Abstract
Retinoblastoma is a rare childhood cancer initiated by RB1 mutation or MYCN amplification, while additional alterations may be required for tumor development. However, the view on single nucleotide variants is very limited. To better understand oncogenesis, we determined the genomic landscape of retinoblastoma. We performed exome sequencing of 71 retinoblastomas and matched blood DNA. Next, we determined the presence of single nucleotide variants, copy number alterations and viruses. Aside from RB1, recurrent gene mutations were very rare. Only a limited fraction of tumors showed BCOR (7/71, 10%) or CREBBP alterations (3/71, 4%). No evidence was found for the presence of viruses. Instead, specific somatic copy number alterations were more common, particularly in patients diagnosed at later age. Recurrent alterations of chromosomal arms often involved less than one copy, also in highly pure tumor samples, suggesting within-tumor heterogeneity. Our results show that retinoblastoma is among the least mutated cancers and signify the extreme sensitivity of the childhood retina for RB1 loss. We hypothesize that retinoblastomas arising later in retinal development benefit more from subclonal secondary alterations and therefore, these alterations are more selected for in these tumors. Targeted therapy based on these subclonal events might be insufficient for complete tumor control.
Retinoblastoma is a childhood cancer of the retina. Although the disease is relatively rare accounting for 2% of childhood cancers1, retinoblastoma is the most common intra-ocular malignancy in children2. From a clinical genetics perspective, three retinoblastoma types can be distinguished: familial (10%), sporadic heritable (30%) and non-heritable (60%). Patients with familial or sporadic heritable retinoblastoma have a germ line mono-allelic RB1 mutation and have acquired a second RB1 hit in the retina. While familial patients have inherited the mutant allele, sporadic heritable patients have acquired a de novo RB1 mutation. The majority of non-heritable retinoblastoma (95%) is also caused by bi-allelic inactivation of RB1 but in this case occurring through two subsequent somatic events in the developing retina. A minority of non-heritable retinoblastoma (2%) is caused by amplification of the oncogene MYCN3. Recently, chromothrypsis of chromosome 13 disrupting the RB1 locus has been described as an alternative mechanism for RB1 inactivation4. Possibly, 13q chromothrypsis accounts for the remaining patients for whom no RB1 or MYCN alterations can be found by Sanger sequencing, Multiplex Ligation-dependent Probe Amplification (MLPA) or RB1 promotor methylation assays.
Yet, while inactivation of RB1 in the developing retina is sufficient for neoplastic onset, it has been suggested that additional genetic alterations are required for malignant progression5. In agreement, based on comprehensive genome-wide next-generation sequencing (NGS) efforts, it was claimed that two to eight genetic alterations are required to drive tumorigenesis6. Throughout the last decade, useful insights about secondary genetic alterations in retinoblastoma have been obtained by studies profiling large (>50 Kb) somatic copy number alterations (SCNAs). Unlike many other cancers, very little is known about smaller genetic alterations (<50 Kb) that are typically identified by genome-wide NGS. To date, only 14 retinoblastoma samples have been profiled for single nucleotide variations (SNVs) and insertions and deletions (INDELs) in a genome-wide fashion7,8.
Our current study set out to determine the prevalence of SNV and INDELs in order to facilitate the identification of the genetic alterations that drive retinoblastoma development. To this end, whole-exome sequencing (WES) of a diverse set of retinoblastoma and matching blood DNA was performed (N = 71). Using dedicated bioinformatics and statistics, this dataset allowed for the identification of SNVs/INDELs, SCNAs including chromothrypsis, loss of heterozygosity (LOH) and viral content. By integrating genomics with clinical and histopathological data, our study aims to provide a detailed molecular landscape, which may help better understand retinoblastoma development.
Results
Identification of significantly mutated genes by somatic SNV/INDEL analysis
To identify genes relevant for retinoblastoma carcinogenesis, we compared SNVs/INDELs observed in 71 primary retinoblastoma samples with a variant database established from blood DNA of the same patients (N = 70, from one bilateral patient, one tumor sample per eye was analyzed). Sequencing, alignment and enrichment statistics are provided (Table S1). The number of exonic variants ranged between 18,510 and 23,772 variants per tumor sample, adding up to a total of 1,386,285 variants in the tumor cohort. By comparing tumor variants with the pooled blood variant database, only 5,797/1,386,285 (0.41%) variants were considered somatically acquired. Using the criteria for possible pathogenic variants (see Materials and methods, chapter 1.4), only 258/5,797 (4.45%) of the somatic variants remained (Table S2). These 258 somatic variants were distributed over 202 unique genes, of which only 3 genes were recurrently (i.e. at least two different patients) mutated; RB1 (51 variants, 44 patients), BCOR (5 variants, 5 patients), and CREBBP (2 variants, 2 patients). For tumors with SNVs/INDELs in RB1, tumor cellularity could be estimated based on the variant allele frequency (VAF). Patients that were compound heterozygous for RB1 (confirmed by conventional DNA diagnostics) had a mean VAF close to 0.5 (mean 0.48) and patients with homozygous RB1 mutations (confirmed by conventional DNA diagnostics) had a mean VAF close to 1 (mean 0.98), indicating high tumor cellularity in our cohort. Variant details for BCOR and CREBBP genes are given in Table 1. VAFs of BCOR and CREBBP were well below 1, indicating that not all alleles were mutated and therefore possibly not all tumor cells harbored these secondary variants. In short, although relatively lenient thresholds were used for pathogenicity and recurrence, aside from RB1, only two genes were identified as recurrently hit by somatic and possibly pathogenic mutations.
Table 1. Somatic and possibly pathogenic variants for genes that showed at least 2 variants, excluding RB1. DP = depth of coverage, VAF = Variant allele frequency.
| Gene | Type | Refseq cDNA annotation | DP | VAF | ID |
|---|---|---|---|---|---|
| BCOR | stopgain SNV | NM_001123384:c.T4472A:p.L1491X | 36 | 0.92 | T66 |
| BCOR | nonsynonymous SNV | NM_001123383:c.G3001C:p.E1001Q | 71 | 0.46 | T63 |
| BCOR | frameshift deletion | NM_001123384:c.3314delA:p.D1105fs | 82 | 0.50 | T61 |
| BCOR | stopgain SNV | NM_001123383:c.C2926T:p.R976X | 56 | 0.38 | T57 |
| BCOR | frameshift deletion | NM_001123384:c.4047_4053del:p.1349_1351del | 48 | 0.19 | T23 |
| CREBBP | nonsynonymous SNV | NM_001079846:c.T4308G:p.C1436W | 81 | 0.17 | T62 |
| CREBBP | nonframeshift deletion | NM_001079846:c.6629_6631del:p.2210_2211del | 11 | 0.27 | T47 |
In parallel to our custom defined filtering strategy MutSigCV9 was used, a tool purposed to find significantly mutated genes based on frequency and adjusting for sequence content and coverage. Somatic variants that were not located in segmental duplication regions, had at least 10x sequencing depth and were flagged “PASS” by the GATK variant filter were used as input for MutSigCV (2085 variants). Only RB1 was significantly mutated (q-value 3.35E-11) (Table S3). Although BCOR was not considered significantly mutated after multiple testing correction (p-value 1.30E-03, q-value 1), notably this gene ranked sixth in the MutSigCV gene list.
