Abstract
A decline in electron transport chain (ETC) activity is associated with many human diseases. Although diminished mitochondrial ATP production is recognized as a source of pathology, the contribution of the associated reduction in the ratio of the amount of oxidized nicotinamide adenine dinucleotide (NAD+) to that of its reduced form (NADH) is less clear. We used a water-forming NADH oxidase from L. brevis (LbNOX) as a genetic tool for inducing a compartment-specific increase of the NAD+/NADH ratio in human cells. We used LbNOX to demonstrate the dependence of key metabolic fluxes, gluconeogenesis, and signaling on the cytosolic or mitochondrial NAD+/NADH ratios. Expression of LbNOX in the cytosol or mitochondria ameliorated proliferative and metabolic defects caused by an impaired ETC. The results underscore the role of reductive stress in mitochondrial pathogenesis and demonstrate the utility of targeted LbNOX for direct, compartment-specific manipulation of redox state.
One Sentence Summary
We developed a genetically encoded tool for raising NAD+/NADH ratios and showed it can complement an impaired electron transport chain in human cells.
A decline in electron transport chain (ETC) activity has been linked to numerous human disorders, ranging from rare genetic syndromes to common diseases such as neurodegeneration, cancer, and diabetes, as well as the aging process itself (1, 2). How a decline in ETC activity gives rise to the spectrum of observed pathology cannot be readily explained by a simple deficiency in adenosine triphosphate (ATP) production (1). A key challenge in deciphering mitochondrial pathogenesis stems from the fact that the ETC performs at least two coupled functions: redox transfer of electrons from NADH [the reduced form of nicotinamide adenine dinucleotide (NAD+)] to oxygen and a simultaneous conversion of the free energy of the electromotive force into a proton gradient across the mitochondrial inner membrane. In principle, pathology could stem from an excess of reducing equivalents (termed reductive stress or pseudohypoxia, which includes stalling of NAD+-coupled reactions) or a reduced proton gradient (impairing pH and voltage-coupled processes, including ATP synthesis by the F1FO-ATPase). Currently there are no methods for dissecting the redox function of the ETC from its proton pumping function.
Here, we report the application of a genetically encoded tool for compartment-specific manipulation of the NAD+/NADH ratio. Our tool is based on the flavin adenine dinucleotide (FAD)-dependent H2O-forming NADH oxidases, which catalyze the four-electron reduction of O2 to two molecules of H2O (Fig. 1A). We focused on bacterial oxidases with specificity for NADH over NADPH (3–7), whose natural function is protection of redox balance and defense against oxygen toxicity (8). Such oxidases have been successfully expressed in bacteria and yeast for biotechnological applications (9–11). We screened several H2O-forming NADH oxidases by expressing their human codon-optimized, epitope-tagged versions in cultured human cancer-derived epithelial (HeLa) cells. The enzyme from L. brevis (LbNOX) was most highly expressed and had the highest oxidase activity when targeted to mitochondria (fig. S1).
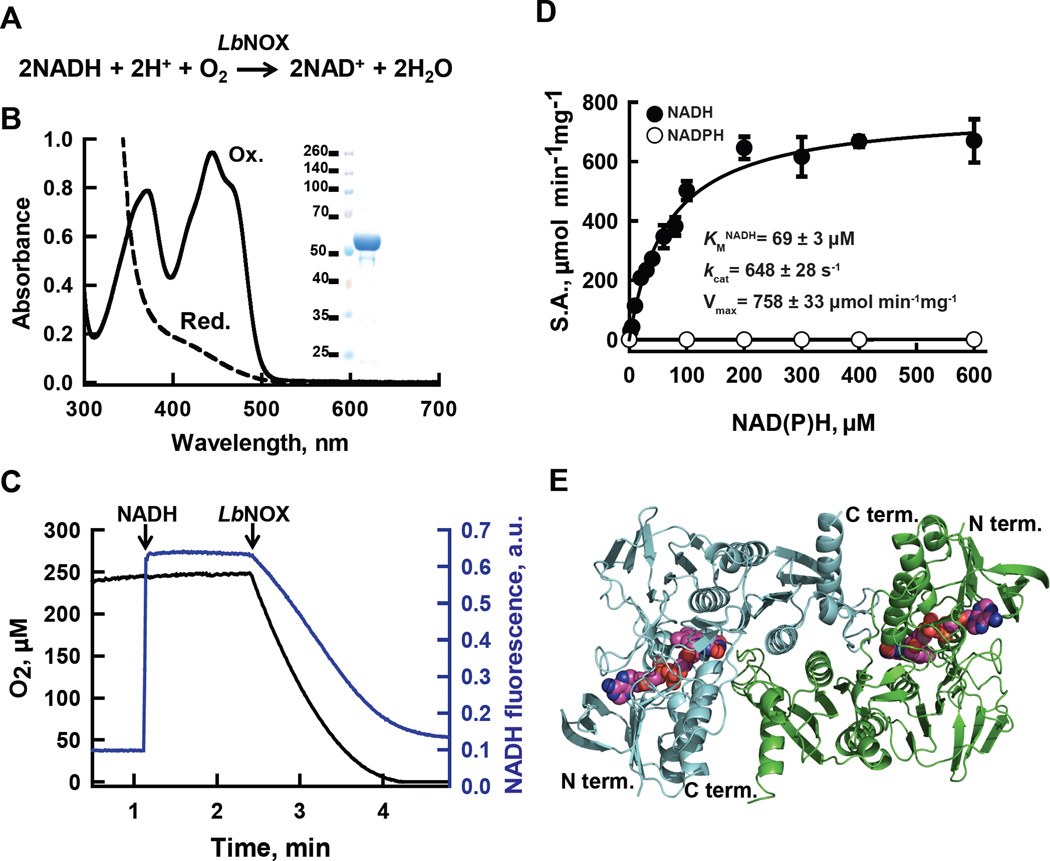
Figure 1. H2O-forming NADH oxidase from L. brevis (LbNOX.
