Abstract
Background
Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae commonly cause infections worldwide. BlaCTX-M-15 has been commonly detected in hospital isolates in Mwanza, Tanzania. Little is known regarding the faecal carriage of ESBL isolates and blaCTX-M-15 allele among humans in the community in developing countries.
Methods
A cross-sectional study involving 334 humans from the community settings in Mwanza City was conducted between June and September 2014. Stool specimens were collected and processed to detect ESBL producing enterobacteriaceae. ESBL isolates were confirmed using disc approximation method, commercial ESBL plates and VITEK-2 system. A polymerase chain reaction and sequencing based allele typing for CTX-M ESBL genes was performed to 42 confirmed ESBL isolates followed by whole genome sequence of 25 randomly selected isolates to detect phylogenetic groups, sequence types plasmid replicon types.
Results
Of 334 humans investigated, 55 (16.5 %) were found to carry ESBL-producing bacteria. Age, history of antibiotic use and history of admission were independent factors found to predict ESBL-carriage. The carriage rate of ESBL-producing Escherichia coli was significantly higher than that of Klebsiella pneumoniae (15.1 % vs. 3.8 %, p = 0.026). Of 42 ESBL isolates, 37 (88.1 %) were found to carry the blaCTX-M-15 allele. Other transferrable resistance genes were aac(6’)Ib-cr, aac(3)-IIa, aac(3)-IId, aadA1, aadA5, strA, strB and qnrS1. Eight multi-locus sequence types (ST) were detected in 25 E. coli isolates subjected to genome sequencing. ST-131 was detected in 6 (24 %), ST-38 in 5 (20 %) and 5 (20 %) clonal complex − 10(ST-617, ST-44) of isolates. The pathogenic phylogenetic groups D and B2 were detected in 8/25 (32 %) and 6/25 (24 %) of isolates respectively. BlaCTX-M-15 was found to be located in multiple IncY and IncF plasmids while in 13/25(52 %) of cases it was chromosomally located.
Conclusion
The overlap of multi-drug resistant bacteria and diversity of the genotypes carrying CTX-M-15 in the community and hospitals requires an overall approach that addresses social behaviour and activity, rationalization of the antibiotic stewardship policy and a deeper understanding of the ecological factors that lead to persistence and spread of such alleles.
Background
Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae are currently a major problem in hospitalized patients worldwide [1–3]. The prevalence of ESBLs among clinical isolates varies between countries and from institution to institution [2, 4]. Tanzania is one of the sub-Saharan African countries facing increasing numbers of health care associated infections due to multi-drug resistant Gram-negative bacteria [5–8]. Data regarding ESBL isolates in Tanzania are limited to tertiary hospital based studies only.
Several studies performed in developed countries have demonstrated ESBL carriage in the community [9–13]. In addition, in many studies from developed and middle-income countries it could be demonstrated that ESBL-producing bacteria are common in domestic and companion animals in the community [14, 15]. Human to animal contact seems to play a role in the transfer of ESBL producing bacteria between both populations [14, 16, 17]. In Thailand, different factors such as better education, history of hospitalization and the use of antibiotics have been found to be independently predictors for ESBL carriage [18]. Despite the potential risk of ESBL acquisition in the community and transfer between humans and animals there is no study which has documented ESBL carriage and associated risk factors in the human community in Tanzania. Therefore this study was done to investigate the magnitude of ESBL carriage and diversity of ESBL genotypes, and to identify factors associated with it among humans in the community in Mwanza.
Methods
Sample size and sampling
A cross-sectional study was conducted between June and September 2014 in three rural districts (Igogo, Mbugani and Kirumba) with squatters in Mwanza city. The characteristics of the rural districts are described in Table 1. Sample size was calculated using the formula by Cochran [19]. A prevalence of 22.1 % from a study performed in Madagascar was used for calculation [20]. The minimum sample size obtained was 297.
Table 1.
Profile of Igogo, Mbugani and Kirumba
| Variables | Igogo | Mbugani | Kirumba |
|---|---|---|---|
| Population size | 27,303 | 39,041 | 29,354 |
| Number of hospitals/Dispensaries | 2 | 5 | 4 |
| Number of pharmacy/Medicine shops | 18 | 10 | 4 |
| Population per pharmacy/medicine shops | 1516.8 | 3904.1 | 7338.5 |
| Industrial activities | • Industrial areas • Car garages • Storages warehouses |
• None | • Car garages |
| Type of toilets | Latrines | Latrines | Latrines |
| Damping ground | Present | Present | Present |
| Distance from Tertiary hospital (Bugando Medical Centre) | 1.56 km | 2.16 km | 3.82 km |
| ESBL rates | 20.5 % | 15.2 % | 11.6 % |
Streets within these rural districts with characteristics of squatter settlements were purposively selected. A total of 3144 households were obtained from these streets. The number of households in each street was obtained from the household registers at the street Executive Officer‘s office. Simple random sampling was used to select 334 households which were included in the study. Using a random number generator, the households to be included in the study were determined. A total of 334 stool samples (one per participant) were collected. From every participant enrolled, additional information (with the use of an interview questionnaire) such as age, gender, size of the family, history of antibiotic use in the past one month and history of admission in the past one year, were collected.
