Abstract
Vemurafenib, a RAF inhibitor, extends survival in patients with BRAFV600-mutant melanoma but activates extracellular signal–regulated kinase (ERK) signaling in RAS-mutant cells. In a patient with a BRAFV600K-mutant melanoma responding to vemurafenib, we observed accelerated progression of a previously unrecognized NRAS-mutant leukemia. We hypothesized that combining vemurafenib with a MAP–ERK kinase (MEK) inhibitor would inhibit ERK activation in the melanoma and prevent ERK activation by vemurafenib in the leukemia, and thus suppress both malignancies. We demonstrate that intermittent administration of vemurafenib led to a near-complete remission of the melanoma, and the addition of the MEK inhibitor cobimetinib (GDC-0973) caused suppression of vemurafenib-induced leukemic proliferation and ERK activation. Antimelanoma and antileukemia responses have been maintained for nearly 20 months, as documented by serial measurements of tumor-derived DNA in plasma in addition to conventional radiographic and clinical assessments of response. These data support testing of intermittent ERK pathway inhibition in the therapy for both RAS-mutant leukemia and BRAF-mutant melanoma.
SIGNIFICANCE
We show that in a patient with simultaneous RAS-mutant leukemia and BRAF-mutant melanoma, intermittent RAF inhibitor therapy induced a near-complete melanoma response, and addition of a MEK inhibitor prevented RAF inhibitor-induced activation of the RAS-mutant leukemia. Intermittent therapy may permit greater pathway inhibition with less toxicity, avoid chronic relief of pathway feedback, and have enhanced effectiveness compared with chronic administration.
INTRODUCTION
Activating mutations at the V600 codon of BRAF are found in 40% to 60% of melanomas. These mutations lead to hyperactivation of the extracellular signal–regulated kinase (ERK) pathway, which causes feedback inhibition of RAS activation and maintains the RAF kinases in a monomeric state. Currently available ATP-competitive RAF inhibitors, such as vemurafenib, bind to BRAFV600E monomer and thus inhibit its catalytic activity and activation of ERK signaling. Vemurafenib leads to clinically significant responses in nearly half of patients with BRAFV600E/K-mutated melanoma and improves progression-free and overall survival (1). This led to U.S. Food and Drug Administration (FDA) approval of vemurafenib in 2011. In contrast, in cells with sufficient levels of RAS activation, RAF forms activated dimers. Binding of vemurafenib and other RAF inhibitors to one member of the dimer pair results in transactivation of the other RAF molecule and causes activation of ERK signaling (2–4). This may stimulate proliferation of tumors with active RAS.
We previously reported a patient with metastatic BRAFV600K-mutant melanoma who, when treated with vemurafenib, experienced dramatic shrinkage of his melanoma but induction of proliferation of a previously unsuspected chronic myelomonocytic leukemia (CMML) that harbored an oncogenic NRASG12R mutation (5). In vitro , vemurafenib induced proliferation of the CMML cells, which could be blocked by concurrent MAP–ERK kinase (MEK) inhibition. We hypothesized that treating this patient with combined therapy with RAF and MEK inhibitors would treat the melanoma and reduce proliferation of the patient’s concurrent CMML.
Here, we report that combined therapy with vemurafenib and the MEK inhibitor GDC-0973 (now called cobimetinib) did indeed prevent proliferation of the CMML while maintaining a near-complete response of BRAFV600K-mutated melanoma. This was achieved and maintained with intermittent dosing of both drugs.
RESULTS
Clinical Case
The patient is a 76-year-old man with stage IV (T3aNxMIb) BRAFV600K -mutant melanoma who was started on therapy with vemurafenib in February 2012 (5). After 2 weeks of treatment, there was already a marked improvement in his melanoma, but his white blood cell (WBC) count increased to 80.9 × 103 /μL. Bone marrow evaluation revealed that the patient also had CMML harboring an NRASG12R mutation. As the CMML regressed quickly upon vemurafenib discontinuation, the patient was subsequently treated with vemurafenib at a dose of 720 mg twice daily on an intermittent dosing schedule and experienced a near-complete melanoma response. Specifically, vemurafenib was held when the patient developed toxicities (fatigue or joint pain) or when the WBC count approached 80 × 103 /μL. Once the toxicities resolved and the WBC count decreased to <20 × 103 /μL, vemurafenib was resumed. The patient was managed with intermittent dosing of single-agent vemurafenib for 49 weeks (Fig. 1A), achieving a near-complete melanoma response.
