesis34 but not for growth in vitro35. Clearly, additional work is required to establish the role of these HAD-type hydrolases. However, the possibility that PlsC is a bifunctional enzyme is particularly satisfying as this would mean that a single operon encodes all of the enzymes required for TAG biosynthesis during vegetative growth.
The transcriptomics data extend our understanding of how rhodococci shift their global metabolism to promote TAG accumulation. Pathways that generate propionyl-CoA and malonyl-CoA, the building blocks for FA biosynthesis, were among the most highly up-regulated including those involved in benzoate catabolism, the TCA cycle, the methylmalonyl-CoA pathway and acetyl-CoA carboxylases. Genes encoding the Entner-Doudoroff pathway and the pentose-phosphate pathway were also up-regulated. Increased flux through these pathways in gluconate-grown cells17 is not surprising considering that gluconate feeds into the Entner-Doudoroff pathway. The latter’s up-regulation during growth on benzoate suggests its primary importance in generating NADPH for FA biosynthesis. By contrast, NADPH does not appear to be generated by an NAD(P)+-dependent glyceraldehyde 3-phosphate dehydrogenase as occurs in PD630 under conditions of TAG accumulation18,25. Other up-regulated processes that provide building blocks for TAGs include glyceroneogeneis, fatty acid biosynthesis, and branched amino acid catabolism. These processes were also up-regulated under N-limiting TAG-accumulation during growth on gluconate17. Branched amino acid catabolism is a source of nitrogen, NADPH and TAG precursors. In glyceroneogenesis, the up-regulation of genes encoding PEP carboxykinase is consistent with the former catalyzing the rate-limiting step26. OAA for glyceroneogenesis may be generated through the TCA cycle as the lower abundance of pycA transcripts suggests that it is not generated through the carboxylation of pyruvate. Genes whose transcripts were less abundant include those involved in TAG degradation and FA oxidation. Interestingly, although eight predicted lipase-encoding genes were down-regulated during TAG accumulation, six were up-regulated (Table S4). Metabolic flux studies would be required to determine whether any turnover of TAGs occurs under these conditions.
Finally, the transcriptomics data highlight important gaps in our knowledge. Foremost, our knowledge of the Kennedy pathway in Rhodococcus is incomplete since, as noted above, the lone plsB homolog is down-regulated under TAG-accumulating conditions. At the same time, it is unclear why there is so much apparent redundancy in central metabolism in Rhodococcus. Most notably, genes encoding four predicted pyruvate dehydrogenases were highly up-regulated in this study. While this is consistent with a dramatic shift toward acetyl-CoA production during TAG accumulation, it is possible that some of these isozymes act on other α-keto acids. Similarly, two nitrite reductases, three putative glutamate dehydrogenases and two glutamine synthases (glnA and glnA3, a glutamate-ammonia ligase) were dysregulated. Although redundancy in rhodococcal catabolic enzymes is well known20, functional characterization of the central metabolic isozymes is required to establish their respective physiological roles.
We demonstrated that the atf8-encoded DGAT is a major determinant of TAG accumulation in RHA1 under N-limiting conditions. This DGAT likely plays the same role in PD630 and other oleaginous rhodococci in which it is conserved. Importantly, targeted disruption of atf8 perturbed TAG accumulation to a greater extent than that reported in any other rhodococcal atf deletion to date. Moreover, overexpressing atf8 significantly increased the production of TAG in benzoate-grown RHA1, establishing the potential of engineering rhodococci to optimize TAG production from different substrates. Finally, we identified a previously overlooked suite of HAD-type hydrolases that appear to be involved in TAG biosynthesis. These findings, together with the transcriptomics data, should facilitate the engineering of rhodococci for the production of TAGs as feedstock for various applications9,10.