Laboratory procedures
A total of 334 non-repetitive stool specimens were obtained from humans. All specimens were cultured on MacConkey agar supplemented with 2 μg/mL cefotaxime (Oxoid, Basingstoke, UK) and plain MacConkey agar plates to isolate lactose fermenting colonies to investigate these for the antimicrobial susceptibility pattern.
Strain selection
One colony from predominant morphologically similar colonies was selected on the MacConkey agar plate with 2 μg/mL cefotaxime for subsequent characterization while a representative of predominant morphologically similar lactose fermenter colonies was also selected from a plain MacConkey agar plate.
ESBL confirmation and susceptibility testing
ESBL isolates were confirmed using disc approximation method as previously described [5], commercial ESBL CHROMagar (CHROMagar Company, Paris, France) and VITEK-2 compact system (AST-card N214 and N248, bioMérieux, Nürtingen, Germany) in case of ambiguous results. Antimicrobial susceptibility testing using ciprofloxacin (5 μg), gentamicin (10 μg), tetracycline (30 μg) and trimethoprim/sulphamethoxazole (1.25/23.75 μg) was performed with all isolates based on Clinical Laboratory Standard Institute (CLSI) Guidelines [21].
Analysis of the CTX-M-Allele
A total of 42 ESBL-producing isolates were available for further characterization. First, the presence of CTX-M was identified using CTX-M-F (sequence: 5’-TCTTCCAGAATAAGGAATCCC-3’) and CTX-M-R (sequence: 5’-CCGTTTCCGCTATTACAAAC-3’) amplifying 909 bp of the blaCTX-M gene. In case of ambiguities and additional set of primers (CTF-F: 5’-GACAGACTATTCATGTTGTTG-3’ and CTF-R: 5’-CGATTGCGGAAAAGCACGTC-3’) was used to differentiate blaCTX-M-15 from blaCTX-M-28. All CTX-M products were sequenced with both forward and reverse primers using the automated sequencer ABI Prism® 3100 (Life technologies, Road Grand Island, USA). The blastN algorithm of NCBI (http://www.ncbi.nlm.nih.gov/blast/) was to identify the ESBL alleles.
SHV-F (5’-GCAAAACGCCGGGTTATTC-3’) and SHV-R (5’-GGTTAGCGTTGCCAGTGCT-3’) were used to amplify 940 bp of the blaSHV gene. PCR using the primers TEM-F (5’-ATGAGTATTCAACATTTCCG-3’) and TEM-R (5-TTAATCAGTGAGGCACCTAT-3’) was used to amplify 851 bp of the blaTEM gene [22].
Whole genome sequencing
Twenty five CTX-M-15-producing Escherichia coli strains were randomly selected for whole genome sequencing (WGS). Genomic DNA was isolated using Purelink Genome DNA Mini kit (Invitrogen, Darmstadt, Germany) according to the manufacturer’s instruction. WGS was carried out on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) with an Illumina Nextera XT library with 2x300bp paired-end reads. The reads were assembled using SPAdes (version 3.0) [23]. The assembled contigs were analysed and examined for the presence of transferrable resistance genes, virulence genes, multi-locus sequence types, and plasmid replicon types using ResFinder, VirulenceFinder, MLST 2.0, and PlasmidFinder software, [24–27], respectively.
The location of blaCTX-M-15 was determined by extracting the contigs harbouring blaCTX-M-15 and studying the genes flanking blaCTX-M-15 gene using NCBI blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Data analysis
Data were double-entered into Microsoft Excel and analysed using STATA version 11. Results were summarized using proportions (%) for categorical data and medians (IQR) for continuous variables. Categorical variables were compared using either Pearson’s Chi–squared or Fisher’s exact test, where appropriate. To determine the predictors of ESBL carriage, univariate followed by multivariate logistic regressions analysis was performed. The predictors tested included age, sex, number of residents in a household, antibiotic use in the last month, admission history and presence of animals at home. Odd ratios with respective 95 % confidence interval (CI) were reported. Predictors with a p-value of less than 0.05 were considered statistically significant.
Limitations of the study
In this study the primers pair used are not for amplification of all CTX-M groups, and might have introduced a great bias towards group 1 to which CTX-M-15 belongs. However, the sequence covered aligned clearly align to the product with CTX-M-15 standard and this was further confirmed for the 25 isolates which underwent WGS. The WGS of 25 randomly selected isolates also confirmed the presence CTX-M-15 in all 25 isolates. Finally, 13 ESBL isolates could not be recovered for PCR and sequencing.
Results
Demographic
Of 334 humans sampled, 196 (58.7 %) were female. The median age was ten years (IQR 5–23). All sampled participants used tap water. The median number of children in a household was three (IQR 2–4). The majority of participants 156/334 (46.7 %) were from Igogo rural district (Table 1).
ESBL carriage and rates of ESBL by isolates
Of 334 individuals from the Mwanza city community, 55 (16.5 %) were found to be colonized by ESBL-producing Enterobacteriaceae. A total of 323 lactose fermenting isolates (270 E. coli and 53 Klebsiella pneumoniae) were obtained from plain MacConkey agar plates. Out of E. coli and Klebsiella pneumoniae isolates, 42/270 (15.5 %) and 2/53 (3.8 %) were ESBL producers, respectively (p = 0.026). E. coli (42/55; 76.3 %) formed the majority of ESBL isolates in this population. A total of eleven ESBL isolates were other Enterobacteriaceae species (Enterobacter spp: n = 4, Proteus mirabilis: n = 5 and Proteus vulgaris: n = 2). Their ESBL rates could not be calculated because these isolates were not targeted for in using plain MacConkey agar plates.