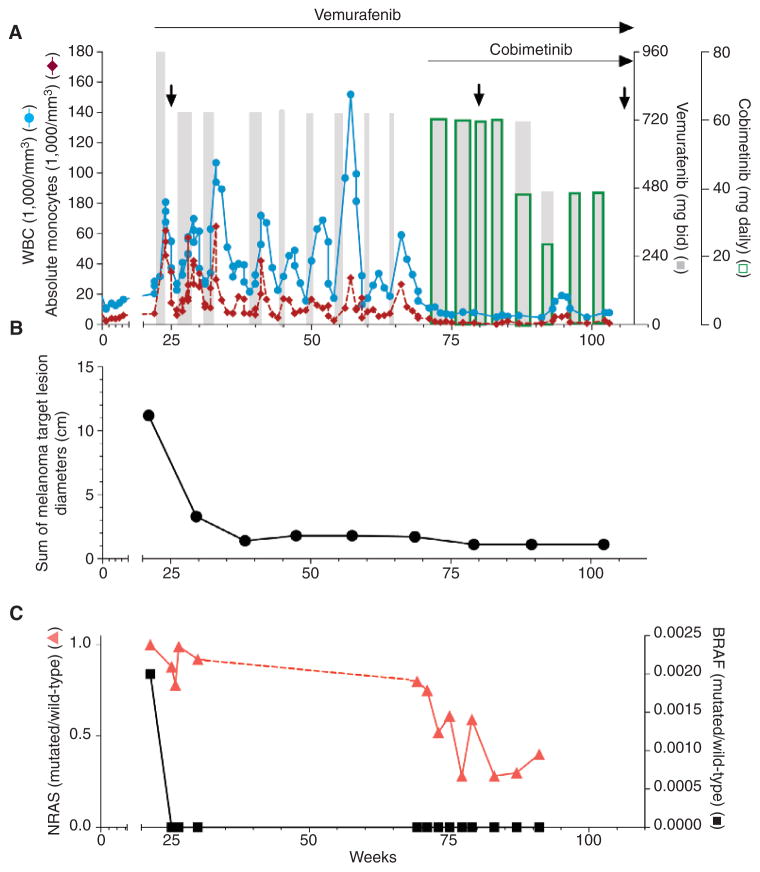
Figure 1.
Treatment and clinical course of a patient with BRAFV600K-mutant melanoma and NRASG12R-mutant CMML treated with vemurafenib and cobimetinib. A, the peripheral WBC (blue lines) and monocyte counts (red lines) throughout the treatment. Gray bars, vemurafenib therapy; open green bars, cobimetinib therapy. The width of the bars indicates duration of treatment. The relative heights of the bars reflect dose level adjustments as indicated on the right Y-axes [960, 720, and 480 mg twice a day (bid) for vemurafenib; 60, 40, and 20 mg every day for cobimetinib]. Arrows indicate times of bone marrow examinations. The antimelanoma response based on radiographic tumor measurements using Response Evaluation Criteria in Solid Tumors (RECIST) 1.1 criteria is shown in B. C, the quantification of circulating CMML-derived (red line) and melanoma-derived (black line) DNA in the plasma measured by digital PCR. Y-axes indicate the ratio of mutated DNA to wild-type DNA circulating in the plasma. No samples were available for analysis between weeks 30 and 69 (indicated by the dotted line).
In November 2012, Genentech agreed to supply cobimetinib under a single-patient use Investigator New Drug application . Cobimetinib is a selective, noncompetitive inhibitor of MEK1/2 that inhibits ERK1/2 phosphorylation in cells and proliferation at IC50 values of 1.8 and 8 nmol/L, respectively (6). The patient was started on the recommended phase II dose for cobimetinib of 60 mg orally daily for 3 weeks, followed by 1 week off the drug. Vemurafenib (720 mg orally twice a day) was administered concurrently with cobimetinib. As shown in Fig. 1A, the addition of the MEK inhibitor resulted in suppression of the CMML, and the patient achieved normal WBC counts for the first time since beginning vemurafenib. The patient initially tolerated 3 weeks of vemurafenib/cobimetinib combination therapy, but subsequently experienced fatigue and anemia that required dose adjustments of both drugs. We found that 40 mg/d of cobimetinib was sufficient to inhibit vemurafenib-induced proliferation of the CMML but that 20 mg/d was not sufficient (Fig. 1A). We ultimately found that 7- to 10-day courses of combination vemurafenib (480 mg twice a day) and cobimetinib (40 mg/d) were tolerable and were not associated with elevation of the peripheral WBC count. Drug holidays of 2 to 3 weeks (or longer) were given as needed for the resolution of adverse symptoms, largely fatigue.
