Abstract
BACKGROUND
The discovery of low-frequency coding variants affecting the risk of coronary artery disease has facilitated the identification of therapeutic targets.
METHODS
Through DNA genotyping, we tested 54,003 coding-sequence variants covering 13,715 human genes in up to 72,868 patients with coronary artery disease and 120,770 controls who did not have coronary artery disease. Through DNA sequencing, we studied the effects of loss-of-function mutations in selected genes.
RESULTS
We confirmed previously observed significant associations between coronary artery disease and low-frequency missense variants in the genes LPA and PCSK9. We also found significant associations between coronary artery disease and low-frequency missense variants in the genes SVEP1 (p.D2702G; minor-allele frequency, 3.60%; odds ratio for disease, 1.14; P = 4.2×10−10) and ANGPTL4 (p.E40K; minor-allele frequency, 2.01%; odds ratio, 0.86; P = 4.0×10−8), which encodes angiopoietin-like 4. Through sequencing of ANGPTL4, we identified 9 carriers of loss-of-function mutations among 6924 patients with myocardial infarction, as compared with 19 carriers among 6834 controls (odds ratio, 0.47; P = 0.04); carriers of ANGPTL4 loss-of-function alleles had triglyceride levels that were 35% lower than the levels among persons who did not carry a loss-of-function allele (P = 0.003). ANGPTL4 inhibits lipoprotein lipase; we therefore searched for mutations in LPL and identified a loss-of-function variant that was associated with an increased risk of coronary artery disease (p.D36N; minor-allele frequency, 1.9%; odds ratio, 1.13; P = 2.0×10−4) and a gain-of-function variant that was associated with protection from coronary artery disease (p.S447⋆; minor-allele frequency, 9.9%; odds ratio, 0.94; P = 2.5×10−7).
CONCLUSIONS
We found that carriers of loss-of-function mutations in ANGPTL4 had triglyceride levels that were lower than those among noncarriers; these mutations were also associated with protection from coronary artery disease. (Funded by the National Institutes of Health and others.)
ALTHOUGH GENOMEWIDE ASSOCIATION studies have identified more than 56 loci associated with the risk of coronary artery disease,1–3 the disease-associated variants are typically common (minor-allele frequency >5%) and located in noncoding sequences; this has made it difficult to pinpoint causal genes and affected pathways. This lack of a causal mechanism has in part hindered the immediate translation of the findings of genomewide association studies into new therapeutic targets. However, the discovery of rare or low-frequency coding-sequence variants that affect the risk of coronary artery disease has facilitated advances in the prevention and treatment of disease. The most recent example of such advances is the development of a new class of therapeutic agents that is based on the discovery of the gene encoding pro-protein convertase subtilisin/kexin type 9 (PCSK9) as a regulator of low-density lipoprotein (LDL) cholesterol4 and the discovery that low-frequency and loss-of-function variants in this gene protect against coronary artery disease.5,6
Recently, low-frequency coding variation across the genome was systematically tabulated with the use of next-generation exome and whole-genome sequencing data from more than 12,000 persons of various ancestries (including a major contribution from the National Heart, Lung, and Blood Institute Exome Sequencing Project). Protein-altering variants (i.e., nonsynonymous, splice-site, and nonsense single-nucleotide substitutions) that were observed at least twice among these 12,000 persons were included in a genotyping array (hereafter referred to as the exome array). In addition, the exome array contains previously described variants from genomewide association studies, a sparse genomewide grid of common markers, markers that are informative with regard to ancestry (i.e., African American, Native American, and European), and some additional content. Additional information on the design of the exome array is provided at http://genome.sph.umich.edu/wiki/Exome_Chip_Design. In this study, we focused on the 220,231 autosomal variants that were present on the array and were expected to alter protein sequence (i.e., missense, nonsense, splice-site, and frameshift variants) and used these to test the contribution of low-frequency coding variation to the risk of coronary artery disease.
METHODS
STUDY DESIGN AND PARTICIPANTS
We performed a discovery study involving 42,335 patients with coronary artery disease and 78,240 controls from 20 individual studies (hereafter referred to as the discovery cohort). For variants with suggestive associations, we sought replication of our findings in an independent study of 30,533 patients and 42,530 controls assembled from 8 individual studies (hereafter referred to as the replication cohort). The names of the individual studies and information on the numbers of participants and phenotypic definitions of patients and controls in the discovery cohort and the replication cohort are provided in Tables S1 and S2, respectively, in the Supplementary Appendix, available with the full text of this article at NEJM.org. The 5755 participants from the Bangladesh Risk of Acute Vascular Events (BRAVE) study and the 22,072 participants from the Pakistan Risk of Myocardial Infarction Study (PROMIS) were of South Asian ancestry; all other participants were of European ancestry.
GENOTYPING AND QUALITY CONTROL
Samples were genotyped on the Illumina HumanExome BeadChip array, version 1.0 or 1.1, or the Illumina OmniExome array (which includes markers from the HumanExome BeadChip) in accordance with the manufacturer's recommended protocol. Our genotyping methods, as well as the quality-control procedures that were used to remove low-quality samples and variants, are described in the Supplementary Appendix.
FOLLOW-UP ANGPTL4 SEQUENCING
The exon sequences of ANGPTL4 were obtained from the exome sequences7 of 6924 persons who had early-onset myocardial infarction and those of 6834 persons who were free from coronary artery disease (see the Methods section in the Supplementary Appendix for details). The names of the individual studies and information on the numbers of participants and phenotypic definitions of patients and controls for ANGPTL4 sequencing are provided in Table S3 in the Supplementary Appendix.
STATISTICAL ANALYSIS
Of the 220,231 variants on the exome array, 54,003 (covering 13,715 genes) were present in our study at sufficient frequency (minor-allele frequency >0.01%) to allow for individual-variant association testing as described in the Supplementary Appendix. In the discovery phase, we defined a suggestive novel association between a variant and the risk of coronary artery disease as one with a meta-analysis P value of 1×10−4 or lower. For variants with suggestive association, we performed association analysis in the replication studies as described in the Supplementary Appendix. We defined significant novel associations as those that were nominally significant (P<0.05) in the replication cohort and that had an overall P value of less than 7.7×10−8 in the discovery and replication cohorts combined (a Bonferroni-corrected threshold that accounted for 54,003 markers with minor-allele frequencies >0.01% being tested initially and 12 markers being tested in the replication cohort).