Susceptibility pattern
A total of 279 (E. coli, 228 and Klebsiella pneumoniae, 51) non-ESBL isolates and 55 ESBL isolates from humans stool specimens were obtained. The 55 ESBL isolates were significantly more resistant to trimethoprim/sulphamethoxazole (SXT), tetracycline (TET), gentamicin (CN) and ciprofloxacin (CIP) than the non-ESBL isolates; Table 2 (p < 0.001). All isolates were sensitive to ertapenem, meropenem, imipenem and tigecycline. The resistance rates of the isolates to TET, SXT, CN and CIP were 48.8 %, 64.9 %, 14.4 % and 12.9 % respectively.
Table 2.
Resistance rates of ESBL and Non-ESBL isolates to TET, CIP, CN and SXT
| Antibiotic | NON-ESBL (n = 279) | ESBL (n = 55) | Total (n = 334) | P value |
|---|---|---|---|---|
| Tetracycline | 119 (42.7 %) | 44 (80 %) | 163 (48.8 %) | p < 0.001 |
| Ciprofloxacin | 6 (2.1 %) | 37 (67.2 %) | 43 (12.9 %) | p < 0.001 |
| Gentamicin | 14 (5.2 %) | 34 (61.8 %) | 48 (14.4 %) | p < 0.001 |
| Co-trimoxazole | 184 (66.6 %) | 52 (94.5 %) | 216 (64.7 %) | p < 0.001 |
SXT Trimethoprim/sulphamethoxazole, TET tetracycline, CN gentamicin and CIP ciprofloxacin
Predictors of ESBL carriage
Higher median age was observed among individuals colonized with ESBLs compared to those without colonization (17 [IQR 6–38] vs. 10 [IQR 5–22], p = 0.028). On univariate analysis, the ESBL carriage significantly increased with the number of children in the household (OR = 1.34, 95 % CI 1.14–1.57, p < 0.001). Humans from Igogo were significantly more colonized by ESBL-producing isolates than those from Kirumba on univariate analysis (OR = 1.9, 95 % CI 1.0–4.2, p = 0.05).
Of 208 individuals who had a history of using antibiotics in the past one month, 44 (21.1 %) were found to be colonized with ESBL isolates as compared to 11 (8.7 %) of those with no history of antibiotic use (OR = 2.88, 95 % CI 1.3–5.66, p = 0.004). In addition, significantly higher rate of colonization was observed among individuals with history of admission in the past one year compared to those with no history (66.6 % vs. 15.1 %; p = 0.001).
On multivariate logistic regression analysis only history of admission, history of antibiotic use and increasing age were independent predictors of ESBL carriage (Table 3).
Table 3.
Predictors of ESBL carriage among 334 humans in the community in Mwanza, city
| Variables | Positive ESBL carriage (55) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| OR (95 % CI) | P value | OR (95 % CI) | P value | ||
| Age(years)a | 17 (IQR 6–17) | 1.02 (1.2–1.3) | 0.028 | 1.07 (1.04–1.10) | <0.001 |
| Number of childrena | 4 (IQR 2–5) | 1.34 (1.14–1.57) | <0.001 | 1.15 (0.95–1.39) | 0.134 |
| Sex | |||||
| Male (138) | 22 (15.9 %) | 1 | |||
| Female (196) | 33 (16.8 %) | 1.06 (0.94–1.72) | 0.828 | 1.33 (0.67–2.63) | 0.410 |
| Location | |||||
| Kirumba (112) | 13 (11.6 %) | 1 | |||
| Mbugani (66) | 10 (15.2 %) | 1.35 (0.49–3.59) | 0.49 | ||
| Igogo (156) | 32 (20.5 %) | 1.9 (1.0–4.2) | 0.05 | 1.33 (0.901–1.97) | 0.149 |
| Antibiotic use | |||||
| Yes (208) | 44 (21.2 %) | 1 | |||
| No (126) | 11 (8.7 %) | 2.8 (1.38–5.6) | 0.004 | 27 (6.63–116) | <0.001 |
| Admission history | |||||
| No (325) | 49 (15.1 %) | 1 | |||
| Yes (9) | 6 (66.7 %) | 11.3 (2.7–46.5) | 0.001 | 7.4 (1.43–38.5) | 0.017 |
amedian
ESBL alleles and resistance genes
Of 42 ESBL isolates available for typing, 37 (88.1 %) were found to carry the blaCTX-M-15 allele. Of these, 34 (91.8 %) were E. coli, two Klebsiella pneumoniae and one Enterobacter spp. The remaining five ESBL isolates (three E. coli isolates and two Enterobacter spp.) were not positive for CTX-M group 1 alleles. Further screening for the presence of SHV- and TEM-type ESBL-genes was negative. The whole genome sequence of 25 randomly selected E. coli strains confirmed all to harbour blaCTX-M-15. Analysis of the sequenced isolates revealed that in 13/25 isolates CTX-M-15 was found to be located in the chromosome.