Control of the peripheral WBC and monocyte counts correlated with a decrease in spleen length as assessed by computed tomography (CT) scan (Supplementary Fig. S1). Bone marrow examinations performed at weeks 57 and 85 while on combination therapy showed persistent CMML without evidence of disease progression. The patient has now been on intermittent treatment for 85 weeks; for the first 50 weeks he received only vemurafenib and for the next 35 weeks he received vemurafenib with cobimetinib. The effect on the melanoma, as assessed by CT scans, is shown in Fig. 1B and Supplementary Fig. S2A–S2C. By week 40, the patient experienced a near-complete response with only a residual 1.1-cm subcarinal lymph node observed on imaging, which has been maintained despite subsequent intermittent treatment with only RAF and MEK inhibitors.
Quantitative Assessment of Circulating Tumor DNA
Antimelanoma and anti-CMML effects were also assessed by quantifying circulating tumor-derived DNA using a digital PCR assay (7, 8). The effect of treatment on melanoma-derived (BRAFV600K) and CMML-derived (NRASG12R) DNA in the plasma throughout the treatment is shown in Fig. 1C. This methodology allows for the detection and quantification of mutant alleles with sensitivity as low as 0.001% (see Methods and Supplementary Fig. S3A and S3B). The effect of combined RAF and MEK inhibition on the levels of melanoma-derived BRAFV600K throughout the treatment was consistent with tumor volume, as assessed by radiographic evaluation shown in Fig. 1B (Supplementary Fig. S2A–S2C). In fact, once the maximal radiographic antimelanoma response was achieved, circulating melanoma-derived BRAFV600K DNA was no longer detectable. Likewise, the level of CMML-derived NRASG12R DNA in the plasma correlated with the peripheral WBC count (Fig. 1C); however, CMML-derived DNA remained detectable throughout the treatment despite normal peripheral WBC counts, consistent with the observation that CMML remained detectable in the bone marrow.
Antileukemic Efficacy of MEK Inhibition in RAS-Mutant Leukemias
Given the effect of combined RAF–MEK inhibition on the patient’s monocytosis and NRASG12R-mutant allele burden, we next assessed the effects of vemurafenib and combined vemurafenib plus cobimetinib on ERK signaling in leukemic cells throughout the treatment. We measured the levels of phosphorylated ERK (pERK) in the CD14+ cells from the peripheral blood of the patient using phosphoprotein flow cytometry. As expected and previously shown (5), activated ERK was increased in monocytes during the time of vemurafenib monotherapy as compared with the same cell population before vemurafenib therapy (Supplementary Fig. S4A). There was also an increase in the frequency of CD14+cells in the peripheral blood at this time. In contrast, monocytes analyzed from the time of combined vemurafenib plus cobimetinib had reduced ERK activation compared with the same cells during a period of observation following vemurafenib withdrawal (Supplementary Fig. S4B). Moreover, the frequency of CD14+ cells among peripheral blood mononuclear cells was lowest in the samples obtained from the time of combined vemurafenib plus cobimetinib therapy.
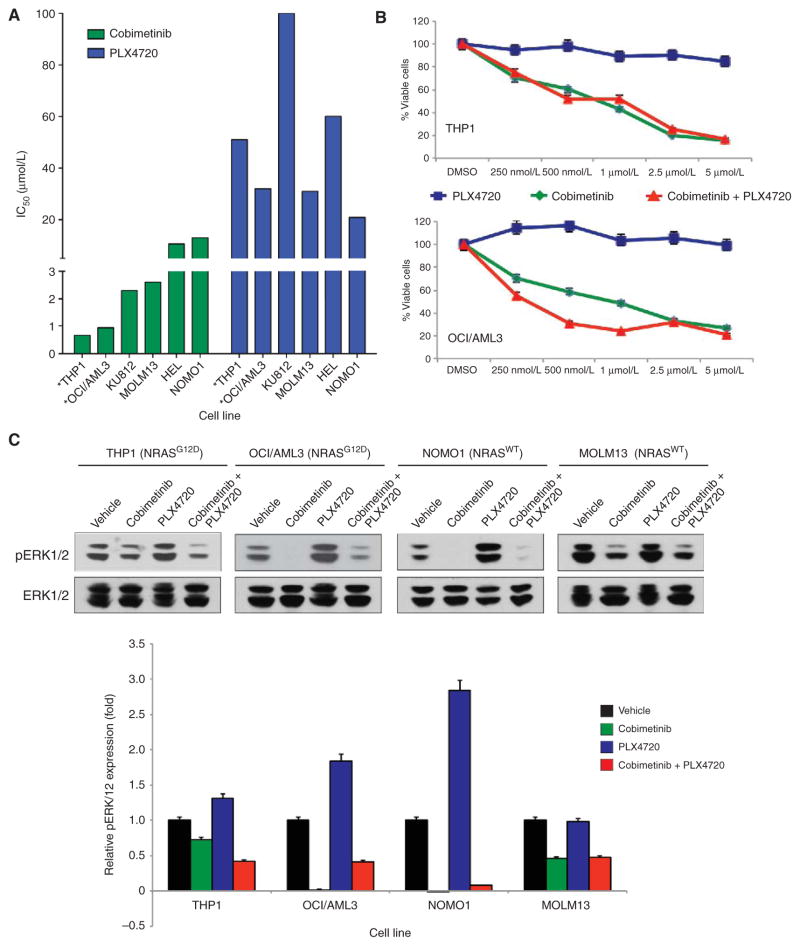
We next sought to compare the efficacy of pharmacologic inhibition of MEK versus inhibition of RAF and combined RAF–MEK inhibition in myeloid leukemia cell lines that were BRAFWT/NRASWT or BRAFWT/NRASG12D mutant. Treatment with PLX4720 alone increased ERK phosphorylation in all of these BRAF wild-type cell lines and had minimal efficacy in inducing cell death, as expected (Fig. 2A–C). In contrast, although MEK inhibition with cobimetinib had a modest and variable inhibitory effect on the NRAS wild-type cell lines (IC50, 2.3–13 μmol/L), the NRAS-mutant lines were highly sensitive to MEK inhibition (IC50, 0.66–0.94 μmol/L; Fig. 2A). Moreover, combined MEK and RAF inhibition was able to suppress NRAS-mutant leukemic proliferation. ERK phosphorylation was greater in three of four cell lines treated with combined RAF and MEK inhibition compared with MEK inhibition alone (Fig. 2C), suggesting that combined use of vemurafenib attenuated the antileukemic effect of cobimetinib.
