Abstract
Polymorphic non-coding variants at the NOS1AP locus have been associated with the common cardiac, metabolic and neurological traits and diseases. Although, in vitro gene targeting-based cellular and biochemical studies have shed some light on NOS1AP function in cardiac and neuronal tissue, to enhance our understanding of NOS1AP function in mammalian physiology and disease, we report the generation of cre recombinase-conditional Nos1ap over-expression transgenic mice (Nos1apTg). Conditional transgenic mice were generated by the pronuclear injection method and three independent, single-site, multiple copies integration event-based founder lines were selected. For heart-restricted over-expression, Nos1apTg mice were crossed with Mlc2v-cre and Nos1ap transcript over-expression was observed in left ventricles from Nos1apTg; Mlc2v-cre F1 mice. We believe that with the potential of conditional over-expression, Nos1apTg mice will be a useful resource in studying NOS1AP function in various tissues under physiological and disease states.
Keywords: Cardiac arrhythmia, Conditional over-expression, GENOME-wide association studies, NOS1AP, QT interval, Sudden cardiac death, Transgenic mouse
Introduction
NOS1AP encodes the C-terminal PDZ domain ligand of neuronal nitric oxide synthase (nNOS) and was originally cloned from a rat hippocampal cDNA library (Jaffrey et al. 1998). NOS1AP has an N-terminal PTB domain and a C-terminal PDZ binding region (Jaffrey et al. 1998). In neuronal tissues, NOS1AP has been shown to (a) compete with PSD95 for interaction with the nNOS PDZ domain through its C-terminus, suggesting that NOS1AP may influence nNOS function at synaptic and post-synaptic structures (Jaffrey et al. 1998), (b) act as an adaptor protein linking nNOS to its downstream targets like Dexras1 (Fang et al. 2000) and synapsins (Jaffrey et al. 2002) through its PTB (phosphotyrosine binding) domain, (c) regulate dendritic spine formation and patterning at synapses through its PTB-domain mediated interactions with carboxypeptidase E (Carrel et al. 2009) and Scribble/Git1 (Richier et al. 2010), and (d) mediate activation of the nNOS-p38MAPK pathway during neuronal death from excitotoxicity (Li et al. 2013). NOS1AP also forms a protein complex with SCRIB and VANGL to regulate cell polarity and migration, and is associated with breast cancer progression (Anastas et al. 2012).
In addition to these cellular and biochemical studies, mainly focused on NOS1AP function in neuronal tissues, genetic association studies have identified common noncoding variants mapping at the locus encompassing NOS1AP to be associated with complex cardiac, metabolic and neuronal traits/diseases including the electrocardiographic QT interval (Arking et al. 2006; Newton-Cheh et al. 2009; Pfeufer et al. 2009), type 2 diabetes (Becker et al. 2008; Prokopenko et al. 2009) and schizophrenia (Brzustowicz et al. 2004). The QT interval-associated variants at the NOS1AP locus are also associated with risk for sudden cardiac death (SCD) in the general population (Eijgelsheim et al. 2009; Kao et al. 2009) with a hazard ratio of ~1.4 and act as genetic modifiers of long QT syndrome (LQTS) phenotype by influencing QT interval duration and enhancing SCD risk up to tenfold (Crotti et al. 2009; Tomas et al. 2010). However, like most other genetic association studies, the identity, function and mechanisms of action of the underlying noncoding sequence variants and genes remain unknown. There is limited knowledge of NOS1AP function in non-neuronal tissues, a function revealed through genetic association studies. Knockdown of nos1ap expression in zebrafish using morpholinos leads to shortened action potential duration (APD) in excised hearts from developing embryos (Milan et al. 2009), and over-expression of Nos1ap in guinea pig ventricular myocytes using in vivo gene transfer leads to shortened APD mediated by inhibition of L-type calcium currents (Chang et al. 2008). These findings suggest that altered NOS1AP expression level influences cardiac cellular electrophysiology and thus is the most likely causal gene underlying trait association and disease risk. Since only noncoding variants at the NOS1AP locus have been associated with common cardiac, metabolic and neuronal traits/diseases in humans, it is likely that expression level of NOS1AP in various cell types is the primary mechanism through which NOS1AP influences disease risk and trait variation.
