Abstract
Larger organisms tend to live longer, have more potentially carcinogenic cells, and undergo more cell divisions. While one might intuitively expect cancer incidence to scale with body size, this assertion does not hold over the range of different mammals. Explaining this lack of correlation, so-called ‘Peto’s paradox’ can likely increase our understanding of how cancer defense mechanisms are shaped by natural selection. Here, we study the occurrence of microsatellite in mammal genomes and observe that animals with expanded body size restrain the number of microsatellite. To take into account of higher mutation rate in the microsatellite region compared to that of genome, limiting the abundance of somatic mutations might explain how larger organisms could overcome the burden of cancer. These observations may serve as the basis to better understand how evolution has modeled protective mechanisms against cancer development.
Cancer is a unique type of genetic disease in which several sequential mutations are necessary, and each mutation drives a wave of cellular proliferation which in turn leads to gradual increases in tumor size, disorganization and malignancy1. As cancer arises through the accumulation of mutations, each proliferating cell is at risk of malignant transformation, assuming all cells have similar chances of mutation2. Cancer risk is thus expected to increase with larger bodies and longer lifespan, but there appears to be disconnect between prediction and observation across species, a phenomenon termed Peto’s paradox3.
There are many hypotheses but limited research efforts to resolve this paradox. Although large bodies evolved independently, some probable and common mechanisms of the effective cancer suppression in large species include lower somatic mutation rates, redundancy of tumor suppressor genes, lower selective advantage of mutant cells, more efficient immune system, shorter telomeres, and fewer reactive oxygen species due to lower basal metabolic rate2. Katzourakis et al. also suggested that lower levels of tumorgenic endogenous retroviruses in larger bodied species could be the result of evolution of mechanisms capable of limiting retroviral activity4. Recently, Varki & Varki provided several explanations for the reported rarity of carcinomas in captive chimpanzees such as differences in diet, their microbiome, and potential environmental factors5. Abegglen et al. reported that elephants, compared with human, appeared to have multiple copies of tumor suppressor gene (TP53) and also increased level of apoptotic response after DNA damage, which are potential molecular mechanisms of cancer resistance6.
Genomes are scattered with numerous simple repeats, and tandem repeats are iterations of repeat units of any size, from a single base pair to thousands of base pairs. The major types of microsatellites are mono-, di-, tri- and tetranucleotide repeats, but units of five or six nucleotides are also classified as microsatellites7. These are among the most variable types of DNA sequence in the genome8, and genetic variation at many microsatellite loci is characterized by high heterozygosity and the presence of multiple alleles7. Notably, the vast number of mutations in cancer cells were directly associated with changes in microsatellites in tumor DNA9. The cancer patients harbor mutations in mismatch repair genes10,11, which leads to failure to correct slippage errors made by DNA polymerases and consequently to give rise to the length changes, microsatellites instability7. It seems evident that repetitive elements are “hot spots” for mutagenesis and may serve as markers for detecting other types of mutations throughout the genome9,12.
In this sense, relating two seemingly disparate contexts, Peto’s paradox and microsatellites across species, may lead to conceptual advances in understanding the mechanisms underlying the animals that have been evolving mechanisms to suppress cancer ever since the origin of multicellularity. In the light of comparative oncology, we explore the hypothesis that differences in microsatellite occurrence across mammalian species have been shaped by natural selection, with larger animals expected to have smaller number of microsatellites in the genome.
Results and Discussion
We investigated the genome-wide microsatellites (defined as di-, tri-, tetra-, penta-, hexa-nucleotide repeats) across 31 mammalian species (Supplementary Table S1) using RepeatMasker13. It is previously suggested that abundance of microsatellite tends to positively correlate with genome size among a variety of eukaryotes, whereas occurrence of microsatellite is negatively correlated with genome size in plants7,14,15,16. In mammals in particular, it was evident that the total number of microsatellite does not correlate with genome size (P-value = 0.13) (Supplementary Fig. S1).