Figure 2.
Viability curves and immunoblots revealing viability and activation of mitogen–activated protein kinase (MAPK) signaling intermediates in NRASWT and NRASG12D AML cell lines (indicated with *) treated with various concentrations of cobimetinib, PLX4720, or both. A, IC 50 values of BRAF WT/ NRASG12D and BRAFWT/NRASWT AML cell lines with exposure to cobimetinib or PLX4720. B, the cell viability of BRAFWT/NRASG12D AML cell lines with exposure to varying concentrations of cobimetinib, PLX4720, or both. DMSO, dimethyl sulfoxide. In A and B, data were derived from 72 hours of exposure to cells followed by CellTiter-Glo Luminescence assessment. C, immunoblots of pERK1/2 and total ERK1/2 (top) in NRASG12D-mutant and NRASWT human leukemia cell lines exposed to vehicle, cobimetinib, PLX4720, or PLX4720 + cobimetinib. Quantification of pERK1/2 relative to total ERK1/2 is displayed at the bottom. Cells were treated with 500 nmol/L of each drug for 24 hours.
Genetic Analysis of the Leukemia and Melanoma
Given the unique history of this patient harboring two simultaneous malignancies, we sought to define genetically both cancers in more detail, identify any potential genetic modifiers of response to RAF and MEK inhibition, and identify the development of new genetic events in the leukemia. The patient’s melanoma and the CMML were subjected to targeted sequencing of 279 genes known to be recurrently mutated across many malignancies using a bait-capture, next-generation sequencing assay, and custom-designed probes generated using the Nimblegen SeqCap system, essentially as previously described (Supplementary Table S1; ref. 9). The melanoma was sequenced from biopsy material before treatment with vemurafenib, whereas the CMML was sequenced from bone marrow mononuclear cells (BM MNC) before treatment with vemurafenib and also before introduction of cobimetinib.
In the melanoma, 28 somatic mutations in 20 separate genes were identified (Table 1 and Supplementary Table S2; DNA from buccal epithelia cells was used as paired normal). Twenty-three of 28 mutations (82%) involved a C → T or G → A transitions, indicative of UV damage. Of note, there were no mutations or copy-number alterations in p16INK4a or genetic changes suggestive of PI3K–AKT pathway activation that could activate a parallel signaling pathway [other than a PREX2A (10) somatic missense mutation of unknown importance]. In the CMML, genetic analysis of DNA from BM MNCs just after the initiation of vemurafenib and after introduction of cobimetinib consistently detected six mutations in four genes (NRAS, EZH2, TET2, and IDH2), all of which have been previously described in this disorder (11). Five of these mutations (83%) were C → T or G → A transitions. As previously observed by prior investigators, neither of the driver mutations (BRAFV600K or NRASG12R) had a UV damage signature, raising the possibility that the driver mutations arose by non-UV mechanisms.
Table 1.