(A) Reaction catalyzed by LbNOX. (B) UV-visible spectrum of purified LbNOX. Protein (83 µM FAD active sites) in oxidized form (solid line) and after addition of excess of sodium dithionite, reduced form (dashed line). Inset: SDS-PAGE of purified LbNOX. (C) Simultaneous measurement of NADH and oxygen consumption by LbNOX. NADH and LbNOX were added as indicated by arrows. (D) Dependence of the specific activity of recombinant LbNOX on the concentration of NADH and NADPH. Reported values for Vmax, kcat and KM for NADH represent the mean ± S.D. from n=4 independent experiments. (E) Crystal structure of the catalytic dimer of LbNOX. Each of the two-fold symmetry related monomers (cyan and green ribbons) contain bound FAD, shown here in sphere (CPK) representation. Details of the catalytic center on the si-face of FAD and of the substrate selectivity loop are shown in fig. S3A–C.
We evaluated the biochemical properties of LbNOX modified to contain a C-terminal FLAG tag and a cleavable N-terminal hexahistidine tag, and overexpressed in E. coli. Purified LbNOX-FLAG has a yellow color in solution and a characteristic UV-visible absorption spectrum (λmax = 371 and 444 nm) consistent with the presence of FAD, which can be reduced upon the addition of sodium dithionite (Fig. 1B). Our recombinant enzyme consumes oxygen and is strictly specific for NADH rather than NADPH with KM for NADH of 69 ± 3 µM, V max of 758 ± 33 µmol min−1 mg−1 and kcat of 648 ± 28 s−1 which is more active than previously reported (3, 12) (Fig. 1 C and D). The molecular size of LbNOX-FLAG was determined to be 197 ± 4 kD, which indicates that the protein is a tetramer in solution. Although enzymes in this family often produce H2O2, LbNOX-FLAG produces amounts of H2O2 that constitute only 1 to 2 % of the amount of H2O produced during its catalytic cycle (fig. S2A) (4, 6, 7). The apparent KM for O2 of LbNOX-FLAG was below 2 µM (~ 0.17 % O2), as estimated from enzyme-monitored turnover experiments (fig. S2B), which is less than one-tenth of the concentration of oxygen in human venous blood (13). Thus, we expect LbNOX to be active in most animal tissues. The enzymatic properties of LbNOX-FLAG in solution were well founded in the 2.4 Å resolution X-ray structure of this protein that we determined (Fig. 1E, Table S1). Our structure is generally similar to the reported structures of H2O-forming NAD(P)H oxidases from L. sanfranciscensis (PDB ID 2CDU) and S. pyogenes (PDB ID 2BC0) (14, 15). However, our structure captures LbNOX in a new state with molecular oxygen (O2) bound and the redox active Cys 42 in a reduced form (Cys 42-SH) (fig. S3, see Supplementary Materials for a detailed discussion of the X-ray structure). In conclusion, the high selectivity for NADH over NADPH, negligible H2O2 production, and very low KM for O2 made LbNOX attractive for additional studies in human cells.