Somatic copy number alterations
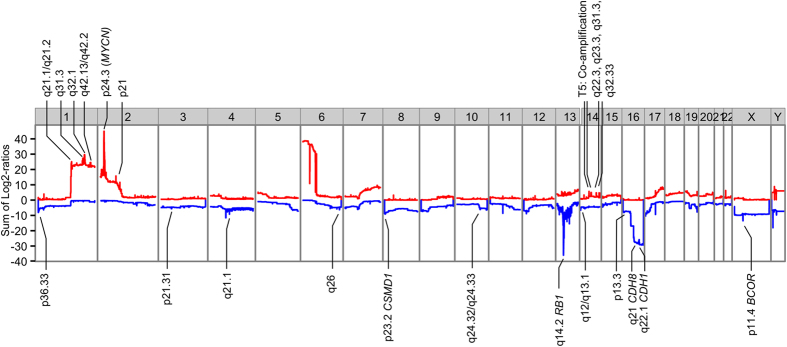
Although WES was primarily designed for the identification of exonic SNVs/INDELs, WES data can also serve well for copy number alteration profiling. Using off-target sequencing reads that are not subject to systematic biases due to bait hybridization efficiencies, we performed SCNA analysis implemented by CopywriteR10. Segmented results are available (Table S4) and when the file suffix is renamed to .seg it can be used for convenient browsing of the SCNA results in Integrative Genome Viewer (IGV)11. For each HGNC approved gene, the sum of Log2-ratios (calculated based on normalized read depth in tumor divided by matched germ line) is plotted along genomic coordinates (Fig. 1). The sum of Log2-ratios is proportional to the total number of acquired DNA copies in all tumor samples (N = 71) for the respective gene. To identify significantly altered genes, which appear as spikes in Fig. 1, GISTIC analysis was performed. Highlighted cytoband labels in Fig. 1 indicate regions identified by GISTIC as significantly altered. Putative target genes are highlighted only if a single protein-coding gene was located at the identified region. All significantly altered regions are given including RefSeq gene annotation (Table S5). One of the significantly altered regions was located at Xp11.4 and included BCOR exclusively. Instead of SNVs, two tumors (T22 and T67) showed focal homozygous loss of BCOR adding up to a mutations frequency of 7/71 (10%) tumors.
Figure 1. Significantly altered copy number regions identified by GISTIC.
A karyogram overview of the cumulative copy number changes (Log2-ratios of tumors versus germ line summed over 71 samples, Y-axis) is shown separately for gains (red) and losses (blue). Based on SCNA focality, amplitude and recurrence, GISTIC identified significantly altered regions, which are highlighted by cytoband labels. For regions that contained a single gene only, the gene symbol is given.
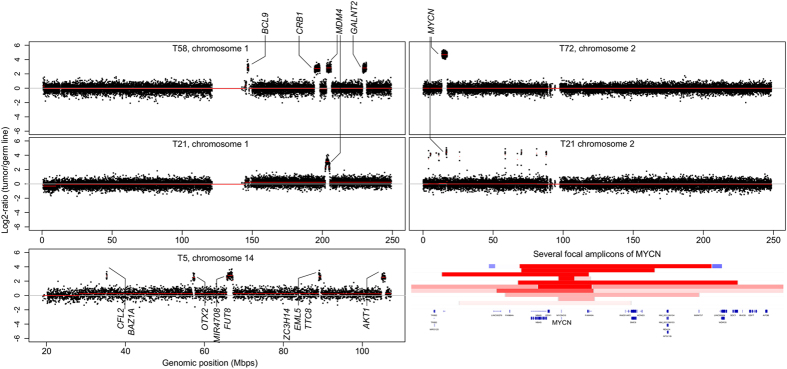
High-level amplifications (in our study defined by >10 copies, Log2-ratio >2.32) were observed at 1q, 2p and 14q and are visualized in more detail in Fig. 2. Amplifications of the four regions at 1q were divided over two tumors, one tumor (T58) with similar copy number for all four regions indicating co-amplification and one tumor (T21) with only 1q32.1 amplification including MDM4 and 26 other genes (Table S5). High-level amplification of 2p24.3 (MYCN) occurred in 6/72 (8%) tumors of which 4/6 (67%) lacked mutations in RB1 (RB1−/−MYCNA)3. The tumor with focal 1q32.1 amplification (T21) also showed focal MYCN-amplification and several additional focal amplifications at 2p with similar copy numbers, indicating co-amplification. One previously described tumor12 had high-level amplification of five focal amplifications at 14q with similar copy numbers as well (Log2-ratio 2.75, ploidy 2*22.75 = 13.5 copies). All high-level focal amplicons showed LOH, indicating that only one of the two alleles was amplified (haplotype amplification) consistent with earlier reports of MYCN amplification in neuroblastoma13.
Figure 2. Examples of high-level gains.
Segmented (orange lines) somatic copy number estimates (black dots, Log2-ratios, Y-axis) are plotted along genomic coordinates (X-axis). Focal and high-level gains were scarce and were restricted to chromosomes 1 (2/71 tumors), 2 (6/71) and 14 (1/71). Established oncogenes contained in the focal gains are highlighted. Only for chromosome 2, the intersection of amplicons included a single gene only (MYCN). Tumors T58, T21, and T5 showed co-amplification of multiple loci at chromosome 1, 2, and 14 respectively.
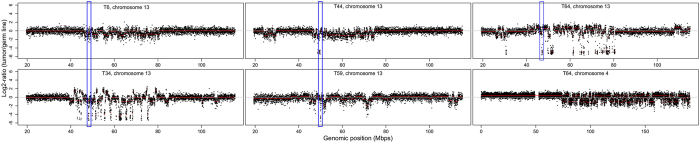
Six cases of chromothrypsis, defined as (sub-) chromosomal shattering, were observed in five different tumors (T6, T34, T44, T59, T64, Fig. 3) for chromosome arm 13q (5/6) and 4q (1/6). For each of the five 13q regions showing chromothrypsis, the RB1 allele was included, indicated by the blue rectangles in Fig. 3. For 2/5 tumors with chromothrypsis at 13q, SCNAs were all losses consistent with initial reports about chromothrypsis14 while for 3/5 tumors SCNAs were both gains and losses. Tumor T64 had chromothrypsis of both 13q and 4q.