Existence of gene targeting methods including a conditional knockout and conditional over-expression design make mice an ideal model system to study gene function. NOS1AP function in neuronal and cardiac tissues has so far been evaluated using in vitroand ex vivo experimental systems. To gain further insights into NOS1AP function at the tissue, organ and organismal levels, here we report the creation of a cre recombinase-conditional, Nos1ap-over-expression transgenic mouse and evaluation of heart-restricted Nos1ap over-expression. In parallel, the International Knockout Mouse Consortium has recently generated the Nos1ap conditional knockout mice (KOMP-CSD ID: 84676).
Materials and methods
Generation of cre recombinase-conditional Nos1ap over-expression transgenic mice
Mouse Nos1ap full length ORF (NM_001109985) was PCR-amplified from a mouse embryonic day 17.5 cDNA library (Clontech, CA) using gene-specific primers (Nos1apORF_XhoIF and Nos1apORF_No-tIR; Supplementary Table 1). The 1.5 kb Nos1ap ORF ampilcon was cloned into the pCLIP vector (George et al. 2007) (gift from Andras Nagy) as a XhoI-NotI fragment for conditional transgene expression. The identity of the conditional clone was confirmed by Sanger sequencing of the entire insert using cloning primers and internal gene-specific sequencing primers Nos1apORF_IntF and Nos1apORF_IntR (Supplementary Table 1). The sequence of loxP sites in the vector backbone was confirmed using the sequencing primers loxP_Seq and Nos1apORF_IntR (Supplementary Table 1). The production of conditional transgenic mice from pronuclear injection of ~12.1 kb long ScaI-linearized Nos1ap-pCLIP plasmid DNA into fertilized mouse eggs (FVB strain) was performed by the Texas A&M Institute for Genomic Medicine.
Characterization of conditional transgenic founders
Conditional transgenic founders were identified by PCR specific to lacZ (LacZ_F and LacZ_R primers; Supplementary Table 1) and mouse Nos1ap cDNA (Nos1ap_cDNA_F and Nos1ap_cDNA_R primers; Supplementary Table 1), and by Southern blotting using lacZ-specific probes (amplified from pCLIP using SouthernProbe_F and SouthernProbe_R primers; Supplementary Table 1). Southern blot analysis of genomic DNA isolated from mouse tail- tips was performed following standard protocols using a 32P-labeled lacZ probe (Perkin Elmer, MA). The transgene DNA has two EcoRV sites that lead to a ~6.8 kb long restriction fragment when probed with a lacZ-specific probe. Transgene insertion sites were identified using fluorescent in situ hybridization (FISH) as described by Dutra et al. (1996). Briefly, metaphase chromosomes were prepared from cultured peripheral blood cells from tail vein bleeds by doing a mitotic arrest with colcemid (0.1 μg/ml, 30 min), followed by hypotonic treatment (0.075 M KCl, 20 min, 37 °C) and fixation with methanol/acetic acid (3:1). On each slide 50 ng Nos1ap-pCLIP plasmid DNA-derived probe was applied along with tenfold excess of Cot1 DNA for repeat blocking. Hybridization mixture (10 μl) containing the labeled probe in 50 % (v/v) formamide, 2× sodium saline citrate, and 10 % (w/v) dextran sulfate was denatured at 75 °C for 10 min and incubated at 37 °C for 30 min for pre-annealing, and then added to denatured slides and hybridized at 37 °C for at least 18 h. After washes slides were counter stained with 4′,6-diamidino-2-phenylindole.