To test for association between variants and risk factors for coronary artery disease, we examined the relationship between low-frequency variants that were significantly associated with disease in the combined (i.e., discovery and replication) analysis and plasma lipid levels in 10,088 samples from the discovery cohort; in order to minimize any effect of ascertainment bias, we limited the analysis to persons without coronary artery disease who had lipid measurements available. We also examined the relationships between the significantly associated low-frequency variants and blood pressure in 146,562 persons from the Cohorts for Heart and Aging Research in Genomic Epidemiology Plus (CHARGE+) consortium. Tables S4 and S5 in the Supplementary Appendix list the individual studies and numbers of participants that contributed to the analyses of plasma lipid levels and blood pressure, respectively. Finally, we queried publicly available exome-array data to explore the relationship between significantly associated low-frequency variants and type 2 diabetes (data from the Type 2 Diabetes Genetic Exploration by Next-Generation Sequencing in Multi-Ethnic Samples [T2D-GENES] Consortium, Genetics of Type 2 Diabetes [GoT2D] Consortium, and the Diabetes Genetics Replication and Meta-Analysis [DIAGRAM] Consortium; accessed in November 2015 at www.type2diabetesgenetics.org/). Additional details of the risk-factor association analyses are provided in the Methods section in the Supplementary Appendix.
From the sequencing data, we used linear regression to test the association between ANGPTL4 loss-of-function alleles and plasma lipid levels in 8085 persons for whom lipid measurements were available, using models described in the Supplementary Appendix. We calculated the significance of the association between ANGPTL4 loss-of-function alleles and the risk of coronary artery disease with the use of 100,000 study-stratified permutations of case–control phenotypes.
RESULTS
LOW-FREQUENCY CODING VARIANTS ASSOCIATED WITH CORONARY ARTERY DISEASE
The discovery cohort comprised 120,575 persons (42,335 patients and 78,240 controls) (Table S1 in the Supplementary Appendix). In the discovery cohort, we found significant associations between low-frequency coding variants in the LPA and PCSK9 genes and coronary artery disease (Table 1). Both gene loci also harbor common noncoding variants associated with coronary artery disease that had previously been discovered through genomewide association studies. These variants were also present on the exome array and had significant associations with coronary artery disease in our study (Table 1). In a conditional analysis, the associations between coronary artery disease and the low-frequency coding variants in both LPA and PCSK9 were found to be independent of the associations between coronary artery disease and the more common variants (Table 1).
Table 1.
Low-Frequency Coding Variations Previously Associated with Coronary Artery Disease.*
| Locus and SNP | Chromosome and Nucleotide Position† | Allele 1/Allele 2 | Frequency of Allele 1 % | Functional Effect | Odds Ratio‡ | P Value | Conditional P Value§ |
|---|---|---|---|---|---|---|---|
| LPA | |||||||
| rs3798220 | 6: 160961137 | C/T | 1.99 | p.I4399M | 1.54 | 9.7×10−24 | 1.9×10−18 |
| rs2048327 | 6: 160863532 | C/T | 37.36 | NA | 1.09 | 8.1×10−21 | 1.8×10−15 |
| PCSK9 | |||||||
| rs11591147 | 1: 55505647 | T/G | 1.52 | p.R46L | 0.78 | 8.0×10−10 | 1.9×10−7 |
| rs11206510 | 1: 55496039 | C/T | 18.23 | NA | 0.93 | 1.7×10−8 | 4.6×10−6 |
NA denotes not applicable, and SNP single-nucleotide polymorphism.
Chromosome numbers and positions refer to genome build GRCh37.
Odds ratios are for the development of disease in carriers of allele 1.
The conditional P value is the P value from an association test conditioning on the other marker listed in the locus.
In addition to the variants in LPA and PCSK9, 12 additional suggestive associations (P<1×10−4) between low-frequency coding variants and coronary artery disease were identified in the discovery study (Table S6 in the Supplementary Appendix). We tested for associations between these variants and coronary artery disease among the 73,063 persons (30,533 patients and 42,530 controls) in the replication cohort (see Tables S2 and S6 in the Supplementary Appendix for details). Variants in two genes showed association in the replication set and reached the genome-wide level of significance (P<7.7×10−8) in the combined data set (Table 2). A missense variant (encoding p.D2702G; minor-allele frequency, 3.60%) in the gene SVEP1 (which encodes sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1) was associated with an increased risk of coronary artery disease, and a low-frequency missense variant (encoding p.E40K; minor-allele frequency, 2.01%) in ANGPTL4, which encodes angiopoietin-like 4, was associated with protection against coronary artery disease (Table 2).
Table 2.
Novel Low-Frequency Coding Variations Showing Significant Association with Coronary Artery Disease.
| Locus and SNP | Chromosome and Nucleotide Position* | Allele 1/Allele 2 | Frequency of Allele 1 % | Functional Effect | Stage | Odds Ratio† | P Value |
|---|---|---|---|---|---|---|---|
| SVEP1 rs111245230 | 9: 113169775 | C/T | 3.60 | p.D2702G | Discovery | 1.14 | 1.1×10−7 |
| Replication | 1.13 | 1.0×10−3 | |||||
| Combined‡ | 1.14 | 4.2×10−10 | |||||
| ANGPTL4 rs116843064 | 19: 8429323 | A/G | 2.01 | p.E40K | Discovery | 0.87 | 3.0×10−5 |
| Replication | 0.86 | 3.4×10−4 | |||||
| Combined‡ | 0.86 | 4.0×10−8 |
Chromosome numbers and positions refer to genome build GRCh37.
Odds ratios are for the development of disease in carriers of allele 1. P values for testing differences between the effects observed in the discovery and replication results were 0.73 for rs111245230 and 0.11 for rs116843064 (Cochran's Q test for heterogeneity).
The combined stage refers to a meta-analysis that included 72,868 patients with coronary artery disease and 120,770 controls free from the disease.
ASSOCIATION BETWEEN VARIANTS AND RISK FACTORS FOR CORONARY ARTERY DISEASE
We next examined the associations of the SVEP1 p.D2702G and ANGPTL4 p.E40K variants with plasma lipid levels, blood pressure, and type 2 diabetes to determine whether their associations with coronary artery disease could potentially be mediated through an effect on these known risk factors. To minimize any potential effect of case–control ascertainment in testing the association with plasma lipid levels, we restricted our analysis to 10,088 controls who were free from coronary artery disease and to population-based samples with available lipid data from the discovery cohort. The tests of association with blood pressure were performed on a set of 146,562 samples from the CHARGE+ consortium, and the results for tests of association with type 2 diabetes were extracted from publicly available databases. Additional details of the sources and samples for these analyses of risk factors are provided in Tables S4 and S5 in the Supplementary Appendix.