Figure 3. Chromothrypsis of chromosome 4 (1/71 tumors) and 13 (5/71 tumors).
Segmented (orange lines) somatic copy number estimates (black dots, Log2-ratios, Y-axis) are plotted along genomic coordinates (X-axis). Chromothrypsis, characterized by clustered chromosomal alterations, was observed for 6 chromosomes in five different tumors. One tumor (T64) showed chromothrypsis at both chromosome 4 and 13. Blue rectangles indicate the RB1 locus.
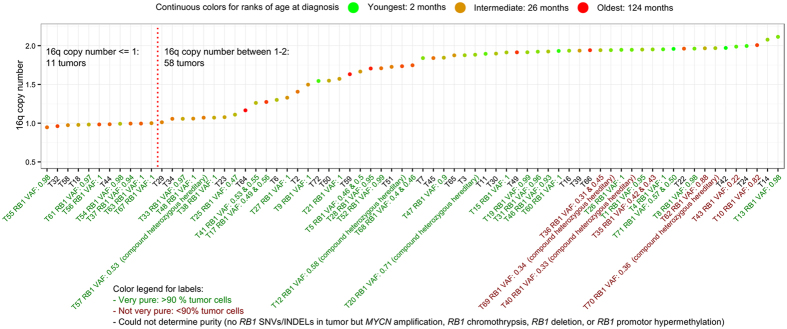
The most frequent large-scale SCNAs were gain of 1q, 2p, and 6p and loss of 16q, in agreement with published studies12,15,16,17,18,19,20,21. This SCNA profile is typical for retinoblastoma and is not observed in other cancers22, indicating that these alterations may confer a growth advantage for retinoblastoma lineages specifically. For the majority of 1p, 2p, 6p, and 16q alterations (91/132, 69%, Table S6), the SCNA did not exceed change of one copy. For illustration, copy numbers of 16q are visualized for each tumor (Fig. 4). 69/71 (97%) of the tumors showed 16q copy number less than two, although only 11/71 (15%) showed copy number below one. VAFs of RB1 are included in the labels and indicate the high tumor cellularity for the majority of the cohort, meaning contamination of non-cancer cells cannot explain incomplete loss of 16q. The remaining explanation is that not all tumor cells showed 16q loss indicating within-tumor heterogeneity for the majority of the cohort. The amplitude of 16q loss may reflect the fraction of cells with 16q loss.
Figure 4. Amplitudes of 16q loss indicate tumor clonality.
For each tumor (dots) the copy number of 16q is plotted, ordered by increasing copy number. A green-to-red color scale was mapped to age at diagnosis, showing a significant positive association between age at diagnosis and 16q loss amplitude (Kendall’s rank correlation test, p-value 1.2E-06). The tumor labels included VAFs of RB1 variants from which contamination with non-tumor cells can be inferred. Although the majority of tumors were considered very pure (>90%, green labels), 16q loss did rarely reach change of one copy (11 tumors with ploidy <=1), suggesting of within-tumor heterogeneity.
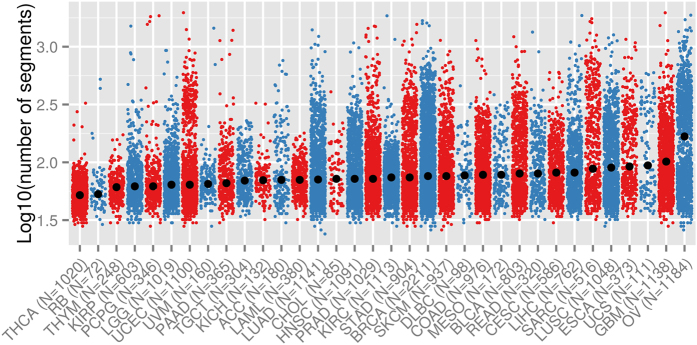
By comparing SCNAs of 46 retinoblastoma samples made by single nucleotide polymorphism (SNP) arrays with 153 high-grade serous ovarian cancer, it was claimed that retinoblastoma genomes are remarkably stable20. To compare the level of SCNAs of retinoblastoma with other cancer types, the total level of genomic disturbance of our cohort was compared with all tumor samples available at The Cancer Genome Atlas (TCGA, N = 22,455 tumors)23. For each individual tumor the total genomic disturbance, represented by the number of DNA segments with equal copy number, is plotted per tumor type (Fig. 5). The tumor types are ordered by the median of total genomic disturbance, confirming that retinoblastoma genomes attained relatively few SCNAs compared to other cancers. It is unclear whether this is a characteristic of pediatric cancer as no cancers types that exclusively occur in children are covered in the TCGA data set.
Figure 5. Number of SCNAs across different cancer types.
For each tumor (dots) the number of contiguous DNA segments (so wild type is 22 + X + Y = 24 segments, log10(24) = 0.38) is plotted for our retinoblastoma cohort and all cancer types available at TCGA in alternating colors. Cancer types are ordered by increasing median of SCNAs, showing that the retinoblastoma genome has relatively few SCNAs compared with cancers available in TCGA.
Validation of identified SNVs/INDELs and SCNAs by RB1
To demonstrate the sensitivity and specificity of the used sequencing methodology for somatic and pathogenic variant detection, WES was compared with standard molecular RB1 diagnostics based on Sanger sequencing and multiplex ligation-dependent probe amplification analysis. For heredity determination, blood DNA diagnostics was performed for all patients, except for T29 (foreign patient). For a subset of patients (36/71, 44%), tumor RB1 diagnostics was performed as well, to aid diagnosis of retinoblastoma heredity. Using a combination of previously described SNV/INDEL and SCNA analysis, the primary disease causing genetic event (RB1 or MYCN alterations) was determined based on WES data and compared with conventional RB1 diagnostics (Supplementary Fig. 1).
For 59/71 (83%) tumors, RB1 inactivation (either by SNV/INDEL or SCNA detection) or high-level focal amplification of MYCN was detected by WES (WES positives). In 31/59 (53%) of WES positives, conventional tumor RB1 diagnostics was performed and the mutation detected by WES was exactly the same as identified by conventional diagnostics, indicating a high true positivity rate. For 28/59 (47%) of WES positives, no tumor DNA diagnostics was available. However, for 6 WES positives without tumor DNA diagnostics, a germ line mutation in RB1 was identified that was always consistent with the somatic RB1 mutation detected by WES, again indicating a high true positivity rate. However, for tumor T7 an RB1 variant was detected by germ line DNA diagnostics in exon that was not detected by WES in the tumor (high GC content, poor coverage), although WES data showed LOH at the RB1 allele. Since, tumor T7 had acquired focal high-level gain of MYCN, based on WES data this amplification would have been falsely considered the primary event.