Several other transferrable resistance genes were detected in the sequenced E. coli genomes. Aminoglycoside resistance genes detected were strA/strB (18/25, 72 %), aadA5 (16/25, 64 %), aac(6’)Ib-cr (17/25, 68 %), aac(3)-IIa (12/25, 48 %), aadA1 (5/25, 20 %) and aac(3)-IId (6/25, 24 %). Quinolone resistance genes detected were aac(6’)Ib-cr (17/25, 68 %) and qnrS1 (6/25, 24 %) (Table 4). All sequenced isolates carried trimethoprim and sulphamethoxazole resistance genes. All but two isolates harboured tetracycline resistance genes.
Table 4.
Resistance genes, sequence types and plasmid replicons of the sequenced isolates
| Isolate | Beta-Lactam genes | Other antibiotic resistance genes | Plasmid replicon type | pMLST | Sequence type | Phylogenetic group |
|---|---|---|---|---|---|---|
| RA005 | a bla CTX-M-15, bla OXA-1, bla TEM-1B | aadA5, aac(6’)Ib-cr, aac(3)-IIa | IncFIA, IncFIB, IncFII | F1:A1:B1 | ST-648 | D |
| RA023 | a bla CTX-M-15 | aadA5, qnrS1 | IncY | - | ST-4450 | A |
| RA025 | a bla CTX-M-15 | aadA5, qnrS1 | IncY | - | ST-4450 | A |
| RA034 | a bla CTX-M-15, bla OXA-1, bla TEM-1B | aadA5, aac(6’)Ib-cr, aac(3)-IIa | IncFIA, IncFIB, IncFII | F1:A1:B1 | ST-648 | D |
| RA043 | bla CTX-M-15 , bla TEM-1B | strB, strA, qnrS1 | IncY | - | ST-2852 | B1 |
| RA045 | a bla CTX-M-15, bla OXA-1 bla TEM-1B | aadA5, aac(6’)Ib-cr, aac(3)-IIa | IncFIA, IncFIB, IncFII | F1:A1:B1 | ST-648 | D |
| RA051 | bla CTX-M-15, bla OXA-1, bla TEM-1B | aadA5,aac(6’)Ib-cr, aac(3)-IIa, strA, strB | IncFIA, IncFIB, IncFII | F31:A4:B1 | ST-617 | A |
| RA061 | a bla CTX-M-15, bla OXA-1 | aadA1, aac(6’)Ib-cr, aac(3)-IIa,strA, strB | IncFIB, IncFII | F1:A-:B33 | ST-38 | D |
| RA073 | a bla CTX-M-15, bla OXA-1 | aadA5, aac(6’)Ib-cr, aac(3)-IIa,strA, strB | IncFIA, IncFIB, IncFII | F1:A1:B16 | ST-131 | B2 |
| RA085 | bla CTX-M-15, bla TEM-1B | strB, strA, qnrS1 | IncFIA, IncFIB, IncFII, IncY | F1:A1:B20 | ST-2852 | B1 |
| RA102 | a bla CTX-M-15, bla OXA-1 | aadA1, aac(6’)Ib-cr, aac(3)-IIa, strA, strB | IncFIB, IncFII | F1:A-:B33 | ST-38 | D |
| RA105 | bla CTX-M-15, bla OXA-1, bla TEM-1B | aadA5, aac(6’)Ib-cr, aac(3)-IId, strA,strB | IncFIA, IncFIB, IncFII | F87:A4:B1 | ST-617 | A |
| RA116 | a bla CTX-M-15, bla OXA-1 | aadA5, aac(6’)Ib-cr,aac(3)-IIa,strA, strB, | IncFIB, IncFIA, IncQ1, IncFII | F1:A1:B16 | ST-131 | B2 |
| RA134 | a bla CTX-M-15 | aadA1, strA, strB | No replicon | ST-205 | B1 | |
| RA166 | bla CTX-M-15, bla OXA-1 | aadA5, aac(6’)Ib-cr, aac(3)-IIa, | IncFIA, IncFIB, | F31:A4:B1 | ST-131 | B2 |
| RA173 | a bla CTX-M-15, bla OXA-1, bla TEM-1B | aadA5, aac(6’)Ib-cr, aac(3)-IId, strA,strB | IncFIB, IncFIA, IncQ1, IncFII | F48:A1:B26 | ST-131 | B2 |
| RA175 | a bla CTX-M-15, bla OXA-1, bla TEM-1B | aadA5, aac(6’)Ib-cr, aac(3)-IId, strA, strB | IncFIA, IncFIB, IncQ1, IncFII | F48:A1:B26 | ST-131 | B2 |
| RA176 | bla CTX-M-15, bla TEM-1B | strB, strA,qnrS1 | IncY | - | ST-38 | D |
| RA194 | bla CTX-M-15, bla OXA-1, bla TEM-1B | aadA5,aac(6’)Ib-cr, aac(3)-IIa, strA, strB | IncFIA, IncFIB, IncFII | F31:A4:B1 | ST-617 | A |
| RA195 | bla CTX-M-15, bla TEM-1B | aadA1, aac(3)-IId, strB, strA | IncFIB, IncFII | F1:A-:B33 | ST-38 | D |
| RA217 | bla CTX-M-15, bla OXA-1 | aadA5, aac(6’)Ib-cr,aac(3)-IIa, strA, strB | IncFIA, IncFIB, IncFII | F31:A4:B1 | ST-44 | A |
| RA218 | bla CTX-M-15, bla OXA-1 | aadA5, aac(6’)Ib-cr,aac(3)-IIa, strA, strB | IncFIB, IncFIA, IncFII | F31:A4:B1 | ST-44 | A |
| RA228 | a bla CTX-M-15, bla OXA-1 | aadA1, aac(6’)Ib-cr, aac(3)-IIa,,strA, strB | IncFIB, IncFII | F1:A-:B33 | ST-38 | D |
| RA246 | bla CTX-M-15, bla OXA-1, bla TEM-1B | aadA5, aac(6’)Ib-cr, aac(3)-IIa, strA,strB | IncFIA, IncFII | F2:A1:B- | ST-131 | B2 |
| RA256 | bla CTX-M-15, bla TEM-1B | strB, strA,qnrS1 | IncY, IncFIB | F46:A-:B24 | ST-2852 | B1 |
ain these isolates CTX-M-15 was located in the chromosome
Multi locus sequence types (MLST), phylogenetic groups and plasmid MLST
Eight different sequence types were observed in the genome-sequenced strains (Table 4). The most common ones were ST131 (6/25, 24 %), ST38 (5/25, 20 %) and ST-648 (3/25, 12 %), ST2852 (3/25, 12 %), and ST617 (3/25, 12 %), Out of 25 isolates, eight (32 %) were members of phylogroup D, seven (28 %) of phylogroup A, six (24 %) of group B2 and four of B1.