Somatic mutations present in the melanoma and in the chronic myelomonocytic leukemia of the patient harboring both disorders simultaneouslya
| Gene symbol | Amino acid change | Tumor variant frequency | Buccal epithelium variant frequency |
|---|---|---|---|
| Melanoma | |||
| EPHA8 | p.S196F | 0.16098 | 0 |
| ERBB4 | p.R106C | 0.14213 | 0.00286 |
| EPHA6 | p.T314I | 0.15333 | 0 |
| EPHA6 | p.P353S | 0.10067 | 0 |
| EPHA6 | p.D535N | 0.19704 | 0 |
| EPHB1 | p.Q927K | 0.1466 | 0 |
| IL7R | p.S184F | 0.14094 | 0.00295 |
| NOTCH4 | p.W306* | 0.10588 | 0.00291 |
| MYB | p.P273S | 0.16 | 0 |
| BRAF | p.V600K | 0.41551 | 0 |
| PREX2 | p.D1072N | 0.13889 | 0.00271 |
| PTPRD | p.G1819R | 0.17842 | 0 |
| PTPRD | p.G1001R | 0.13208 | 0 |
| PTPRD | p.R427Q | 0.15676 | 0.00395 |
| PTPRD | p.G285E | 0.16185 | 0 |
| MLL2 | p.T698L | 0.18919 | 0 |
| FLT1 | p.G1086E | 0.15175 | 0 |
| DICER1 | p.I445fs | 0.2598 | 0 |
| CDH11 | p.A342T | 0.21212 | 0 |
| KEAP1 | p.P549L | 0.19764 | 0.0027 |
| NOTCH3 | p.S2262F | 0.14976 | 0 |
| NOTCH3 | p.G824D | 0.19421 | 0.00297 |
| NOTCH3 | p.P609S | 0.20567 | 0 |
| MEF2B | p.G290S | 0.12745 | 0 |
| CBLC | p.G312E | 0.16667 | 0 |
| PAK7 | p.E518K | 0.18779 | 0 |
| PAK7 | p.P424S | 0.15909 | 0 |
| ERG | p.M109I | 0.16814 | 0 |
|
| |||
| Leukemia | |||
| NRAS | p.G12R | 0.49153 | 0.065 |
| EZH2 | p.R63* | 0.45905 | 0.09259 |
| IDH2 | p.R140Q | 0.06089 | 0 |
| TET2 | p.R1465* | 0.439 | 0.084 |
| TET2 | p.Q1553* | 0.49 | 0.123 |
Mutational analysis performed using IMPACT assay, a high-throughput next-generation sequencing assay of 279 genes known to be mutated in cancer. Melanoma tumor biopsy, bone marrow aspirate mononuclear cells from 2 time points in the patient’s course, and DNA from buccal epithelia cells were sequenced.
DISCUSSION
In this patient, the simultaneous presence of an NRAS-mutant leukemia in the setting of metastatic BRAF-mutant melanoma provided a dramatic example of the effects of activation of ERK by vemurafenib in RAS-mutant cells. Tumor induction has been noted in patients treated with RAF inhibitors, but most cases have been squamous cell cancers of the skin which are easily managed. In our case, acceleration of a previously undiagnosed mutant NRAS CMML occurred. Both the melanoma and the CMML were well controlled with intermittent inhibition of ERK signaling. In the first year, intermittent treatment with the RAF inhibitor alone reduced the melanoma to a near-complete response while mitigating the drug’s proliferative effects on the leukemia. Here, we describe that intermittent combined therapy with MEK and RAF inhibitors counteracted the paradoxical ERK activation induced by the RAF inhibitor in the CMML cells, leading to effective therapy for both malignancies.
The improved efficacy of combining inhibition of RAF and MEK in BRAFV600-mutant metastatic melanoma has been previously described in a phase I/II trial of dabrefenib and trametinib. Within this trial, 162 patients were randomized to one of three treatment arms: dabrafenib monotherapy or dabrafenib with one of two doses of trametinib (1 mg or 2 mg/d). Concomitant treatment with 2 mg daily of MEK inhibitor was associated with increased response rate (76% vs. 54%), increased complete response rate (9% vs. 4%), and prolonged median progression-free survival (9.4 months vs. 5.8 months; ref. 12). In addition to increasing progression-free survival and response rate, combined MEK and RAF inhibition was associated with a decreased frequency of squamous cell carcinomas compared with monotherapy. Inhibitors of MEK and RAF are traditionally administered on schedules designed to inhibit ERK signaling continuously with the presumption that this is required for effective antitumor activity. There are rationales for intermittent therapy, however. The degree of pathway inhibition with MEK inhibitors given daily is limited by toxicity; higher doses and greater pathway inhibition might be achievable with intermittent dosing. Moreover, continuous ERK pathway inhibition causes chronic reactivation of feedback inhibition and other adaptations of the tumor (2, 13). Although the results here come from a single patient, in 2 years of intermittent therapy with RAF inhibition followed by combined RAF–MEK inhibition, no clinical evidence of resistance was apparent. These data suggest that intermittent therapy avoids constant selection for vemurafenib-resistant cells seen with continuous drug administration. In support of this, recent preclinical data with RAF inhibitors suggest that intermittent dosing schedules can delay resistance (13). The experience of this patient strongly supports testing the efficacy of inhibitors of ERK signaling on intermittent schedules in both preclinical and clinical models. The ability to increase dosage and pathway inhibition while reducing feedback could increase the efficacy of these regimens in tumors with mutant RAS and mutant RAF.