Methods
Strains and culture conditions
E. coli DH5α was used for DNA propagation and manipulation. E. coli strains were grown at 37 °C (200 rpm) in LB broth unless otherwise indicated. Bacto agar (1.5% [w/v]; Difco) was used for solid medium. RHA1 and derivative strains were cultivated at 30 °C under aerobic conditions in M9 minimal medium supplemented with trace elements, thiamin and 20 mM benzoate as growth substrate in shake flasks. For nitrogen-limiting conditions, the concentration of ammonium chloride in the M9 medium was reduced from 1 g l−1 to 0.05 g l−1. Expression from the thiostrepton-inducible promoter (PtipA) of pTip-QC2 was induced by adding 10 μg ml−1 of thiostrepton to the culture. Growth was measured as the change in optical density at 600 nm. For lipid analyses, cells were harvested, washed with 150 mM NaCl and frozen at −80 °C. As appropriate, antibiotics were used at the following final concentrations: 100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol.
DNA manipulation
DNA was manipulated using standard protocols36. Oligonucleotides were from Integrated DNA Technologies. The sequences of primers and probes used in this study are listed in Table S1. DNA sequencing was performed at Genewiz Inc. E. coli and RHA1 were transformed with plasmid DNA by electroporation using a MicroPulser with GenePulser cuvettes (Bio-Rad). The Δatf8 mutant was constructed using the sacB counter selection system37. The flanking regions of atf8 were amplified from RHA1 genomic DNA using the atf8-up and atf8-dn oligonucleotides (Table S1). The two amplicons of ~500 bp and were cloned into pK18mobsacB using restriction enzymes XbaI, BamHI and HindIII to yield pK18MSatf8. Kanamycin-sensitive/sucrose-resistant colonies were screened using PCR. The nucleotide sequence of the targeted region was determined in the mutant strain to confirm the gene deletion. For complementation and over-expression experiments, atf8 was amplified from RHA1 genomic DNA using primers atf8-ex-F and atf8-ex-R (Table S1) together with GoTaq DNA polymerase (Promega). The resulting amplicon was digested with NdeI and EcoRI and cloned in pTip-QC238,39 yielding pTipatf8. The nucleotide sequence of the cloned gene was verified and the vector was used to complement the mutant and to overexpress atf8 in WT RHA1.
RNA isolation
Total RNA was extracted using TRIzol® Reagent (Invitrogen) according to the manufacturer’s instructions. The RNA was treated with TURBOTM DNase (Invitrogen) and extracted with phenol-chloroform. The quality and concentration of RNA were assessed using a 2100 Bioanalyzer and the RNA 6000 Nano kit (Agilent). Three μg of total RNA was heated at 65 °C for 5 min, quickly chilled on ice and then mixed with Streptavidin magnetic beads (New England Biolabs) with an equimolar mixture of biotin-labelled probes specific for RHA1 16S and 23S rRNA to deplete rRNA (rRNA-16S, rRNA-23Sa, and rRNA-23Sb (Table S1)). The supernatant was collected and subjected to further rRNA-depletion using the Ribominus™ Transcriptome Isolation Kit (Invitrogen).
RNA-seq analysis
One hundred ng of the rRNA-depleted RNA was used for library construction using NEBNext® Ultra Directional RNA Library Prep Kit for Illumina® and NEBNext® Multiplex Oligos for Illumina® (New England Biolabs) according to the manufacturer’s instructions. The samples were analyzed using a Genome Analyzer IIx (Illumina®). Data were processed using the Sequence Control Software (Illumina®). Single-ended sequence reads were scanned 3′ to 5′. Those with a Phred quality value below 20 were trimmed using the FASTQ Quality Trimmer tool of the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Reads of less than 35 bases after trimming were discarded from further analysis. Trimmed reads were aligned to the RHA1 genome40 using Bowtie 2 in end-to-end mode with the “very sensitive” option41. Reads of rDNA sequences were omitted in subsequent analyses. A custom Perl script was developed to create a GTF file using the recently updated RHA1 annotation by the NCBI Prokaryotic Genome Annotation Pipeline. Mapped read counts per gene were obtained using the HTSeq package42. Differential expression (>2-fold change with a false discovery rate-adjusted p value < 0.05) was analyzed using the DESeq2 package43. The RNA-seq data set was deposited in Gene Expression Omnibus44 under accession number of GSE77158.