We found a significant association between SVEP1 p.D2702G and blood pressure (Table 3, and Table S7 in the Supplementary Appendix). The allele associated with an increased risk of coronary artery disease was also associated with higher systolic blood pressure (0.94 mm Hg higher for each copy of the allele among allele carriers, P = 3.0×10−7) and higher diastolic blood pressure (0.57 mm Hg higher for each copy of the allele among allele carriers, P = 4.4×10−7). We did not find an association between SVEP1 p.D2702G and any plasma lipid trait. In contrast, ANGPTL4 p.E40K was not associated with blood pressure but instead was found to be associated with significantly lower levels of triglycerides (approximately 0.3 standard deviation units lower for each copy of the allele among allele carriers, P = 1.6×10−13) (Table 3) and with higher levels of high-density lipoprotein (HDL) cholesterol (approximately 0.3 standard deviation units higher for each copy of the allele among allele carriers, P = 8.2×10−11) (Table 3). In a conditional analysis, these effects appeared to be at least partially independent of each other (Table S8 in the Supplementary Appendix). We did not observe any significant association between ANGPTL4 p.E40K and LDL cholesterol level (Table 3). Both SVEP1 p.D2702G and ANGPTL4 p.E40K were nominally associated with type 2 diabetes in a direction concordant with the associated risk of coronary artery disease.
Table 3.
Association between Low-Frequency Variants and Traditional Risk Factors.*
| Variant and Risk Factor | Effect (95% CI)† | P Value |
|---|---|---|
| SVEP1 rs111245230 C allele | ||
| LDL cholesterol | 0.011 (−0.066 to 0.088) | 0.78 |
| HDL cholesterol | 0.023 (−0.054 to 0.099) | 0.56 |
| Triglycerides | 0.050 (−0.025 to 0.125) | 0.19 |
| Systolic blood pressure | 0.94 (0.58 to 1.3) | 3.0×10−7 |
| Diastolic blood pressure | 0.57 (0.35 to 0.79) | 4.4×10−7 |
| Type 2 diabetes | 1.13 (1.10 to 1.16) | 0.0062 |
| ANCPTL4 rs116843064 A allele | ||
| LDL cholesterol | −0.064 (−0.153 to 0.025) | 0.16 |
| HDL cholesterol | 0.295 (0.206 to 0.384) | 8.2×10−11 |
| Triglycerides | −0.335 (−0.424 to −0.246) | 1.6×10−13 |
| Systolic blood pressure | −0.18 (−0.66 to 0.30) | 0.46 |
| Diastolic blood pressure | −0.13 (−0.43 to 0.16) | 0.38 |
| Type 2 diabetes | 0.90 (0.87 to 0.92) | 0.028 |
Shown in the table are low-frequency variants associated with coronary artery disease that are outside of known loci that were previously identified in genome-wide association studies. Cl denotes confidence interval, HDL high-density lipoprotein, and LDL low-density lipoprotein.
The effect is the difference per each copy of the allele in units of standard deviation change (in LDL cholesterol level, HDL cholesterol level, and the natural logarithm of triglyceride level), millimeters of mercury (for systolic and diastolic blood pressure), or the odds ratio for disease (for type 2 diabetes) for carriers as compared with noncarriers.
ANGPTL4 LOSS-OF-FUNCTION MUTATIONS, PLASMA LIPID LEVELS, AND CORONARY ARTERY DISEASE
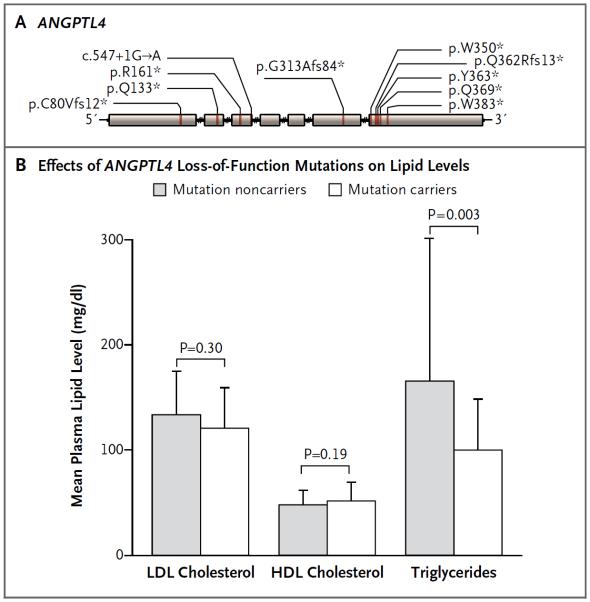
The finding that a missense allele in ANGPTL4 reduced the risk of coronary artery disease, potentially by reducing triglyceride levels, raised the possibility that complete loss-of-function variants in ANGPTL4 may have an even more dramatic effect on triglyceride concentrations and the risk of coronary artery disease. We therefore examined sequence data for the seven protein-coding exons of ANGPTL4 in 6924 patients with early-onset myocardial infarction and 6834 controls free from coronary artery disease (details of the patients and controls are provided in Table S3 in the Supplementary Appendix). We discovered a total of 10 variants that were predicted to lead to loss of gene function (Fig. 1A, and Table S9 in the Supplementary Appendix), carried by 28 heterozygous persons; no homozygous or compound heterozygous persons were discovered. Carriers of loss-of-function alleles had significantly lower levels of triglycerides than did noncarriers (a mean of 35% lower among carriers, P = 0.003) (Fig. 1B, and Table S10 in the Supplementary Appendix), with no significant difference in LDL or HDL cholesterol levels. Moreover, we found a lower risk of coronary artery disease among carriers of loss-of-function alleles (9 carriers among 6924 patients vs. 19 carriers among 6834 controls; odds ratio for disease, 0.47; P = 0.04) (Table S11 in the Supplementary Appendix). A similar investigation was performed for the 48 protein-coding exons of SVEP1; however, only 3 loss-of-function allele carriers were discovered (2 carriers among 6924 patients vs. 1 carrier among 6834 controls).
Figure 1. Loss-of-Function Alleles in ANGPTL4 and Plasma Lipid Levels.