WGS-based phylogenetic tree grouped the isolates into three large clusters, all pathogenic E. coli (group B2 and D) were grouped into one cluster with 14 (56 %) isolates. The other two clusters were clonal complex (CC) ST-10 (ST-617, ST-44, all phylogenetic group A) and another cluster with a mixture of phylogroup A (ST-4450) and B1 (ST-2852 and ST-205) (Fig. 1).
Fig. 1.
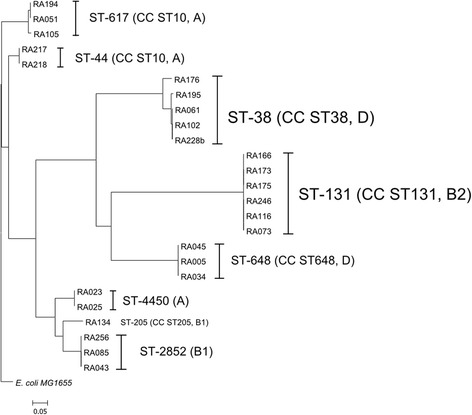
Phylogenetic tree of 25 ESBL-producing E. coli based on the whole genome rooted from E. coli MG1655 genome. The tree was produced using Harvest Suite and drawn by MEGA5 software
A total of 20 (80 %) isolates were found to carry IncF plasmids which were characterized using plasmid-based multi-locus sequence typing (pMLST) to give six different pMLST (F1:A1:B1 (3/25), F31:A4:B1 (5/25), F1:A-:B33 (4/25), F48:A1:B26 (2/25), F1:A1:B16 (2/25), F1:A1:B20, F2:A1:B-, F87:A4:B1 and F46:A-:B24 variants (Table 4).
In most cases, isolates exhibiting an identical sequence type were found to carry different plasmid types.
Virulence genes
Isolates belonging to the phylogenetic group B2 harboured the most virulence genes followed by phylogroup D isolates. Glutamate decarboxylase (gad) that confers resistance to bile salts and the increased serum survival genes (iss) were detected in 13 (52 %) of the isolates. All phylogroup B2 isolates harboured the sat and Iha genes encoding for a serine-protease autotransporter and an iron-dependent adhesion protein, respectively (Table 5).
Table 5.
Virulence factors in relation to ST and phylogenetic group
| Isolate | Sequence type | Phylogroup | Virulence gene |
|---|---|---|---|
| RA005 | ST-648 | D | iha, sat, Ipfa, nfaE, gad |
| RA023 | ST-4450 | A | gad, IpfA |
| RA025 | ST-4450 | A | gad, IpfA |
| RA034 | ST-648 | D | iha, sat, Ipfa, nfaE |
| RA043 | ST-2852 | B1 | gad, IpfA |
| RA045 | ST-648 | D | iha, sat, Ipfa, nfaE |
| RA051 | ST-617 | A | gad, iss |
| RA061 | ST-38 | D | gad, iss |
| RA073 | ST-131 | B2 | iha, sat, Ipfa, nfaE, gad, senB |
| RA085 | ST-2852 | B1 | gad, IpfA |
| RA102 | ST-38 | D | gad, iss |
| RA105 | ST-617 | A | gad, iss |
| RA116 | ST-131 | B2 | iha, sat, Ipfa, nfaE, gad, senB |
| RA134 | ST-205 | B1 | gad, IpfA |
| RA166 | ST-131 | B2 | iha, sat, cnf1, iss, gad, senB |
| RA173 | ST-131 | B2 | iha, sat, Ipfa, nfaE, gad, ireA |
| RA175 | ST-131 | B2 | iha, sat, iss, nfaE, gad, ireA |
| RA176 | ST-38 | D | gad, iss |
| RA194 | ST-617 | A | gad, iss |
| RA195 | ST-38 | D | gad, iss |
| RA217 | ST-44 | A | gad, iss |
| RA218 | ST-44 | A | gad, iss |
| RA228 | ST-38 | D | gad, iss |
| RA246 | ST-131 | B2 | nfaE, Iha, sat, gad, iss |
| RA256 | ST-2852 | B1 | gad, IpfA |
gad, Glutamate decarboxylase; iss, increased serum survival; iha, adherence protein; sat, secreted autotransporter toxin; IpfA, long polar fimbriae; nfaE, diffuse adherence fimbriae; ireA, siderophore receptor; senB, plasmid encoded enterotoxin; cnf1, cytotoxic necrotizing factor
Discussion
The presented study identifies a high proportion of faecal carriage of ESBL-producing E. coli in the Mwanza community. The prevalence of 21 % in Igogo rural district is almost the same as that of 25 % obtained in E. coli clinical isolates in the same town [5]. Compared to a previous study [28] which investigated ESBL carriage in women and neonates admitted at Bugando Medical Centre, similar findings are observed with the magnitude of carriage observed in women. However, the carriage is significantly lower than that obtained in neonates. Our findings are within the range of the ESBL carriage reported in Africa which has been found to range from 10 % in Senegal to 31 % in Niger [20, 29–31]. The high carriage in Niger could be attributed to the fact that the population studied suffered from malnutrition with majority of individuals having previous antibiotic exposure.
