Key Points
Normal ABL1 is a tumor suppressor in BCR-ABL1–induced leukemia.
Allosteric stimulation of the normal ABL1 kinase activity enhanced the antileukemia effect of ABL1 tyrosine kinase inhibitors.
Abstract
Leukemias expressing constitutively activated mutants of ABL1 tyrosine kinase (BCR-ABL1, TEL-ABL1, NUP214-ABL1) usually contain at least 1 normal ABL1 allele. Because oncogenic and normal ABL1 kinases may exert opposite effects on cell behavior, we examined the role of normal ABL1 in leukemias induced by oncogenic ABL1 kinases. BCR-ABL1-Abl1−/− cells generated highly aggressive chronic myeloid leukemia (CML)-blast phase–like disease in mice compared with less malignant CML-chronic phase–like disease from BCR-ABL1-Abl1+/+ cells. Additionally, loss of ABL1 stimulated proliferation and expansion of BCR-ABL1 murine leukemia stem cells, arrested myeloid differentiation, inhibited genotoxic stress-induced apoptosis, and facilitated accumulation of chromosomal aberrations. Conversely, allosteric stimulation of ABL1 kinase activity enhanced the antileukemia effect of ABL1 tyrosine kinase inhibitors (imatinib and ponatinib) in human and murine leukemias expressing BCR-ABL1, TEL-ABL1, and NUP214-ABL1. Therefore, we postulate that normal ABL1 kinase behaves like a tumor suppressor and therapeutic target in leukemias expressing oncogenic forms of the kinase.
Introduction
The ABL1 protein is a ubiquitously expressed nonreceptor tyrosine kinase markedly influenced by subcellular localization and posttranslational modifications.1-3 Cytoplasmic expression of ABL1 leads to increased cell proliferation and survival. In response to genotoxic stress, ABL1 is translocated into the nucleus and/or mitochondria where its activity contributes to modulation of DNA repair, induction of apoptosis/necrosis, and inhibition of cell growth.
Normal ABL1 kinase activity is essential for B- and T-cell development, but expendable in hematopoietic stem cells (HSCs) and the myeloid compartment.4-6 Constitutively activated oncogenic mutants of the ABL1 tyrosine kinase play a central role in the pathogenesis of acute and chronic leukemias. Activation usually occurs as a consequence of chromosomal translocations (BCR-ABL1, TEL-ABL1, and others) or episomal amplification (NUP214-ABL1).1
The BCR-ABL1 fusion oncogene, the product of t(9;22)(q34;q11) is found in all patients with chronic myeloid leukemia (CML), in ∼25% of pre-B acute lymphocytic leukemia (ALL) and occasionally in de novo acute myeloid leukemia (AML).7 BCR-ABL1 kinase is leukemogenic only when expressed in an HSC with self-renewal capacity, thereby transforming it to a leukemia stem cell (LSC).8 In CML–chronic phase (CML-CP), LSCs are capable of generating large numbers of leukemia early progenitor cells (LPCs): leukemia common myeloid (LCMPs) and leukemia granulocyte/macrophage (LGMPs), which cannot self-renew and eventually differentiate to mature cells. Thus, CML-CP is a stem cell–derived but progenitor-driven disease.8 Transition of a relatively benign CML-CP to the aggressive and fatal blast phase (CML–blast phase [CML-BP]) is associated with expansion of LSCs, enhanced proliferation, arrested differentiation, drug resistance, and accumulation of additional genetic and epigenetic aberrations.9,10
NUP214-ABL1 fusion is generated by circularization of the 500-kb genomic region from ABL1 to NUP214 and subsequent extrachromosomal (episomal) amplification.11 The NUP214-ABL1 gene is found in ∼4% of all cases of adult ALL. Other ABL1 fusion genes have been described but are uncommon. For example, the ETV6(TEL)-ABL1 fusion gene is the product of a t(9;12)(q34;p13) and is found occasionally in patients with acute leukemias or myeloproliferative disorders. EML1, ZMIZ1, and RCSD1 were identified as ABL1 partners in ALLs.1
Leukemias expressing oncogenic forms of the ABL1 kinase usually contain the nonmutated allele encoding normal ABL1 kinase which may play an important role in pathogenesis of disease and/or in response to treatment, given its prominent role in regulation of cell motility, adhesion, autophagy, response to DNA damage, apoptosis, and proliferation.1-3 This possibility is supported by previous observations that loss of normal ABL1 expression resulting from interstitial deletion in the normal chromosome 9 [del(9q34)] and/or transcriptional silencing of the alternative ABL1 promoter within BCR-ABL1 translocation occurs during progression of CML-CP to CML-BP.12,13 Of note, in the absence of ABL1, BCR-ABL1 cells displayed reduced sensitivity to tyrosine kinase inhibitors (TKIs) such as imatinib.14 Therefore, we hypothesized that normal ABL1 is a tumor suppressor in CML-CP and therapeutic target in leukemias induced by oncogenic forms of ABL1 kinase.
Materials and methods
BCR-ABL1–positive Abl1−/− and Abl1+/+ cells
BCR-ABL1–positive Abl1−/− and Abl1+/+ bone marrow cells (BMCs) expressing YFP-ABL1 fusion protein or yellow fluorescent protein (YFP) only were obtained and maintained as described in supplemental Methods (see supplemental Data available at the Blood Web site).
Leukemogenesis in vivo
Green fluorescent protein (GFP)-positive or GFP/YFP-positive cells were injected into the tail vein of sublethally irradiated NOD/SCID mice. Animals were killed when first signs of disease were apparent and leukemia development was confirmed at necropsy. These studies were approved by the Temple University institutional animal care and use committee.
Immunostaining
LSCs and LPCs were identified as described before15 and detailed in supplemental Methods.
Colony formation assay
Freshly transfected Lin−c-Kit+Sca-1+ BCR-ABL1 Abl1+/+ and BCR-ABL1 Abl1−/− cells were cultured for 5 weeks in vitro and simultaneously plated in MethoCult H4230 (StemCell Technologies, Vancouver, BC, Canada) in absence of growth factors. Colonies were scored after 5 to 7 days, and replated in fresh Methocult and scored again after 5 to 7 days. Three rounds of serial replating (representing 5 weeks in culture) were performed. Five-week-old tissue-cultured BCR-ABL1 Abl1+/+ and BCR-ABL1 Abl1−/− cells were also plated in Methocult. Colonies were scored after 5 to 7 days.
Competitive growth assay
A mixture of GFP-positive BCR-ABL1 Abl1−/− and GFP/YFP-positive BCR-ABL1 Abl1−/− cells restored with YFP-ABL1 was maintained in Iscove modified Dulbecco medium (IMDM) supplemented with fetal bovine serum (FBS), stem cell factor (SCF), and interleukin-3 (IL-3) and also simultaneously injected into the tail vein of NOD/SCID mice. After 5 weeks, the in vitro and in vivo cell mixtures were analyzed by flow cytometry to determine the percentage of GFP and GFP/YFP double-positive cells.
Histologic and cytologic analysis
Tissue sections of spleen from leukemia-sick mice were fixed in 10% formalin, paraffin embedded, cut into 0.4-mm sections, transferred to glass slides, and stained with hematoxylin and eosin. Cytospin preparations of cells cultured for 7 days in the presence of pretested threshold concentrations of IL-3 + SCF or granulocyte colony-stimulating factor (G-CSF; 10 ng/mL) were stained with Wright-Giemsa.
Apoptosis induced by genotoxic treatment
Indicated concentrations of cisplatin (Platinol-AQ; Bristol-Myers Squibb Co, Princeton, NJ), mitomycin C (Sigma-Aldrich, St. Louis, MO), etoposide (Bedford Laboratories, Bedford, OH), methylnitronitrosoguanidine (MNNG; Sigma-Aldrich), and hydrogen peroxide (Sigma-Aldrich) were added to cells growing in MethoCult H4230 (103/mL) supplemented with IL-3. Colonies were scored after 7 days. Results are represented as the percentage of colony-forming cells after drug treatment in comparison with the untreated control group.