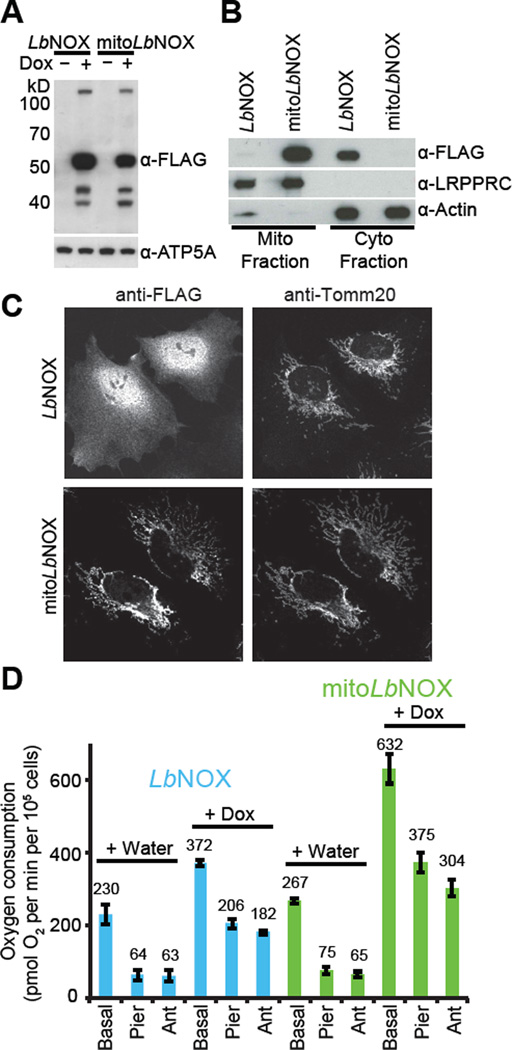
To determine if we could express LbNOX-FLAG safely and efficaciously in various compartments of human cells we used lentiviral infection to generate HeLa cells that expressed untargeted or mitochondria-targeted human codon-optimized LbNOX-FLAG (referred to as LbNOX and mitoLbNOX henceforth) under the control of a doxycycline-inducible promoter (TRE3G) (Fig. 2A, fig. S1A). We used fluorescence microscopy and cell fractionation to confirm diffuse localization of LbNOX and mitochondrial localization of mitoLbNOX (Fig. 2B and C). Cells appeared grossly normal without any impact on cell proliferation or reactive oxygen species (ROS) production (fig. S4A, B). Expression of LbNOX and mitoLbNOX in HeLa cells increased oxygen consumption by 1.6- and 2.4-fold, respectively (Fig. 2D, fig. S4C). The increase was resistant to ETC inhibitors, which indicates that it resulted from LbNOX oxidase activity and not from the increased ETC activity. Despite similar expression levels (Fig. 2A), mitoLbNOX induced a larger increase in oxygen consumption than LbNOX (Fig. 2D), likely because of the higher concentration of NADH within mitochondria (16–18). It is important to remember that in converting NADH to NAD+, LbNOX also consumes protons and oxygen and, therefore, could affect cellular pH or oxygen levels depending on experimental context.
Figure 2. Expression and activity of LbNOX in human cells.
(A) Western blot of LbNOX and mitoLbNOX in HeLa cells after 24-hour induction with water or doxycycline (300 ng/ml). Representative gel from one of three independent experiments. (B) Subcellular localization of LbNOX and mitoLbNOX in HeLa cells determined by cell fractionation. LRPPRC is a mitochondrial marker and Actin is a cytosolic marker. Representative gel from one of three independent experiments. (C) Subcellular localization of LbNOX and mitoLbNOX in HeLa cells determined using fluorescence microscopy. Tomm20 is a marker of mitochondria. (D) Effect of LbNOX and mitoLbNOX expression in HeLa cells on basal, piericidin-resistant and antimycin-resistant oxygen consumption measured with a XF24 extracellular flux analyzer. Mean ± S.E., n=3 independent experiments.
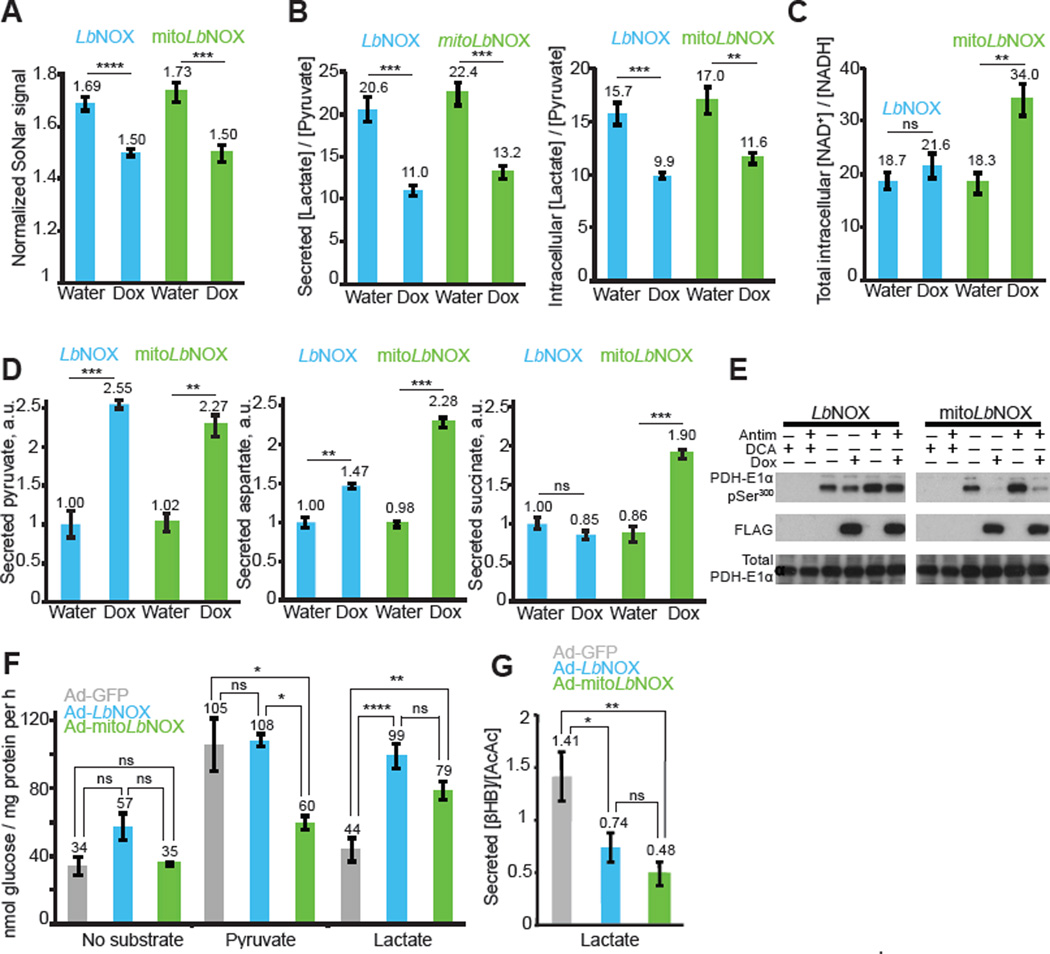
We determined the impact of expressing LbNOX or mitoLbNOX on cellular concentrations of NAD+ and NADH (Fig. 3 and fig. S5). We used a genetic sensor, SoNar (19), to measure cytosolic NADH. SoNar is a fusion of circularly permuted Yellow Fluorescent Protein and a modified NADH binding protein Rex from Thermus aquaticus. Binding of NADH to SoNar leads to an increase in fluorescence. Expression of LbNOX or mitoLbNOX decreased the fluorescence signal from SoNar indicating that both LbNOX and mitoLbNOX decrease cytosolic NADH (Fig. 3A, fig. S5B). Consistent with this