RT-qPCR
Primers and FAM-labelled TaqMan probes (Table S1) were designed using IDT PrimeTime qPCR design tools. The reference gene was sigA (RHA1_RS33345), which encodes the major sigma factor in RHA1. cDNA was synthesized from DNase-treated, Trizol-extracted total RNA using the SuperScript® VILO cDNA synthesis kit according to manufacturer instructions. The cDNA samples were diluted as appropriate for use. Reactions were performed in duplicate using an ABI stepOnePlus real-time PCR system and the following conditions: 20 s at 95 °C followed by 40 cycles of 1 s at 95 °C and 20 s at 60 °C for extension. Standard curves for each target based on genomic DNA were included in each run to quantify absolute target cDNA in the samples. Concentrations were normalized to those of the reference gene under each condition tested. Relative fold differences were calculated using conditions of nitrogen excess as a reference.
Isolation of LDs
LDs were isolated essentially as previously reported5. Briefly, RHA1 cells were cultivated in shake flasks in 50 ml M9 medium, harvested by centrifugation (4,000 × g for 10 min) and washed twice with 20 ml Buffer A (25 mM tricine, 250 mM sucrose, pH 7.8). The washed cells were suspended in 20 ml Buffer A, incubated for 20 min on ice and disrupted by passing them three times through a French pressure cell operated at 100 MPa, 4 °C. The sample was centrifuged at 4,000 × g for 10 min to remove cell debris. Eight ml of the supernatant was loaded into an SW40 tube (Beckman), layered with 2 ml Buffer B (20 mM HEPES, 100 mM KCl, 2 mM MgCl2, pH 7.4) on top, and centrifuged at 180,000 × g for 1 h at 4 °C. The LD fraction on top of the sucrose gradient was collected and transferred to a 1.5 ml Eppendorf tube. LDs were washed with 200 μl Buffer B. To dissolve the lipids and to precipitate proteins, 1 ml chloroform-acetone (1:1, v/v) was added. The sample was vortexed and centrifuged (20,000 g × 10 min). Finally, the organic phase was collected, dried under a nitrogen steam, and suspended in 100 μl chloroform for lipid analysis.
Lipid extraction and thin-layer chromatography (TLC)
For whole RHA1 lipid content analysis, cells were harvested during the transition from exponential to stationary growth phases, washed with a 150 mM NaCl and suspended in 50 mM Tris-HCl, pH 7.0, and 300 mM NaCl buffer. Cells were subjected to four rounds of 60 s of bead beating using a FastPrep-24 bead beater (MP Biomedicals, Solon, OH) set to 5.5. The sample was incubated for 5 min on ice between rounds. To identify lipids, samples of whole cells or isolated LDs were extracted with chloroform-methanol (2:1, v/v). Aliquots were analyzed by thin layer chromatography (TLC) on 60F254 silica gel plates (Merck) using hexane/diethyl ether/acetic acid (80:20:1, v/v/v) as a solvent system. Triolein (Sigma-Aldrich, T7140) was used as the TAG standard. The plates were dried at room temperature for 10 min and immediately sprayed with a 10% cupric sulfate in 8% phosphoric acid solution. The plates were then incubated in an oven at 150 °C for 10 min.
Fatty acid profiling
FA profiles were determined by subjecting 5 mg of freeze-dried whole cells or LD extracts to methanolysis in the presence of 15% (v/v) sulfuric acid as described by Alvarez et al.28. The resulting methylesters were analyzed using an Agilent 6890 series gas chromatograph (GC) equipped with an HP-5 MS 30 m × 0.25 mm capillary column (Hewlett-Packard) and an HP 5973 mass-selective detector. The GC was operated with an injector temperature of 280 °C, a transfer line temperature of 290 °C, and a flow rate of 1 ml min−1 with helium. The temperature program of the oven was 90 °C for 5 min, increased to 240 °C at a rate of 6 °C per min, and then held at 240 °C for 17 min. The mass spectrometer was operated in electron emission scanning mode at 40 to 800 m/z and 1.97 scans per second. Peaks were quantified and identified using the software packages GCsolution Analysis version 2.32 and GCMS Solutions version 2.53 (Shimadzu Scientific Instruments, Columbia, MD) and the NIST08 Library.