ROS, oxidative DNA damage, and genomic instability
Reactive oxygen species (ROS), 8-oxoguanine (8-oxoG), γ-H2AX nuclear foci, and chromosomal aberrations were detected as described before16 and detailed in supplemental Methods.
DPH treatment
CML-CP cells from freshly diagnosed patients, B-ALL xenograft cells, Baf3-TEL-ABL1 cell lines, and murine BMCs expressing NUP214-ABL1 oncogene were treated with 5-[3-(4-fluorophenyl)-1-phenylpyrazol-4-yl]imidazolidine-2,4-dione (DPH; Sigma-Aldrich), imatinib and ponatinib (both from Selleck Chemicals) and evaluated as described in supplemental Methods. These studies were approved by the Temple University institutional review board.
Western analyses
Total, cytoplasmic, and nuclear cell lysates were obtained as described before17 and analyzed as detailed in supplemental Methods.
Genome-wide expression array
The Affymetrix Mouse gene 1.0ST array containing 28 815 probe sets (Affymetrix) was used to measure messenger RNA (mRNA) expression levels. Affymetrix arrays were processed and analyzed as described in supplemental Methods.
Statistical analysis
Results are presented as mean ± standard deviation (SD) and were analyzed by the 2-tailed paired Student t test and the Mann-Whitney rank-sum test when appropriate. Median survival time (MST) of the mice was calculated by Kaplan-Meier log-rank survival analysis. P < .05 was considered statistically significant.
Results
Normal ABL1 kinase plays a tumor suppressor role in BCR-ABL1–induced leukemia
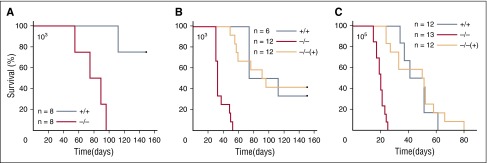
To determine the role of ABL1 in CML, BCR-ABL1 kinase was expressed in Abl1−/− and Abl1+/+ murine BMCs (BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ cells, respectively); in addition, ABL1 kinase expression was restored in BCR-ABL1 Abl1−/− cells by ectopic expression of YFP-ABL1 kinase as described before.14 Although 100% of NOD/SCID mice injected IV with BCR-ABL1 Abl1−/− cells succumbed to leukemia, only 25% of mice injected with BCR-ABL1 Abl1+/+ cells developed lethal disease (MST = 78.8 ± 5.9 days and 140.5 ± 8.2 days, respectively; P < .001) (Figure 1A). When BCR-ABL1–transduced cells were cultured in limited growth factor conditions for 5 weeks to achieve growth factor independence (5-week-old cells), all (100%) mice inoculated with 103 BCR-ABL1 Abl1−/− cells and only 67% of those injected with BCR-ABL1 Abl1+/+ cells developed deadly disease (Figure 1B; MST = 36.2 ± 2.4 days and 105.7 ± 15.3 days, respectively; P < .001). Even though injection of 105 of 5-week-old BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ cells resulted in a deadly leukemia in all mice, the disease was accelerated in the absence of ABL1 (Figure 1C; MST = 19.7 ± 0.9 and 57.4 ± 5.5, respectively; P < .001). Injection of 103 or 105 of BCR-ABL1 Abl1−/− cells reconstituted with YFP-ABL1 [BCR-ABL1 Abl1−/−(+)] induced deadly leukemias in 58% and 100% mice, respectively, with latency (MST = 102.8 ± 12.7 and 45.8 ± 5.1, respectively) similar to that in animals injected with corresponding numbers of BCR-ABL1 Abl1+/+ counterparts (Figure 1B-C).
Figure 1.
Normal ABL1 kinase plays a tumor suppressor role in BCR-ABL1–induced leukemogenesis. BCR-ABL1 Abl1−/− [−/−], BCR-ABL1 Abl1+/+ [+/+], and BCR-ABL1 Abl1−/− leukemia cells reconstituted with YFP-ABL1 [−/−(+)] were inoculated IV into NOD/SCID mice. Kaplan-Meier survival curve of mice injected with (A) 103 freshly transfected cells [(−/−) and (+/+): n = 8] and (B) 103 [(+/+): n = 6, (−/−) and (−/−)+: n = 12] or (C) 105 [(−/−): n = 13, (+/+) and (−/−)+: n = 12] 5-week-old tissue-cultured cells; n = number of mice per group.
ABL1 inhibits expansion of BCR-ABL1–expressing LSCs
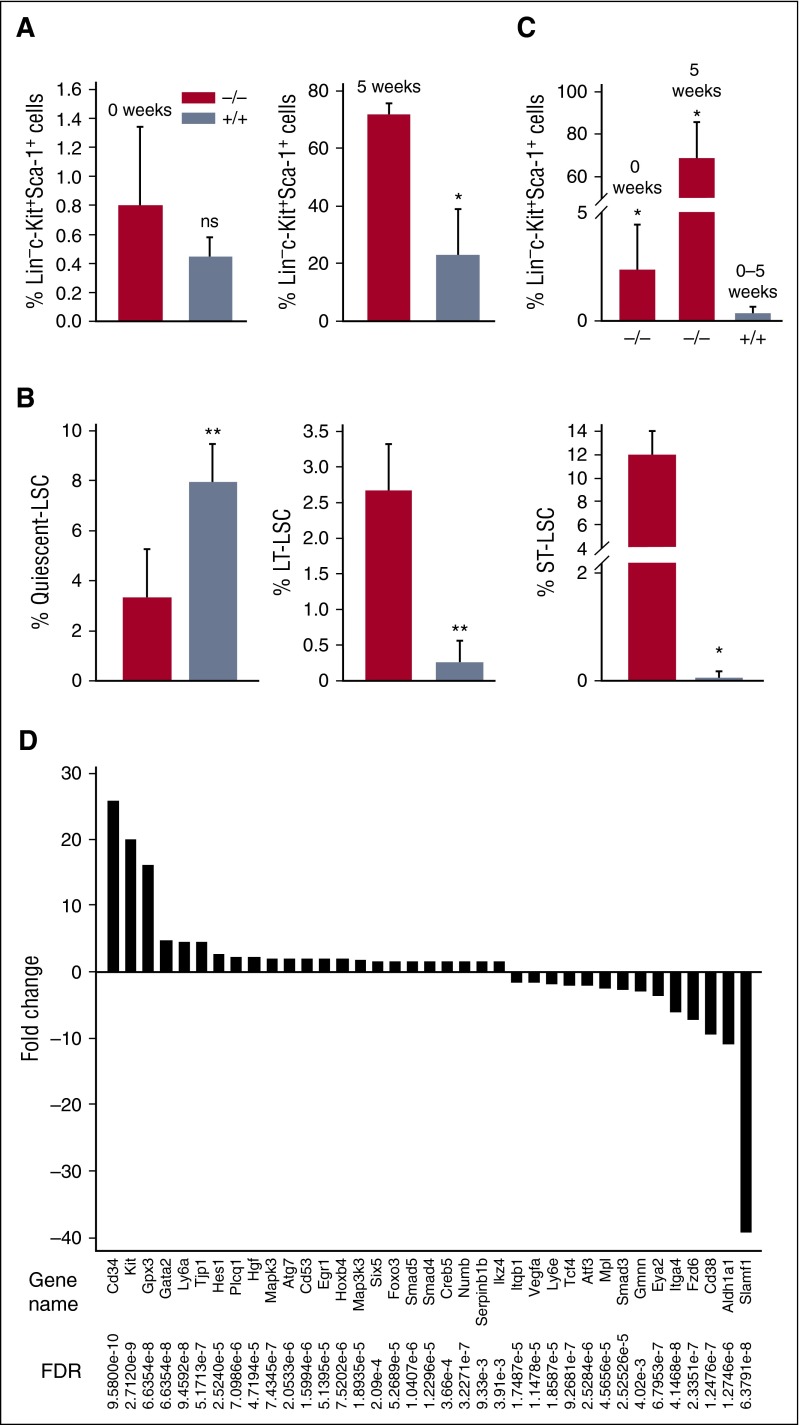
Because uncontrolled outgrowth of LSCs is associated with CML-BP progression and/or therapeutic resistance,18 we sought to determine whether ABL1 affects the accumulation of stem cells in the BCR-ABL1–transformed BMC population. We found no significant difference in the percentage of Lin−c-Kit+Sca-1+ HSCs in freshly harvested Abl1−/− (1.8 ± 0.5) and Abl1+/+ (1.9 ± 0.6) bone marrow. In concordance, the percentage of Lin−c-Kit+Sca-1+ in freshly transduced GFP+ BCR-ABL1 Abl1−/− and GFP+ BCR-ABL1 Abl1+/+ cells did not differ (Figure 2A 0 weeks). However, after 5 weeks of in vitro culture, the percentage of LSCs derived from GFP+ BCR-ABL1 Abl1−/− cells was over threefold higher than those derived from GFP+ BCR-ABL1 Abl1+/+ cells (Figure 2A 5 weeks). Next, we analyzed GFP+ BCR-ABL1–positive Lin−c-Kit+Sca-1+ LSCs cultured for 5 weeks to quantify quiescent (Lin−c-Kit+Sca-1+eFluor670high), long-term (LT) (Lin−c-Kit+Sca-1+CD34−) and short-term (ST) (Lin−c-Kit+Sca-1+CD34+) LSC subpopulations.15 An approximate threefold reduction of the percentage of quiescent LSCs was accompanied by ninefold and >100-fold increase in LT-LSCs and ST-LSCs, respectively, derived from GFP+ BCR-ABL1 Abl1−/− cells in comparison with GFP+ BCR-ABL1 Abl1+/+ cells (Figure 2B).