Panel A shows the loss-of-function mutations discovered by sequencing the seven exons of ANGPTL4 in 9731 persons of European ancestry. The locations of the individual mutations are depicted along the length of the ANGPTL4 gene, starting at the 5′ end (left), along with the predicted functional effect. An asterisk indicates the introduction of a premature stop codon. Panel B shows the mean plasma lipid levels according to loss-of-function allele carrier status. T bars indicate standard deviations. P values were calculated from a linear regression (with the use of a logarithm transformation in the case of triglycerides) with covariates of age, sex, and study. HDL denotes high-density lipoprotein, and LDL low-density lipoprotein.
CODING VARIATION IN LPL AND THE RISK OF CORONARY ARTERY DISEASE
On the basis of the fact that a loss of ANGPTL4 function was associated with reduced risk of coronary artery disease and that ANGPTL4 inhibits lipoprotein lipase (LPL), one would expect a gain of LPL function to also be associated with a lower risk of coronary artery disease, whereas a loss of LPL function would be expected to be associated with a higher risk. In observations consistent with these expectations, we found a low-frequency missense variant in LPL on the exome array that was associated with an increased risk of coronary artery disease (p.D36N; minor-allele frequency, 1.9%; odds ratio for disease, 1.13; P = 2.0×10−4) (Table S12 in the Supplementary Appendix); previous studies have shown that this allele (also known as p.D9N) is associated with LPL activity that is 20% lower in allele carriers than in noncarriers.8 We also identified a nonsense mutation in LPL on the exome array that was significantly associated with a reduced risk of coronary artery disease (p.S447⋆; minor-allele frequency, 9.9%; odds ratio, 0.94; P = 2.5×10−7) (Table S12 in the Supplementary Appendix). Contrary to most instances in which the premature introduction of a stop codon leads to loss of gene function, this nonsense mutation, which occurs in the penultimate codon of the gene, paradoxically induces a gain of LPL function.9
DISCUSSION
Through large-scale exomewide screening, we identified a low-frequency coding variant in ANGPTL4 that was associated with protection against coronary artery disease and a low-frequency coding variant in SVEP1 that was associated with an increased risk of the disease. Moreover, our results highlight LPL as a significant contributor to the risk of coronary artery disease and support the hypothesis that a gain of LPL function or loss of ANGPTL4 inhibition protects against the disease.
ANGPTL4 has previously been found to be involved in cancer pathogenesis and wound healing.10 Previous functional studies also revealed that ANGPTL4 regulates plasma triglyceride concentration by inhibiting LPL.11 The minor allele at p.E40K has previously been associated with lower levels of triglycerides and higher levels of HDL cholesterol.12 We now provide independent confirmation of these lipid effects. In vitro and in vivo experimental evidence suggests that the lysine allele at p.E40K results in destabilization of ANGPTL4 after its secretion from the cell in which it was synthesized. It may be that the p.E40K variant leads to increases in the enzymatic activity of LPL because of this destabilization.13 Previous, smaller studies produced conflicting results regarding p.E40K and the risk of coronary artery disease14,15; we now provide robust support for an association between p.E40K and a reduced risk of coronary artery disease.
To provide confirmatory orthogonal evidence that a loss of ANGPTL4 function is associated with a decreased risk of coronary artery disease, we searched for loss-of-function mutations in this gene. We found that ANGPTL4 loss-of-function mutations were associated with substantially lower triglyceride levels (35% lower than in persons who were not carriers of a loss-of-function mutation), and we also found that these loss-of-function alleles were associated with a 53% lower risk of coronary artery disease. The identification by Dewey et al.16 of additional ANGPTL4 inactivating mutation carriers, now reported in the Journal, provides further evidence of the association between a loss of ANGPTL4 function and lower triglyceride levels and a reduced risk of coronary artery disease.
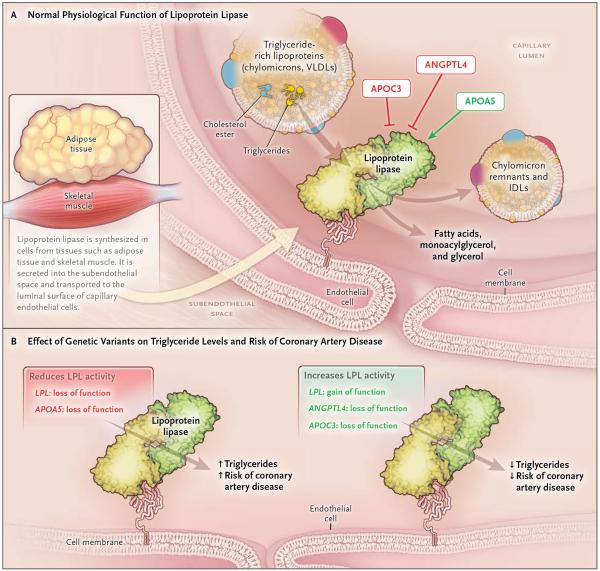
Because ANGPTL4 inhibits LPL, we examined coding variants in LPL that were present on the exome array. A gain-of-function mutation in LPL was found to be associated with protection against coronary artery disease, whereas a partial loss-of-function mutation was associated with increased risk. These data corroborate evidence that has connected the risk of myocardial infarction to coding-sequence mutations in two other genes encoding proteins that modulate LPL activity: apolipoprotein A57 and apolipoprotein C3.17,18 Thus, the available human genetic evidence, including our current findings, is coalescing around the conclusion that, in addition to elevated levels of LDL cholesterol, disordered metabolism of triglyceride-rich lipoproteins through the LPL pathway contributes substantively to coronary artery disease (Fig. 2). These data also support the hypothesis that therapeutic modulation of this pathway — either by direct enhancement of LPL activity or by blocking of the effects of natural LPL inhibitors, such as ANGPTL4 (Fig. 2B) — should reduce both triglyceride levels and the risk of coronary artery disease. Studies involving inhibitors of APOC3,19 of ANGPTL4,16,20 or of other inhibitory regulators in the LPL pathway will be needed to address this possibility.
Figure 2. Genetic Variants Affecting the Lipoprotein Lipase Pathway and the Risk of Coronary Artery Disease.