It is worth noting that in this patient, the leukemia was undiagnosed until the patient was treated with the RAF inhibitor. This suggests that the NRAS mutation alone did not elevate ERK output to sufficient levels to cause clinically overt leukemia. It is very likely that the RAF inhibitor enhanced tumor growth by activating ERK signaling. Suppressing ERK activity, by either discontinuing the RAF inhibitor or treating with nontoxic doses of a MEK inhibitor, was sufficient to suppress leukemia growth. Although the patient did not achieve complete remission of the CMML with MEK inhibition, it is possible that the concomitant vemurafenib administration may have blunted the antileukemic effects of cobimetinib by inducing some activation of ERK in the RAS-mutant leukemia cells (as demonstrated in acute myeloid leukemia cell lines; Fig. 2C).
As described earlier, in vitro drug testing suggested that combined RAF and MEK inhibition limited the efficacy of MEK inhibitor to suppress ERK signaling. In this case, it was not possible to give the MEK inhibitor without vemurafenib because of the necessity to treat both tumors. However, two clinical studies of MEK inhibition in myeloid malignancies have been reported (14, 15). An ongoing phase I/II evaluation of trametinib has noted clinical activity in patients with RAS-mutant myeloid malignancies using the recommended phase II dosing of 2 mg daily (14). An overall response rate of 28% of patients with RAS-mutant leukemia has been observed with 11 of 57 RAS-mutant patients experiencing a marrow complete remission. We believe that the data support testing regimens that effectively inhibit ERK signaling as treatment of RAS-mutant leukemias, which account for 15% to 30% of patients with CMML and other myeloid malignancies (11 , 16). Experience from the treatment of this patient here suggests that intermittent MEK inhibitor administration may enhance pathway inhibition and improve therapeutic efficacy without incurring increased toxicity compared with continuous administration.
We also used quantitative measurements of tumor-derived DNA in plasma to monitor tumor burden dynamically of both malignancies throughout treatment. The burden of BRAFV600K and NRASG12R DNA in plasma correlated with conventional radiographic and hematologic laboratory parameters and confirmed the advantageous effects of combined RAF–MEK inhibition in this patient. In the future, monitoring of tumor-derived DNA in plasma could obviate the need for frequent radiographic monitoring of melanoma with defined genetic alterations present in the bulk melanoma clone.
This case presents an example of ERK activation in RAS-mutant cells induced by vemurafenib and evidence that combined RAF–MEK inhibition offers the potential to treat RAS-mutant disease arising during RAF inhibitor therapy. The observations from the clinical management of this patient present a rationale for intermittent dosing of RAF and MEK inhibitors in the management of BRAF-mutant melanoma and the therapeutic potential of MEK inhibition in refractory RAS-mutant myeloid leukemias.
METHODS
Cell-Free Quantitative Digital PCR of Plasma BRAFV600K and NRASG12R
A TaqMan assay was designed to amplify the region of interest (BRAFV600K or NRASG12R) and distinguish between the wild-type and mutant target using a pair of competitive fluorophore-labeled probes (available upon request). PCR reaction mixtures (20 μL), containing a limited template dilution, were partitioned into approximately 20,000 1-nL droplets using the Bio-Rad QX200 droplet generator system according to the manufacturer’s specifications. The limited template dilution ensured that the estimated number of templates per partition (λ) was within the dynamic range of the instrument. The Poisson correction factor was applied at the analysis stage to account for the eventuality of multiple template occupancy. Following the endpoint PCR amplification, carried under conducive conditions, the individual droplets were analyzed using the Bio-Rad QX200 Droplet Reader system and the proprietary analysis software QuantaLife. The mutant to wild-type ratio and the 95% confidence interval (CI) were calculated as previously described (17). To determine the limit of detection for the designed assays, we measured the lowest detectable amount of mutant target within a large excess of wild-type genomic DNA (gDNA). The observed number of mutant target copies was in close linear relationship with the expected quantities (R2 > 0.99), and a single copy of mutant DNA resolved within 105 copies of wild-type DNA (Supplementary Fig. S3).