Figure 2.
ABL1 inhibits expansion of BCR-ABL1–positive LSCs. (A) Mean percentage ± SD of GFP+Lin−c-Kit+Sca-1+ LSCs in freshly established (0 weeks) and 5-week-old (5 weeks) BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ cells. (B) Mean percentage ± SD of quiescent LSCs (CPDmax Lin−c-Kit+Sca-1+), LT-LSCs (Lin−c-Kit+Sca-1+CD34−Flt3−), and ST-LSCs (Lin−c-Kit+Sca-1+CD34+Flt3−) in BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ leukemia cell populations. (C) Bars represent mean percentage ± SD of GFP+Lin−c-Kit+Sca-1+ LSCs in BMCs isolated from moribund SCID mice transplanted with freshly established (0 weeks) and 5-week-old cultured (5 weeks) BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ cells (see Figure 1). **P < .05, *P < .001 as determined by the unpaired Student t test; ns = not significant. (D) Statistically significant (false discovery rate [FDR] < 0.05) fold changes (>1.5) of expression of indicated genes regulating “stemness” in BCR-ABL1 Abl1−/− vs BCR-ABL1 Abl1+/+ leukemia cells maintained with SCF + IL-3.
GFP+ BMCs obtained from mice with advanced leukemia originally injected with either freshly transduced or 5-week-old GFP+ BCR-ABL1 Abl1+/+ cells contained 0.4% ± 0.3% GFP+Lin−c-Kit+Sca-1+ LSCs, whereas those inoculated with GFP+ BCR-ABL1 Abl1−/− counterparts accumulated 2.4% ± 2.0% and 68.7% ± 16.9% GFP+Lin−c-Kit+Sca-1+ LSCs demonstrating sixfold and 60-fold expansion, respectively (Figure 2C).
Transcriptome analysis by microarrays comparing BCR-ABL1 Abl1−/− vs BCR-ABL1 Abl1+/+ cells revealed differential expression of 33 genes potentially involved in regulating stem cell–like characteristics (Figure 2D). In general, genes positively regulating “stemness” including Cd34, Kit, Ly6a, Hoxb, FoxO3, Smad5, and Smad4 were upregulated and genes inhibiting “stemness” such as Ly6e, Fzd6, Eya2, and Slamf1 were downregulated in BCR-ABL1 Abl1−/− vs BCR-ABL1 Abl1+/+ cells. Moreover, Ingenuity Pathway Analysis (IPA) suggests that upregulation of Smad4/5 stimulatory activity and downregulation of Smad3 inhibitory activity downstream of the transforming growth factor β (TGFβ) superfamily signaling (supplemental Figure 1) combined with overexpression of the receptor tyrosine kinase Kit may contribute to abundant expansion of BCR-ABL1 Abl1−/− Lin−c-Kit+Sca-1+ LSCs.19
ABL1 inhibits proliferation of BCR-ABL1–expressing leukemia cells
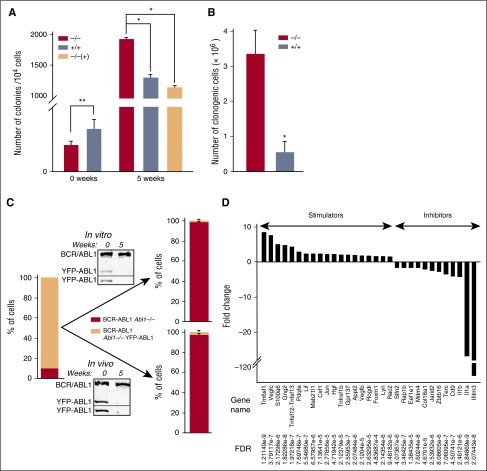
Although BCR-ABL1 stimulates proliferation, ABL1 is known to both promote as well as inhibit cell proliferation depending on the cellular context.1,20 Therefore, we examined whether the tumor-suppressive function of ABL1 is mediated through regulation of cell proliferation. Freshly transduced Lin−c-Kit+Sca-1+ BCR-ABL1 Abl1+/+ cells displayed 1.5 times higher clonogenic potential than BCR-ABL1 Abl1−/− counterparts (Figure 3A 0 weeks), in concordance with other reports indicating that cytoplasmic ABL1 promoted cell cycle progression in CML-CP cells.21 Conversely, 5-week-old cultured BCR-ABL1 Abl1−/− cells formed ∼1.5 times more colonies when compared with BCR-ABL1 Abl1+/+ and BCR-ABL1/YFP-ABL1 Abl1−/− counterparts (Figure 3A 5 weeks). Overall, during 5 weeks of continuous culture, BCR-ABL1 Abl1−/− cells generated ∼6 times more clonogenic cells than BCR-ABL1 Abl1+/+ cells (Figure 3B).
Figure 3.
ABL1 inhibits proliferation of BCR-ABL1 leukemia cells. (A) Mean number of colonies ± SD from BCR-ABL1 Abl1−/−, BCR-ABL1 Abl1+/+, and BCR-ABL1/YFP-ABL1 Abl1−/− [−/−(+)] leukemia cells; *P < .001, **P < .05. (B) Mean number of total clonogenic cells ± SD generated by 106 BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ cells during 5 weeks in vitro culture; *P < .001. (C) The composition of initial cell mixture (left bar) and those after 5 weeks of in vitro and in vivo expansion (right bars). Western blot analyses using anti-ABL1 and anti-GFP/YFP antibodies (top and bottom boxes in each panel, respectively) show expression of BCR-ABL1 as well as YFP-ABL1 protein in the initial cell mixture (0) and after 5 weeks (5). (D) Statistically significant (FDR < 0.05) fold changes (>1.5) of expression of indicated genes regulating cell proliferation in BCR-ABL1 Abl1−/− vs BCR-ABL1 Abl1+/+ leukemia cells maintained with SCF + IL-3.
