Abstract
Background
Staphylococcus aureus, a gram-positive pathogen, causes many human infections. Methicillin-resistant S. aureus (MRSA) is the most common drug-resistance bacteria. Nearly all MRSA bacteria are resistant to several drugs. Specific antibodies are the main components of the host’s humoral immunity, and play a significant role in the process of the host’s resistance to bacterial infection.
Results
A single-chain variable fragment (scFv) library was constructed using mRNA from the peripheral blood mononuclear cells of S. aureus infected volunteers. After the scFv library DNA was transformed into Escherichia coli TG1, ~1.7 × 107 independent clones with full-length scFv inserts. The scFv library was screened by phage display for three rounds using S. aureus as an antigen. The single clones were chosen at random and the scFvs were expressed for enzyme-linked immunosorbent assay (ELISA) assessment. Approximately 50 % of the clones were positive with good binding activity to S. aureus. To improve the stability of scFvs, scFv-fragment crystallizable regions (-Fcs) were constructed and expressed in E. coli DH5α. The expressed scFv-Fcs were purified and identified by western blot. These antibodies were further characterized and analyzed for bioactivity. The results showed that the expression level and folding of scFv-Fcs induced at 25 °C without isopropyl β-D-1-thiogalactopyranoside (IPTG) were higher than that induced at 32 °C with 1.0 mmol/L IPTG. scFv-Fcs had good bioactivity and could specifically bind with S. aureus.
Conclusion
scFv-Fcs against S. aureus were successfully constructed and are good candidates for the development of future adjunctive therapy for severe S. aureus infections.
Keywords: Staphylococcus aureus, Single-chain variable fragment, Fragment crystallizable regions, scFv-Fc, Phage display
Background
Staphylococcus aureus, a gram-positive pathogen, causes an large array of diverse human infections, ranging from relatively minor skin and wound infections to more serious life-threatening diseases, such as deep-tissue infections, pneumonia, and bacteremia [1, 2]. Based on data from the U.S Centers for Disease Control and Prevention, S. aureus infection has been deemed the most lethal of all infectious diseases. The emergence and spread of multidrug-resistant strains in communities and even in hospitals are making therapeutic intervention increasingly difficult and expensive [3–5]. Meticillin-resistant S. aureus (MRSA) is the most common drug-resistant bacteria and causes clinical diseases such as skin infections, pneumonia, and septicemia. Nearly all MRSAs are also multidrug resistant and are not sensitive to β-lactam antibiotics, such as penicillin and cephalosporins, or to chloromycetin, lincomycin, aminoglycoside antibiotics, tetracycline antibiotics, and macrolide antibiotics. Some S. aureus are also not sensitive to vancomycin [6–9]. With only a few new antibiotics in development, considerable interest and efforts have been directed toward exploring active and passive immune-mediated therapeutic approaches to prevent and treat staphylococcal infections.
Specific antibodies are the main component of a host’s humoral immunity, and play a significant role in the process of creating a host’s resistance to bacterial infection. Antibodies can bind with the antigens of bacteria and kill or eliminate the bacteria through neutralization, activating complements, promoting phagocytosis, and antibody-dependent cellular cytotoxicity (ADCC). For nearly 30 years, antibodies have gone through four stages of development—murine antibodies, human-mouse chimeric antibodies, humanized antibodies, and fully human antibodies. Fully human antibodies are used to reduce the rejection reaction when treating human diseases [10, 11].
Traditional monoclonal antibodies were murine. If used to treat human diseases, the human anti-mouse antibody (HAMA) reaction would occur; therefore, fully human antibodies were perfected to treat human diseases. A fully human antibody is prepared in one of two ways—using transgenic mice or constructing a human phage single-chain variable fragment (scFv) library. The molecular weight of the fully human antibody is too high to use phage display for selection. scFv has the property of low molecular weight, which is suitable for phage display, and has antigen-binding properties; therefore, scFvs is the best block for constructing other types of antibodies. However, scFvs are not stable and do not have the properties of fragment crystallizable regions (Fcs), such as ADCC, so Fcs were fused with scFvs, creating scFv-Fc, to increase stability and recover Fc function.
In this study, an scFv library was constructed using mRNA from peripheral blood mononuclear cells (PBMCs) of volunteers infected with S. aureus and specific fully human scFv-Fvs against S. aureus were developed with the hope that an adjunctive therapy is developed for severe S. aureus infections.
Methods
PBMCs of the five volunteers enrolled in this study who were infected with S. aureus were used for the construction of a scFv library. The criterion for selecting volunteers was only that they were infected with S. aureus, which was confirmed by tests from the clinical laboratory in the first affiliated hospital of Sichuan medical university, China. The experiment was approved by the ethics committee of Sichuan medical university (No. 5105025012142). All volunteers were adults and provided written informed consent.