Leukemia Cell Line Drug Studies
Cell lines were originally obtained from DSMZ or the American Type Culture Collection, and all cell lines were authenticated by Promega short-tandem repeat analysis. A total of 10,000 viable cells were plated in 96-well microtiter plates in 200 μL of RPMI media with different concentrations of PLX4720, cobimetinib, or both in triplicate. The 48-hour proliferation was assessed using the Cell Viability Luminescent Assay Kit (CellTiter-Glo; Promega). Results were normalized to growth of cells in media containing an equivalent volume of dimethyl sulfoxide (DMSO). The concentration at which 50% inhibition in proliferation occurred was determined using GraphPad Prism 5.0 software. For Western blot analysis of signaling pathways, cell lines were exposed to different concentrations of PLX4720, cobimetinib, or both for 4 hours. Cells were then collected and lysed in lysis buffer and separated by electrophoresis. Nitrocellulose membrane was blocked in TBST/5% milk and incubated with antibodies.
Flow Cytometric Analysis of pERK in Primary Peripheral Blood Mononuclear Cells
Cryopreserved peripheral blood mononuclear cells were thawed, washed with PBS, fixed, and stained for surface markers to distinguish myeloid cell populations (CD14+) along with intracellular pERK as previously described (5).
Genetic Analysis
We used standard techniques to extract gDNA from the melanoma tumor, leukemic BM MNC, and saliva specimens. Barcoded, massively parallel sequencing libraries were prepared (New England Biolabs, Kapa Biosystems), and exon capture was performed on barcoded pools (Nimblegen SeqCap) according to the manufacturer’s directions. Briefly, we designed and synthesized synthetic DNA probes complementary to the coding sequence of 279 genes known to undergo somatic genomic alterations in cancer (Supplementary Table S1). gDNA libraries were subjected to solution-phase hybrid capture using the DNA probes, followed by massively parallel sequencing on the Illumina HiSeq 2500. We sequenced 100 bases from both ends of library DNA fragments, achieving approximately 15 million purity filtered reads per sample. This yielded target gene haploid coverage of 209-, 429-, 388-, and 271-fold from the melanoma, BM MNC number #1 (prevemurafenib therapy), BM MNC number #2 (on vemurafenib and cobimetinib), and saliva samples, respectively. Paired reads were aligned to the reference human genome using the Burrows–Wheeler Alignment tool (18) and post-processed using the Genome Analysis Toolkit according to best practices (19). Single-nucleotide variants were called using muTect (20), and indels were called using SomaticIndelDetector (19). Because somatic mutations in the leukemia were present at low levels in the saliva sample, we additionally retained mutations and indels if the variant allele frequency in the tumor was >5 times that in the matched normal. All alterations were manually reviewed using the Integrative Genomics Viewer (21).
Supplementary Material
Supplementary Table 1: List of genes sequenced in melanoma, saliva, and bone marrow mononuclear cells.
Supplementary Table 2. Somatic mutations detected in melanoma and bone marrow mononuclear cells.
Supplementary Figure 1: Effect of treatment on spleen length as measured by CT scan.
Specified amounts of DNA containing BRAFV600K (Panel A) or NRASG12R (Panel B) mutations were placed into a large excess of 5ng of BRAF/NRAS wildtype genomic DNA followed by detection of mutant target DNA. Displayed is the correlation between the detected amounts of mutant DNA and input mutant DNA.
Effect of treatment on the subcarinal mass (Panel A). Currently, only a 1.1 cm lymphoid mass remains detectable. After 11 months of treatment with vemurafenib alone, a pericardial nodule appeared (red circle) which regressed upon addition of cobimetinib (Panel B). Size of the spleen (asterisk) increased on vemurafenib but shrunk once cobimetinib was added (Panel C).
(A) Flow cytometric staining of peripheral blood mononuclear cells (PBMCs) for CD14+ cells before (week 3.3 according to Figure 1) and after treatment with vemurafenib (week 4.6) shows an increase in the number of CD14+ cells consistent with stimulation by vemurafenib (top row) . Phospho-flow analysis (bottom row) shows an increase in pERK levels in CD14+ cells. (B) In contrast, combined vemurafenib plus cobimetinib therapy (collected on week 73 according to Figure 1) resulted in a decrease in both the frequency of CD14+ cells amongst PBMCs (top row) as well as a decrease in pERK expression in CD14+ cells (bottom row) compared to PBMC collected off all treatment 2 weeks earlier (week 71).
Acknowledgments
The authors thank Drs. Sarang Abhyankar and Mika Sovak at Genentech for supplying cobimetinib and, along with Dr. Alice Chung, for helpful suggestions on the article.