To test directly whether loss of ABL1 provides a growth advantage, competition assays were performed in which 105 freshly transduced cells consisting of 10% GFP-positive BCR-ABL1 Abl1−/− cells and 90% GFP/YFP-double positive BCR-ABL1/YFP-ABL1 Abl1+/+ cells were maintained for 5 weeks either in liquid culture or in NOD/SCID mice. Flow cytometry detected ∼95% of GFP-positive and 5% of GFP/YFP-positive cells in the mixtures obtained in vitro and in vivo, indicating that the absence of ABL1 provided ∼200-fold growth advantage for BCR-ABL1–transformed cells (Figure 3C bars). Overgrowth of BCR-ABL1 Abl1−/− cells was confirmed by western blot analysis demonstrating undetectable YFP-ABL1 expression in cell mixtures harvested after 5 weeks (Figure 3C western blot insets).
Transcriptome analysis by microarrays was consistent with a growth advantage of BCR-ABL1 Abl1−/− cells; 19 proproliferative genes (eg, Vegfc, Vegfb, Pdgfa, Lif, Hgf, Lyn, Rac2) were upregulated and 12 antiproliferative genes (eg, Ifitm3, Il1a, Il1b) were downregulated in these cells as compared with BCR-ABL1 Abl1+/+ cells (Figure 3D). In addition, IPA suggests that enhanced signaling from phospholipase Cγ1 (PLCγ1) and upregulation of cytoplasmic tyrosine kinases such as Lyn, Fyn, and Syk may contribute to accelerated proliferation rate of BCR-ABL1 Abl1−/− cells overexpressing receptor tyrosine kinase Kit (supplemental Figure 2).22-25 Moreover, upregulation of Pdgfa, Vegfb, Vegfc, and Csf1 expression in BCR-ABL1 Abl1−/− cells implicates autocrine stimulation of cell growth.
ABL1 promotes myeloid differentiation of BCR-ABL1–positive leukemia cells
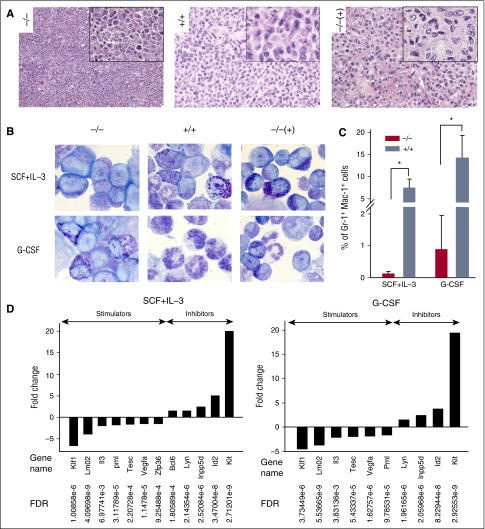
Because moderate expression levels of BCR-ABL1 in the presence of ABL1 promote differentiation in CML-CP and differentiation arrest in CML-BP is associated with enhanced BCR-ABL1 expression and downregulation of ABL1, we sought to determine the role of normal ABL1 in the maturation of the leukemic cells.26,27 Although all moribund mice injected with BCR-ABL1 cells developed splenomegaly (supplemental Figure 3), hematoxylin/eosin staining of spleen sections obtained from mice injected with BCR-ABL1 Abl1−/− cells displayed myeloblastic morphology (Figure 4A −/−). Conversely, spleens harvested from moribund animals injected with BCR-ABL1 Abl1+/+ and BCR-ABL1/YFP-ABL1 Abl1−/− cells revealed moderate to complete myeloid differentiation of leukemic cells ranging from promyelocytes to myelocytes, metamyelocytes, and mature neutrophils (Figure 4A +/+ and −/−(+), respectively).
Figure 4.
ABL1 promotes myeloid differentiation of BCR-ABL1 leukemia cells. (A) Representative hematoxylin-eosin–stained spleen sections from moribund SCID mice injected with BCR-ABL1 Abl1−/− [−/−], BCR-ABL1 Abl1+/+ [+/+] and BCR-ABL1/YFP-ABL1 Abl1−/− [−/−(+)] cells; magnification, ×20, inset, ×40. (B) Wright-Giemsa staining of BCR-ABL1 Abl1+/+ [+/+], BCR-ABL1 Abl1−/− [−/−] and BCR-ABL1/YFP-ABL1 Abl1−/− [−/−(+)] leukemia cells maintained in the presence of SCF + IL-3 (top panels) or G-CSF (bottom panels). Representative images are shown; magnification, ×100. (C) Mean percentages of Gr-1+/Mac-1+ cells ± SD in BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ leukemia cells maintained in the presence of SCF + IL-3 or G-CSF; *P < .002. (D) Statistically significant (FDR < 0.05) fold changes (>1.5) of expression of genes regulating myeloid differentiation in BCR-ABL1 Abl1−/− vs BCR-ABL1 Abl1+/+ leukemia cells maintained with SCF + IL-3 or G-CSF.
In vitro, in the presence of IL-3 + SCF, cultures of either BCR-ABL1 Abl1−/− or BCR-ABL1 Abl1+/+ cells were composed almost exclusively of myeloid precursors having deep blue cytoplasm, but the BCR-ABL1 Abl1+/+ population contained more mature myeloid cells displaying abundance of cytoplasmic granules including seemingly fully differentiated neutrophils with hypersegmented multilobed nuclei (Figure 4B SCF + IL-3). In the presence of G-CSF, cultures of BCR-ABL1 Abl1+/+ cells were predominantly composed of mature neutrophils with very few myeloid precursors whereas cultures of BCR-ABL1 Abl1−/− cells contained mostly myeloid precursors and few neutrophils (Figure 4B G-CSF). Reconstitution of ABL1 expression in BCR-ABL1/YFP-ABL1 Abl1−/− leukemia cells facilitated accumulation of myeloid precursors and/or mature neutrophils in the presence of SCF + IL-3 and G-CSF, respectively (Figure 4B, −/−(+)). Flow cytometry detected <1% of Gr-1+Mac-1+ cells in BCR-ABL1 Abl1−/− population whereas >7% and >14% of BCR-ABL1 Abl1+/+ cells were Gr-1+Mac-1+ in the presence of SCF + IL-3 and G-CSF, respectively (Figure 4C).
Transcriptome microarray analysis supported a myeloid differentiation arrest profile of BCR-ABL1 Abl1−/− cells when compared with BCR-ABL1 Abl1+/+ cells; 7 genes promoting differentiation were inhibited (Klf1, Lmo2, Il3, Pml, Tesc, Vegfa, and Zfp36) whereas 5 genes associated with differentiation arrest were preferentially expressed (Bcl6, Lyn, Id2, Inpp5d, and Kit) (Figure 4D).
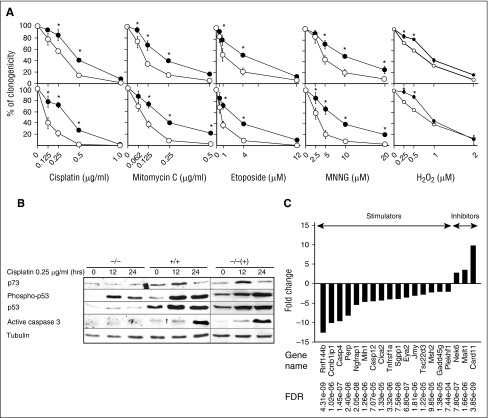
ABL1 promotes DNA damage-induced apoptosis in BCR-ABL1–expressing leukemic cells
BCR-ABL1 kinase promotes cell survival after genotoxic stress whereas ABL1 facilitates apoptosis by exerting opposite effects on caspase 9–mediated activation of caspase 3.28,29 To elucidate the impact of ABL1 loss in BCR-ABL1 cells, BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ cells were exposed to genotoxic agents such as cisplatin, mitomycin C, etoposide, MNNG, or hydrogen peroxide. Results of the clonogenic assay demonstrated that ABL1 enhanced the toxicity of DNA-damaging agents (Figure 5A). ABL1 has been reported to modulate survival/apoptosis by regulation of p53 and p73.30-33 Western blot followed by densitometry analysis show more abundant expression of p53 (11- to 16-fold), phospho-serine 15 p53 (1.5-fold to twofold), and p73 (fivefold), resulting in caspase 3 activation in cisplatin-treated BCR-ABL1 Abl1+/+ when compared with BCR-ABL1 Abl1−/− leukemic cells (Figure 5B). BCR-ABL1 Abl1−/− cells reconstituted with YFP-ABL1 [BCR-ABL1 Abl1−/−(+)] display a similar pattern of expression of these proteins to BCR-ABL1 Abl1+/+ cells.