Mouse husbandry, breeding and genotyping
All protocols for animal care, use and euthanasia were reviewed and approved by the Institutional Animal Care and Use Committee of Johns Hopkins University (Protocol MO12M412) and were in accordance with Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines. All animals were fed a standard rodent chow ad libitum. Genomic DNA was isolated from the tail-tips of 3 weeks old mice following standard methods. FVB mice were obtained from The Jackson Laboratory (FVB/NJ; 001800). The conditional transgenic mice were maintained on a pure inbred background by breeding to FVB mice (FVB-Nos1apTg) and were genotyped by PCR specific to lacZ (LacZ_F and LacZ_R primers; Supplementary Table 1) and mouse Nos1ap cDNA (Nos1ap_cDNA_F and Nos1ap_cD-NA_R primers; Supplementary Table 1). Mice carrying targeted knock-in of cre at the myosin light chain 2v gene (Mlc2v-cre, 129/SvJ and Black Swiss mixed background) (Chen et al. 1998) were provided by Kenneth R. Chien (Massachusetts General Hospital, Boston), and were genotyped for the knock-in cre allele using cre-specific primers (Mlc2v-cre_F and Mlc2c-cre_R; Supplementary Table 1). Mlc2v-cre mice were maintained on a mixed background by breeding to FVB mice. For heart-restricted over-expression, FVB-Nos1apTg mice were crossed with Mlc2v-cre mice to generate Nos1apTg; Mlc2v-cre F1 mice. All molecular analyses of heart-restricted Nos1ap over-expression were performed in these adult (3–4 months) F1 mice and their wild-type control littermates.
The cre-recombinase conditional Nos1ap over-expression transgenic mice described here will be available to the research community upon acceptance of the manuscript.
RNA expression
Adult mice were euthanized using inhaled isoflurane in a closed chamber. Left ventricles were dissected and snap-frozen using liquid N2 and stored at −80 °C. Total RNA was isolated from ~10 mg dry tissue (frozen tissue) using TRIzol. DNase digestion and RNA clean-up was performed using RNeasy Mini kit and RNase-Free DNase set (Qiagen, CA), following manufacturer’s instructions. cDNA was synthesized by oligo-dT primed reverse transcription performed on 1 μg total RNA using SuperScript III First-Strand Synthesis System (Invitrogen, NY), following manufacturer’s instructions. Quantitative expression analysis of Nos1ap was performed using mouse-specific Taq-Man Gene Expression assay (Mm01290688_m1) (Applied Biosystems, NY). Real-time quantitative PCR (qPCR) was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems, NY) and analyzed using Sequence Detection System Software v.2.1 (Applied Biosystems, NY). Expression was measured in technical triplicates and the averages of the threshold cycle (Ct) values were used for analysis. Actb expression, assessed using mouse Actb Endogenous Control TaqMan Gene Expression assay (Applied Biosystems, NY), was used for normalization.
Statistical analyses
All comparisons between two groups of mice were performed by comparing means using Student’s t test. We used an alpha level of 0.05 for all statistical tests.
Results and discussion
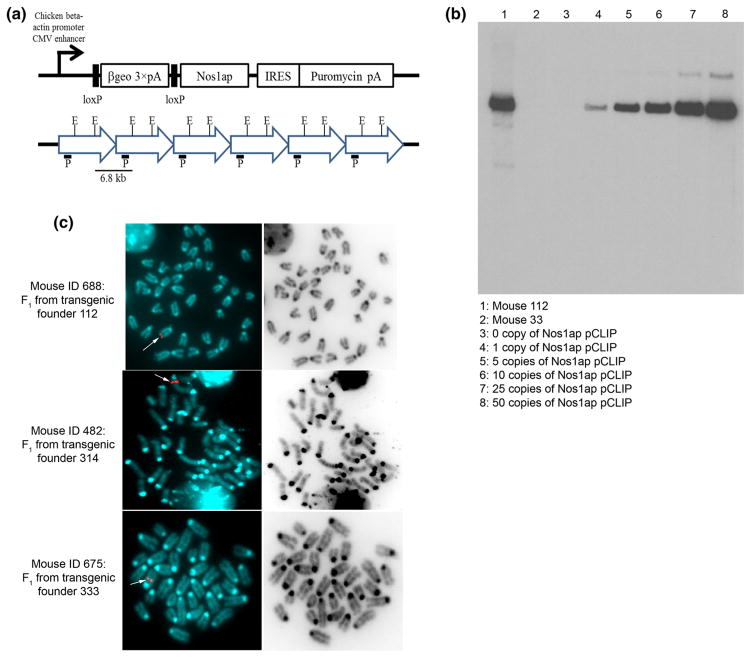
For generation of cre recombinase-conditional Nos1ap over-expression transgenic mice, Nos1ap ORF (NM_001109985), with start and stop codons, was cloned as an XhoI-NotI fragment into the pCLIP vector (George et al. 2007) downstream of the loxP-flanked (floxed) lacZ-neomycin fusion gene (βgeo) (Fig. 1). In this construct, a CMV enhancer combined with a chicken β-actin promoter (pCAGG) (Okabe et al. 1997) drives the expression of floxed βgeo followed by three polyadenylation (pA) signals. These components are followed by the Nos1ap ORF, which is then followed by an IRES-puromycin-pA cassette. We used the pCAGG (Okabe et al. 1997) to drive the expression of a conditional transgene since the Nos1ap endogenous promoter is not characterized. We used pCLIP as the vector for its ability to generate cre recombinase-conditional over-expression as in the presence of cre recombinase, the floxed βgeo cassette is deleted, leaving the gene of interest under the control of a strong constitutive enhancer-promoter element. We utilized the cre-loxP system (Sauer and Henderson 1988; Sternberg et al. 1981) for creating conditional mice which can be used to study the effects of both tissue-restricted as well as ubiquitous over-expression of Nos1ap. Although pCLIP has two selection markers, neomycin and puromycin, we did not use these selection markers in generation of transgenic mice. ~12.1 kb long ScaI-linearized Nos1ap-pCLIP plasmid DNA was used for pronuclear injection of fertilized mouse eggs (FVB) to create the conditional transgenic founders.