Ammonium determination
Ammonium concentration was quantified in culture media using indophenol blue45. Briefly, media samples were diluted 50–1000-fold to 10 ml using distilled water. To this were added: 0.4 ml of reagent (10% w/v phenol solution in ethanol), 0.4 ml catalyst (0.5% w/v nitroprusside in water) and 1 ml oxidizing solution (20% w/v trisodium citrate and 1% w/v sodium hydroxide solution, mixed with 5% sodium hypochlorite 4:1 v/v). Samples were incubated at 37 °C with shaking for 2 h in the dark. Absorbance was read at 630 nm and concentrations were calculated using a standard curve of 0.2–2 mg ml−1 ammonium chloride.
Protein production, purification and analysis
Protein extracts were prepared from RHA1 by suspending cells in 50 mM Tris-HCl, pH 7.0 containing 300 mM NaCl and 1 mg ml−1 CHAPS. Cells were subjected to four rounds of 60 s of bead beating as described above. Protein extracts were analyzed by SDS-PAGE.
DGAT activity
DGAT activity was measured using a spectrophotometric assay that monitors the reaction of dithionitrobenzoate (DTNB) with the CoASH liberated from the DGAT-catalyzed esterification of acyl-CoA and diglyceride4. Briefly, assays were performed in a total volume of 1 ml and absorbance was recorded at 412 nm. Ten μl DTNB solution (18 mg ml−1 in dimethyl sulfoxide) and 15 μl each of 1 mM palmitoyl-CoA and 1 mM diolein were added to 950 μl 50 mM Tris-HCl, 150 mM NaCl, pH 8.0. The base line was recorded, the reaction was initiated with 10 μl of lysate, and the absorbance was recorded for 1 min. Initial reaction rates were calculated from the data using Excel and an ε412 of 14,150 M−1cm−1 for the nitrothiobenzoate dianion.
Computational analysis
Database searches and alignments were carried out using BLAST 2.2.1746 and CLUSTALW47. Reference protein sequences were retrieved from the NCBI database.
Additional Information
How to cite this article: Amara, S. et al. Characterization of key triacylglycerol biosynthesis processes in rhodococci. Sci. Rep. 6, 24985; doi: 10.1038/srep24985 (2016).
Supplementary Material
Acknowledgments
This research was supported by Large Scale Applied Research Project 2108 from Genome Canada, US Department of Energy award DE-EE0006112, and a Canada Research Chair (to L. D. E.).
Footnotes
Author Contributions S.A., N.S. and L.D.E. designed and conceived the experiments. L.D.E. supervised the execution of the experiments. S.A. and N.S. performed the growth experiments and lipids analyses. S.A. also performed protein purification and activity assays. N.S. constructed the atf8 mutant constructions with the help of J.L. and performed the transcriptomics. H.O. and S.A. analysed the transcriptomic data. C.D.S. performed the RT-qPCR experiments. S.A. and L.D.E. wrote the paper with contributions from H.O. and C.D.S. All authors read and approved the manuscript.