Panel A shows normal physiological function of lipoprotein lipase (LPL) and regulation of LPL by gene products of ANGPTL4, APOC3, and APOA5. LPL, which is both transported across and anchored to capillary endothelial cells by the protein GPIHBP1, normally hydrolyzes the triglycerides that are present in circulating lipoproteins and reduces the plasma triglyceride level. Its activity is reduced by ANGPTL4 and APOC3 and increased by APOA5. Not shown here are other important regulators of LPL activity, including APOC2 and ANGPTL3. Green arrows indicate enhancers, and red blocked arrows indicate inhibitors. IDL denotes intermediate-density lipoprotein, and VLDL very-low-density lipoprotein. Panel B shows altered function of LPL in mutation carriers. Mutations affecting LPL and proteins interacting with LPL are shown, along with expected effect on LPL activity, plasma triglyceride levels, and risk of coronary artery disease. LPL loss of function refers to p.D36N, and gain of function refers to p.S477* (see Table S12 in the Supplementary Appendix). ANGPTL4 loss of function refers to both p.E40K and loss-of-function mutations (Tables S9, S10, and S11 in the Supplementary Appendix). APOC317,18 and APOA57 loss of function refers to multiple loss-of-function mutations.
The identification of a disease-associated missense variant in SVEP1 points to a potentially novel genetic mechanism leading to atherosclerosis. SVEP1 is a cell-adhesion molecule that acts as a ligand for integrin α9β121; knockdown of SVEP1 alters inflammatory signaling in a cellular model of sepsis.22 Although we observed that the low-frequency variant that was associated with an increased risk of coronary artery disease was also associated with significantly higher blood pressure, the latter effect was modest as compared with that found in a previous study, in which a genetically mediated 1.6–mm Hg increase in systolic blood pressure was associated with a 10% increased risk of coronary artery disease23 (in our study, SVEP1 p.D2702G was associated with an approximately 1–mm Hg increase in systolic blood pressure but a 14% increased risk of coronary artery disease). Thus, the exact mechanism by which SVEP1 contributes to coronary artery disease and the extent to which hypertension and diabetes mediate this risk remain to be clarified.
Several limitations of our study deserve consideration. In our samples, approximately three quarters of the variants on the array were either monomorphic (i.e., only the human reference allele was observed among the persons in the discovery cohort) or very rare (frequency below 0.1%); therefore, we cannot make any estimates of their contribution to the risk of coronary artery disease. Moreover, we estimate that the Illumina HumanExome BeadChip, version 1.0, covers only approximately 80% of coding variants in persons of European ancestry that have an allele frequency of 0.1% or higher (Fig. S1 in the Supplementary Appendix). Furthermore, despite a combined sample size of more than 120,000 persons in the discovery cohort, we had only 80% power to detect an odds ratio of approximately 1.5 for variants with a minor-allele frequency of 0.5% (or an odds ratio of approximately 2.0 for variants with a minor-allele frequency of 0.1%) at the required stringent level of significance (Fig. S2 in the Supplementary Appendix). Hence, analyses of even larger data sets and more comprehensive coverage of exonic variants may reveal further coding variants associated with the risk of coronary artery disease.
In summary, through large-scale exomewide screening, we identified several low-frequency coding variants that are associated with either an increased risk of or protection against coronary artery disease. Our results highlight the LPL pathway as a significant contributor to the risk of coronary artery disease and support speculation that therapeutic modulation of this pathway might be protective against coronary artery disease.
Supplementary Material
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supported by a career development award from the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH) (K08HL114642 to Dr. Stitziel) and by the Foundation for Barnes–Jewish Hospital. Dr. Peloso is supported by the National Heart, Lung, and Blood Institute of the NIH (award number K01HL125751). Dr. Kathiresan is supported by a Research Scholar award from the Massachusetts General Hospital, the Donovan Family Foundation, grants from the NIH (R01HL107816 and R01HL127564), a grant from Fondation Leducq, and an investigator-initiated grant from Merck. Dr. Merlini was supported by a grant from the Italian Ministry of Health (RFPS-2007-3-644382). Drs. Ardissino and Marziliano were supported by Regione Emilia Romagna Area 1 Grants. Drs. Farrall and Watkins acknowledge the support of the Wellcome Trust core award (090532/Z/09/Z), the British Heart Foundation (BHF) Centre of Research Excellence. Dr. Schick is supported in part by a grant from the National Cancer Institute (R25CA094880). Dr. Goel acknowledges EU FP7 & Wellcome Trust Institutional strategic support fund. Dr. Deloukas's work forms part of the research themes contributing to the translational research portfolio of Barts Cardiovascular Biomedical Research Unit, which is supported and funded by the National Institute for Health Research (NIHR). Drs. Webb and Samani are funded by the British Heart Foundation, and Dr. Samani is an NIHR Senior Investigator. Dr. Masca was supported by the NIHR Leicester Cardiovascular Biomedical Research Unit (BRU), and this work forms part of the portfolio of research supported by the BRU. Dr. Won was supported by a postdoctoral award from the American Heart Association (15POST23280019). Dr. McCarthy is a Wellcome Trust Senior Investigator (098381) and an NIHR Senior Investigator. Dr. Danesh is a British Heart Foundation Professor, European Research Council Senior Investigator, and NIHR Senior Investigator. Drs. Erdmann, Webb, Samani, and Schunkert are supported by the FP7 European Union project CVgenes@target (261123) and the Fondation Leducq (CADgenomics, 12CVD02). Drs. Erdmann and Schunkert are also supported by the German Federal Ministry of Education and Research e:Med program (e:AtheroSysMed and sysINFLAME), and Deutsche Forschungsgemeinschaft cluster of excellence “Inflammation at Interfaces” and SFB 1123. Dr. Kessler received a DZHK Rotation Grant. The analysis was funded, in part, by a Programme Grant from the BHF (RG/14/5/30893 to Dr. Deloukas). Additional funding is listed in the Supplementary Appendix.