result, intracellular and secreted lactate/pyruvate ratios, traditionally used proxies for the cytosolic NADH/NAD+ ratio (17), decreased in cells expressing LbNOX or mitoLbNOX (Fig. 3B, fig. S5D). The ratio of total cellular NAD+ to total NADH, based on HPLC measurements, was increased 2-fold by mitoLbNOX, whereas LbNOX did not have a significant effect (Fig. 3C, fig. S5C). Perturbation of the total NAD+/NADH ratio likely reflects changes in amounts of mitochondrial NADH because most of the effect on the ratio resulted from changes in NADH concentration (fig. S5A), and most of the NADH inside the cell is present in mitochondria. The latter is supported by fractionation experiments (20) and by the observation that the majority of NAD(P)H autofluorescence in cells comes from mitochondria (21). In summary, LbNOX and mitoLbNOX can be used to perturb the NAD+/NADH ratio and our compartment-specific measurements of HeLa cells (Fig. 3A–C) indicate that although perturbation of the mitochondrial NAD+/NADH ratio leads to changes in the cytosolic NAD+/NADH ratio, the converse is not true.
Figure 3. Effect of LbNOX and mitoLbNOX on NAD+/NADH ratios, metabolic fluxes, PDH phosphorylation and gluconeogenesis.
(A–C) Effect of LbNOX and mitoLbNOX expression in HeLa cells on (A) cytoplasmic NADH concentrations determined with fluorescence microscopy using SoNar expressing cells (n=7), (B) intracellular and secreted lactate/pyruvate ratio determined by LC-MS (n=4), and (C) intracellular NAD+/NADH ratios determined by HPLC (n=4). Student’s t-test. ns P > 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. Mean ± S.E. (D) Effect of LbNOX and mitoLbNOX expression in HeLa cells on release rate of pyruvate, aspartate and succinate, determined by comparing concentrations in spent versus fresh media. Student’s t-test. ns P > 0.05, ** P < 0.01, *** P < 0.001. Mean ± S.E., n=3 replicates from one experiment. (E) Effect of LbNOX and mitoLbNOX expression in HeLa cells on PDH phosphorylation. Representative gel from one of three independent experiments. (F) Effect of adenoviral transduction of GFP, LbNOX or mitoLbNOX on primary rat hepatocyte gluconeogenesis in DMEM containing no glucose, no glutamine and no pyruvate using either no substrate, 5mM pyruvate, or 5mM lactate. One-way ANOVA followed by Tukey’s multiple comparisons test. ns P > 0.05, * P < 0.05, ** P < 0.01, **** P < 0.0001. Mean ± S.E., n=3 (no substrate, pyruvate) or n=7 (lactate) independent experiments. (G) Effect of LbNOX and mitoLbNOX on secreted β-hydroxybutyrate/acetoacetate ratio in rat hepatocytes performing gluconeogenesis from lactate as a substrate. Metabolite levels determined using LC-MS. One-way ANOVA followed by Tukey’s multiple comparisons test. ns P > 0.05, * P < 0.05, ** P < 0.01. Mean ± S.E., n=10 independent experiments.