Construction of the scFv library
The scFv library was constructed by referring to previous reports by our research group [12]. Briefly, PBMCs from volunteers infected with S. aureus were isolated by Ficoll Paque Plus (Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA) according to the manufacturer’s instructions. mRNA was extracted from the PBMCs using the Dynabeads mRNA DIRECT kit (Invitrogen, USA) and was used to synthesize full-length cDNA using the SMART cDNA library construction kit (Clontech, USA). The variable regions of heavy-chain (VH) and light-chain (VL) immunoglobulin (Ig) were amplified by four subsequent polymerase chain reactions (PCRs) using a set of primers and following Qing Yuan [12] and the VH and VL gene repertoires were linked by overlapping extension PCR [12].
The phage display vector pCANTAB-5E, Escherichia coli strain TG1, and helper phage M13K07 (Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA) were used to create the phage library. The scFv library DNA (VH-linker-VL) and phagemid pCANTAB-5E vector were digested with Sfi I and Not I. The digested scFv fragments were inserted into the vector to generate a scFv-gene III fusion library using T4 DNA ligase (New England BioLabs, UK) at 4.0 °C overnight. The ligated products were transformed into E. coli TG1 by electroporation and grown at 37 °C on culture plates containing lysogeny broth (LB) medium supplemented with 100 μg/mL ampicillin and 2.0 % glucose. Thirty clones from the library were selected at random and identified by PCR to estimate the proportion of full-length scFv clones. The PCR products were digested by BstN I for producing a fingerprint map to estimate the diversity of scFvs. All clones on the plates were scraped and suspended in LB containing 15 % glycerol.
Phage amplification
Eighty microliters of scraped bacterial cells or frozen cell suspensions under glycerol were incubated in 40 mL LB containing 100 μg/mL ampicillin and 2.0 % glucose until the optical density at a wavelength of 600 nm (OD600) = 0.2 while shaking at 37 °C. The bacteria were collected by centrifugation and suspended in 40 mL LB with ampicillin without glucose. Approximately 6 × 109 transducing unit (TU) of helper M13K07 (Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA) were added to each milliliter of cell suspension, incubated for 15 min at 37 °C without agitation, and incubated for another 2.0 h with agitation. Kanamycin was added to obtain a final concentration of 20 μg/mL, after which the cells were incubated overnight at 32 °C. The phage was subsequently precipitated with polyethylene glycol (PEG)-NaCl (20 % PEG, 2.5 mol/L NaCl) and resuspended in 0.01 mol/L phosphate-buffered saline (PBS) buffer.
Affinity selection
Immunotubes were coated with ~1.0 × 108 cfu/mL S. aureus in coating buffer (0.1 mol/L Na2CO3/NaHCO3, pH 9.6) and left overnight at 4.0 °C; these were then sequentially blocked with 4.0 % fat-free milk for 1.0 h at 37 °C and gentle agitation. Approximately 1 × 1012 TU from the freshly amplified scFv libraries were added to the blocked immunotubes and incubated at room temperature for 2.0 h. The immunotubes were rinsed 10 times with 0.01 mol/L PBST (0.05 % Tween-20 in 0.01 mol/L PBS) and 0.01 mol/L PBS separately, then eluted with 0.1 mol/L glycine-HCl (pH 2.2) and amplified as above. In the second and third rounds of affinity selection, 1 × 107 cfu/mL and 1 × 106 cfu/mL bacteria antigen, respectively, were coated and the immunotubes were washed separately 15 to 20 times with 0.01 mol/L PBST and 0.01 mol/L PBS.
Phage enzyme-linked immunosorbent assay
Following affinity selection of the scFv library in three rounds, individual clones were randomly chosen and grown at 37 °C. The phage was amplified according to the previously described protocols. The amplified phage preparation (1012 TU) was blocked with 4.0 % fat-free milk in 0.01 mol/L PBS for 30 min and added to an immunoassay plate coated with S. aureus for enzyme-linked immunosorbent assay (ELISA) and the negative control (0.01 mol/L PBS) was set up. Plates were incubated for 1.0 h, washed three times with washing buffer PBST, and finally incubated with horseradish peroxidase (HRP)-conjugated anti-M13 mouse monoclonal antibody (Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA). The immunoreactions were developed by incubating in TMB liquid substrate (Sigma-Aldrich Company, Inc., St. Louis, MO, USA) for 15 min. The reactions were stopped by adding 2.0 mol/L H2SO4. Absorbance at 450 nm was recorded and used to select the phage that expressed scFv for recognizing the target antigens.