Grant Support
O. Abdel-Wahab is supported by an NIH K08 Clinical Investigator Award (1K08CA160647-01) and the Josie Robertson Investigator Program. P.B. Chapman is supported in part by the John F. Figge Research Fund.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
Margaret K. Callahan has received a commercial research grant from Bristol-Myers Squibb and is a consultant/advisory board member of GlaxoSmithKline and Bristol-Myers Squibb. N. Rosen has received a commercial research grant from Chugai and is a consultant/advisory board member of Chugai and AstraZeneca. P.B. Chapman is a consultant/advisory board member of Genentech. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: O. Abdel-Wahab, V.M. Klimek, R. Rampal, M.K. Callahan, D.B. Solit, P.B. Chapman
Development of methodology: A.A. Gaskell, M.K. Callahan, T. Merghoub, D.B. Solit, R.L. Levine
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): O. Abdel-Wahab, V.M. Klimek, A.A. Gaskell, A. Viale, E. Kim, R. Rampal, J.J. Harding, M.K. Callahan, T. Merghoub, M.F. Berger, P.B. Chapman
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): O. Abdel-Wahab, V.M. Klimek, A.A. Gaskell, D. Cheng, E. Kim, R. Rampal, M. Bluth, J.J. Harding, T. Merghoub, M.F. Berger, N. Rosen, R.L. Levine, P.B. Chapman
Writing, review, and/or revision of the manuscript: O. Abdel- Wahab, V.M. Klimek, A.A. Gaskell, R. Rampal, J.J. Harding, M.K. Callahan, D.B. Solit, N. Rosen, R.L. Levine, P.B. Chapman
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J.J. Harding, M.F. Berger
Study supervision: O. Abdel-Wahab, P.B. Chapman
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joseph E, Pratilas C, Poulikakos P, Tadi M, Wang W, Taylor B, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci U S A. 2010;107:14903–8. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulikakos P, Zhang C, Bollag G, Shokat K, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan MK, Rampal R, Harding JJ, Klimek VM, Chung YR, Merghoub T, et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med. 2012;367:2316–21. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo EF, Belvin M, Boggs J, Deng Y, Hoeflich KP, Ly J, et al. Pre-clinical disposition of GDC-0973 and prospective and retrospective analysis of human dose and efficacy predictions. Drug Metab Dispos. 2012;40:919–27. doi: 10.1124/dmd.111.043778. [DOI] [PubMed] [Google Scholar]
- 7.Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. Guidelines for minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 8.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagle N, Berger M, Davis M, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–5. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31:2428–36. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 12.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–5. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borthakur G, Popplewell L, Boyiadzis M, Foran JM, Platzbecker U, Vey N, et al. Phase I/II trial of the MEK1/2 inhibitor trametinib (GSK1120212) in relapsed/refractory myeloid malignancies: evidence of activity in patients with RAS mutation-positive disease [abstract]. Proceedings of the 54th ASH Annual Meeting and Exposition; 2012 Dec 8–11; Atlanta, GA. p. Abstract nr 677. [Google Scholar]
- 15.Jain N, Curran E, Iyengar NM, Diaz-Flores E, Kunnavakkam R, Popplewell L, et al. Phase II study of the oral MEK inhibitor selumetinib (AZD6244) in advanced acute myeloid leukemia (AML) J Clin Oncol. 2012;30(suppl):abstr 6582. [Google Scholar]
- 16.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whale AS, Huggett JF, Cowen S, Speirs V, Shaw J, Ellison S, et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012;40:e82. doi: 10.1093/nar/gks203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cibulskis K, Lawrence M, Carter S, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorvaldsdóttir H, Robinson J, Mesirov J. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: List of genes sequenced in melanoma, saliva, and bone marrow mononuclear cells.
Supplementary Table 2. Somatic mutations detected in melanoma and bone marrow mononuclear cells.
Supplementary Figure 1: Effect of treatment on spleen length as measured by CT scan.
Specified amounts of DNA containing BRAFV600K (Panel A) or NRASG12R (Panel B) mutations were placed into a large excess of 5ng of BRAF/NRAS wildtype genomic DNA followed by detection of mutant target DNA. Displayed is the correlation between the detected amounts of mutant DNA and input mutant DNA.
Effect of treatment on the subcarinal mass (Panel A). Currently, only a 1.1 cm lymphoid mass remains detectable. After 11 months of treatment with vemurafenib alone, a pericardial nodule appeared (red circle) which regressed upon addition of cobimetinib (Panel B). Size of the spleen (asterisk) increased on vemurafenib but shrunk once cobimetinib was added (Panel C).
(A) Flow cytometric staining of peripheral blood mononuclear cells (PBMCs) for CD14+ cells before (week 3.3 according to Figure 1) and after treatment with vemurafenib (week 4.6) shows an increase in the number of CD14+ cells consistent with stimulation by vemurafenib (top row) . Phospho-flow analysis (bottom row) shows an increase in pERK levels in CD14+ cells. (B) In contrast, combined vemurafenib plus cobimetinib therapy (collected on week 73 according to Figure 1) resulted in a decrease in both the frequency of CD14+ cells amongst PBMCs (top row) as well as a decrease in pERK expression in CD14+ cells (bottom row) compared to PBMC collected off all treatment 2 weeks earlier (week 71).