For 12/71 (17%) tumor samples, no RB1 mutations or MYCN amplification could be found by WES (WES negatives). For 4/12 WES negatives, tumor DNA diagnostics was performed. For one tumor (T16), indeed no second RB1 hit or MYCN amplification was found by conventional DNA diagnostics (true negative). For the other 3 WES negatives, RB1 mutations were found by conventional diagnostics including a tumor with promotor hypermethylation followed by LOH, one tumor with deletion of the promotor region and subsequent LOH and one tumor with compound heterozygous splice-site INDELs that were not annotated by Annovar in the WES-data. For the remaining 8/12 WES negatives, no tumor DNA diagnostics was performed but for 6 of them, a germ line mutation was found. All these 6 patients that were WES negative but had a germ line RB1 mutation showed clear LOH at the RB1 allele in the WES data. Apparently, somatic mutations of RB1 were missed by WES for these 6 tumors. One patient had a pathogenic deep intronic mutation (intron 18 inversion and splice site mutation), three had complicated insertions and two had mutations that were missed by WES because of thresholds for calling variants homozygous (RB1 variant in T39 VAF 81% and in T45 VAF 78%) and therefore somatic.
In short, all RB1 variants identified by WES that passed criteria to be called somatic and pathogenic were confirmed by conventional methods. Assuming that all tumors in our cohort either had RB1-inactivation or MYCN amplification, the sensitivity of WES was 83% (59/71). Incomplete sensitivity can be explained by epigenetic events, complex insertions, variants outside WES target areas and used thresholds for calling variants somatic.
Quantification of viral traces in retinoblastoma tumor and blood
After an HIV outbreak in Zambia, the incidence of retinoblastoma was increased, suggesting a role for oncoviruses in retinoblastoma development24. It was also shown that the use of barrier methods of contraception, associated with decreased risk for human papilloma virus (HPV) infection, significantly decreased the risk of having a child affected by retinoblastoma25. Since HPV is known to inhibit the RB1 pathway26,27,28, HPV infection is a plausible driver for retinoblastoma development alternative to - or in concert with RB1 mutations. However, controversies exist surrounding the presence of viruses in retinoblastoma tissue29,30. To test whether viral DNA was present in our tumor cohort, we aligned all reads to all available viral reference genomes (2547 viruses) and counted the number of well-mapped reads per virus. As a positive control for the detection of viruses in our WES data cohort, the Enterobacteria phage phiX174 sensu lato (phiX174) virus was used. The phiX174 virus DNA was spiked into sequencing libraries to guarantee enough library complexity required for proper Illumina sequencing. Virtually all reads that mapped to viral reference genomes (99.8%) mapped to phiX174. The remaining viral reads (0.2%) mapped to 26 viruses, which have not been associated with cancer previously (Table S7). In all, no evidence could be found for the presence of tumor viruses in our retinoblastoma cohort.
Testing for mutual exclusivity of identified somatic events and correlation with phenotype data
The identification of negative correlations between genetic events can give important insights into the molecular signaling pathways that underlie carcinogenesis. For example, the RB1 pathway is inactivated in all glioblastoma tumors but is caused either by somatic RB1 mutations, CDKN2A/B mutations, or CDK4 amplification31. The power of analyses aimed to identify negative correlations depends on the overall sample size and the frequency of occurrence of the event pairs. To be more specific, if two events rarely occur, a relatively big sample size is required to detect a significant negative correlation. Since retinoblastoma is a rare disease and identified genetic events beyond RB1 inactivation appear to be rare except for large SCNAs, identification of mutually exclusive events might be challenging.
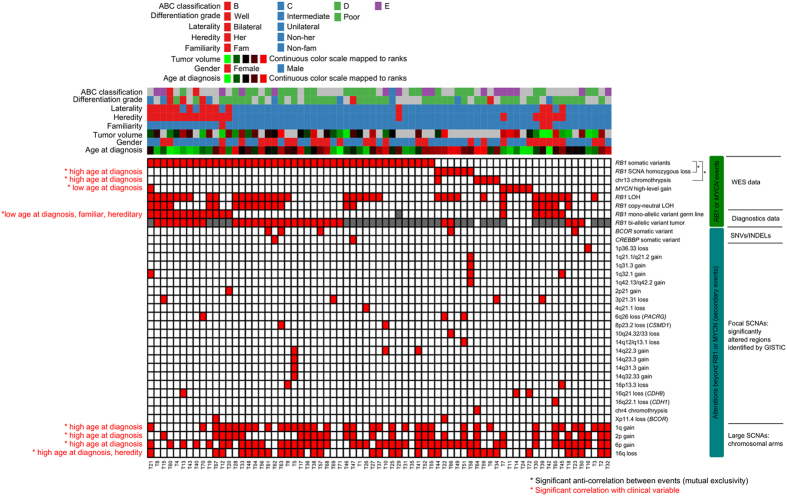
Nevertheless, to identify negative correlations and ultimately signaling pathways, a binary event matrix was compiled from the identified SNV/INDEL and SCNA analysis (Fig. 6). For each pair-wise combination of somatic events, the phi correlation coefficient was calculated and tested for significance. After correcting for multiple hypothesis testing, significant negative correlations were only found between RB1 SNVs/INDELs with either RB1 loss (FDR = 0.01) or chromothrypsis at chromosome 13 (FDR = 0.03). High-level focal amplification of MYCN and RB1 mutations by was not considered significantly inversely correlated because tumor T21 acquired both alterations (single nucleotide frameshift deletion in exon 15 of RB1; NM_000321:c.1397delA:p.E466fs). Also for tumor T7, from a non-familial unilateral patient, both high-level amplification of MYCN and RB1 inactivation was observed (20 nucleotides frameshift insertion in exon 1; NM_000321:c.25ins20:pT9fs), although RB1 inactivation was missed by WES because of low exon 1 coverage. However, LOH at the RB1 locus was clearly identified through WES which is a strong indication of RB1 inactivation.
Figure 6. Landscape of somatic alterations in retinoblastoma.
A binary event matrix (red: event occurred, white: event did not occur, grey: data not available) for SNVs/INDELs and SCNA, aligned with color-coded sample information. Columns represent tumor samples and rows represent events. For focal SCNAs containing a single gene only, the gene symbol is given in parentheses. Events that showed significant negative correlation are paired with black lines highlighted on the right of the matrix. Significant correlations between events and clinical variables are indicated by red labels on the left of the matrix. For large SCNAs (1p, 2p, 6p, and 16q), tumors with a mean ploidy more than 2.3 or less than 1.7 were called altered (red boxes).