Construction and expression of scFv-Fcs
pLZ16 was designed based on pUC vector for soluble scFv-Fc antibody production by introducing leader peptide-encoding sequencing (PhoA leader), cloning sites (HindIII, BamHI, NotI) and FLAG tag. The Fc was inserted into the plasmid (Fig. 1). The selected scFvs with good binding activity and specificity were ligated with the pLZ16 vector and transformed into DH5α cells of E. coli. The positive clones were cultured to OD600 = 0.5 at 37 °C in LB with 100 μg/mL ampicillin, 2.0 % glucose (LBAG) and then centrifuged at 5000 r/min for 10 min. One of the following two techniques was used to induce scFv-Fcs expression. One technique was to resuspend the pellets with LBA containing 2.0 mmol/L MgCl2 and culture at 25 °C for 50 h [13]. The other was to resuspend the pellets with LBA and 1.0 mmol/L IPTG and culture at 32 °C for 5.0 h. After centrifugation, the pellet was collected and resuspended in 0.01 mol/L PBS. The suspended cells were sonicated and centrifuged, and the supernatant was detected by ELISA and western blot.
Fig. 1.
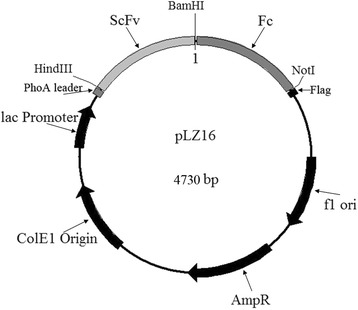
Plasmid map of pLZ16. The vector was constructed based on the pUC vector and used for soluble single-chain variable fragment- fragment crystallizable region (scFv-Fc) production
scFv-Fc functionality
The binding activity of and specificity of scFv-Fcs to S. aureus was tested by dot blot and ELISA using the anti-human IgG1 Fc antibody. Briefly, S. aureus, S. albus, S. citreus, Salmonella typhi, Bacillus anthracis,, Shigella Castellani, Proteusbacillus vulgaris, Monilia albican, α-hemolytic streptococcus and β-hemolytic streptococcus was suspended in Na2CO3/NaHCO3 coating buffer at a concentration of 107 cfu/mL and the ELSIA plates were coated at 4.0 °C overnight or the nitrocellulose membrane was dotted for dot blot analysis. The scFv-Fcs were purified and 1 μg/mL purified scFv-Fcs were used as first antibody and the anti-human IgG1 Fc antibody was used as second antibody for test. The dot blot immunoreactions was developed by chemiluminscence and the ELISA immunoreactions was developed by TMB substrate.
The binding activity of scFv-Fcs were also analyzed by BIAcore (BLITZ). The scFv-Fcs were immobilized on the anti-hIgG Fc capture (AHC) biosensor. Then the S. aureus with different concentration (1 × 107 cfu/mL, 2 × 107 cfu/mL, 4 × 107 cfu/mL) were added and the association and dissociation of scFv-Fcs and S. aureus were detected. The 0.01 mol/L PBS were used as control.
The opsonization of scFv-Fcs was researched by testing the phagocytosis of antibody-opsonized pathogens. The peritoneal macrophages of mice were isolated. The macrophages and S. aureus (1 × 107 cfu) was mixed at the ratio 1:20 in RPMI-1640 medium containing 10 % fetal calf serum (FCS). Ten microliter of scFv-Fc (100 μg/mL) was added into the mixture and incubated at 37 °C for 2 h with gentle agitation. The mouse IgG1 was used as control and each group was repeat for three times. Then the 200 μL of mixture was spread the LB plates for overnight culture. The following day, the clone numbers were calculated.
Statistical method
Background noise correction was performed from ELISA by subtracting the absorbance. All the data were repeated for three times and the data sets the mean and the standard error were calculated. Data were presented as means ± SEM. Differences between groups were determined by the two-tailed t-test. P < 0.05 was considered statistically significant. *p < 0.05, **P < 0.01, ***P < 0.001.