As observed previously [18, 28], antibiotic exposure, history of admission, or increase in age were found to predict ESBL carriage on multivariate logistic regression analysis in our setting as well.
As in previous studies [16, 29], the blaCTX-M-15 was the commonest allele observed in the community in Mwanza, Tanzania. This allele has also been found to be the commonest allele in E. coli and Klebsiella pneumoniae isolates from Bugando Medical Centre [6, 32]. This could possibly be explained by contacts between individuals admitted to the hospital, and due to the fact that the Bugando Medical Centre is the only tertiary hospital in the region. Also environmental contamination by hospital sewages and transfer through the food-chain in the city such as the fish-consumption could play a significant role [33]. This is further supported by the fact that, as in hospital E. coli isolates from a previous study [32], ST-131 was commonly found in the community. However, more studies are needed to establish the transmission pathways especially the role of food-chain and environment in the persistence and spread of ESBL isolates in the city.
Though marginally significant on univariate analysis, people from Igogo were 1.9 times more likely to carry ESBL isolates than those from Kirumba. The population of these two rural districts are almost equal. However, Igogo rural district is nearer to the Bugando Medical Centre and has more industrial and garage activities than Kirumba. These could contribute to more environmental contamination that might lead to bacteria being resistant to various metals and chemicals. In addition, the population per pharmacy/medicine shops is low at Igogo rural district as compared to that in Kirumba rural district; this might contribute to an easy accessibility to antibiotics and more environmental contamination. More research to explore human’s activities, environmental contamination and the role of antibiotic usage are needed. A geographical factor worth studying regarding the influence on ESBL carriage is the fact that in all rural communities studied a majority of the population lives in the hills, and use latrines that are not connected to city sewage.
As compared to hospital isolates [5, 32], E. coli isolated in the community were more resistant to gentamicin, sulphamethoxazole/trimethoprim and tetracycline. These results necessitate an urgent review whether these antibiotics are of value for empirical treatment of infections caused by E. coli such as urinary tract infections.
This study further confirms the role of IncF plasmids with multiple resistance genes to be responsible in transmitting resistance genes among Enterobacteriaceae isolates [32, 34]. An important finding in this study is the detection of IncY plasmids in 20 % of isolates carrying quinolone resistance gene (qnrS1) and aminoglycosides resistance genes (aadA5, strA and strB) in addition to blaCTX-M-15. Similar plasmids have been recently detected in Nigeria among healthy pregnant women [35]. In addition, 57 % (13/25) of the isolates displayed a chromosomally located CTX-M-15, thereby enabling a vertical transfer of the resistance. The findings underscore the importance to investigate the epidemiology of ESBL-producing bacteria on the African continent to ascertain transmission pathways and factors associated to the persistence.
Conclusion
High ESBL carriage, especially of blaCTX-M-15, is observed in the community among Escherichia coli in Mwanza city. Predictors for ESBL carriage are the use of antibiotic, history of hospital admission and increase in age. This information is useful for introducing pre-admission screening, planning empirical treatment by identifying high risk ESBL patients and adapting empirical treatment of infections towards covering ESBL isolates in patients with urosepsis. The findings that hospital clones and plasmids are also in the community isolates require more studies using a One Health approach to determine the role of human’s activities in relation to the persistence, circulation, spread and transmission of CTX-M-15-producing E. coli strains in Mwanza city in order to provide appropriate recommendations to control this public health threat.
Ethical considerations
The protocol to perform this study was approved by Catholic University of Health and Allied Sciences and Bugando Medical Centre (CUHAS/BMC) ethics review board (CREC/043/2014). In addition, all participants signed written informed consent for participation in the study. Where participants were children, a parent or guardian signed the consent.
Availability of data and materials
The raw data of the 25 sequenced E. coli are available at the European Nucleotide Archive (ENA) under the project number PRJEB12376.
Acknowledgements
The authors acknowledge the support provided by their respective universities and encouragement to undertake this study. We appreciate the technical assistance by Christina Gerstmann and Hiren Ghosh at the Institute of Medical Microbiology in Giessen.