Figure 5.
ABL1 promotes apoptosis in BCR-ABL1 leukemia cells in response to genotoxic agents. (A) Results represent mean percentage of clonogenic activity ± SD of BCR-ABL1 Abl1−/− (●) and BCR-ABL1 Abl1+/+ (○) leukemia cells after treatment with indicated DNA damage-inducing agents in the presence (top row) or absence (bottom row) of SCF + IL-3; *P < .05. (B) Western blot analysis of p73, p53, phospho-serine 15 of p53 (phospho-p53) and activated caspase 3 in BCR-ABL1 Abl1−/− [−/−], BCR-ABL1 Abl1+/+ [+/+], and BCR-ABL1/YFP-ABL1 Abl1−/− [−/−(+)] leukemia cells treated with cisplatin (0.25 μg/mL); tubulin served as loading control. (C) Statistically significant (FDR < 0.05) fold changes (>2.0) of expression of indicated genes in BCR-ABL1 Abl1−/− vs BCR-ABL1 Abl1+/+ leukemia cells maintained with SCF + IL-3.
The proapoptotic gene expression signature of BCR-ABL1 Abl1+/+ cells was also detected by transcriptional microarrays. Several proapoptotic genes including Rnf144b, Ccnb1ip1, Casp4, Casp12, Eya2, and Gadd45g were downregulated and 3 antiapoptotic genes Nek6, Malt1, and Card11 were upregulated in BCR-ABL1–positive Abl1−/− cells compared with BCR-ABL1 Abl1+/+ cells (Figure 5C). In addition, IPA suggests that proapoptotic signaling involving JNK1-dependent phosphorylation of p53 and BCL2 is downregulated in BCR-ABL1–positive Abl1−/− cells (supplemental Figure 4).34,35
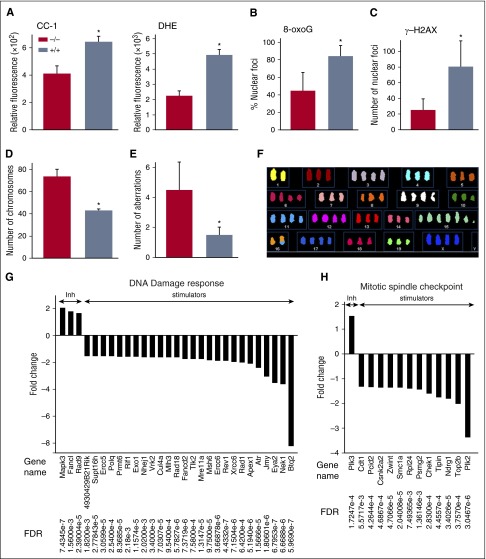
ABL1 prevents accumulation of chromosomal aberrations in BCR-ABL1–expressing leukemic cells
BCR-ABL1–positive LSCs and LPCs contain high levels of ROS-induced oxidative DNA damage such as 8-oxoG and DNA double strand breaks (DSBs), which contribute to accumulation of chromosomal aberrations.16,36 By contrast, ABL1 prevents genomic instability.37 To evaluate the role of ABL1 in modulating ROS-induced oxidative DNA damage in BCR-ABL1 leukemia cells, endogenous ROS were measured with RedoxSensor Red CC-1 dye which detects superoxide anion (⋅O2−) and hydrogen peroxide (H2O2) and with dihydroethidium (DHE) which detects ⋅O2−. BCR-ABL1 Abl1−/− cells contained 1.5 and 2.5 times less ROS detected by CC-1 and DHE, respectively, when compared with BCR-ABL1 Abl1+/+ cells (Figure 6A). Transcriptional microarray analysis suggests that lower levels of H2O2 and ⋅O2− in BCR-ABL1 Abl1−/− cells were probably caused by 15-fold upregulation of glutathione peroxidase 3 (Gpx3) responsible for detoxification of H2O2 and 1.7-fold downregulation of mitochondrial electron transfer flavoprotein-ubiquinone oxidoreductase (Etfdh) which facilitates production of superoxide.38,39 As predicted by previous reports, decrease in ROS levels in BCR-ABL1 Abl1−/− cells was accompanied by twofold lower oxidative DNA damage measured by 8-oxoG as well as threefold decrease in the number of DSBs quantitated by γ-H2AX when compared with BCR-ABL1 Abl1+/+ cells (Figure 6B-C).16,40
Figure 6.
ABL1 prevents accumulation of chromosomal aberrations in BCR-ABL1 leukemia cells. (A) Mean ± SD of relative fluorescence of ROS; CC-1 (left panel) and DHE (right panel) in BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ leukemia cells; *P < .001. (B-C) Mean percentage ± SD of (B) 8-oxoG foci and (C) γ-H2AX foci in BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ leukemia cells assessed by immunofluorescence in 4,6 diamidino-2-phenylindole (DAPI)-counterstained nuclei; *P < .001. (D-F) SKY analysis of BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ leukemia cells based on the (D) number of chromosomes and (E) number of aberrations; results represent mean ± SD; *P < .001. (F) Representative image showing chromosome analysis of metaphase spread of BCR-ABL1 Abl1−/− leukemia cell using SKY. (G-H) Statistically significant (FDR < 0.05) fold changes (>1.5) of expression of indicated genes regulating (G) DNA damage response and (H) mitotic spindle assembly checkpoint in BCR-ABL1 Abl1−/− vs BCR-ABL1 Abl1+/+ leukemia cells maintained with SCF + IL-3.
These unexpected findings prompted us to examine whether ABL1 regulated the accumulation of chromosomal aberrations in BCR-ABL1 leukemia cells. BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ cells were cultured for 10 weeks followed by spectral karyotype analysis (SKY) to detect acquired chromosomal aberrations. Results from metaphase spreads revealed a nearly twofold increase in chromosome numbers and ∼2.5-fold higher number of chromosomal aberrations (whole chromosome and intrachromosomal gains/losses, as well as nonrecurrent chromosomal translocations) in BCR-ABL1 Abl1−/− cells compared with BCR-ABL1 Abl1+/+ cells (Figure 6D-F), consistent with overall high levels of genomic instability in the former cells.
The absence of ABL1 did not affect cell cycle distribution of BCR-ABL1 cells in response to DNA damage (supplemental Figure 5), but transcriptional microarray analysis revealed that the presence of ABL1 in BCR-ABL1 cells is associated with expression of numerous genes whose products regulate DNA damage response and mitotic spindle assembly checkpoint (Figure 6G-H). The majority of genes downregulated in BCR-ABL1 Abl1−/− cells in comparison with BCR-ABL1 Abl1+/+ cells are involved in promotion of DNA repair, for example, Ercc5, Fancd2, Polg, Exo1, Nhej1, Rif1, Rad18, Mre11, Msh6, Xrcc6, Top2b, and Atr. IPA suggested that simultaneous downregulation of Fancd2, and Xrcc6 and Nhej1 (involved in faithful homologous recombination and relatively faithful classical nonhomologous end-joining [C-NHEJ], respectively) may favor highly unfaithful PARP1-mediated alternative NHEJ resulting in facilitated accumulation of additional chromosomal aberrations in BCR-ABL1 Abl1−/− cells (supplemental Figure 6).41 Genes involved in kinetochore/spindle/centrosome regulation (Pcid, Csnk2a2, Zwint, Plk2, Nek1) and sister chromatid segregation (Smc1a) are downregulated in BCR-ABL1 Abl1−/− cells compared with BCR-ABL1 Abl1+/+ cells, which may be responsible for abundant aneuploidy in the former cells.