We performed metabolic profiling on medium in which cells expressing LbNOX or mitoLbNOX had been grown (Fig. 3D and fig. S6A–B). We identified pyruvate, aspartate and succinate as three metabolites whose consumption or release was changed more than two-fold (Student’s t-test; P < 0.01) by either enzyme. These changes are attributable to compartment-specific changes of NAD+/NADH by LbNOX or mitoLbNOX (see Supplementary Materials for discussion). It is notable that LbNOX and mitoLbNOX did not have a significant effect on the uptake of glucose and release of lactate (fig. S6C).
In vitro phosphorylation of mitochondrial pyruvate dehydrogenase (PDH) is regulated by the NAD+/NADH ratio (22), but this has never been shown in intact cells. Treatment of HeLa cells with dichloroacetate (DCA), an inhibitor of pyruvate dehydrogenase kinases (PDKs), inhibits phosphorylation of PDH, and antimycin treatment, which blocks the ETC and decreases the mitochondrial NAD+/NADH ratio, increases PDH phosphorylation. In agreement with in vitro studies, PDH was almost completely dephosphorylated in the presence of mitoLbNOX but not LbNOX (Fig. 3E and fig. S6D). The data on PDH phosphorylation are consistent with our observation that mitoLbNOX, but not LbNOX, increases mitochondrial NAD+/NADH ratio in HeLa cells (Fig. 3C).
We expressed LbNOX and mitoLbNOX in primary rat hepatocytes to study gluconeogenesis. The cytosolic NAD+/NADH ratio has been reported to affect gluconeogenesis, though classical approaches relied on indirect methods for manipulating the redox state (23, 24). In our hepatocyte system, rates of gluconeogenesis were significantly higher if pyruvate rather than lactate was used as a substrate, which we attribute to the NAD+/NADH ratio-dependent inhibition of lactate to pyruvate conversion (23). Consistent with this hypothesis rates of gluconeogenesis from lactate were increased to those seen with pyruvate when primary hepatocytes expressed LbNOX, whereas gluconeogenesis from pyruvate was not affected (Fig. 3F). Gluconeogenesis from lactate was also increased by mitoLbNOX. Gluconeogenesis from pyruvate, however, was inhibited by mitoLbNOX, perhaps because strong oxidation of mitochondrial NADH prevents formation of malate (25). In assays of gluconeogenesis from lactate expression of either LbNOX or mitoLbNOX decreased the ratio of secreted β- hydroxybutyrate/acetoacetate (Fig. 3G), the classical proxy for the mitochondrial NADH/NAD+ ratio (17). Although the NAD+/NADH ratio appears to be increased we cannot exclude the possibility that LbNOX or mitoLbNOX induce hypoxia. LbNOX did increase mitochondrial NAD+/NADH ratio in rat hepatocytes (Fig. 3G) but not in HeLa cells (Fig. 3C), which might reflect differences in cell type or media conditions.