Funding
This study was supported by a grant from the Wellcome Trust (WT087546MA) to SACIDS and also by grants from the Bundesministerium fuer Bildung und Forschung (BMBF, Germany) within the framework of the RESET research network (contract no. 01KI1313G) and the German Center for Infection research (DZIF/grant number 8000 701–3 [HZI] to TC and CI and TI06.001 to TC).
Abbreviations
- BMC
Bugando Medical Centre
- CC
clonal complex
- CTX
cefotaxime
- CUHAS
Catholic University of Health and Allied Sciences
- ESBL
extended spectrum beta-lactamases
- gad
glutamate decarboxylase
- iha
adherence protein
- Inc
incompatibility
- IQR
interquartile range
- iss
increased serum survival
- OR
odd ratio
- PCR
polymerase chain reaction
- sat
secreted autotransporter
- SHV
sulfhydryl variable
- ST
sequence type
- Str
streptomycin resistant
- TEM
temoniera
- Tet
tetracycline resistant
- WGS
whole genome sequence
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SEM, MMM, MM and TC designed the study. SEM, MFM, NM, RJ and JS collected the data and performed laboratory testing. CI, LF, TC performed molecular testing, whole genome sequencing and sequence analysis. SEM, MMM, RJ and LF analysed and interpreted the data. SEM, MMM and LF prepared the first draft of the manuscript. All authors read and approved the final manuscript.
References
- 1.Bell JM, Turnidge JD, Gales AC, Pfaller MA, Jones RN, Group SAS. Prevalence of extended spectrum β-lactamase (ESBL)-producing clinical isolates in the Asia-Pacific region and South Africa: regional results from SENTRY Antimicrobial Surveillance Program (1998–99) Diagn Microbiol Infect Dis. 2002;42(3):193–198. doi: 10.1016/S0732-8893(01)00353-4. [DOI] [PubMed] [Google Scholar]
- 2.Mshana SE, Matee M, Rweyemamu M. Antimicrobial resistance in human and animal pathogens in Zambia, Democratic Republic of Congo, Mozambique and Tanzania: an urgent need of a sustainable surveillance system. Ann Clin Microbiol Antimicrob. 2013;12(1):28. doi: 10.1186/1476-0711-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peirano G, Pitout JD. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25: H4. Int J Antimicrob Agents. 2010;35(4):316–321. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Aibinu I, Ohaegbulam V, Adenipekun E, Ogunsola F, Odugbemi T, Mee B. Extended-spectrum β-lactamase enzymes in clinical isolates of Enterobacter species from Lagos, Nigeria. J Clin Microbiol. 2003;41(5):2197–2200. doi: 10.1128/JCM.41.5.2197-2200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mshana SE, Kamugisha E, Mirambo M, Chakraborty T, Lyamuya EF. Prevalence of multiresistant gram-negative organisms in a tertiary hospital in Mwanza, Tanzania. BMC research notes. 2009;2(1):49. doi: 10.1186/1756-0500-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mshana SE, Hain T, Domann E, Lyamuya EF, Chakraborty T, Imirzalioglu C. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis. 2013;13(1):466. doi: 10.1186/1471-2334-13-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christopher A, Mshana SE, Kidenya BR, Hokororo A, Morona D. Bacteremia and resistant gram-negative pathogens among under-fives in Tanzania. Ital J Pediatr. 2013;39(1):27. doi: 10.1186/1824-7288-39-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manyahi J, Matee MI, Majigo M, Moyo S, Mshana SE, Lyamuya EF. Predominance of multi-drug resistant bacterial pathogens causing surgical site infections in Muhimbili national hospital. Tanzania BMC Res Notes. 2014;7(1):500. doi: 10.1186/1756-0500-7-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khanfar HS, Bindayna KM, Senok AC, Botta GA. Extended spectrum beta-lactamases (ESBL) in Escherichia coli and Klebsiella pneumoniae: trends in the hospital and community settings. J. Infect. Dev. Ctries. 2009;3(04):295–299. doi: 10.3855/jidc.127. [DOI] [PubMed] [Google Scholar]
- 10.Strömdahl H, Tham J, Melander E, Walder M, Edquist P, Odenholt I. Prevalence of faecal ESBL carriage in the community and in a hospital setting in a county of Southern Sweden. Eur J Clin Microbiol Infect Dis. 2011;30(10):1159–1162. doi: 10.1007/s10096-011-1202-5. [DOI] [PubMed] [Google Scholar]
- 11.Reuland E, Overdevest I, Al Naiemi N, Kalpoe J, Rijnsburger M, Raadsen S, Ligtenberg-Burgman I, van der Zwaluw K, Heck M, Savelkoul P. High prevalence of ESBL-producing Enterobacteriaceae carriage in Dutch community patients with gastrointestinal complaints. Clin Microbiol Infect. 2013;19(6):542–549. doi: 10.1111/j.1469-0691.2012.03947.x. [DOI] [PubMed] [Google Scholar]
- 12.Wickramasinghe NH, Xu L, Eustace A, Shabir S, Saluja T, Hawkey PM. High community faecal carriage rates of CTX-M ESBL-producing Escherichia coli in a specific population group in Birmingham, UK. J Antimicrob Chemother. 2012;67(5):1108–1113. doi: 10.1093/jac/dks018. [DOI] [PubMed] [Google Scholar]
- 13.Valenza G, Nickel S, Pfeifer Y, Eller C, Krupa E, Lehner-Reindl V, Höller C. Extended-spectrum-β-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob Agents Chemother. 2014;58(2):1228–1230. doi: 10.1128/AAC.01993-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, Clabots C, Kuskowski MA, Trott DJ. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob Agents Chemother. 2011;55(8):3782–3787. doi: 10.1128/AAC.00306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, Stubbe M, Doi Y, Zong Z, Kola A. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemotherapy. 2014;69(5):1224–30. doi: 10.1093/jac/dkt516. [DOI] [PubMed] [Google Scholar]
- 16.Meunier D, Jouy E, Lazizzera C, Kobisch M, Madec J-Y. CTX-M-1-and CTX-M-15-type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int J Antimicrob Agents. 2006;28(5):402–407. doi: 10.1016/j.ijantimicag.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Hammerum AM, Heuer OE. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin Infect Dis. 2009;48(7):916–921. doi: 10.1086/597292. [DOI] [PubMed] [Google Scholar]
- 18.Luvsansharav U-O, Hirai I, Nakata A, Imura K, Yamauchi K, Niki M, Komalamisra C, Kusolsuk T, Yamamoto Y. Prevalence of and risk factors associated with faecal carriage of CTX-M β-lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob Chemother. 2012;67(7):1769–1774. doi: 10.1093/jac/dks118. [DOI] [PubMed] [Google Scholar]
- 19.Cochran WG. Sampling techniques. New York: John Wiley & Sons; 2007.