References
- Goodfellow M. In The Prokaryotes – Actinobacteria (ed Rosenberg E. et al.) 595–650 (Springer-Verlag 2014). [Google Scholar]
- Hernandez M. A. et al. Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genomics 9, 600, doi: 10.1186/1471-2164-9-600 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez H. M. In Microbiology Monographs Vol. 16 (ed Steinbüchel Alexander) (Springer, 2010). [Google Scholar]
- Barney B. M., Wahlen B. D., Garner E., Wei J. & Seefeldt L. C. Differences in substrate specificities of five bacterial wax ester synthases. Appl Environ Microbiol 78, 5734–5745, doi: 10.1128/AEM.00534-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. et al. Identification of the major functional proteins of prokaryotic lipid droplets. J Lipid Res 53, 399–411, doi: 10.1194/jlr.M021899 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvachúa D., Karp E. M., Nimlos C. T., Vardonab D. R. & Beckham G. T. Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem in press (2015). [Google Scholar]
- Kurosawa K., Plassmeier J., Kalinowski J., Ruckert C. & Sinskey A. J. Engineering l-arabinose metabolism in triacylglycerol-producing Rhodococcus opacus for lignocellulosic fuel production. Metab Eng 30, 89–95, doi: 10.1016/j.ymben.2015.04.006 (2015). [DOI] [PubMed] [Google Scholar]
- Ahmad M. et al. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50, 5096–5107, doi: 10.1021/bi101892z (2011). [DOI] [PubMed] [Google Scholar]
- Gouda M. K., Omar S. H. & Aouad L. M. Single cell oil production by Gordonia sp. DG using agro-industrial wastes. World J Microbiol Biotechnol 24, 1703–1711 (2008). [Google Scholar]
- Liang M. H. & Jiang J. G. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog Lipid Res 52, 395–408, doi: 10.1016/j.plipres.2013.05.002 (2013). [DOI] [PubMed] [Google Scholar]
- Alvarez H. M., Silva R. A., Herrero M., Hernández M. A. & Villalba M. S. Metabolism of triacylglycerols in Rhodococcus species: insights from physiology and molecular genetics. Journal of Molecular Biochemistry 2, 69–78 (2012). [Google Scholar]
- Hernandez M. A., Comba S., Arabolaza A., Gramajo H. & Alvarez H. M. Overexpression of a phosphatidic acid phosphatase type 2 leads to an increase in triacylglycerol production in oleaginous Rhodococcus strains. Appl Microbiol Biotechnol 99, 2191–2207, doi: 10.1007/s00253-014-6002-2 (2015). [DOI] [PubMed] [Google Scholar]
- Hernandez M. A., Arabolaza A., Rodriguez E., Gramajo H. & Alvarez H. M. The atf2 gene is involved in triacylglycerol biosynthesis and accumulation in the oleaginous Rhodococcus opacus PD630. Appl Microbiol Biotechnol 97, 2119–2130, doi: 10.1007/s00253-012-4360-1 (2012). [DOI] [PubMed] [Google Scholar]
- Holder J. W. et al. Comparative and functional genomics of Rhodococcus opacus PD630 for biofuels development. PLoS Genet 7, e1002219, doi: 10.1371/journal.pgen.1002219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez A. F., Alvarez H. M., Kalscheuer R., Waltermann M. & Steinbuchel A. Cloning and characterization of a gene involved in triacylglycerol biosynthesis and identification of additional homologous genes in the oleaginous bacterium Rhodococcus opacus PD630. Microbiology 154, 2327–2335, doi: 10.1099/mic.0.2008/016568-0 (2008). [DOI] [PubMed] [Google Scholar]
- Chen Y. et al. Integrated omics study delineates the dynamics of lipid droplets in Rhodococcus opacus PD630. Nucleic Acids Res 42, 1052–1064, doi: 10.1093/nar/gkt932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila Costa J. S., Herrero O. M., Alvarez H. M. & Leichert L. Label-free and redox proteomic analyses of the triacylglycerol-accumulating Rhodococcus jostii RHA1. Microbiology 161, 593–610, doi: 10.1099/mic.0.000028 (2015). [DOI] [PubMed] [Google Scholar]
- MacEachran D. P., Prophete M. E. & Sinskey A. J. The Rhodococcus opacus PD630 heparin-binding hemagglutinin homolog TadA mediates lipid body formation. Appl Environ Microbiol 76, 7217–7225, doi: 10.1128/AEM.00985-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba M. S. & Alvarez H. M. Identification of a novel ATP-binding cassette transporter involved in long-chain fatty acid import and its role in triacylglycerol accumulation in Rhodococcus jostii RHA1. Microbiology 160, 1523–1532, doi: 10.1099/mic.0.078477-0 (2014). [DOI] [PubMed] [Google Scholar]