Each event in the binary event matrix was tested for statistical association with the phenotypic variables aligned on top of the event matrix (Fig. 6). The phenotypic variable that correlated with most of the events was age at diagnosis. While focal high-level MYCN amplification and RB1 germ line mutations by DNA diagnostics were associated with young age at diagnosis, homozygous RB1 loss, chromothrypsis of chromosome 13, and alterations of chromosomal arms were associated with old age at diagnosis. By definition, RB1 germ line mutations detected by DNA diagnostics were associated with familial and heritable retinoblastoma. No significant relations between SNVs/INDELs of BCOR or CREBBP with clinical variables were found.
Discussion
The introduction of NGS enabled researchers to study genetics at unprecedented speed and dramatically reduced costs32. In particular, cancer research has benefited from the NGS development and as a result, genetic defects underlying many cancer types were revealed33. However, retinoblastoma is largely underrepresented in whole-exome/genome sequencing efforts with only 14 retinoblastoma samples profiled to date4,20. Our study is the first WES study that determined the somatic genomic landscape of retinoblastoma. Considering the low incidence of retinoblastoma, the sample size of our study was relatively large and therefore enabled powerful analyses.
Genes recurrently hit by SNVs/INDELs aside from RB1 were restricted to BCOR and CREBBP and were only observed in the minority (5/71 (7%) and 2/71 (3%) tumors respectively) of tumors. Somatic pathogenic BCOR mutations were reported in retinoblastoma before with similar frequency (6/46 tumors, 13%)20. For the BCOR gene, next to 4 truncating (two stop-gain mutations, two frameshift deletions) and one non-synonymous SNV, homozygous somatic copy number losses were observed in two tumors (T22, and T67). This indicates that the observed BCOR mutations are loss-of-function mutations, in agreement with reports about deleterious BCOR mutations in acute myeloid leukemia34. Germ line mutations in BCOR can give rise to two X-linked syndromes characterized by microphthalmia49 and other malformations: Lenz microphthalmia and oculofaciocardiodental (OFCD) syndrome35, characterized by retarded eye growth. The function of BCOR appears to be crucial during early development, since knock-down of BCOR expression during embryogenesis in drosophila caused severe developmental perturbations of the eye, skeleton and central nervous system. Given that BCOR gene function is associated with proper eye development, it seems plausible that inactivation of this gene can be related to retinoblastoma.
For an even smaller fraction of our cohort (2/72, 3% tumors), CREBBP mutations were found. Heterozygous germ line mutations in CREBBP are known to cause the Rubinstein-Taybi syndrome (RSTS)36, characterized by mental retardation, growth retardation and distinct dysmorphology. There have been several reports of RTST patients with rare pediatric neural tumors37. Analysis of CREBBP mutations in RSTS patients showed that the mutations were concentrated (44.7% of RSTS patients) in the histone acyltransferase (HAT) domain (295/2442 (12%) of amino acids make up the HAT domain in CREBBP)38, indicating that the HAT domain is crucial for proper CREBBP function. The somatic CREBBP mutation in sample T62 is located in the most highly conserved (11 organisms, up to frog) region of the HAT domain. The CREBBP mutation in tumor T47 was not located in any of the CREBBP protein domains. Assuming CREBBP mutations were heterozygous (expected VAF 0.5) similarly to RSTS patients, given the VAFs (0.17 and 0.27) a quarter to about half of the cells were mutated for CREBBP. In addition to SNVs, tumor T48 had a heterozygous focal SCNA loss at 16p13.3 covering CREBBP exclusively. Focal SCNA losses of CREBBP in retinoblastoma tissue were also recognized previously at low frequency (2/94, 2% tumors)4. Since this event is so rare and amplitudes of loss are not very high (1/72, 1%), GISTIC analysis (Fig. 1, Table S5) did not consider this locus as significantly altered.
Apart from the three genes (RB1, BCOR, CREBBP) that showed somatic mutations in at least 2 different tumors, there were 199 other genes that were mutated in a single tumor only. Since these genes were mutated so infrequently, the molecular and clinical significance of these genes for retinoblastoma carcinogenesis is not apparent. Yet, in case these genes comprise different key components of a particular molecular signaling pathway, they could be very valuable for understanding retinoblastoma biology. Enrichment analysis of gene ontologies, including biological processes, pathways, diseases and many more, did not result in any significantly overrepresented ontology. This doesn’t necessarily mean all infrequently mutated genes are irrelevant but means no coherence could be found and therefore their relevance could not be demonstrated. Possibly, this gene list is polluted with false positives that are introduced due to artifacts in local INDEL realignment (GATK) PCR, sequencing, mapping and/or stochastic processes that influenced genotype calling. The possibility of false positive variants that can randomly affect genes signifies the importance to focus on recurrently mutated genes.
The number of SCNAs in our retinoblastoma samples was relatively low compared with other cancer types from the TCGA data set. Since loss of RB1 under certain conditions may lead to genomic instability through missegregation of chromosomes eventually causing aneuploidy39,40,41,42, the relatively few SCNAs might appear surprising. Although our data showed that retinoblastoma is the second least disrupted cancer out of all TCGA cancer types, it is of note that no pediatric cancer types are covered in the TCGA data set. It is known that pediatric tumors have lower mutation rates than adult tumors6 and therefore, comparison with other pediatric cancers like neuroblastoma (unfortunately unavailable in TCGA) might provide another perspective. Also, although the genomic fraction hit by SCNAs in retinoblastoma might be relatively low compared with adult tumors, SCNAs in retinoblastoma were concentrated at specific genomic regions indicating their significance. For example, gain of chromosome arm 6p was observed in the majority of tumors (48/71, 68%) and only the minority (12/71, 17%) did not show any SCNA at 1p, 2p, 6p or 16q. Although most of the samples had little non-cancer contamination, losses of 16q often did not exceed loss of one copy, indicating that 16q loss is subclonal. Similarly, VAFs of BCOR and CREBBP and amplitudes of most of the large SCNAs were consistent with intra-tumor heterogeneity.
Although SCNAs were not detected in all tumors, we hypothesize that all tumors were aneuploid although with varying degrees. For example, in 58/69 (84%) tumor samples with 16q copy number decrease, the 16q copy number was between one and two copies, even in samples with very high tumor cellularity. We hypothesize that previously described RB1 loss induced genomic instability can affect any chromosome but selection pressure of growth favoring SCNAs is required for SCNAs to exceed our detection thresholds. Since SCNAs were less abundant in tumors of patients diagnosed at young age, we hypothesize that clonal selection was less persevere in these patients. Possibly, tumors that arose during early stages of retina development originated from more undifferentiated precursors. The intrinsic proliferative capacity of these cells probably was sufficient for development of full-blown tumors and therefore there was no window for clonal selection on SCNAs. Lesions that arose at later stages of retinal development might have been neoplastic, but specific SCNAs favored growth of these cells and therefore cells with favorable SCNAs were selected for.