- 20.Andriatahina T, Randrianirina F, Hariniana ER, Talarmin A, Raobijaona H, Buisson Y, Richard V. High prevalence of fecal carriage of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a pediatric unit in Madagascar. BMC Infect Dis. 2010;10(1):204. doi: 10.1186/1471-2334-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CLSI . Perfomance standards for antimicrobial susceptibility testing; twenty first information supplement vol. CLSI document M100-S21. Wayne: Clinical and Laboratory Standards Institute; 2011. [Google Scholar]
- 22.Schmiedel J, Falgenhauer L, Domann E, Bauerfeind R, Prenger-Berninghoff E, Imirzalioglu C, Chakraborty T. Multiresistant extended-spectrum beta-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol. 2014;14(1):187. doi: 10.1186/1471-2180-14-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Pontén TS, Ussery DW, Aarestrup FM. Multilocus sequence typing of total genome sequenced bacteria. J Clin Microbiol. 2012;50:1355-361. [DOI] [PMC free article] [PubMed]
- 25.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52(5):1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson E, Kayega J, Seni J, Mushi MF, Kidenya BR, Hokororo A, Zuechner A, Kihunrwa A, Mshana SE. Evaluation of existence and transmission of extended spectrum beta lactamase producing bacteria from post-delivery women to neonates at Bugando Medical Center, Mwanza-Tanzania. BMC Res Notes. 2014;7(1):279. doi: 10.1186/1756-0500-7-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruppé E, Woerther P-L, Diop A, Sene A-M, Da Costa A, Arlet G, Andremont A, Rouveix B. Carriage of CTX-M-15-producing Escherichia coli isolates among children living in a remote village in Senegal. Antimicrob Agents Chemother. 2009;53(7):3135–3137. doi: 10.1128/AAC.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lonchel CM, Meex C, Gangoué-Piéboji J, Boreux R, Assoumou M-CO, Melin P, De Mol P. Proportion of extended-spectrum ß-lactamase-producing Enterobacteriaceae in community setting in Ngaoundere, Cameroon. BMC Infect Dis. 2012;12(1):53. doi: 10.1186/1471-2334-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woerther P-L, Angebault C, Jacquier H, Hugede H-C, Janssens A-C, Sayadi S, El Mniai A, Armand-Lefèvre L, Ruppé E, Barbier F. Massive increase, spread, and exchange of extended spectrum β-lactamase–encoding genes among intestinal Enterobacteriaceae in hospitalized children with severe acute malnutrition in Niger. Clin Infect Dis. 2011;53(7):677–685. doi: 10.1093/cid/cir522. [DOI] [PubMed] [Google Scholar]
- 32.Mshana S, Imirzalioglu C, Hain T, Domann E, Lyamuya E, Chakraborty T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect. 2011;17(8):1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x. [DOI] [PubMed] [Google Scholar]
- 33.Leverstein-van Hall M, Dierikx C, Cohen Stuart J, Voets G, Van Den Munckhof M, van Essen-Zandbergen A, Platteel T, Fluit A, van de Sande-Bruinsma N, Scharinga J. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011;17(6):873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 34.Rafaï C, Frank T, Manirakiza A, Gaudeuille A, Mbecko J-R, Nghario L, Serdouma E, Tekpa B, Garin B, Breurec S. Dissemination of IncF-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from surgical-site infections in Bangui, Central African Republic. BMC Microbiol. 2015;15(1):15. doi: 10.1186/s12866-015-0348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortini D, Fashae K, Villa L, Feudi C, García-Fernández A, Carattoli A. A novel plasmid carrying bla CTX-M-15 identified in commensal Escherichia coli from healthy pregnant women in Ibadan, Nigeria. J Global Antimicrob Res. 2015;3(1):9–12. doi: 10.1016/j.jgar.2014.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data of the 25 sequenced E. coli are available at the European Nucleotide Archive (ENA) under the project number PRJEB12376.