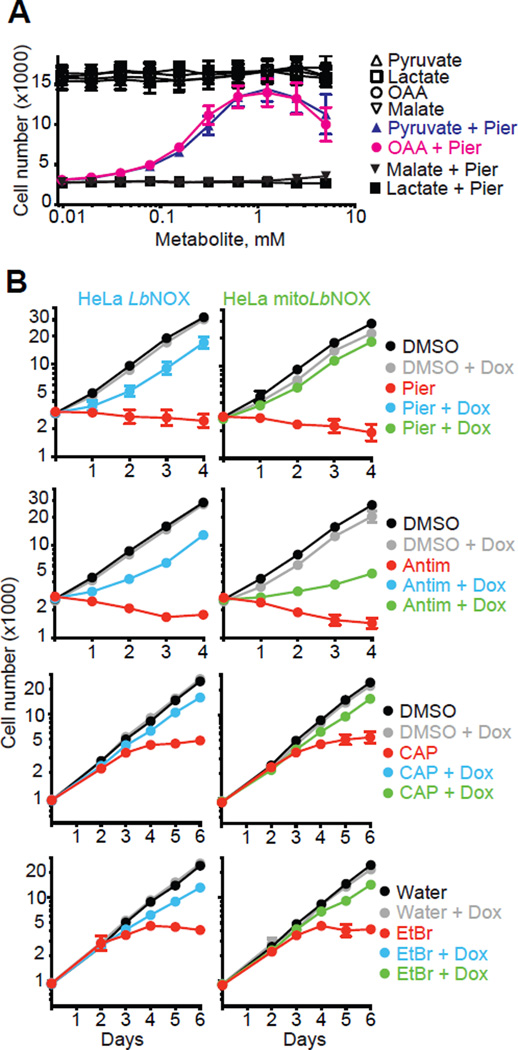
Mammalian cells lacking a functional ETC require the addition of exogenous pyruvate and uridine for cell proliferation (26–28). Uridine is required because one of the enzymes in de novo uridine biosynthesis (dihydroorotate dehydrogenase) is coupled to the ETC through coenzyme Q (CoQ). The requirement of pyruvate, however, has been less clear because it participates in many reactions but has been proposed to rescue cell growth by recycling NAD+ from NADH through cytosolic lactate dehydrogenase (26, 29). If the NAD+ recycling hypothesis is correct then supplementation with oxaloacetate should have the same effect as pyruvate because it can be reduced by malate dehydrogenase while recycling NAD+. Oxaloacetate, like pyruvate, rescued the proliferation defect induced by piericidin whereas malate and lactate did not (Fig. 4A). Alpha-ketobutyrate also rescues the proliferative defect induced by ETC inhibition (30). Furthermore, a large number of α-keto acids can rescue pyruvate dependence of proliferation in cells with intact ETC (31). These findings support the NAD+ recycling hypothesis, though they are still indirect as α-keto acids have many metabolic roles.
Figure 4. NAD+ recycling rescues proliferation in cells with impaired ETC.
(A) Effect of pyruvate, oxaloacetate, lactate and malate addition on proliferation of HeLa Tet3G Luciferase cells in the presence of 200 µM uridine and in the presence or absence of 1 µM piericidin. Mean ± S.E., n=5 independent experiments. (B) Effect of LbNOX and mitoLbNOX expression in HeLa cells on inhibition of cell proliferation by 1 µM piericidin, 1 µM antimycin, 10 µg/ml chloramphenicol and 30 ng/ml ethidium bromide in the presence of 200 µM uridine. Mean ± S.E., n=3 independent experiments.
We used LbNOX to directly test whether NAD+ recycling is an essential function of the ETC that is required for cell proliferation. We inhibited ETC function, with piericidin (a complex I inhibitor), antimycin (a complex III inhibitor), ethidium bromide (a mtDNA replication inhibitor) and chloramphenicol (an inhibitor of mitochondrial translation) in HeLa cells supplemented with uridine but lacking pyruvate. HeLa cells cannot proliferate in these conditions (Fig. 4B and fig. S7). Expression of either LbNOX or mitoLbNOX rescued cell proliferation in the presence of these ETC inhibitors indicating that regeneration of NAD+ in either cytosol or mitochondria is sufficient to complement ETC activity that is required for cell proliferation (Fig. 4B). Metabolic profiling showed that of the nine metabolites whose uptake or release is affected greater than two-fold by antimycin (Student’s t-test; P < 0.01), all could be reversed by either LbNOX or mitoLbNOX, reflecting a metabolic rescue (fig. S8, see Supplementary Materials for discussion). Our metabolic profiling data is complementary to recent studies demonstrating an inhibition of aspartate biosynthesis in cells with dysfunctional ETC (30, 32, 33). As a control, we also showed that the rescue by LbNOX or mitoLbNOX was not attributable to an effect on mitochondrial membrane potential (fig. S9A–C), nor was it due to a rescue of ETC-derived ATP synthesis (fig. S9D–G).
Collectively, these studies (Fig. 4, fig. S7–9) underscore the importance of NAD+ recycling by the ETC to support proliferation. In healthy cells, the ETC produces ATP and simultaneously recycles mitochondrial NADH to NAD+, with a secondary oxidation of cytosolic NADH via shuttles. In the absence of a functional ETC, glycolysis is capable of compensating for the lack of ATP production, but it is net redox neutral. NAD+ recycling is likely key for cell proliferation because many biosynthetic pathways produce NADH as a byproduct (34). These insights confirm the longstanding hypothesis (26, 29) that pyruvate supplementation rescues proliferation in cells with disrupted ETC by restoring NAD+/NADH balance via the LDH reaction.
In the future, LbNOX and engineered or naturally occurring variants may become valuable tools for studying compartmentalization of redox metabolism. These constructs will allow for a dissection of the relative contributions of redox imbalance and ATP insufficiency to mitochondrial disease pathogenesis. If a substantial amount of the organ pathology of mitochondrial disease stems from reductive stress or pseudohypoxia, then expression of this single polypeptide holds promise as a “protein prosthesis” for the large number of disorders characterized by ETC dysfunction.