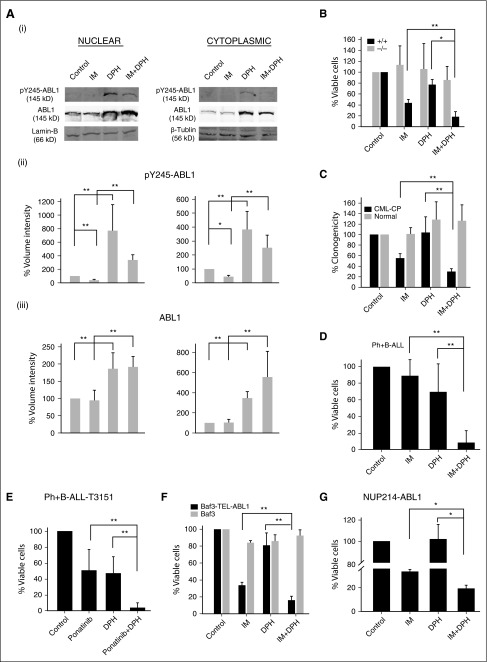
Activation of native ABL1 kinase enhances the efficacy of TKIs in leukemias expressing oncogenic ABL1 kinase mutants
Inhibition of intracellular ABL1 kinase may require threefold to 10-fold higher concentrations of imatinib than required to effectively inhibit BCR-ABL1 kinase42 (supplemental Figure 7). For example, 1 to 2 µM imatinib, the concentration typically achievable in steady state in patients,43,44 inhibits cytoplasmic BCR-ABL1, but it is suboptimal for inhibition of nuclear and mitochondrial ABL1.45,46 Because ABL1 kinase enhanced the sensitivity of CML cells to imatinib,14 we hypothesized that stimulation of residual ABL1 activity may enhance the effect of TKIs in CML-CP treatment.
ABL1 allosteric activator DPH has been reported to stimulate ABL1 kinase by displacing the myristate moiety from the myristoyl-binding site, thereby preventing the autoinhibitory conformation of ABL1.47 To determine whether DPH was able to induce and/or maintain the activity of ABL1 kinase under imatinib treatment, ABL1 was overexpressed in Phoenix cells followed by treatment with 10 μM DPH, 1 μM imatinib, or a combination of the 2 agents. DPH induced approximately eightfold and fourfold increase of phospho-Y245-ABL1, indicative of ABL1-kinase activation, in nuclear and cytoplasmic fractions, respectively (Figure 7A). Moreover, ∼50% to 70% of DPH-mediated activation of ABL1 kinase persisted in the presence of imatinib.
Figure 7.
Activation of native ABL1 enhances the efficacy of TKIs against leukemias expressing oncogenic ABL1 kinase mutants. (A) Phoenix cells overexpressing ABL1 were treated with dimethylsulfoxide (DMSO) (control), DPH, imatinib (IM), and a combination of imatinib followed by DPH (IM+DPH). (i) Representative western blot analysis of phospho-tyrosine 245-ABL1 (pY245-ABL1), ABL1, lamin-B and β-tubulin in nuclear (left panel) and cytoplasmic (right panel) cell lysates. Quantification of normalized pY245-ABL1 (ii) and ABL1 (iii) levels to lamin-B and β-tubulin. Bars represent mean percentage volume intensity ± SD. (B-G) Percentage of viable cells or colonies ± SD from cells treated for 72 hours with diluent (Control), imatinib (IM), ponatinib, DPH, and combinations: (B) BCR-ABL1 Abl1−/− and BCR-ABL1 Abl1+/+ cells, (C) Lin−CD34+ cells from 6 CML-CP patients and 3 healthy donors, (D) xenograft cells from 3 freshly diagnosed BCR-ABL1 B-ALL patients, (E) xenograft cells from 3 relapsed B-ALL patients carrying BCR-ABL1(T315I) mutation, (F) Baf3 and Baf3-TEL-ABL1 cells, (G) NUP214-ABL1–positive murine cells; *P < .001, **P < .05 as determined by the unpaired Student t test.
To determine whether ABL1 kinase could be a therapeutic target, we examined the effect of imatinib and/or DPH on BCR-ABL1 Abl1+/+ and BCR-ABL1 Abl1−/− cells. We found that treatment with 0.125 µM imatinib in combination with 10 µM DPH for 3 consecutive days reduced the number of BCR-ABL1 Abl1+/+ leukemia cells when compared with single agents (Figure 7B). In contrast, BCR-ABL1 Abl1−/− cells were not sensitive to the combination treatment thereby confirming the ABL1-specific effect of DPH. In addition, the combination of 1 µM imatinib and 10 µM DPH reduced clonogenic growth of CML-CP Lin−CD34+ cells by more than twofold in comparison with either agent alone, at the same time normal counterparts were not affected by the treatments (Figure 7C). Moreover, BCR-ABL1–positive B-ALL primary human xenograft cells and those harboring imatinib-resistant BCR-ABL1 (T315I) mutation were extremely sensitive (>10-fold) to the combination of imatinib (1 µM) + DPH (10 µM) and ponatinib (12.5 nM) + DPH, respectively, when compared with single-agent treated cells (Figure 7D-E).
Leukemia cells expressing TEL-ABL1 and NUP214-ABL1 oncogenic kinases were also more sensitive (more than twofold) to the combination of imatinib + DPH (Figure 7F-G).
Discussion
We showed here that ABL1 exerts a tumor suppressor function in CML-CP by restricting the expansion of LSCs, inhibiting clonogenic activity of LSCs/LPCs, promoting myeloid differentiation and apoptosis, and reducing genomic instability. In concordance, loss of ABL1 facilitated development of highly malignant CML-BP–like disease in mice inoculated with BCR-ABL1 Abl1−/− BMCs in contrast to CML-CP–like disease that arose from BCR-ABL1–transformed Abl1+/+ cells.
The tumor suppressor function of ABL1 contrasting the oncogenic function of BCR-ABL1 is supported by reports that BCR-ABL1 and ABL1 can exert opposite effects on a variety of cellular functions.48 We have shown that BCR-ABL1 kinase, which localizes exclusively to the cytoplasm,49 protects leukemic cells from genotoxic stress, and facilitates genomic instability by promoting unfaithful DNA repair and deregulation of mitotic spindle.50-55 Conversely, ABL1 which is targeted to the nucleus upon DNA damage can inhibit or stimulate DNA repair and stabilize mitotic spindle.37,56-61 BCR-ABL1 inhibits apoptosis,62 conversely nuclear and mitochondrial ABL1 kinase may facilitate apoptosis or necrosis.28,31-33,63 Activation of the nuclear ABL1 inhibits cell proliferation but cytoplasmic BCR-ABL1 stimulates cell cycle progression.64,65 These striking differences between ABL1 and BCR-ABL1 functions may explain the dramatic changes in proliferation, apoptosis, and genomic instability of BCR-ABL1 leukemic cells upon the loss of ABL1.