Fig. 1.
Generation of cre recombinase-conditional Nos1ap over-expression transgenic mice. a Nos1ap pCLIP transgene construct (adapted from George et al. 2007), and representation for multiple copy head-to-tail insertion of the transgene and the expected 6.8 kb Southern band with lacZ probe (P) and EcoRV (E) digestion; βgeo β-galactosidase-neomycin fusion gene, pA signal, IRES internal ribosome entry site; b identification of transgenic founder by Southern blotting. Southern blotting performed on mouse tail genomic DNA digested with EcoRV and probed using lacZ specific probe. Lanes 3–8 have wild type mouse tail genomic DNA spiked with zero or multiple copies, as indicated, of Nos1ap pCLIP plasmid DNA per diploid genome. Mouse 112 was selected as a founder line for further experiments; c single site transgene integration in cre recombinase-conditional Nos1ap over-expression transgenic lines. Metaphase FISH using Nos1ap pCLIP plasmid specific probe performed in cultured peripheral blood cells of F1 mice derived from cross between 112, 314, and 333 transgenic founders and FVB mice. Red dots in the left panel (white arrow) indicate site of integration on chromosome 4 (top), chromosome 12 (middle) and chromosome 2 (bottom) for 112, 314 and 333 lines, respectively
Out of a total of 35 live pups, three transgenic founders (Nos1apTg, IDs 112, 314 and 333) were identified by PCR on genomic DNA using primers specific to lacZ and Nos1ap cDNA, and were confirmed by Southern blotting using a lacZ-specific probe (Fig. 1, data not shown). These three founder mice were crossed with FVB mice to generate FVB-Nos1apTg F1 mice for maintenance of individual lines (henceforth referred to as the 112, 314 and 333 lines). We then analyzed FVB-Nos1apTg F1 mice to identify the site of transgene integration by FISH using a Nos1ap-pCLIP plasmid DNA-derived probe. All three lines were found to have a single site of transgene integration, in chromosome 4, chromosome 12 and chromosome 2 for the 112, 314 and 333 lines, respectively (Fig. 1). Normal Mendelian segregation of the transgenic locus was observed for all 3 lines and FVB-Nos1apTg F1 mice were indistinguishable in appearance and behavior from wild type littermates.