Microsatellites can be found anywhere in the genome, both in protein-coding and noncoding regions. Due to their high mutability, microsatellites are thought to play a major role in genome evolution by creating and maintaining quantitative genetic variation14,17. To understand the selective landscapes in which species evolved in terms of occurrence of microsatellite, we used linear regression to test association between number of microsatellites and body mass. As our surrogate measure of relative level of total number of cells present in each organism, we followed previous studies in the use of body mass4,18. We observed a significant negative correlation (slope = −0.042, P-value = 2.0E-04 and R2 = 0.36), indicating that the number of microsatellites in the whole genome is smaller in species with larger body size (Fig. 1 and Table 1). As multicellular organism expanded the body size, the challenge of suppressing somatic evolution dramatically increases; however, that challenge was successfully tackled in terms of abundance of microsatellite, in which mutation rate is higher than in genome.
Figure 1. Number of microsatellites in genome against body mass in 31 mammalian species.
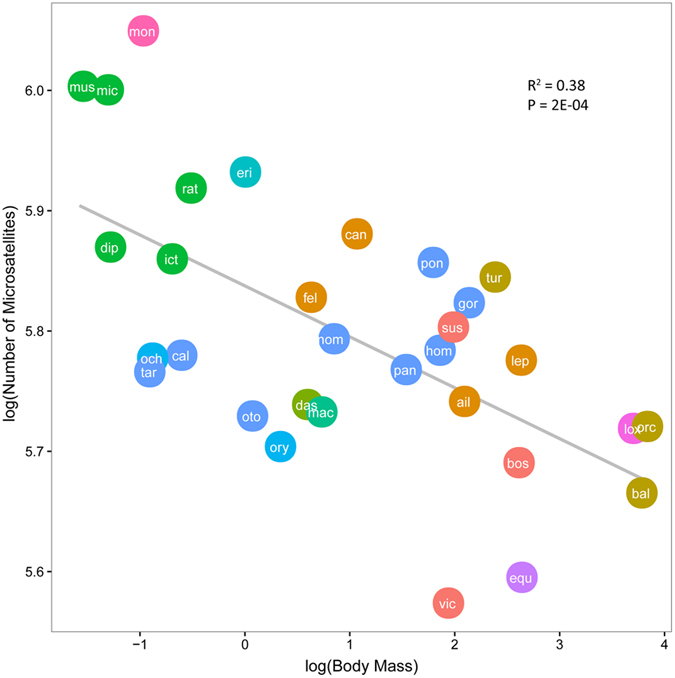
Both traits are log-transformed, and different colors denote different orders.
Table 1. Relationships between number of microsatellites and life history traits in non-phylogenetic models.
| Dependent variable | Simple linear regression |
Multiple linear regression |
||||
|---|---|---|---|---|---|---|
| df1 | slope(beta) | R2 | P-value | slope (beta) | P-value | |
| body mass | 29 | −0.042 | 0.38 | <0.001 | −0.042 | 0.023 |
| 2temperature-corrected mass-specific basal metabolic rate | 29 | 0.047 | 0.25 | <0.01 | 0.00077 | 0.97 |
1df denotes degree of freedom
2activation energy of E = 0.65 was used to correct for temperature. Results from other E ranges are shown in Supplementary Table S2.
As life history traits are often correlated each other, it remains possible that the apparent correlation of body mass with microsatellite occurrence could be confounded by other life history traits. There is increasing evidence that variations in rates of nucleotide substitution show relationships with body size18,19,20 and metabolic rate17. Higher metabolic rates, associated with reactive oxygen species (ROS) and metabolic stress along with other by-products of metabolism, can lead to tumorigenesis and appear to be inversely proportional to animal body size21. Hence, clarifying if other traits have played a role in determining the number of microsatellites is very crucial in assessing the effect of body mass correctly.