Remarkably, even though ABL1 did not affect myeloid differentiation of HSCs,5,6 and BCR-ABL1 seemed to promote myeloid differentiation,26 loss of ABL1 in BCR-ABL1 leukemic cells resulted in dramatic arrest of myeloid differentiation. Similarly, although ABL1 does not regulate the number of HSCs, BCR-ABL1 promotes rather mild expansion of LSCs in CML-CP4,66 (and present study); nonetheless, rapid expansion of LSCs was observed in the absence of ABL1. It is plausible to speculate that a combined effect of enhanced proliferation and differentiation arrest resulted in rapid expansion of BCR-ABL1 Abl1−/− LSCs.
The opposite effects of BCR-ABL1 and ABL1 may result from differences in their substrate preferences; in addition, their intracellular substrate repertoire may differ due to modification of kinase-substrate interactions and/or intracellular localization.67,68 Accordingly, the exclusively cytoplasmic localization of BCR-ABL1 may allow it to escape the modulatory effect of intranuclear proteins such as retinoblastoma (Rb) tumor suppressor which negatively regulates ABL1.69 Moreover, BCR-ABL1 demonstrated more abundant binding and modification of actin microfilaments than ABL1, which plays an important role in leukemogenesis.70
Considering their opposite, pro- and antioncogenic properties, the BCR-ABL1-to-ABL1 ratio may play a key role in shaping the ultimate malignant features of the BCR-ABL1–transformed cells. It has been reported that CML-CP to CML-BP progression is associated with hypermethylation of the ABL1 promoter embedded in the translocation, which may be accompanied by clonal loss of expression of ABL1 transcribed from nontranslocated chromosome 9.13,14,71-73 That would tip the balance toward BCR-ABL1 oncogene in individual clonogenic cells, which in conjunction with decreased sensitivity to imatinib14 might promote malignant progression. Moreover, patients undergoing imatinib treatment with yet undetected clones carrying BCR-ABL1 kinase mutations may be at greater risk of clonal evolution, eventually leading to CML-BP because even partial inhibition of ABL1 kinase (especially in patients treated with higher doses of imatinib) in the presence of intact BCR-ABL1 kinase mutant (eg, T315I) would promote chromosomal instability (supplemental Figure 8).
The fact that ABL1 kinase not only opposes BCR-ABL1 (present study) but also sensitizes CML cells to TKIs14 suggests that ABL1 activity could be explored therapeutically. The standard clinical dose of imatinib (400 mg per day) corresponds to trough plasma levels of ∼1 to 2 μM which effectively inhibited BCR-ABL1,43,44 whereas ABL1 kinase required 3 to 10 μM of the drug to suppress its activity45,46 (and this work). The tumor suppressor function of the intact ABL1 creates therapeutic opportunity for a combination of TKI and ABL1 activator. We showed here that allosteric activation of normal ABL1 kinase activity by DPH “hypersensitized” leukemic cells expressing oncogenic variants of ABL1 to TKIs in vitro. The beneficial antileukemia effect of imatinib + DPH was detected only in leukemia cells expressing ABL1, which supports the notion that DPH exerted its activity by activating ABL1, but not by displacing HSP90 from BCR-ABL1.74
Optimization of DPH or other ABL1 activators may generate new drugs useful for development of novel therapeutic modalities against tumors expressing oncogenic mutants of ABL1.75,76 For example, combination of TKI and allosteric ABL1 activator may enhance the therapeutic effect of TKIs in a cohort of CML-CP “poor responders” (no major cytogenetic response in 12 months, BCR-ABL1 transcript levels >10% after 3 months) and also in a majority of more aggressive BCR-ABL1–positive B-ALLs, which display unfavorable prognosis on TKIs.77,78
In summary, we showed here that the proto-oncogene ABL1 acts as a tumor suppressor in CML-CP cells expressing its oncogenic variant, BCR-ABL1. Moreover, analysis of the CGAP Mitelman database revealed that actual del(9q34) or deletions encompassing this region resulting in loss of ABL1 have been detected in other hematologic malignancies (see supplemental Table 1), thus suggesting that ABL1 may play a tumor suppressor role not only in CML. On the other hand, normal ABL1 kinase could be targeted to enhance the therapeutic effect of TKIs in leukemias driven by BCR-ABL1 and other oncogenic forms of ABL1 kinase. It is possible that the enhanced cytotoxic effect of TKI in the context of activation of ABL1 may not be limited to tumors expressing oncogenic forms of ABL1 but it could be a more general paradigm as supported by our findings of a similar effect in leukemia cells harboring the FLT3-ITD mutation when treated with the FLT3 inhibitor AC220 in combination with DPH (supplemental Figure 8).
Acknowledgments
The authors thank Dr John Tobias from the Molecular Profiling Core of the University of Pennsylvania (Philadelphia, PA) for the assistance with microarray analyses.
This work was supported by National Institutes of Health/National Cancer Institute CA134458 (T. Skorski), HARMONIA grant 2014/14/M/NZ5/00441 from the Polish National Science Center (T. Stoklosa), and by the Austrian Science Fund (FWF) F4704-B20 (P.V.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.D. performed in vitro and ex vivo experiments for stemness, proliferation, differentiation, DNA damage response, DPH treatment, and microarray analysis, and wrote the first draft of the manuscript; M.K. performed in vitro and in vivo experiments for proliferation and in vitro experiments for stemness, myeloid differentiation, apoptosis, and DNA damage response; G.H. performed in vivo experiments; M.N.-S. assisted with experiments using primary cells; K.K., D.R., M.A.W., and B.G. ran histopathological analyses and differentiation tests; E.B.-G. assisted with flow cytometry data analysis; C.R. performed the SKY assay; S.C.-R., P.V., T. Stoklosa, and M.M. provided patient samples and primary leukemia xenografts, and performed CGAP Mitelman database analysis; O.H. provided NUP214-ABL1–transformed cells; H.v.d.K. performed the experiments in supplemental Figure 1; and T. Skorski conceived the idea, supervised the project, and wrote the final version of the manuscript.
Conflict-of-interest disclosure: P.V. served as a consultant and/or obtained honoraria and research funding from Novartis, Ariad, Bristol-Myers Squibb, Pfizer, and Celgene. The remaining authors declare no competing financial interests.
Correspondence: Tomasz Skorski, Department of Microbiology and Immunology and Fels Institute for Cancer Research and Molecular Biology, School of Medicine, Temple University, 3400 N. Broad St, MRB 548, Philadelphia, PA 19140; e-mail: tskorski@temple.edu.