The significance of regions and genes hit by SCNAs was determined by GISTIC which makes use of SCNA focality, amplitude and recurrence. For few GISTIC regions, a single gene was included (e.g. MYCN, RB1, BCOR), while most GISTIC regions harbored multiple genes complicating target gene identification. Focal gains at chromosome 1q in human retinoblastoma are rare but in murine retinoblastoma, focal gains of MDM2 were reported43. Human paralog MDM4 has been proposed as putative retinoblastoma driver5,44 and was one of the 27 genes included in the significantly altered 1q31.2 region (Figs 1 and 2, Table S5). By inhibiting p53-induced apoptosis, gain of MDM2/4 is suggested to prevent major cell death in RB1 inactivated retina45. However, cone photoreceptor precursor cells, the alleged retinoblastoma cells of origin, show intrinsic p53-inhibition by high expression of MDM2 that might leave gain of MDM2/4 redundant. Furthermore, since TP53 mutations in retinoblastoma never have been found, inactivation of p53 seems not to be a requirement for retinoblastoma development. Opposed to glioma where minimal regions of 1q gain exclusively included MDM4, target gene identification of 1q gain in retinoblastoma is not conclusive yet. Possibly, focal or single gene gains at 1q are rare because multiple genes at 1q are relevant for retinoblastoma development, illustrated by tumor T21 that showed focal gains at four separate 1q regions. Similarly, focal alterations for 6p in retinoblastoma were not found in our cohort, indicating multiple 6p loci could be relevant for retinoblastoma carcinogenesis. Although E2F3 has been presented as 6p candidate gene18,46, the neighboring gene SOX4 was gained with similar frequency in our cohort and was the exclusively gained gene in urothelial carcinoma47. Also for 16q loss, identification of the target gene is not completely conclusive. In our cohort, two GISTIC regions at 16q were identified exclusively including CDH1 and CDH8 respectively. However, CDH11 which is located in between CDH1 and CDH8 was identified as the most frequently lost gene of 16q previously48. It was shown that inactivation of CDH11 in murine retinoblastoma accelerated tumor growth validating its retinoblastoma suppressive function49. However, this does not directly discriminate CDH1 and CDH8 as retinoblastoma suppressors, and these discrepancies signify the difficulty of pinpointing a single gene as driver. We conclude that, unless the intersection of SCNAs exclusively covers a single gene like for RB1, MYCN or BCOR, identification of SCNA target genes remains challenging.
This study aimed to identify recurrent genetic alterations subsequent to RB1 loss that drive tumor progression. Targeted disruption of gain-of-function alterations that are crucial for retinoblastoma cells to survive may lead to cell death. Our analyses of enucleated retinoblastoma showed that SNVs/INDELs beyond RB1 inactivation were extremely rare. Also, since the VAFs of the secondary variants were well below 1, these variants most likely were not present in all tumor cells. Therefore, targeted treatment based on these secondary alterations, possibly can only disrupt parts of the tumor in parts of the retinoblastoma population. For several tumors, inactivation of RB1 alone was sufficient to develop tumors that were so advanced they had to be enucleated. In agreement with the drastic consequences of RB1 loss during early childhood, only 1% of the children who carry an RB1 mutation remain unaffected50, indicating not much additional factors are involved. Given the high penetrance and the requirement for bi-allelic RB1 inactivation, the second hit almost inevitably occurs. Possibly, the high chance of acquiring a second RB1 inactivation can be explained by RB1 haploinsufficiency. It was shown that heterozygous inactivation of RB1 in RPE cells resulted in increased incidence of chromosome missegregation during mitosis51. Possibly, heterozygous RB1 inactivation increases the risk for (copy neutral) LOH and partly explains the high disease penetrance.
In contrast to SNVs/INDELs, larger genetic alterations concentrated at specific genomic regions were more common, in particular for patients diagnosed at a later age. Only when RB1 inactivation occurred at later age, additional genetic alterations might have had sufficient proliferative value to get selected for. Although SCNAs might be relevant for tumor growth in at least a subset of the retinoblastoma population, the clinical relevance remains to be determined. Furthermore, SCNAs might be associated with increased tumor progression but inhibition of these genetic events does not necessarily reverts the phenotype. Also, similarly to secondary SVNs/INDELs, the majority of SCNAs were subclonal and therefore targeting these alterations specifically might be insufficient for complete control of retinoblastoma.
Materials and Methods
Tissue collection
Tumor samples were obtained from retinoblastoma patients after primary enucleation and peripheral blood samples were collected at initial presentation before treatment. Tumor samples were snap frozen in liquid nitrogen and stored at −80 °C until further analysis. All patient samples and clinical and histopathological features were collected and stored according to local ethical regulations. All patients gave consent verbally, as this was the standard in the time the included patients were diagnosed. Since genetic analyses of our study focused on tumor DNA and not germ line DNA, waiver of informed consent was specifically given for genetic analyses by The Medical Ethics Review Committee of the VU University Medical Center which is registered with the USA OHRP as IRB00002991. The FWA number assigned to VU University Medical Center is FWA00017598. It was required that germ line variants were anonymized and pooled during somatic small variant identification. All experimental protocols used in this study were approved by the Medical Ethics Review Committee of the VU University Medical Center and all methods were carried out in accordance with the approved guidelines.
Sample preparation for whole-exome sequencing
Genomic DNA from frozen tumor retinoblastoma specimens (N = 72) and matching white blood cell DNA (N = 71, 1 bilateral patient: one tumor sample per affected eye, 1 blood sample) was isolated with the NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany) or Wizard Genomic DNA Purification Kit (Promega, Madison, USA). DNA quality was analyzed for high molecular bands >20 kb by agarose gel electrophoresis. DNA concentrations were determined by Qubit 3.0 Fluorometer (Life technologies, Bleiswijk, The Netherlands). DNA yields and quality were within the same range for all samples. Genomic DNA was sheared using Covaris Focused-ultrasonicator (Covaris, Woburn, USA). Quality control of fragmented DNA was performed with Genomic DNA ScreenTape (Agilent technologies, Santa Clara, USA). DNA end-repair and ligation of sequencing and indexing adapters was done using Truseq Nano DNA library prep kit (Illumina, San Diego, USA). Exon enrichment was performed using SeqCapEZ v3.0 (Roche Nimblegen, Madison, USA). Sample preparation was performed in two batches, the first batch comprised tumor samples T1-T24, the second batch T25-T72. Within batches, tumor IDs were randomly assigned. Six samples were pooled per sequencing library and sequenced with 125 bps paired-end sequencing, HiSeq 2500 HT v3/4 (Illumina) yielding on average 40.6 million read pairs (range 28.6–56.8 million read pairs, N = 143).