Supplementary Material
Acknowledgments
We thank Dr. Victor Vitvitsky for technical support with HPLC. We thank members of the Mootha lab for valuable discussions and feedback. This work was supported by a T-R01 R01GM099683 from the National Institutes of Health. D.V.T. was supported by Tosteson and Fund for Medical Discovery Postdoctoral Fellowship Award. R.P.G. was supported by a T32DK007191 from the National Institutes of Health. V.K.M. is an Investigator of the Howard Hughes Medical Institute. The Massachusetts General Hospital has filed a patent application on the technology described in this paper. Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession number 5ER0.
Footnotes
References and Notes
- 1.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012 Nov 15;491:374. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual review of genetics. 2005;39:359. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hummel W, Riebel B. Isolation and biochemical characterization of a new NADH oxidase from Lactobacillus brevis. Biotechnology letters. 2003 Jan;25:51. doi: 10.1023/a:1021730131633. [DOI] [PubMed] [Google Scholar]
- 4.Lopez de Felipe F, Hugenholtz J. Purification and characterisation of the water forming NADH-oxidase from Lactococcus lactis. International Dairy Journal. 2001;11:37. [Google Scholar]
- 5.Yu J, et al. Characterization of the Streptococcus pneumoniae NADH oxidase that is required for infection. Microbiology. 2001 Feb;147:431. doi: 10.1099/00221287-147-2-431. [DOI] [PubMed] [Google Scholar]
- 6.Higuchi M, et al. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. Journal of general microbiology. 1993 Oct;139:2343. doi: 10.1099/00221287-139-10-2343. [DOI] [PubMed] [Google Scholar]
- 7.Stanton TB, Jensen NS. Purification and characterization of NADH oxidase from Serpulina (Treponema) hyodysenteriae. Journal of bacteriology. 1993 May;175:2980. doi: 10.1128/jb.175.10.2980-2987.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higuchi M, Yamamoto Y, Kamio Y. Molecular biology of oxygen tolerance in lactic acid bacteria: Functions of NADH oxidases and Dpr in oxidative stress. Journal of bioscience and bioengineering. 2000;90:484. [PubMed] [Google Scholar]
- 9.Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2007 Feb 13;104:2402. doi: 10.1073/pnas.0607469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Applied and environmental microbiology. 2006 May;72:3653. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heux S, Cachon R, Dequin S. Cofactor engineering in Saccharomyces cerevisiae: Expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metabolic engineering. 2006 Jul;8:303. doi: 10.1016/j.ymben.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Kuzu M, Niefind K, Hummel W, Schomburg D. Crystallization and preliminary crystallographic analysis of a flavoprotein NADH oxidase from Lactobacillus brevis. Acta crystallographica. Section F, Structural biology and crystallization communications. 2005 May 1;61:528. doi: 10.1107/S174430910501153X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray JF, Mason RJ. Murray and Nadel's textbook of respiratory medicine. 5th. Philadelphia, PA: Saunders/Elsevier; 2010. [Google Scholar]
- 14.Lountos GT, et al. The crystal structure of NAD(P)H oxidase from Lactobacillus sanfranciscensis: insights into the conversion of O2 into two water molecules by the flavoenzyme. Biochemistry. 2006 Aug 15;45:9648. doi: 10.1021/bi060692p. [DOI] [PubMed] [Google Scholar]
- 15.Wallen JR, et al. Structural Analysis of Streptococcus pyogenes NADH Oxidase: Conformational Dynamics Involved in Formation of the C(4a)-Peroxyflavin Intermediate. Biochemistry. 2015 Nov 17;54:6815. doi: 10.1021/acs.biochem.5b00676. [DOI] [PubMed] [Google Scholar]
- 16.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell metabolism. 2011 Oct 5;14:545. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. The Biochemical journal. 1967 May;103:514. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, et al. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell metabolism. 2011 Oct 5;14:555. doi: 10.1016/j.cmet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, et al. SoNar, a Highly Responsive NAD+/NADH Sensor, Allows High-Throughput Metabolic Screening of Anti-tumor Agents. Cell metabolism. 2015 May 5;21:777. doi: 10.1016/j.cmet.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sies H. Metabolic compartmentation. London ; New York: Academic Press; 1982. [Google Scholar]
- 21.Eng J, Lynch RM, Balaban RS. Nicotinamide adenine dinucleotide fluorescence spectroscopy and imaging of isolated cardiac myocytes. Biophysical journal. 1989 Apr;55:621. doi: 10.1016/S0006-3495(89)82859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettit FH, Pelley JW, Reed LJ. Regulation of pyruvate dehydrogenase kinase and phosphatase by acetyl-CoA/CoA and NADH/NAD ratios. Biochemical and biophysical research communications. 1975 Jul 22;65:575. doi: 10.1016/s0006-291x(75)80185-9. [DOI] [PubMed] [Google Scholar]