APPENDIX
The authors' full names and academic degrees are as follows: Nathan O. Stitziel, M.D., Ph.D., Kathleen E. Stirrups, Ph.D., Nicholas G.D. Masca, Ph.D., Jeanette Erdmann, Ph.D., Paola G. Ferrario, Dr. rer. nat., Inke R. König, Dr. rer. biol. hum., Peter E. Weeke, M.D., Ph.D., Thomas R. Webb, Ph.D., Paul L. Auer, Ph.D., Ursula M. Schick, Ph.D., Yingchang Lu, M.D., Ph.D., He Zhang, Ph.D., Marie-Pierre Dube, Ph.D., Anuj Goel, M.Sc., Martin Farrall, F.R.C.Path., Gina M. Peloso, Ph.D., Hong-Hee Won, Ph.D., Ron Do, Ph.D., Erik van Iperen, M.Sc., Stavroula Kanoni, Ph.D., Jochen Kruppa, Ph.D., Anubha Mahajan, Ph.D., Robert A. Scott, Ph.D., Christina Willenborg, Ph.D., Peter S. Braund, Ph.D., Julian C. van Capelleveen, M.D., Alex S.F. Doney, M.D., Ph.D., Louise A. Donnelly, Ph.D., Rosanna Asselta, Ph.D., Piera A. Merlini, M.D., Stefano Duga, Ph.D., Nicola Marziliano, Ph.D., Josh C. Denny, M.D., M.S., Christian M. Shaffer, B.S., Nour Eddine El-Mokhtari, M.D., Andre Franke, Ph.D., Omri Gottesman, M.D., Stefanie Heilmann, Ph.D., Christian Hengstenberg, M.D., Per Hoffmann, Ph.D., Oddgeir L. Holmen, M.D., Kristian Hveem, M.D., Ph.D., Jan-Håkan Jansson, M.D., Ph.D., Karl-Heinz Jöckel, Ph.D., Thorsten Kessler, M.D., Jennifer Kriebel, Ph.D., Karl L. Laugwitz, M.D., Eirini Marouli, Ph.D., Nicola Martinelli, M.D., Ph.D., Mark I. McCarthy, M.D., Ph.D., Natalie R. Van Zuydam, Ph.D., Christa Meisinger, M.D., M.P.H., Tõnu Esko, Ph.D., Evelin Mihailov, M.Sc., Stefan A. Escher, Ph.D., Maris Alver, M.Sc., Susanne Moebus, Ph.D., Andrew D. Morris, M.D., Martina Müller-Nurasyid, Ph.D., Majid Nikpay, Ph.D., Oliviero Olivieri, M.D., Louis-Philippe Lemieux Perreault, Ph.D., Alaa AlQarawi, B.Sc., Neil R. Robertson, M.Sc., Karen O. Akinsanya, Ph.D., Dermot F. Reilly, Ph.D., Thomas F. Vogt, Ph.D., Wu Yin, Ph.D., Folkert W. Asselbergs, M.D., Ph.D., Charles Kooperberg, Ph.D., Rebecca D. Jackson, M.D., Eli Stahl, Ph.D., Konstantin Strauch, Ph.D., Tibor V. Varga, M.Sc., Melanie Waldenberger, Ph.D., Lingyao Zeng, M.Sc., Aldi T. Kraja, D.Sc., Ph.D., Chunyu Liu, Ph.D., Georg B. Ehret, M.D., Christopher Newton-Cheh, M.D., M.P.H., Daniel I. Chasman, Ph.D., Rajiv Chowdhury, M.D., Ph.D., Marco Ferrario, M.D., Ian Ford, Ph.D., J. Wouter Jukema, M.D., Ph.D., Frank Kee, M.D., Kari Kuulasmaa, Ph.D., Børge G. Nordestgaard, M.D., D.M.Sc., Markus Perola, M.D., Ph.D., Danish Saleheen, M.B., B.S., Ph.D., Naveed Sattar, F.R.C.P., Ph.D., Praveen Surendran, Ph.D., David Tregouet, Ph.D., Robin Young, Ph.D., Joanna M.M. Howson, Ph.D., Adam S. Butterworth, Ph.D., John Danesh, F.R.C.P., D.Phil., Diego Ardissino, M.D., Erwin P. Bottinger, M.D., Raimund Erbel, M.D., Paul W. Franks, Ph.D., Domenico Girelli, M.D., Ph.D., Alistair S. Hall, M.D., Ph.D., G. Kees Hovingh, M.D., Ph.D., Adnan Kastrati, M.D., Wolfgang Lieb, M.D., Thomas Meitinger, M.D., William E. Kraus, M.D., Svati H. Shah, M.D., M.P.H., Ruth McPherson, M.D., Ph.D., Marju Orho-Melander, Ph.D., Olle Melander, M.D., Ph.D., Andres Metspalu, M.D., Ph.D., Colin N.A. Palmer, Ph.D., Annette Peters, Ph.D., Daniel J. Rader, M.D., Muredach P. Reilly, M.B., B.Ch., M.S.C.E., Ruth J.F. Loos, Ph.D., Alex P. Reiner, M.D., Dan M. Roden, M.D., Jean-Claude Tardif, M.D., John R. Thompson, Ph.D., Nicholas J. Wareham, M.B., B.S., Ph.D., Hugh Watkins, M.D., Ph.D., Cristen J. Willer, Ph.D., Sekar Kathiresan, M.D., Panos Deloukas, Ph.D., Nilesh J. Samani, F.R.C.P., and Heribert Schunkert, M.D.
The authors' affiliations are as follows: Cardiovascular Division, Departments of Medicine and Genetics, and the McDonnell Genome Institute (N.O.S.), and the Division of Statistical Genomics, Department of Genetics and Center for Genome Sciences and Systems Biology (A.T.K.), Washington University School of Medicine, St. Louis; William Harvey Research Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London (K.E.S., S. Kanoni, E. Marouli, P.D.), and Institute of Cardiovascular Science, Faculty of Population Health Sciences, University College London (F.W.A.), London, Department of Haematology (K.E.S.), Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care (R.C., P.S., R.Y., J.M.M.H., A.S.B., J.D.), and National Institute of Health Research Blood and Transplant Research Unit in Donor Health and Genomics (J.D.), University of Cambridge, MRC Epidemiology Unit, Institute of Metabolic Science, Addenbrooke's Hospital (R.A.S., N.J.W.), and Wellcome Trust Sanger Institute (J.D.), Cambridge, Departments of Cardiovascular Sciences (N.G.D.M., T.R.W., P.S.B., N.J.S.) and Health Sciences (J.R.T.), University of Leicester, and NIHR Leicester Cardiovascular Biomedical Research Unit, Glenfield Hospital (N.G.D.M., T.R.W., P.S.B., J.R.T., N.J.S.), Leicester, Division of Cardiovascular Medicine, Radcliffe Department of Medicine (A.G., M. Farrall, H.W.), the Wellcome Trust Centre for Human Genetics (A.G., M. Farrall, A. Mahajan, M.I.M., N.R.R., H.W.), and Oxford Centre for Diabetes, Endocrinology and Metabolism (M.I.M., N.R.V.Z., N.R.R.), University of Oxford, and Oxford National Institute for Health Research Biomedical Research Centre, Churchill Hospital (M.I.M.), Oxford, Medical Research Institute, University of Dundee, Ninewells Hospital and Medical School, Dundee (A.S.F.D., L.A.D., C.N.A.P.), School of Molecular, Genetic and Population Health Sciences, University of Edinburgh Medical School, Edinburgh (A.D.M.), Robertson Centre for Biostatistics (I.F.) and British Heart Foundation, Glasgow Cardiovascular Research Centre (N.S.), University of Glasgow, Glasgow, and Leeds Institute of Genetics, Health and Therapeutics, University of Leeds, Leeds (A.S.H.) — all in the United Kingdom; Institute for Integrative and Experimental Genomics, University of Lübeck (J.E., C.W.), DZHK (German Center for Cardiovascular Research), partner site Hamburg/Lübeck/Kiel (J.E., P.G.F., I.R.K., C.W.), and Institut für Medizinische Biometrie und Statistik, Universität zu Lübeck (P.G.F., I.R.K., J. Kruppa), Lübeck, Department of Medical Statistics, University Medical Center Göttingen, Göttingen (J. Kruppa), Klinik für Innere Medizin, Kreiskrankenhaus Rendsburg, Rendsburg (N.E.E.