- Patrauchan M. A. et al. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. J Bacteriol 187, 4050–4063, doi: 187/12/4050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Llorens J. M. et al. Phenylacetate catabolism in Rhodococcus sp. strain RHA1: a central pathway for degradation of aromatic compounds. J Bacteriol 187, 4497–4504, doi: 187/13/4497 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago G., Diacovich L., Arabolaza A., Tsai S. C. & Gramajo H. Fatty acid biosynthesis in actinomycetes. FEMS Microbiol Rev 35, 475–497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Witkowski A. & Joshi A. K. Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res 42, 289–317. (2003). [DOI] [PubMed] [Google Scholar]
- Sutcliffe I. C., Brown A. K. & Dover L. G. In In Biology of Rhodococcus (ed Alvarez H. M.) 30–60 (Microbiology Monographs Series, Springer, 2010). [Google Scholar]
- MacEachran D. P. & Sinskey A. J. The Rhodococcus opacus TadD protein mediates triacylglycerol metabolism by regulating intracellular NAD(P)H pools. Microb Cell Fact 12, 104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson R. W. & Reshef L. Glyceroneogenesis revisited. Biochimie 85, 1199–1205 (2003). [DOI] [PubMed] [Google Scholar]
- Kuznetsova E. et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem 281, 36149–36161, doi: M605449200 (2006). [DOI] [PubMed] [Google Scholar]
- Alvarez H. M., Mayer F., Fabritius D. & Steinbuchel A. Formation of intracytoplasmic lipid inclusions by Rhodococcus opacus strain PD630. Arch Microbiol 165, 377–386 (1996). [DOI] [PubMed] [Google Scholar]
- Alvarez H. M., Kalscheuer R. & Steinbüchel A. Accumulation of storage lipids in species of Rhodococcus and Nocardia and effect of inhibitors and polyethylene glycol. Fett/Lipid 99, 239–246 (1997). [Google Scholar]
- Feisthauer S. et al. Differences of heterotrophic 13CO2 assimilation by Pseudomonas knackmussii strain B13 and Rhodococcus opacus 1CP and potential impact on biomarker stable isotope probing. Environ Microbiol 10, 1641–1651, doi: 10.1111/j.1462-2920.2008.01573.x (2008). [DOI] [PubMed] [Google Scholar]
- Alvarez H. M. & Steinbuchel A. Triacylglycerols in prokaryotic microorganisms. Appl Microbiol Biotechnol 60, 367–376, doi: 10.1007/s00253-002-1135-0 (2002). [DOI] [PubMed] [Google Scholar]
- Alvarez H. In Triglycerides and cholesterol research Vol. 6 (ed Welson L. T.) 159–176 (Nova Science, New York, 2006). [Google Scholar]
- Alvarez H. M. Relationship between b-oxidation pathway and the hydrocarbon-degrading profile in actinomycets bacteria. Int Biodeter Biodegradation 52, 35–42 (2003). [Google Scholar]
- Sassetti C. M. & Rubin E. J. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA 100, 12989–12994, doi: 10.1073/pnas.2134250100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. E. et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7, e1002251, doi: 10.1371/journal.ppat.1002251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. & Russell D. W. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY 2001). [Google Scholar]
- van der Geize R., Hessels G. I., van Gerwen R., van der Meijden P. & Dijkhuizen L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Delta1-dehydrogenase, In Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol Lett 205, 197–202, doi: S0378109701004645 (2001). [DOI] [PubMed] [Google Scholar]
- Dong L., Nakashima N., Tamura N. & Tamura T. Isolation and characterization of the Rhodococcus opacus thiostrepton-inducible genes tipAL and tipAS: application for recombinant protein expression in Rhodococcus. FEMS Microbiol Lett 237, 35–40, doi: 10.1016/j.femsle.2004.06.012 (2004). [DOI] [PubMed] [Google Scholar]
- Nakashima N. & Tamura T. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl Environ Microbiol 70, 5557–5568 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod M. P. et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA 103, 15582–15587, doi: 10.1073/pnas.0607048103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. & Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M. & Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleff F. In Methods of Seawater Analysis (ed In Grasshoff K.) 126–133 (Verlag Chemie, 1976). [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402, doi: 10.1093/nar/25.17.3389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Higgins D. & Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.