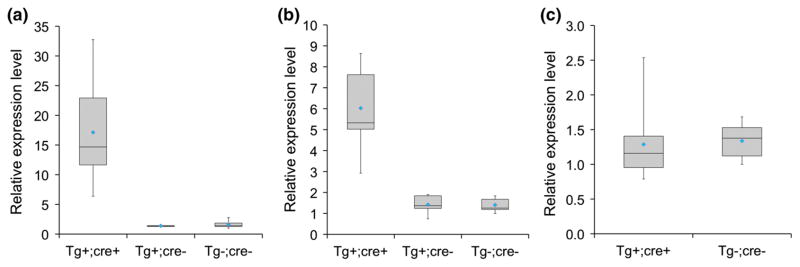
For heart-restricted over-expression, FVB-Nos1apTg cre-conditional mice from the 112, 314 and 333 lines were crossed with Mlc2v-cre driver mice (Chen et al. 1998) to generate Nos1apTg; Mlc2v-cre F1 mice. The FVB-Nos1apTg cre-conditional transgenic mice could also be used to study the effects of global Nos1ap over-expression by crossing with a ubiquitous cre line such as CMV-cre (Schwenk et al. 1995). cre expression in the knock-in Mlc2v-cre mice recapitulates endogenous Mlc2v expression (Chen et al. 1998) and is expressed in heart starting as early as embryonic day 9.5 (Robson et al. 2010). Expression of Nos1ap transcript was evaluated in left ventricles excised from adult (3–4 months old) F1 mice. Nos1ap transcript was found to be overexpressed in left ventricles of Nos1apTg; Mlc2v-cre mice from the 112 and 333 lines as compared to wild type littermates, t(18) = 5.77, p <0.001 for the 112 line and t(12) = 5.98, p < 0.001 for the 333 line (Fig. 2). On average Nos1apTg; Mlc2v-cre mice from the 112 line had higher expression levels as compared to mice from the 333 line. Nos1ap expression in left ventricles from mice carrying the Nos1apTg locus, but not the Mlc2v-cre locus, was comparable to wild type littermates in the 112 and the 333 lines, t(11) = 0.66, p = 0.52 for the 112 line and t(10) = 0.07, p = 0.94 for the 333 line, indicating that over-expression from Nos1apTg locus is not “leaky” and is dependent on the presence of cre recombinase (Fig. 2). No over-expression of Nos1ap transcript was observed in left ventricles of Nos1apTg; Mlc2v-cre mice from the 314 line, t(13) = 0.22, p = 0.83, indicating that the Nos1apTg locus is transcriptionally silenced (Fig. 2).
Fig. 2.
Nos1apTg; Mlc2v-cre (Tg+;cre+) mice from two transgenic lines overexpress Nos1ap transcript in left ventricles. a Box-and-Whisker plots showing relative expression levels for Nos1ap transcript in left ventricles from Tg+;cre+ (n = 10), Tg+;cre− (conditional transgene only, n = 3) and wild type control littermates (Tg−;cre−, n = 10) mice derived from transgenic founder 112. Expression of Nos1ap transcript was higher in Tg+;cre+ mice as compared to Tg−;cre− mice, t(18) = 5.77, p <0.001 and expression of Nos1ap transcript was not significantly different between Tg+;cre− and Tg−;cre−mice, t(11) = 0.66, p = 0.52; b same as a, mice derived from transgenic founder 333 (Tg+;cre+ n = 7, Tg+;cre− n = 5, Tg−;cre− n = 7). Expression of Nos1ap transcript was higher in Tg+;cre+ mice as compared to Tg−;cre− mice, t(12) = 5.98, p <0.001 and expression of Nos1ap transcript was not significantly different between Tg+;cre− and Tg−;cre− mice, t(10) = 0.07, p = 0.94; c Same as a, mice derived from transgenic founder 314 (Tg+;cre+ n = 8, Tg−;cre− n = 7). No significant difference in expression level was observed between the two groups of mice derived from transgenic founder 314, t(13) = 0.22, p = 0.83
We chose to generate Nos1ap over-expression transgenic mice because of our interests in understanding cardiac repolarization, cardiac arrhythmias and SCD. Of the nearly dozen genes associated with QT interval (Newton-Cheh et al. 2009; Pfeufer et al. 2009), a quantitative and reproducible measure of ventricular repolarization, we chose Nos1ap because: (a) it is the single largest contributor to QT interval variation, (b) not much is known about its cardiac function in physiology and disease, and (c) genetic variations at NOS1AP are also associated with increased risk for SCD (Eijgelsheim et al. 2009; Kao et al. 2009), act as modifiers of LQTS (Crotti et al. 2009; Tomas et al. 2010), and are associated with other common complex traits including schizophrenia (Brzustowicz et al. 2004) and glucose metabolism (Becker et al. 2008; Prokopenko et al. 2009). In fact, Nos1ap is the first gene to be associated with electrical propagation defects in the cardiac system. The cre-conditional design of the transgenic line can be used to overexpress Nos1ap in a variety of tissue types, both in adults and embryos, and we thus believe that these mice would be a useful resource. And to the best of our knowledge this is the first report of creation of a Nos1ap over-expression allele in mice.