We evaluated the correlation between microsatellite abundance and life history traits with multiple regression model to account for their simultaneous contributions while controlling for potential confounders. In addition, as temperature is known to affect metabolic rate22, following the previous study, we considered temperature-corrected mass-specific metabolic rate23 as a confounding variable in the model. Body size still remained as the only significant variable confirming that it is the most significant predictor of microsatellite density, which in turn indicates that observed correlation between abundance of microsatellite and body mass is robust against variations in temperature and metabolic rate (Table 1). The result was consistent when mass-specific metabolic rate (not temperature-corrected) was included in the model (Supplementary Table S2).
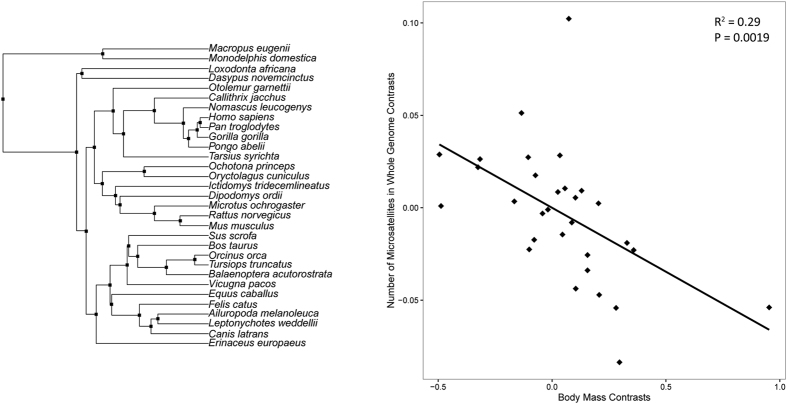
However, this conclusion still may be premature without phylogenetic comparative analyses of evolution in microsatellites occurrence. When species are used as data points, relationships between raw values of any traits are difficult to interpret, because shared phylogenetic history means that assumptions of statistical independence are likely to be violated24,25,26. It had been demonstrated that such approaches may lead to overestimation, excessively high type I error rates and inaccurate estimations of correlations or slopes27,28. The correlation was thus re-evaluated in a phylogenetic context. After correcting for phylogenetic proximity, the independent contrasts of body mass versus number of microsatellites were correlated significantly and negatively (slope = −0.069, P-value = 0.0019) (Fig. 2). Although this method is also limited for loss of statistical power and its reliance on the assumption of constant rates of trait evolution through time29,30, a consistent result supports the evidence that microsatellite abundance is significantly associated with body size.
Figure 2. Phylogenetic independent contrasts of body mass versus number of microsatellites in whole genome region.
(a) Rectangle indicates the contrasts in the phylogeny (Canis familiaris was replaced with Canis latrans in this analysis) (b) Relationship between phylogenetically independent contrasts of body mass and contrasts of number of microsatellites in whole genome region.
Comparing genomic regions of interest for cancer research such as proto-oncogenes, tumor suppressor genes or whole protein-coding genes, widespread in mammalian genomes can provide important insights into how these classes of genes have been in subject to natural selection31. We first observed that body mass still contributed significantly (P-value < 0.05) to the microsatellite occurrence within genic region under both simple (Supplementary Fig. S2) and multiple regression models (Supplementary Table S3) but not under comparative phylogenetic analysis. As we focus our attention to proto-oncogene and tumor suppressor genes, a negative trend was observed between microsatellite abundance and body mass, but the correlation was not highly significant (slope = −0.059, R2 = 0.12, P-value = 0.076) (Supplementary Fig. S3). More complete results can be expected with better quality of genome annotation and better definition of proto-oncogene and tumor suppressor genes across species (for example, different numbers of copies of the genes can also alter the level of cancer resistance6,32). Interestingly, common minke whale characterized itself as a stricter regulator than any other species. Common minke whale seemed to very extremely suppress the occurrence of microsatellites in genic region, proto-oncogene and tumor suppressor gene regions where the accumulated mutations can cause the cancer developments at relatively higher chance.