-M.), Institute of Clinical Molecular Biology (A.F.) and Institute of Epidemiology and Biobank popgen (W.L.), Christian-Albrechts-University Kiel, Kiel, Institute of Human Genetics (S.H., P.H.) and Department of Genomics, Life and Brain Center (S.H., P.H.), University of Bonn, Bonn, Deutsches Herzzentrum München (C.H., T.K., L.Z., A.K., H.S.) and Institute of Human Genetics (T.M.), Technische Universität München, DZHK partner site Munich Heart Alliance (C.H., K.L.L., M.M.-N., T.M., A.P., H.S.), I. Medizinische Klinik und Poliklinik, Klinikum rechts der Isar der Technischen Universität München (K.L.L.), Department of Medicine I (M.M.-N.), and Institute for Medical Informatics, Biometry and Epidemiology, Chair of Genetic Epidemiology (K.S.), Ludwig-Maximilians-Universität, Munich, Institute for Medical Informatics, Biometry and Epidemiology, University Hospital Essen, Essen (K.-H.J., S.M., R.E.), and Research Unit of Molecular Epidemiology (J. Kriebel, M.W.) and Institutes of Epidemiology II (J. Kriebel, C.M., M.W., A.P.), Genetic Epidemiology (M.M.-N., K.S.), and Human Genetics (T.M.), Helmholtz Zentrum München–German Research Center for Environmental Health, and German Center for Diabetes Research (J. Kriebel), Neuherberg — all in Germany; Departments of Medicine (P.E.W., J.C.D., C.M.S., D.M.R.), Biomedical Informatics (J.C.D.), and Pharmacology (D.M.R.), Vanderbilt University Medical Center, Nashville; Laboratory for Molecular Cardiology, Department of Cardiology, Copenhagen University Hospital Rigshospitalet (P.E.W.), and Copenhagen University Hospital and Faculty of Health and Medical Sciences, University of Copenhagen (B.G.N.), Copenhagen; School of Public Heath, University of Wisconsin–Milwaukee, Milwaukee (P.L.A.); Fred Hutchinson Cancer Research Center (U.M.S., C.K., A.P.R.), and Department of Epidemiology, University of Washington (A.P.R.), Seattle; the Genetics of Obesity and Related Metabolic Traits Program (Y.L., R.J.F.L.), the Charles Bronfman Institute for Personalized Medicine (U.M.S., Y.L., R.D., O.G., E.P.B., R.J.F.L.), the Center for Statistical Genetics and Institute for Genomics and Multiscale Biology, Department of Genetics and Genomic Sciences, Zena and Michael A. Weiner Cardiovascular Institute (R.D.), and the Department of Psychiatry (E.S.), and Mindich Child Health and Development Institute (R.J.F.L.), Icahn School of Medicine at Mount Sinai, New York; Department of Internal Medicine, Division of Cardiovascular Medicine (H.Z., C.J.W.), and Departments of Computational Medicine and Bioinformatics (C.J.W.) and Human Genetics (C.J.W.), University of Michigan, Ann Arbor; Université de Montréal, Faculté de médecine, Département de médecine (M.-P.D., J.-C.T.), and the Montreal Heart Institute (M.-P.D., L.-P.L.P., J.-C.T.), Montreal, and Ruddy Canadian Cardiovascular Genetics Centre, University of Ottawa Heart Institute, Ottawa (M.N., R.M.) — both in Canada; Center for Human Genetic Research and Cardiovascular Research Center (G.M.P., H.-H.W., C.N.-C., S. Kathiresan) and the Cardiology Division (C.N.-C., S. Kathiresan), Massachusetts General Hospital, the Departments of Medicine (G.M.P., H.-H.W., C.N.-C., S. Kathiresan) and Genetics (T.E.), Harvard Medical School, Division of Endocrinology, Boston Children's Hospital (T.E.), Division of Preventive Medicine, Brigham and Women's Hospital (D.I.C.), and Department of Nutrition, Harvard School of Public Health (P.W.F.), Boston, and the Program in Medical and Population Genetics, Broad Institute (G.M.P., H.-H.W., C.N.-C., S. Kathiresan), and Broad Institute of the Massachusetts Institute of Technology and Harvard (T.E.), Cambridge, and the Framingham Heart Study, Framingham (C.L.) — all in Massachusetts; Samsung Advanced Institute for Health Sciences and Technology (SAIHST), Sungkyunkwan University, Samsung Medical Center, Seoul, South Korea (H.-H.W.); Departments of Biostatistics (E.I.) and Vascular Medicine (J.C.C., G.K.H.), Academic Medical Center, Amsterdam, Department of Cardiology, Division of Heart and Lungs, UMC Utrecht, and Durrer Center for Cardiogenetic Research, ICIN-Netherlands Heart Institute (F.W.A.), and Interuniversity Cardiology Institute of the Netherlands (J.W.J.), Utrecht, and Department of Cardiology, Leiden University Medical Center, Leiden (J.W.J.) — all in the Netherlands; Department of Biomedical Sciences, Humanitas University, and Humanitas Clinical and Research Center (R.A., S.D.), and Division of Cardiology Niguarda Hospital (P.A.M.), Milan, Division of Cardiology, Azienda Ospedaliero–Universitaria di Parma, Parma (N. Marziliano, D.A.), Associazione per lo Studio della Trombosi in Cardiologia, Pavia (N. Marziliano, D.A.), Department of Medicine, Section of Internal Medicine, University of Verona, Verona (N. Martinelli, O.O., D.G.), and EPIMED Research Center, Department of Clinical and Experimental Medicine, University of Insubria, Varese (M. Ferrario) — all in Italy; Division of Medical Genetics, Department of Biomedicine, University of Basel, Basel (P.H.), and Cardiology Division, Department of Medicine, Geneva University Hospital, Geneva (G.B.E.) — both in Switzerland; HUNT Research Center, Department of Public Health and General Practice, Norwegian University of Science and Technology (O.L.H., K.H.), and Department of Medicine, Levanger Hospital, Nord-Trøndelag Health Trust Norway (K.H.), Levanger, and St. Olav Hospital, Trondheim University Hospital, Trondheim (O.L.H.) — all in Norway; Department of Public Health and Clinical Medicine, Research Unit Skellefteå, Umeå University (J.