Results and discussion
Construction of the scFv library
mRNA was extracted from PBMCs of volunteers infected S. aureus. The quality of the mRNA was the key to synthesize the full-length cDNA. The integrity of the extracted mRNA was determined by testing the OD260/OD280 and electrophoresis. The synthesized ds cDNA appeared as a smear of from 0.1 to 8.0 kb on the gel and the size of distribution, yield, and intensity of cDNA were satisfied (data not shown). The full-length cDNA was used as a template to amplify the regions of VH and VL, including Vκ and Vλ. The VH, Vκ, and Vλ gene repertoires were amplified with 42, 16, and 18 optimized primer sets, respectively. Nearly all PCR reactions were effective and the correct PCR products were obtained from the gel picture; the expected size of the amplified VH and VL (including Vκ and Vλ) was ~350 bp. A part of amplified VH, Vκ, and Vλ were shown in Fig. 2. The VH-linker gene and VL-linker gene repertories were amplified and the VH-linker-VL gene repertories (750 bp) were obtained by overlapping extension PCR. The DNA library of human scFvs was developed.
Fig. 2.
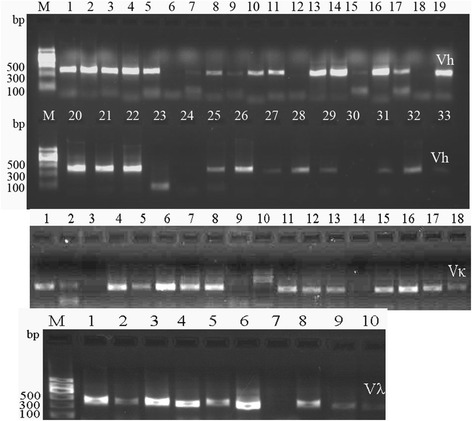
Amplification of VH, Vκ, and Vλ gene repertoires. Lane M: DNA Marker II (Tiangen, CN). The expected size of the amplified VH and VL (including Vκ and Vλ) was ~350 bp. A part of amplified VH, Vκ, and Vλ were shown in Fig. 2
The DNA of the scFv library was ligated with phagemid vector pCANTAB-5E and transformed into TG1 cells of E. coli. Thirty clones were randomly chosen and the inserted scFvs were identified by PCR. The results showed that all clones contained full-length scFv genes. By BstN I digestion of the scFv DNA, the fingerprint maps showed that almost all scFvs were different (Fig. 3); For further analysis of the diversity of the library, the 10 clones were picked randomly for sequencing. The results showed that the sequences of those scFvs were diverse (Fig. 4). The library was estimated to contain 1.7 × 107 unique scFv members.
Fig. 3.
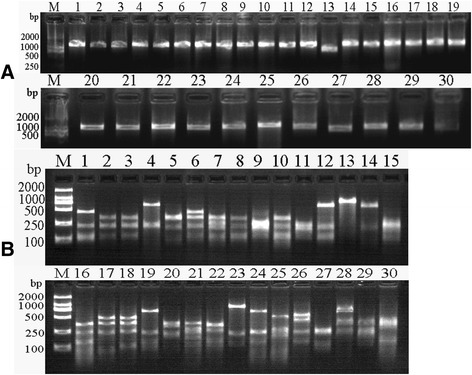
a Thirty clones were chosen randomly from the single-chain variable fragment (scFv) library and the positive clones inserted into full scFv genes were identified by polymerase chain reaction (PCR). Lane M: 2000 bp DNA Marker (TaKaRa, JP). Lanes 1–30: amplified scFvs from different clones randomly picked. b PCR products of scFvs were digested by BstN I. 2000 bp DNA Marker (TaKaRa, JP). Lane M: 2000 bp DNA Marker (TaKaRa, JP). Lanes 1–30: Fingerprints of scFvs digested by BstN I
Fig. 4.
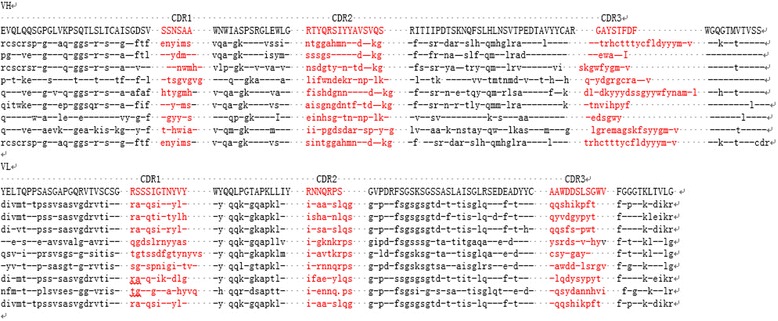
Amino acid sequences of 10 clones which were randomly picked from the library and analyzed by DNA sequence
Affinity selection
The phage library was amplified and for each round of affinity selection, ~1012 TU freshly amplified scFv antibody library were used for screening. Approximately 1.1 × 106, 3.1 × 106, and 1.2 × 107 phages were recovered after the first, second, and third cycles, respectively. Phage pools before and after each round of affinity selection were tested by ELISA, which was performed by using the anti-M13 HRP-conjugated secondary antibody. The phage pool before affinity selection had no signal with the antigen, but after three rounds of screening, there was strong binding activity between the library phage and antigen (data not shown). Hundreds of clones were randomly chosen from the phage pool after three rounds of affinity selection. The phages amplified from those clones were tested by ELISA and about 50 % showed positive whose absorbance of OD450 was two-fold those of the negative controls.