The extension of evolutionary thinking into cancer biology has contributed to realization that cancer defenses both between tissues within individual and between species have been influenced by natural selection33. Our results indicate that larger mammals tend to exert more effective control over microsatellite occurrence throughout the genome. We suggest that a driving force for this restraint in larger and longer-lived animals is their higher expected cancer rates given the number of cells and number of cell divisions that occur.
Methods
Data and identifications of microsatellite
We downloaded 30 complete mammalian genomes from the NCBI and USCS databases and the assembled whole genome of common minke whale34 to finally retain 31 species. Microsatellites were identified (masked as simple repeats) using RepeatMasker version 4.0.5 (http://repeatmasker.genome.washington.edu)13 with the “no_is” parameter to skip bacterial insertion element check. The human proto-oncogene and tumor suppressor gene were retrieved from the UniprotKB (KW-0656 and KW-0043 respectively)35 and found the orthologous genes across 27 publically available species using Ensembl database36. We then used BLAST to search the genomic sequence of the orthologous gene for common minke whale to finally retain the lowest common number of 27 gene set throughout 28 mammalian species37.
Multiple linear regression analysis and Phylogenetic comparative analyses
Life history traits correlate with each other, and thus body size could in principle be a surrogate measure of a different life history trait, as has been previous shown for body temperature and metabolic rate. Mammalian life history data (Supplementary Table S1) was mainly taken from PanTHERIA database38 and the phylogenetic tree from TimeTree39. We used the phylogenetically independent contrasts (PIC) approach as implemented by the Analysis of Phylogenetic and Evolution (APE)40 package in R version 3.2.2 (http://cran.r-project.org/) to control for shared ancestry26. To correct for the effect of temperature in metabolic rate, mass-specific metabolic rate of each species was transformed to 25 °C, following the previous study23. A range of average activation energy (E = 0.4, 0.65 and 0.8) was considered, and the result was robust (Supplementary Tables S2 and S3). The relationship between the standardized independent contrasts were then investigated through ordinary least squares regression analysis, with regression lines constrained to pass through the origin28. Traits were log-transformed in all regression analyses. The lm function in R was used to perform regression analyses.
Additional Information
How to cite this article: Park, J. Y. et al. Evolutionary constraints over microsatellite abundance in larger mammals as a potential mechanism against carcinogenic burden. Sci. Rep. 6, 25246; doi: 10.1038/srep25246 (2016).
Supplementary Material
Acknowledgments
This work was funded by a grant from the National Fisheries Research and Development Institute (NFRDI), contribution number RP-2015-BT-009. This research was also supported by a grant from Marine Biotechnology Program (PJT200620, Genome Analysis of Marine Organisms and Development of Functional Applications) funded by Ministry of Oceans and Fisheries, Korea.
Footnotes
Author Contributions J.P., H.K., S.C. and J.K. conceived this study and participated in the study design. Y.A., C.A., J.K. and E.K. conducted the sample selection and collected data. J.P., S.C. and J.K. performed statistical analysis and bioinformatics investigation. All the authors contributed to the final version of the manuscript.