Supplementary Material
Acknowledgments
We are grateful to Dr. Andras Nagy (Mount Sinai Hospital, Toronto) for providing the pCLIP vector and to Dr. Kenneth Chien (Massachusetts General Hospital, Boston) for providing the Mlc2v-cre mice. We are thankful to Dr. Gordon Tomaselli, Dr. David Kass and Gary Steel (Johns Hopkins University, Baltimore) for critical discussions. This work was supported in part by funds from the Donald R. Reynolds Foundation and the National Institutes of Health.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10529-014-1473-x) contains supplementary material, which is available to authorized users.
Conflict of interest Aravinda Chakravarti is on the Scientific Advisory Board of Biogen Idec and this potential competing interest is managed by the policies of the Johns Hopkins University School of Medicine.
Contributor Information
Dallas R. Auer, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, 733 N. Broadway, Baltimore, MD 21205, USA
Polina Sysa-Shah, Email: psysa1@jhmi.edu, Department of Molecular and Comparative Pathobiology, Johns Hopkins University School of Medicine, 733 N. Broadway, Baltimore, MD 21205, USA.
Djahida Bedja, Email: dbedja1@jhmi.edu, Department of Molecular and Comparative Pathobiology, Johns Hopkins University School of Medicine, 733 N. Broadway, Baltimore, MD 21205, USA.
Jessica L. Simmers, Email: jlsimmers@gmail.com, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, 733 N. Broadway, Baltimore, MD 21205, USA
Evgenia Pak, Email: epak@mail.nih.gov, National Human Genome Research Institute, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892, USA.
Amalia Dutra, Email: adutra@mail.nih.gov, National Human Genome Research Institute, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892, USA.
Ronald Cohn, Email: ronald.cohn@sickkids.ca, The Hospital for Sick Children, University of Toronto, 555 University Avenue, Toronto, ON M5G 1X8, Canada.
Kathleen L. Gabrielson, Email: kgabriel@jhmi.edu, Department of Molecular and Comparative Pathobiology, Johns Hopkins University School of Medicine, 733 N. Broadway, Baltimore, MD 21205, USA
Aravinda Chakravarti, Email: aravinda@jhmi.edu, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, 733 N. Broadway, Baltimore, MD 21205, USA.
Ashish Kapoor, Email: ashish@jhmi.edu, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, 733 N. Broadway, Baltimore, MD 21205, USA.
References
- Anastas JN, Biechele TL, Robitaille M, Muster J, Allison KH, Angers S, Moon RT. A protein complex of SCRIB, NOS1AP and VANGL1 regulates cell polarity and migration, and is associated with breast cancer progression. Oncogene. 2012;31:3696–3708. doi: 10.1038/onc.2011.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Pfeufer A, Post W, Kao WH, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- Becker ML, Aarnoudse AJ, Newton-Cheh C, Hofman A, Witteman JC, Uitterlinden AG, Visser LE, Stricker BH. Common variation in the NOS1AP gene is associated with reduced glucose-lowering effect and with increased mortality in users of sulfonylurea. Pharmacogenet Genom. 2008;18:591–597. doi: 10.1097/FPC.0b013e328300e8c5. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Simone J, Mohseni P, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS. Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am J Hum Genet. 2004;74:1057–1063. doi: 10.1086/420774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel D, Du Y, Komlos D, Hadzimichalis NM, Kwon M, Wang B, Brzustowicz LM, Firestein BL. NOS1AP regulates dendrite patterning of hippocampal neurons through a carboxypeptidase E-mediated pathway. J Neurosci. 2009;29:8248–8258. doi: 10.1523/JNEUROSCI.5287-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KC, Barth AS, Sasano T, Kizana E, Kashiwakura Y, Zhang Y, Foster DB, Marban E. CAPON modulates cardiac repolarization via neuronal nitric oxide synthase signaling in the heart. Proc Natl Acad Sci USA. 2008;105:4477–4482. doi: 10.1073/pnas.0709118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kubalak SW, Minamisawa S, Price RL, Becker KD, Hickey R, Ross J, Jr, Chien KR. Selective requirement of myosin light chain 2v in embryonic heart function. J Biol Chem. 1998;273:1252–1256. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- Crotti L, Monti MC, Insolia R, Peljto A, Goosen A, Brink PA, Greenberg DA, Schwartz PJ, George AL., Jr NOS1AP is a genetic modifier of the long-QT syndrome. Circulation. 2009;120:1657–1663. doi: 10.1161/CIRCULATIONAHA.109.879643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra AS, Mignot E, Puck JM. Gene localization and syntenic mapping by FISH in the dog. Cytogenet Cell Genet. 1996;74:113–117. doi: 10.1159/000134395. [DOI] [PubMed] [Google Scholar]