scFv specificity test
From the hundreds of clones those with higher absorbance had better binding activity to the antigen and were selected for testing scFv specificity. The S. aureus, S. albus, S. citreus, S. typhi, B. anthracis, S. castellani, P. vulgaris, M. albicans, α-H.streptococcus and β-H. streptococcus (1 × 107 cfu/ml) were diluted in coating buffer and coated in wells of ELISA plates at 4.0 °C overnight. The ELISA results showed that 14 clones had good specificity with S. aureus and no binding reaction with the control bacteria (Fig. 5). Number 15 and 16 (0.01 mol/L PBS was added, not scFvs) were used as the negative controls. Each test was repeated for three times, the value of the blank was removed, and the standard errors were calculated. The scFvs with better binding activity and specificity (S78, S117, S128, S182) were inserted into the pLZ16 vector with human IgG1 Fc.
Fig. 5.
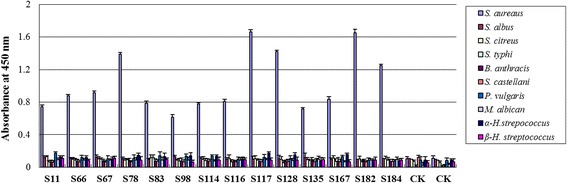
The specificity of selected single-chain variable fragment (scFvs) detected by enzyme-linked immunosorbent assay. Number 15 and 16 were negative controls
Construction and expression of scFv-Fcs
To overcome the instability of scFvs, scFv-Fcs were constructed. The Fc region comprises the CH2 and CH3 domains and the hinge region of the human IgG1. The hinge serves as a flexible spacer between the two parts of the Fc-fusion protein, allowing each part of the molecule to function independently. After transformation, the clones were identified by PCR amplification. The results showed that the bands at 1.5 kb were amplified (Fig. 6a). The sequencing results showed that the scFv-Fcs were correct. Next, the scFv-Fcs were induced to express at 25 °C for 50 h without IPTG or 32 °C for 5.0 h with 1.0 mmol/L IPTG. The expressed scFv-Fcs were coated on the ELISA plates and detected with goat anti-human IgG1 Fc HRP-conjugated antibody. The results showed that OD450 of scFv-Fcs expressed at 25 °C was higher than that expressed at 32 °C (Fig. 6b). The positive results of detection with human IgG1 Fc HRP-conjugated antibody showed that the Fc regions were successfully fused into scFv. The expressed scFv-Fc (S78, S117) under the two conditions were purified. From sodium dodecyl sulfate polyacrylamide gel electrophoresis staining (SDS-PAGE) (Fig. 7a), the scFv-Fcs was pure and not degraded after 50 h expression at lower temperatures (25 °C). The lower temperature will degrade the aggregation of expressed protein and stabilize the bioactivity of scFv-Fc. MgCl2 was added to the medium to maintain the cell wall. Later, the expressed scFv-Fcs (S78, S117, S128, S182) at 25 °C were detected by western blot with the anti-FLAG HRP-conjugated antibody and a single band of ~67 kDa appeared (Fig. 7b). Based on this, it was concluded that a lower temperature was beneficial to the expression and folding of scFv-Fcs and that the system used is suitable for the production of Fc-fused antibody fragments.
Fig. 6.
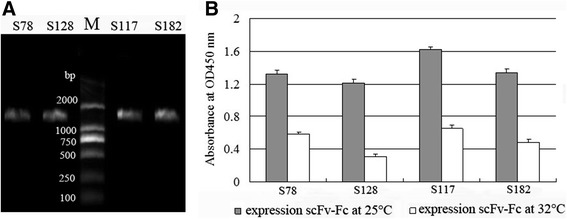
a Constructed scFv-Fcs (S78, S128, S117, S182) were amplified and their sizes were ~1.5 kb. Lane M: 2000 bp DNA Marker (TaKaRa, JP). b Expression level of scFv-Fcs (S78, S128, S117, S182) expressed at 25 °C and 32 °C, respectively, were tested by enzyme-linked immunosorbent assay
Fig. 7.