-H.J.), and Department of Public Health and Clinical Medicine, Umeå University Hospital (P.W.F.), Umeå, Genetic and Molecular Epidemiology Unit, Lund University Diabetes Center, Department of Clinical Sciences (S.A.E., T.V.V., P.W.F.), and Department of Clinical Sciences in Malmo, Lund University Diabetes Center, Lund University, Clinical Research Center, Skåne University Hospital (M.O.-M., O.M.), Malmo — all in Sweden; Estonian Genome Center, University of Tartu (T.E., E. Mihailov, M.A., A. Metspalu), and Institute of Molecular and Cell Biology, (M.A., A. Metspalu), Tartu, Estonia; Princess Al-Jawhara Al-Brahim Center of Excellence in Research of Hereditary Disorders (PACER-HD), King Abdulaziz University, Jeddah, Saudi Arabia (A.A., P.D.); Merck Sharp & Dohme, Rahway, NJ (K.O.A., D.F.R., T.F.V., W.Y.); Division of Endocrinology, Diabetes, and Metabolism, Department of Medicine, Ohio State University, Columbus (R.D.J.); the Population Sciences Branch, National Heart, Lung, and Blood Institute, Bethesda (C.L.), and Center for Complex Disease Genomics, McKusick–Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore (G.B.E.) — both in Maryland; Queen's University of Belfast, Belfast, Northern Ireland (F.K.); National Institute for Health and Welfare, Helsinki (K.K., M.P.); Department of Biostatistics and Epidemiology (D.S.), Department of Genetics, Cardiovascular Institute and Institute of Translational Medicine and Therapeutics (D.J.R.), and the Cardiovascular Institute (M.P.R.), Perelman School of Medicine, University of Pennsylvania, Philadelphia; Center for Noncommunicable Diseases, Karachi, Pakistan (D.S.); Sorbonne Université, UPMC Univ Paris 06, ICAN Institute for Cardiometabolism and Nutrition, Paris (D.T.); and Duke Molecular Physiology Institute and Division of Cardiology, Department of Medicine, Duke University, Durham, NC (W.E.K., S.H.S.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.The CARDIoGRAMplusC4D Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schunkert H, König IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikpay M, Goel A, Won HH, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–30. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abifadel M, Varret M, Rabès JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–6. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 6.Kathiresan S. A PCSK9 missense variant associated with a reduced risk of early-onset myocardial infarction. N Engl J Med. 2008;358:2299–300. doi: 10.1056/NEJMc0707445. [DOI] [PubMed] [Google Scholar]
- 7.Do R, Stitziel NO, Won HH, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–6. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailly F, Tugrul Y, Reymer PW, et al. A common variant in the gene for lipoprotein lipase (Asp9→Asn): functional implications and prevalence in normal and hyperlipidemic subjects. Arterioscler Thromb Vasc Biol. 1995;15:468–78. doi: 10.1161/01.atv.15.4.468. [DOI] [PubMed] [Google Scholar]
- 9.Kozaki K, Gotoda T, Kawamura M, et al. Mutational analysis of human lipoprotein lipase by carboxy-terminal truncation. J Lipid Res. 1993;34:1765–72. [PubMed] [Google Scholar]
- 10.Tan MJ, Teo Z, Sng MK, Zhu P, Tan NS. Emerging roles of angiopoietin-like 4 in human cancer. Mol Cancer Res. 2012;10:677–88. doi: 10.1158/1541-7786.MCR-11-0519. [DOI] [PubMed] [Google Scholar]
- 11.Lafferty MJ, Bradford KC, Erie DA, Neher SB. Angiopoietin-like protein 4 inhibition of lipoprotein lipase: evidence for reversible complex formation. J Biol Chem. 2013;288:28524–34. doi: 10.1074/jbc.M113.497602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romeo S, Pennacchio LA, Fu Y, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–6. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin W, Romeo S, Chang S, Grishin NV, Hobbs HH, Cohen JC. Genetic variation in ANGPTL4 provides insights into protein processing and function. J Biol Chem. 2009;284:13213–22. doi: 10.1074/jbc.M900553200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folsom AR, Peacock JM, Demerath E, Boerwinkle E. Variation in ANGPTL4 and risk of coronary heart disease: the Atherosclerosis Risk in Communities Study. Metabolism. 2008;57:1591–6. doi: 10.1016/j.metabol.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talmud PJ, Smart M, Presswood E, et al. ANGPTL4 E40K and T266M: effects on plasma triglyceride and HDL levels, post-prandial responses, and CHD risk. Arterioscler Thromb Vasc Biol. 2008;28:2319–25. doi: 10.1161/ATVBAHA.108.176917. [DOI] [PubMed] [Google Scholar]
- 16.Dewey FE, Gusarova V, O'Dushlaine C, et al. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl J Med. 2016;374:1123–33. doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 19.Gaudet D, Alexander VJ, Baker BF, et al. Antisense inhibition of apolipoprotein C-III in patients with hypertriglyceridemia. N Engl J Med. 2015;373:438–47. doi: 10.1056/NEJMoa1400283. [DOI] [PubMed] [Google Scholar]
- 20.Desai U, Lee EC, Chung K, et al. Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid pheno-type found in angiopoietin-like 4 knockout mice. Proc Natl Acad Sci U S A. 2007;104:11766–71. doi: 10.1073/pnas.0705041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato-Nishiuchi R, Nakano I, Ozawa A, et al. Polydom/SVEP1 is a ligand for integrin α9β1. J Biol Chem. 2012;287:25615–30. doi: 10.1074/jbc.M112.355016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakada TA, Russell JA, Boyd JH, Thair SA, Walley KR. Identification of a non-synonymous polymorphism in the SVEP1 gene associated with altered clinical outcomes in septic shock. Crit Care Med. 2014;43:101–8. doi: 10.1097/CCM.0000000000000604. [DOI] [PubMed] [Google Scholar]
- 23.International Consortium for Blood Pressure Genome-Wide Association Studies Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.