- Eijgelsheim M, Newton-Cheh C, Aarnoudse AL, van Noord C, Witteman JC, Hofman A, Uitterlinden AG, Stricker BH. Genetic variation in NOS1AP is associated with sudden cardiac death: evidence from the Rotterdam study. Hum Mol Genet. 2009;18:4213–4218. doi: 10.1093/hmg/ddp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- George SH, Gertsenstein M, Vintersten K, Korets-Smith E, Murphy J, Stevens ME, Haigh JJ, Nagy A. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:4455–4460. doi: 10.1073/pnas.0609277104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Snowman AM, Eliasson MJ, Cohen NA, Snyder SH. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron. 1998;20:115–124. doi: 10.1016/s0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Benfenati F, Snowman AM, Czernik AJ, Snyder SH. Neuronal nitric-oxide synthase localization mediated by a ternary complex with synapsin and CAPON. Proc Natl Acad Sci USA. 2002;99:3199–3204. doi: 10.1073/pnas.261705799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao WH, Arking DE, Post W, Rea TD, Sotoodehnia N, Prineas RJ, Bishe B, Doan BQ, Boerwinkle E, Psaty BM, Tomaselli GF, Coresh J, Siscovick DS, Marban E, Spooner PM, Burke GL, Chakravarti A. Genetic variations in nitric oxide synthase 1 adaptor protein are associated with sudden cardiac death in US white community-based populations. Circulation. 2009;119:940–951. doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LL, Ginet V, Liu X, Vergun O, Tuittila M, Mathieu M, Bonny C, Puyal J, Truttmann AC, Courtney MJ. The nNOS-p38MAPK pathway is mediated by NOS1AP during neuronal death. J Neurosci. 2013;33:8185–8201. doi: 10.1523/JNEUROSCI.4578-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, Arking DE, Amsterdam AH, Sabeh KM, Mably JD, Rosenbaum DS, Peterson RT, Chakravarti A, Kaab S, Roden DM, MacRae CA. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120:553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- Pfeufer A, Sanna S, Arking DE, Muller M, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko I, Zeggini E, Hanson RL, Mitchell BD, et al. International Type 2 Diabetes 1q Consortium (2009) Linkage disequilibrium mapping of the replicated type 2 diabetes linkage signal on chromosome 1q. Diabetes. 58:1704–1709. doi: 10.2337/db09-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richier L, Williton K, Clattenburg L, Colwill K, O’Brien M, Tsang C, Kolar A, Zinck N, Metalnikov P, Trimble WS, Krueger SR, Pawson T, Fawcett JP. NOS1AP associates with Scribble and regulates dendritic spine development. J Neurosci. 2010;30:4796–4805. doi: 10.1523/JNEUROSCI.3726-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson A, Allinson KR, Anderson RH, Henderson DJ, Arthur HM. The TGFbeta type II receptor plays a critical role in the endothelial cells during cardiac development. Dev Dyn. 2010;239:2435–2442. doi: 10.1002/dvdy.22376. [DOI] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acid Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Hamilton D, Hoess R. Bacteriophage P1 site-specific recombination. II. Recombination between loxP and the bacterial chromosome. J Mol Biol. 1981;150:487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]
- Tomas M, Napolitano C, De Giuli L, Bloise R, Subirana I, Malovini A, Bellazzi R, Arking DE, Marban E, Chakravarti A, Spooner PM, Priori SG. Polymorphisms in the NOS1AP gene modulate QT interval duration and risk of arrhythmias in the long QT syndrome. J Am Coll Cardiol. 2010;55:2745–2752. doi: 10.1016/j.jacc.2009.12.065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.