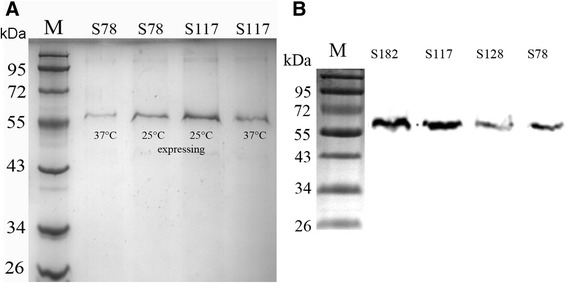
a The scFv-Fcs (S78, S117) expressed at 25 °C and 32 °C respectively were purified and run SDS-PAGE. Lane M: prestained protein ladder (Fermentas, USA); b The scFv-Fcs (S78, S128, S117, S182) were identified by western blot. Lane M: prestained protein ladder (Fermentas, USA)
scFv-Fc functionality
The dot blot results showed that the scFv-Fcs had good binding activity with the S. aureus using the goat anti-human IgG1 Fc HRP-conjugated antibody (Fig. 8a). In addition, the specificity of scFv-Fcs remained and had no cross reaction with S. aureus, S. albus, S. citreus, S. typhi, B. anthracis, S. castellani, P. vulgaris, M. albicans, α-H. streptococcus or β-H streptococcus (Fig. 8b). The BIAcore (BLITZ) results also showed that the scFv-Fcs (S78, S117, S128, S182) could bind S. aureus well (Data not shown). The real-time association and dissociation of S117 and S. aureus were showed in Fig. 9, and the equilibrium dissociation constant (KD), the rate constants for association (ka) and dissociation (kd) between S. aureus and selected scFv-Fcs were listed in Table 1, which were determined by BIAcore (BLITZ).
Fig. 8.
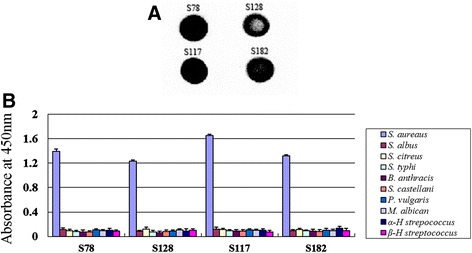
a Binding activity of scFv-Fcs (S78, S128, S117, S182) and S. aureus were detected by dot blot. The immunoreactions was developed by chemiluminscence (b). Detection of the specificity of scFv-Fcs (S78, S128, S117, S182) by enzyme-linked immunosorbent assay
Fig. 9.

Analysis of the association and dissociation of S117 and S. aureus. The S117 was immobilized on AHC biosensor, the different concentration of S. aureus were tested. (run1: 1 × 107 cfu/mL, run 2: 2 × 107 cfu/mL, run 3: 4 × 107 cfu/mL)
Table 1.
The kinetic constants determined by BIAcore (BLITZ) analysis for scFvs
| scFv | ka (1/Ms) | kd (1/s) | kd/ka(KD, M) |
|---|---|---|---|
| S78 | 4.937e4 | 5.731e-4 | 1.161e-8 |
| S128 | 2.123e4 | 8.231e-4 | 3.877e-8 |
| S117 | 5.539e5 | 4.804e-4 | 8.673e-9 |
| S182 | 2.213e4 | 7.648e-4 | 3.456e-8 |
The opsonization of scFv-Fcs was detected. Compared with mouse IgG controls, the plates spread with the mixture containing scFv-Fc (S78, S117) had less clones (Table 2) and it has statistically significant for S78 (P < 0.01) and S117 (P < 0.001). The results showed the scFv-Fcs (S78, S117) against S. aureus could play the role of opsonization, and promoting the function of phagocytosis of macrophage.
Table 2.
Test of the opsonization of scFv-Fcs
| clones | clones | ||
|---|---|---|---|
| S. aureus + macrophage + S78 | (42 ± 5) × 104 ** | S. aureus + macrophage + IgG | (78 ± 7) × 104 |
| S. aureus + macrophage + S128 | (62 ± 6) × 104 | S.aureus + macrophage | (82 ± 8) × 104 |
| S. aureus + macrophage + S117 | (26 ± 5)104*** | ||
| S. aureus + macrophage + S182 | (56 ± 6) × 104 |
**P <0.01, ***P <0.001
Discussion
Commercial murine IgG against S. aureus has been used in research but no fully human antibody against S. aureus has been commercially available for treatment of the infection of S. aureus. Our scFv-Fc against S. aureus was a fully human antibody, and the affinity of scFv was not evolved in vivo; therefore, after the functional scFv-Fc was selected, the affinity of scFv-Fc will be improved by affinity maturation in vitro and the antigenic determinant of S. aureus also will be researched. We hope to establish an adjunctive therapy for severe S. aureus infections using those antibodies.