Pre-processing of sequencing reads and quality control
Adapter removal and 5′-end quality trimming was performed using Trimmomatic52 using default parameters. Read quality control of cleaned data was done with FastQC. Clean sequencing reads were mapped to UCSC genome version hg19 with Burrows-Wheeler aligner (BWA)53 in paired-end mode. Post-mapping quality control was done by Picard CalculateHsMetrics. Sequencing targets are considered all exons in RefSeq release 70. Genomic locations of the baits are defined in the specification of the SeqCap EZ v3.0 documentation.
Gender tests for sample quality control
Based on the number of reads mapped to chromosome Y relative to all mapped reads, gender estimations were made. Tumor T53 was obtained from a female patient according to the patient information records but based on WES data its gender was estimated to be male (Supplementary Fig. 2). This observation questions the identity of the T53 sample, and therefore it cannot be guaranteed that the correct germ line sample was included. Therefore T53 was discarded in all further analyses.
Somatic and pathogenic SNVs/INDELs
For variant calling, GATK54 was used to recalibrate base call scores, to re-align reads around INDELs and to call variants using the haplotype caller. Variants with low coverage (depth <5 reads), low GATK variant quality (GATK variant QUAL <50) and/or strand bias (FisherStrandBias >60) were discarded and remaining variants were annotated with ANNOVAR55. According to the rules of the local medical and ethical committee, genome-wide variants of germ line samples were anonymized and were merged into a single retinoblastoma germ line variant database. To identify somatic variants, only tumor variants that were not recorded in the retinoblastoma germ line variant data base were considered if they were at least covered by 10 reads. Two variants were considered identical when they had similar genomic coordinates, reference and mutant allele sequences, and the same GATK genotype calls (AA, AB, BB). To filter for somatic pathogenic variants, the somatic variants were required to be infrequent (<0.1%) in Exome Sequencing Project version 6500 and 1000 genomes 2014Oct. Variants were also were required to be marked “PASS” by GATK and were truncating (stop-gain/loss, frameshift-gain/loss, and INDELs) or splice site mutations or amino acid substitutions that were scored pathogenic by at least 2 out of 4 pathogenicity predicting programs (SIFT, LJB_LRT_Pred, PolyPhen2, and MutationTaster).
Validation of RB1 mutations by conventional DNA diagnostics
Validation of somatic and possibly pathogenic mutations was done by comparing RB1 mutations identified by WES with results from diagnostics testing on tumor and germ line DNA by conventional methods described previously56. Germ line RB1 genetic testing was performed for all patients but one (T29, bilateral patient). Tumor DNA diagnostics was available for 36 patients. 12/35 patients for whom no tumor DNA diagnostics was performed, a germ line RB1 mutation was found. The remaining 23 patients without detectable germ line mutation and no available DNA diagnostics were all non-heritable unilateral patients diagnosed before 2007.
Somatic copy number alterations
For somatic copy number alteration detection, the depth of coverage for 20 Kbps genomic windows was summarized using off-target reads only. The percentage of off-target reads per sample was on average 18% (range 11.9–23.4%, N = 141). Genomic windows that are hard to quantify with NGS (low mappability regions) according to the uniqueness of the ENCODE reference genome were discarded. Depth of coverage was normalized for GC content and mappability and log2-transformed ratios of tumor and matching germ line (Log2-R-ratios, LRR) were used for segmentation with DNAcopy under default parameters. These analyses were implemented by CopywriteR10.
Allelic imbalance and loss of heterozygosity
All variants detected by GATK for all samples (N = 143) were pooled in a single database using VCFtools57. For each unique variant position and each sample, the B/variant allele frequency (BAF or VAF) and genotypes were calculated by SAMtools mpileup. Variant positions with at least 20x depth of coverage in at least 80% of the samples were considered, adding up to 208,822 unique markers. Using BAFsegmentation58, mirrored BAF (mBAF) were computed and mBAF segmentation was performed using allelic imbalance threshold 0.6 and a minimum of 5 consecutive markers for calling. Allelic imbalance was called loss of heterozygosity when segmented mBAF values exceeded 0.8.
Additional Information
Data availability: Sequencing data is stored at the European Genomics Archive under accession number EGAS00001001690 and access is controlled by Data Access Committee EGAC00001000431.
How to cite this article: Kooi, I. E. et al. Somatic genomic alterations in retinoblastoma beyond RB1 are rare and limited to copy number changes. Sci. Rep. 6, 25264; doi: 10.1038/srep25264 (2016).
Supplementary Material
Acknowledgments
We would like to thank all members of the VU University Medical Center Retinoblastoma team for feedback and discussions. The research was supported by grants from Stichting VUmc CCA (VU University Medical Center Cancer Center Amsterdam foundation) grant number CCA20083-05 and Stichting KIKA (Dutch Children Cancer-Free foundation) grant number 98. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions I.E.K. wrote the manuscript. B.M.M., S.E.vM. and Y.dV performed the experiments. E.I.K., M.P.G.M. and N.A. analyzed the data. I.E.K., B.M.M., C.J.D. and A.C.M. collected the samples. C.J.D. and A.H.vdH. provided RB1 genetic testing results. H.tR., J.C. and J.C.D. designed the study. H.M.H. and G.J.L.K. provided clinical feedback. All authors read and reviewed the manuscript.