References
- Vogelstein B. & Kinzler K. W. The multistep nature of cancer. Trends Genet. 9, 138–141 (1993). [DOI] [PubMed] [Google Scholar]
- Caulin A. F. & Maley C. C. Peto’s Paradox: evolution’s prescription for cancer prevention. Trends Ecol. Evol. 26, 175–182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R., Roe F., Lee P., Levy L. & Clack J. Cancer and ageing in mice and men. Br. J. Cancer 32, 411 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A. et al. Larger mammalian body size leads to lower retroviral activity. PLoS Pathog 10, e1004214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki N. M. & Varki A. On the apparent rarity of epithelial cancers in captive chimpanzees. Phil. Trans. R. Soc. B 370, 20140225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abegglen L. M. et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 314, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nature reviews genetics 5, 435–445 (2004). [DOI] [PubMed] [Google Scholar]
- Weber J. L. Informativeness of human (dC-dA)n (dG-dT)n polymorphisms. Genomics 7, 524–530 (1990). [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Loeb K. R. & Anderson J. P. Multiple mutations and cancer. Proceedings of the National Academy of Sciences 100, 776–781 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionov Y., Peinado M. A., Malkhosyan S., Shibata D. & Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363, 558–561 (1993). [DOI] [PubMed] [Google Scholar]
- Fishel R. et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75, 1027–1038 (1993). [DOI] [PubMed] [Google Scholar]
- Boland C. R. et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 58, 5248–5257 (1998). [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth G., Gáspári Z. & Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10, 967–981 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. M. Simple sequences and the expanding genome. Bioessays 18, 421–425 (1996). [DOI] [PubMed] [Google Scholar]
- Morgante M., Hanafey M. & Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30, 194–200 (2002). [DOI] [PubMed] [Google Scholar]
- Kashi Y., King D. & Soller M. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 13, 74–78 (1997). [DOI] [PubMed] [Google Scholar]
- Amos W. & Filipe L. N. Microsatellite frequencies vary with body mass and body temperature in mammals, suggesting correlated variation in mutation rate. PeerJ 2, e663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. P. & Palumbi S. R. Body size, metabolic rate, generation time, and the molecular clock. Proceedings of the National Academy of Sciences 90, 4087–4091 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly J. F., Allen A. P., West G. B. & Brown J. H. The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proceedings of the National Academy of Sciences of the United States of America 102, 140–145 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V. A metabolic perspective of Peto’s paradox and cancer. Phil. Trans. R. Soc. B 370, 20140223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly J. F., Brown J. H., West G. B., Savage V. M. & Charnov E. L. Effects of size and temperature on metabolic rate. science 293, 2248–2251 (2001). [DOI] [PubMed] [Google Scholar]
- Takemoto K. & Kawakami Y. The proportion of genes in a functional category is linked to mass-specific metabolic rate and lifespan. Scientific Reports 5, 10008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney K. D. & Garland T. Did genetic drift drive increases in genome complexity. PLos genet 6, e1001080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (1985). [DOI] [PubMed] [Google Scholar]
- Garland T., Bennett A. F. & Rezende E. L. Phylogenetic approaches in comparative physiology. J. Exp. Biol. 208, 3015–3035 (2005). [DOI] [PubMed] [Google Scholar]
- Garland T. & Janis C. M. Does metatarsal/femur ratio predict maximal running speed in cursorial mammals? J. Zool. 229, 133–133 (1993). [Google Scholar]
- Garland T., Midford P. E. & Ives A. R. An introduction to phylogenetically based statistical methods, with a new method for confidence intervals on ancestral values. Am. Zool. 39, 374–388 (1999). [Google Scholar]
- Griffith O., Moodie G. & Civetta A. Genome size and longevity in fish. Experimental gerontology 38, 333–337 (2003). [DOI] [PubMed] [Google Scholar]
- Freckleton R., Harvey P. & Pagel M. Phylogenetic dependence and ecological data: a test and review of evidence. Am. Nat 160, 716–726 (2002). [DOI] [PubMed] [Google Scholar]
- Roche B. et al. Natural resistance to cancers: a Darwinian hypothesis to explain Peto’s paradox. BMC Cancer 12, 387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi V. A. et al. The origins and evolution of the p53 family of genes. Cold Spring Harbor perspectives in biology 2, a001198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney L., Maley C. C., Breen M., Hochberg M. E. & Schiffman J. D. Peto’s paradox and the promise of comparative oncology. Phil. Trans. R. Soc. B 370, 20140177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. Y. et al. Cetaceans evolution: insights from the genome sequences of common minke whales. BMC Genomics 16, 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A. et al. The universal protein resource (UniProt). Nucleic Acids Res. 33, D154–D159 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T. et al. The Ensembl genome database project. Nucleic Acids Res. 30, 38–41 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. E. et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals: Ecological Archives E090-184. Ecology 90, 2648–2648 (2009). [Google Scholar]
- Zimin A. V. et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol 10, R42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J. & Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.