- 23.Sistare FD, Haynes RC., Jr The interaction between the cytosolic pyridine nucleotide redox potential and gluconeogenesis from lactate/pyruvate in isolated rat hepatocytes. Implications for investigations of hormone action. The Journal of biological chemistry. 1985 Oct 15;260:12748. [PubMed] [Google Scholar]
- 24.Krebs HA, Freedland RA, Hems R, Stubbs M. Inhibition of hepatic gluconeogenesis by ethanol. The Biochemical journal. 1969 Mar;112:117. doi: 10.1042/bj1120117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Kalhan SC, Hanson RW. What is the metabolic role of phosphoenolpyruvate carboxykinase? The Journal of biological chemistry. 2009 Oct 2;284:27025. doi: 10.1074/jbc.R109.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989 Oct 27;246:500. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 27.Morais R, Gregoire M, Jeannotte L, Gravel D. Chick embryo cells rendered respiration-deficient by chloramphenicol and ethidium bromide are auxotrophic for pyrimidines. Biochemical and biophysical research communications. 1980 May 14;94:71. doi: 10.1016/s0006-291x(80)80189-6. [DOI] [PubMed] [Google Scholar]
- 28.Harris M. Pyruvate blocks expression of sensitivity to antimycin A and chloramphenicol. Somatic cell genetics. 1980 Nov;6:699. doi: 10.1007/BF01538969. [DOI] [PubMed] [Google Scholar]
- 29.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods in enzymology. 1996;264:304. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan LB, et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015 Jul 30;162:552. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKeehan WL, McKeehan KA. Oxocarboxylic acids, pyridine nucleotide-linked oxidoreductases and serum factors in regulation of cell proliferation. Journal of cellular physiology. 1979 Oct;101:9. doi: 10.1002/jcp.1041010103. [DOI] [PubMed] [Google Scholar]
- 32.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015 Jul 30;162:540. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardaci S, et al. Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nature cell biology. 2015 Oct;17:1317. doi: 10.1038/ncb3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakker BM, et al. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS microbiology reviews. 2001 Jan;25:15. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 35.Votyakova TV, Reynolds IJ. Detection of hydrogen peroxide with Amplex Red: interference by NADH and reduced glutathione auto-oxidation. Archives of biochemistry and biophysics. 2004 Nov 1;431:138. doi: 10.1016/j.abb.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 36.Gibson QH, Swoboda BE, Massey V. Kinetics and Mechanism of Action of Glucose Oxidase. The Journal of biological chemistry. 1964 Nov;239:3927. [PubMed] [Google Scholar]
- 37.Tapley TL, Eichner T, Gleiter S, Ballou DP, Bardwell JC. Kinetic characterization of the disulfide bond-forming enzyme DsbB. The Journal of biological chemistry. 2007 Apr 6;282:10263. doi: 10.1074/jbc.M611541200. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Bunkoczi G, et al. Phaser.MRage: automated molecular replacement. Acta crystallographica. Section D, Biological crystallography. 2013 Nov;69:2276. doi: 10.1107/S0907444913022750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010 Feb;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011 Apr;67:235. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004 Dec;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- 44.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamentsky L, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011 Apr 15;27:1179. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012 May 25;336:1040. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008 Jul 11;134:112. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nature protocols. 2007;2:287. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 49.Claiborne A, Mallett TC, Yeh JI, Luba J, Parsonage D. Structural, redox, and mechanistic parameters for cysteine-sulfenic acid function in catalysis and regulation. Advances in protein chemistry. 2001;58:215. doi: 10.1016/s0065-3233(01)58006-7. [DOI] [PubMed] [Google Scholar]
- 50.Yeh JI, Claiborne A, Hol WG. Structure of the native cysteine-sulfenic acid redox center of enterococcal NADH peroxidase refined at 2.8 A resolution. Biochemistry. 1996 Aug 6;35:9951. doi: 10.1021/bi961037s. [DOI] [PubMed] [Google Scholar]
- 51.Mallett TC, Parsonage D, Claiborne A. Equilibrium analyses of the active-site asymmetry in enterococcal NADH oxidase: role of the cysteine-sulfenic acid redox center. Biochemistry. 1999 Mar 9;38:3000. doi: 10.1021/bi9817717. [DOI] [PubMed] [Google Scholar]
- 52.Seo BB, Matsuno-Yagi A, Yagi T. Modulation of oxidative phosphorylation of human kidney 293 cells by transfection with the internal rotenone-insensitive NADH-quinone oxidoreductase (NDI1) gene of Saccharomyces cerevisiae. Biochimica et biophysica acta. 1999 May 26;1412:56. doi: 10.1016/s0005-2728(99)00051-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.