scFv comprised only Ig VH and VL connected by a soluble and flexible oligopeptide. scFv use is easy for the design and construction of a phage display antibody library, and phage display technology makes possible the direct isolation of monovalent scFv antibody fragments; however, scFv has disadvantages of being unstable, having a short serum half-life, and lacking avidity because of monovalent binding, all of which could limit its effectiveness. Reducing these disadvantages would be useful in making scFv more successful as an antibody. The scFv-Fc format has certain properties of IgGs, such as bivalency, tag-free detection, and Fc-mediated effector functions. Moreover, scFv-Fc antibodies have been successfully used to neutralize viral, bacterial, and fungal pathogens in vitro and in vivo, which suggests its potential uses in therapy [14–16].
In this study, the scFv against S aureus was screened and merged with Fc. The scFv-Fc was expressed with the vector pLZ16 and the better expression condition was incubated at 25 °C for 50 h without IPTG. According to Hiroyoshi (2008) [13], optimum conditions were established for bacterial cultivation and protein expression, utilizing an unusually long cultivation time (>50 h), very low temperature (25 °C), and no IPTG, thereby leading to the production and extracellular secretion of fragment antigen-binding (Fab) proteins in a high-yield using the vector pComb43C9 with lactose promoter.
Based on the report of Hiroyoshi [13], the vector pLZ16 was designed in this study for expressing the soluble scFv-Fc antibody. The results showed that the overall gene expression level under the conditions of 25 °C for 50 h and without IPTG induction was higher than the expression under the conditions of 32 °C for 5.0 h with IPTG. The production yield of the functional scFv-Fc at 25 °C was highly dependent on the following points: (1) freshness of a single colony of the phagemid-harboring E. coli cells; (2) temperature controls, including pre-warming of the culture medium in the inoculation steps; and (3) medium exchange to one with no glucose at the induction step for protein expression [13].
Another study reported that auto-induction without IPTG allows efficient screening many clones in parallel for expression and solubility, as cultures need only to be inoculated and grown to saturation, and yields of target protein are typically several-fold higher than that obtained by conventional IPTG induction [17].
The expressed and purified scFv-Fcs have good bioactivity and specificity with S. aureus. In subsequent studies, the antigenic determinant mechanisms of action will be researched.
Conclusions
In this work, a human scFv library with a volume of 1.7 × 107 was constructed. From the library, specific scFvs against S. aureus were selected using three rounds of phage display. The scFv-Fcs with stable structure developed and expressed in E. coli DH5α, and the suitable expression system induced at 25 °C was beneficial for the correct folding of scFv-Fcs. The purified scFv-Fcs also had the functionality of good binding activity and specificity to S. aureus. These antibodies could be used as candidates for the development of future adjunctive therapy for severe S. aureus infections.
Availability of data and materials
Considering the data has not been used for applying patent, the data will not be shared.
Acknowledgments
This work was supported by the Sichuan Youth Science and Technology Foundation (No. 2012JQ0018) and the Luzhou City Bureau of science and technology (2013LZLY-K77).
Abbreviations
- ADCC
antibody-dependent cellular cytotoxicity
- ELISA
enzyme-linked immunosorbent assay
- Fc
fragment crystallizable region
- FCS
fetal calf serum
- Ig
immunoglobulin
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- HAMA
human anti-mouse antibody
- MRSA
meticillin resistant Staphylococcus aureus
- PBMC
peripheral blood mononuclear cell
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PEG
polyethylene glycol
- scFv
single-chain variable fragment
- VH
variable regions of heavy-chain
- VL
variable regions of light-chain
Footnotes
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
SJ made the main contributions to this study, and acquired, analyzed, and interpreted the data. TW participated in the design and coordination of the study and in the construction of the vector for expressing scFv-Fcs. YY participated in the screening of scFvs. XW constructed the scFv library. WX participated in the immunoassays. QY contributed to the conception and design of the study, the analysis and interpretation of the data, and drafting and revising the manuscript. All authors read and approved the final manuscript.