References
- Linabery A. M. & Ross J. A. Trends in childhood cancer incidence in the U.S. (1992–2004). Cancer 112, 416–32 (2008). [DOI] [PubMed] [Google Scholar]
- Parkin D. M., Stiller C. A., Draper G. J. & Bieber C. A. The international incidence of childhood cancer. Int. J. Cancer 42, 511–520 (1988). [DOI] [PubMed] [Google Scholar]
- Rushlow D. E. et al. Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet. Oncol. 14, 327–34 (2013). [DOI] [PubMed] [Google Scholar]
- McEvoy J. et al. RB1 gene inactivation by chromothripsis in human retinoblastoma. Oncotarget 5, 438–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaras H. et al. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 17, 1363–1372 (2008). [DOI] [PubMed] [Google Scholar]
- Vogelstein B. et al. Cancer genome landscapes. Science 339, 1546–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy J. et al. RB1 gene inactivation by chromothripsis in human retinoblastoma. Oncotarget 5, 438–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481, 329–34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T. et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 16, 49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol B. M. et al. High resolution SNP array profiling identifies variability in retinoblastoma genome stability. Genes. Chromosomes Cancer 53, 1–14 (2014). [DOI] [PubMed] [Google Scholar]
- Yamada M. et al. Amplified allele of the human n- myc oncogene in neuroblastomas. Japanese J. Cancer Res. 79, 670–673 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wal J. E. et al. Comparative genomic hybridisation divides retinoblastomas into a high and a low level chromosomal instability group. J. Clin. Pathol. 56, 26–31 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillington D. M. et al. Comparative genomic hybridization of 49 primary retinoblastoma tumors identifies chromosomal regions associated with histopathology, progression, and patient outcome. Genes Chromosom. 36, 121–128 (2003). [DOI] [PubMed] [Google Scholar]
- Zielinski B. et al. Detection of chromosomal imbalances in retinoblastoma by matrix-based comparative genomic hybridization. Genes. Chromosomes Cancer 43, 294–301 (2005). [DOI] [PubMed] [Google Scholar]
- Chen D., Gallie B. L. & Squire J. A. Minimal regions of chromosomal imbalance in retinoblastoma detected by comparative genomic hybridization. Cancer Genet. 129, 57–63 (2001). [DOI] [PubMed] [Google Scholar]
- Ganguly A., Nichols K. E., Grant G., Rappaport E. & Shields C. Molecular karyotype of sporadic unilateral retinoblastoma tumors. Retina 29, 1002–1012 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481, 329–34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooi I. E. et al. Loss of photoreceptorness and gain of genomic alterations in retinoblastoma reveal tumor progression, EBioMedicine 2, doi: 10.1016/j.ebiom.2015.06.022 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J. N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintu C., Athale U. H. & Patil P. S. Childhood cancers in Zambia before and after the HIV epidemic. Arch. Dis. Child. 73, 100–105 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunin G. R. et al. Pre- and Postconception Factors Associated with Sporadic Heritable and Nonheritable Retinoblastoma. Cancer Res. 49, 5730–5735 (1989). [PubMed] [Google Scholar]
- Decaprio J. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54, 275–283 (1988). [DOI] [PubMed] [Google Scholar]
- Dyson N., Howley P. M., Münger K. & Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243, 934–7 (1989). [DOI] [PubMed] [Google Scholar]
- Whyte P. et al. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334, 124–9 (1988). [DOI] [PubMed] [Google Scholar]
- Orjuela M. et al. Presence of Human Papilloma Virus in Tumor Tissue from Children with Retinoblastoma: An Alternative Mechanism for Tumor Development. Clin. Cancer Res. 6, 4010–4016 (2000). [PubMed] [Google Scholar]
- Gillison M. L. et al. Human retinoblastoma is not caused by known pRb-inactivating human DNA tumor viruses. Int. J. Cancer 120, 1482–90 (2007). [DOI] [PubMed] [Google Scholar]
- Ciriello G., Cerami E., Sander C. & Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 22, 398–406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E. L., Auger H., Jaszczyszyn Y. & Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 30, 418–426 (2014). [DOI] [PubMed] [Google Scholar]
- Offit K. Decade in review—genomics: A decade of discovery in cancer genomics. Nat. Rev. Clin. Oncol. 11, 632–634 (2014). [DOI] [PubMed] [Google Scholar]
- Tiacci E. et al. The corepressors BCOR and BCORL1: two novel players in acute myeloid leukemia. Haematologica 97, 3–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton E. et al. BCOR analysis in patients with OFCD and Lenz microphthalmia syndromes, mental retardation with ocular anomalies, and cardiac laterality defects. Eur. J. Hum. Genet. 17, 1325–35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F. et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376, 348–51 (1995). [DOI] [PubMed] [Google Scholar]
- Miller R. W. & Rubinstein J. H. Tumors in Rubinstein-Taybi syndrome. Am. J. Med. Genet. 56, 112–5 (1995). [DOI] [PubMed] [Google Scholar]
- Sharma N., Mali A. M. & Bapat S. A. Spectrum of CREBBP mutations in Indian patients with Rubinstein-Taybi syndrome. J. Biosci. 35, 187–202 (2010). [DOI] [PubMed] [Google Scholar]
- Manning A. L., Longworth M. S. & Dyson N. J. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 24, 1364–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A., Lentini L., Schillaci T., Iovino F. & Di Leonardo A. RNAi mediated acute depletion of retinoblastoma protein (pRb) promotes aneuploidy in human primary cells via micronuclei formation. BMC Cell Biol. 10, 79 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Harn T. et al. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 24, 1377–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Vasconcellos I. et al. Rb1 haploinsufficiency promotes telomere attrition and radiation-induced genomic instability. Cancer Res. 73, 4247–55 (2013). [DOI] [PubMed] [Google Scholar]
- Conkrite K., Sundby M., Mu D., Mukai S. & MacPherson D. Cooperation between Rb and Arf in suppressing mouse retinoblastoma. J. Clin. Invest. 122, 1726–1733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampieri K. et al. Genomic differences between retinoma and retinoblastoma. Acta Oncol. 47, 1483–1492 (2008). [DOI] [PubMed] [Google Scholar]
- Laurie N. a et al. Inactivation of the p53 pathway in retinoblastoma. Nature 444, 61–66 (2006). [DOI] [PubMed] [Google Scholar]
- Grasemann C. et al. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene 24, 6441–9 (2005). [DOI] [PubMed] [Google Scholar]
- Eriksson P. et al. Detailed Analysis of Focal Chromosome Arm 1q and 6p Amplifications in Urothelial Carcinoma Reveals Complex Genomic Events on 1q, and SOX4 as a Possible Auxiliary Target on 6p. Plos One 8, e67222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchong M. N. et al. Minimal 16q Genomic Loss Implicates Cadherin-11 in Retinoblastoma. Mol. Cancer Res. 2, 495–503 (2004). [PubMed] [Google Scholar]
- Marchong M. N. et al. Cdh11 acts as a tumor suppressor in a murine retinoblastoma model by facilitating tumor cell death. Plos Genet. 6, e1000923 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaras H. et al. Retinoblastoma. Lancet (London, England) 379, 1436–46 (2012). [DOI] [PubMed] [Google Scholar]
- Coschi C. H. et al. Haploinsufficiency of an RB-E2F1-Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 4, 840–53 (2014). [DOI] [PubMed] [Google Scholar]
- Bolger A. M. Lohse M. & Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, doi: 10.1093/bioinformatics/btu170 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dommering C. J. et al. RB1 mutation spectrum in a comprehensive nationwide cohort of retinoblastoma patients. J. Med. Genet. 51, 366–74 (2014). [DOI] [PubMed] [Google Scholar]
- Danecek P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staaf J. et al. Segmentation-based detection of allelic imbalance and loss-of-heterozygosity in cancer cells using whole genome SNP arrays. Genome Biol. 9, R136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.