References
- 1.Pezato R, Bottura L, de Paula SR, Voegels RL, Bachi AL, Gregório LC. Bone: the final frontier for Staphylococcus aureus penetration in chronic rhinosinusitis. J Otolaryngol Head Neck Surg. 2013;42(1):45. doi: 10.1186/1916-0216-42-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamada K, Wanchun J, Ohkura T, Murai A, Hayakawa R, Kinoshita K, Mizutani M, Okamoto A, Namikawa T, Ohta M. Detection of methicillin-resistant Staphylococcus aureus using a specific anti-PBP2a chicken IgY antibody. Jpn J Infect Dis. 2013;66(2):103–8. [DOI] [PubMed]
- 3.Faghri J, Shahbazzadeh D, Pooshang Bagheri K, Moghim S, Ghase mian Safaei H, Nasr Esfahani B, Fazeli H, Yazdani R, Mirmohammad Sadeghi H. Two Dimensional Structural Analysis and Expression of a New Staphylococcus aureus Adhesin Based Fusion Protein. Iran J Basic Med Sci. 2012;15(2):725–38. [PMC free article] [PubMed]
- 4.Liang SY, Khair HN, McDonald JR, Babcock HM, Marschall J. Daptomycin versus vancomycin for osteoarticular infections due to methicillin-resistant Staphylococcus aureus (MRSA): a nested case–control study. Eur J Clin Microbiol Infect Dis. 2013;33(4):659–6. doi: 10.1007/s10096-013-2001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulido Pérez A, Baniandrés Rodríguez O, Ceballos Rodríguez MC, Mendoza Cembranos MD, Campos Domínguez M, Suárez Fernández R. Skin Infections Caused by Community-Acquired Methicillin-Resistant Staphylococcus aureus: Clinical and Microbiological Characteristics of 11 Cases. Actas Dermosifiliogr, 2013, 29. pii: S0001-7310(13)00321-9. doi: 10.1016/j.ad.2013.09.002. [DOI] [PubMed]
- 6.Kali A, Stephen S, Umadevi S, Kumar S, Joseph NM, Srirangaraj S. Changing Trends in Resistance Pattern of Methicillin Resistant Staphylococcus aureus. J Clin Diagn Res. 2013;7(9):1979–82. doi: 10.7860/JCDR/2013/6142.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jurke A, Kock R, Becker K, Thole S, Hendrix R, Rossen J, Daniels-Haardt I, Friedrich A. Molecular epidemiology of meticillin-resistant Staphylococcus aureus (MRSA): think regionally but use globally uniform typing languages. Euro Surveill. 2013;18(43):20617. [DOI] [PubMed]
- 8.Krakauer T, Stiles BG, Krakauer T, Stiles BG. The staphylococcal enterotoxin (SE) family: SEB and siblings. Virulence. 2013;4(7):19. doi: 10.4161/viru.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salgado-Pabón W, Case-Cook LC, Schlievert PM. Molecular analysis of staphylococcal superantigens. Methods Mol Biol. 2014;1085:169–85. doi: 10.1007/978-1-62703-664-1_10. [DOI] [PubMed] [Google Scholar]
- 10.Chen W, Gong R, Ying T, Prabakaran P, Zhu Z, Feng Y, Dimitrov DS. Discovery of Novel Candidate Therapeutics and Diagnostics Based on Engineered Human Antibody Domains. Curr Drug Discov Technol. 2013;11(1):28–40. [DOI] [PMC free article] [PubMed]
- 11.Mandrup OA, Friis NA, Lykkemark S, Just J, Kristensen P. A novel heavy domain antibody library with functionally optimized complementarity determining regions. PLoS One. 2013;8(10):e76834. doi: 10.1371/journal.pone.0076834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qing Y, Li H, Xu W, Yuchuan W, Yan G, Chengwen L, Siji N. Construction of human non-immune library and selection of scFvs against IL-33. Appl Biochem Biotechnol. 2012;167:498–509. [DOI] [PubMed]
- 13.Kuba H, Furukawa A, Okajima T, Furukawa K. Efficient bacterial production of functional antibody fragments using a phagemid vector. Protein Expr Purif. 2008;58:292–300. doi: 10.1016/j.pep.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Moutel SEI, Marjou A, Vielemeyer O, Nizak C, Benearoch P, Dubel S, Perez F. A multi-Fc-species system for recombinant antibody production. BMC Biotechnol. 2009;9:14. [DOI] [PMC free article] [PubMed]
- 15.Rülker T, Voß L, Thullier P, O’ Brien LM, Pelat T, Perkins SD, Langermann C, Schirrmann T, Dübel S, Marschall HJ, Hust M, Hülseweh B. Isolation and characterization of a human-like antibody fragment (scFv) that inactivates VEEV in vitro and in vivo. Plos one. 2012;7:e37242. [DOI] [PMC free article] [PubMed]
- 16.West AP, Jr, Galimidi RP, Gnanapragasam PNP, Bjorkman PJ. Single-chain Fv-based anti-HIV proteins: potential and limitations. J Virol. 2012;86:195–202. doi: 10.1128/JVI.05848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studier William F. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Considering the data has not been used for applying patent, the data will not be shared.