References
- 1.Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nat Rev Cancer. 2013;13(8):559–571. doi: 10.1038/nrc3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang JY. The capable ABL: what is its biological function? Mol Cell Biol. 2014;34(7):1188–1197. doi: 10.1128/MCB.01454-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity [published correction appears in Sci Signal. 2011;4(188):er4]. Sci Signal. 2010;3(139):re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardin JD, Boast S, Schwartzberg PL, et al. Bone marrow B lymphocyte development in c-abl-deficient mice. Cell Immunol. 1995;165(1):44–54. doi: 10.1006/cimm.1995.1185. [DOI] [PubMed] [Google Scholar]
- 5.Schwartzberg PL, Stall AM, Hardin JD, et al. Mice homozygous for the ablm1 mutation show poor viability and depletion of selected B and T cell populations. Cell. 1991;65(7):1165–1175. doi: 10.1016/0092-8674(91)90012-n. [DOI] [PubMed] [Google Scholar]
- 6.Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65(7):1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 7.Walz C, Cross NC, Van Etten RA, Reiter A. Comparison of mutated ABL1 and JAK2 as oncogenes and drug targets in myeloproliferative disorders. Leukemia. 2008;22(7):1320-1334. [DOI] [PMC free article] [PubMed]
- 8.Marley SB, Gordon MY. Chronic myeloid leukaemia: stem cell derived but progenitor cell driven. Clin Sci (Lond) 2005;109(1):13–25. doi: 10.1042/CS20040336. [DOI] [PubMed] [Google Scholar]
- 9.Penserga ET, Skorski T. Fusion tyrosine kinases: a result and cause of genomic instability. Oncogene. 2007;26(1):11-20. [DOI] [PubMed]
- 10.Amabile G, Di Ruscio A, Müller F, et al. Dissecting the role of aberrant DNA methylation in human leukaemia. Nat Commun. 2015;6:7091. doi: 10.1038/ncomms8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graux C, Cools J, Melotte C, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36(10):1084-1089. [DOI] [PubMed]
- 12.Brahmbhatt MM, Trivedi PJ, Patel DM, Shukla SN, Patel PS. Location of the BCR/ABL fusion genes on both chromosomes 9 in Ph negative young CML patients: an Indian experience. Indian J Hematol Blood Transfus. 2014;30(4):241–246. doi: 10.1007/s12288-013-0316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zion M, Ben-Yehuda D, Avraham A, et al. Progressive de novo DNA methylation at the bcr-abl locus in the course of chronic myelogenous leukemia. Proc Natl Acad Sci USA. 1994;91(22):10722–10726. doi: 10.1073/pnas.91.22.10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virgili A, Koptyra M, Dasgupta Y, et al. Imatinib sensitivity in BCR-ABL1-positive chronic myeloid leukemia cells is regulated by the remaining normal ABL1 allele. Cancer Res. 2011;71(16):5381–5386. doi: 10.1158/0008-5472.CAN-11-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolton-Gillespie E, Schemionek M, Klein HU, et al. Genomic instability may originate from imatinib-refractory chronic myeloid leukemia stem cells. Blood. 2013;121(20):4175–4183. doi: 10.1182/blood-2012-11-466938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieborowska-Skorska M, Kopinski PK, Ray R, et al. Rac2-MRC-cIII-generated ROS cause genomic instability in chronic myeloid leukemia stem cells and primitive progenitors. Blood. 2012;119(18):4253–4263. doi: 10.1182/blood-2011-10-385658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slupianek A, Dasgupta Y, Ren SY, et al. Targeting RAD51 phosphotyrosine-315 to prevent unfaithful recombination repair in BCR-ABL1 leukemia. Blood. 2011;118(4):1062–1068. doi: 10.1182/blood-2010-09-307256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrotti D, Jamieson C, Goldman J, Skorski T. Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest. 2010;120(7):2254–2264. doi: 10.1172/JCI41246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Söderberg SS, Karlsson G, Karlsson S. Complex and context dependent regulation of hematopoiesis by TGF-beta superfamily signaling. Ann N Y Acad Sci. 2009;1176:55–69. doi: 10.1111/j.1749-6632.2009.04569.x. [DOI] [PubMed] [Google Scholar]
- 20.Szczylik C, Skorski T, Nicolaides NC, et al. Selective inhibition of leukemia cell proliferation by BCR-ABL antisense oligodeoxynucleotides. Science. 1991;253(5019):562–565. doi: 10.1126/science.1857987. [DOI] [PubMed] [Google Scholar]
- 21.Rosti V, Bergamaschi G, Ponchio L, Cazzola M. c-abl function in normal and chronic myelogenous leukemia hematopoiesis: in vitro studies with antisense oligomers. Leukemia. 1992;6(1):1–7. [PubMed] [Google Scholar]
- 22.Markova B, Albers C, Breitenbuecher F, et al. Novel pathway in Bcr-Abl signal transduction involves Akt-independent, PLC-gamma1-driven activation of mTOR/p70S6-kinase pathway. Oncogene. 2010;29(5):739–751. doi: 10.1038/onc.2009.374. [DOI] [PubMed] [Google Scholar]
- 23.Singh MM, Howard A, Irwin ME, et al. Expression and activity of Fyn mediate proliferation and blastic features of chronic myelogenous leukemia. PLoS One. 2012;7(12):e51611. doi: 10.1371/journal.pone.0051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourgne C, Janel A, Berger J, et al. Phosphorylation of spleen tyrosine kinase at tyrosine 348 (pSyk348) may be a marker of advanced phase of chronic myeloid leukemia (CML). Leuk Res. 2015;39(3):329–334. doi: 10.1016/j.leukres.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Gioia R, Leroy C, Drullion C, et al. Quantitative phosphoproteomics revealed interplay between Syk and Lyn in the resistance to nilotinib in chronic myeloid leukemia cells. Blood. 2011;118(8):2211–2221. doi: 10.1182/blood-2010-10-313692. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi S, Kimura F, Ikeda T, et al. BCR-ABL promotes neutrophil differentiation in the chronic phase of chronic myeloid leukemia by downregulating c-Jun expression. Leukemia. 2009;23(9):1622–1627. doi: 10.1038/leu.2009.74. [DOI] [PubMed] [Google Scholar]
- 27.Chang JS, Santhanam R, Trotta R, et al. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNP-E2 dependent suppression of C/EBPalpha-driven myeloid differentiation. Blood. 2007;110(3):994-1003. [DOI] [PMC free article] [PubMed]
- 28.Raina D, Pandey P, Ahmad R, et al. c-Abl tyrosine kinase regulates caspase-9 autocleavage in the apoptotic response to DNA damage. J Biol Chem. 2005;280(12):11147-11151. [DOI] [PubMed]
- 29.Deming PB, Schafer ZT, Tashker JS, Potts MB, Deshmukh M, Kornbluth S. Bcr-Abl-mediated protection from apoptosis downstream of mitochondrial cytochrome c release. Mol Cell Biol. 2004;24(23):10289–10299. doi: 10.1128/MCB.24.23.10289-10299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan ZM, Huang Y, Whang Y, et al. Role for c-Abl tyrosine kinase in growth arrest response to DNA damage. Nature. 1996;382(6588):272–274. doi: 10.1038/382272a0. [DOI] [PubMed] [Google Scholar]
- 31.Yuan ZM, Shioya H, Ishiko T, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399(6738):814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 32.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399(6738):809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 33.Gong JG, Costanzo A, Yang HQ, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399(6738):806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 34.Oleinik NV, Krupenko NI, Krupenko SA. Cooperation between JNK1 and JNK2 in activation of p53 apoptotic pathway. Oncogene. 2007;26(51):7222–7230. doi: 10.1038/sj.onc.1210526. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhary P, Vishwanatha JK. c-Jun NH2-terminal kinase-induced proteasomal degradation of c-FLIPL/S and Bcl2 sensitize prostate cancer cells to Fas- and mitochondria-mediated apoptosis by tetrandrine. Biochem Pharmacol. 2014;91(4):457–473. doi: 10.1016/j.bcp.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koptyra M, Cramer K, Slupianek A, Richardson C, Skorski T. BCR/ABL promotes accumulation of chromosomal aberrations induced by oxidative and genotoxic stress. Leukemia. 2008;22(10):1969–1972. doi: 10.1038/leu.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumura S, Hamasaki M, Yamamoto T, et al. ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat Commun. 2012;3:626. doi: 10.1038/ncomms1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigues JV, Gomes CM. Mechanism of superoxide and hydrogen peroxide generation by human electron-transfer flavoprotein and pathological variants. Free Radic Biol Med. 2012;53(1):12–19. doi: 10.1016/j.freeradbiomed.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Herault O, Hope KJ, Deneault E, et al. A role for GPx3 in activity of normal and leukemia stem cells. J Exp Med. 2012;209(5):895–901. doi: 10.1084/jem.20102386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koptyra M, Falinski R, Nowicki MO, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108(1):319–327. doi: 10.1182/blood-2005-07-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wray J, Williamson EA, Singh SB, et al. PARP1 is required for chromosomal translocations. Blood. 2013;121(21):4359–4365. doi: 10.1182/blood-2012-10-460527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zipfel PA, Zhang W, Quiroz M, Pendergast AM. Requirement for Abl kinases in T cell receptor signaling. Curr Biol. 2004;14(14):1222–1231. doi: 10.1016/j.cub.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 43.Larson RA, Druker BJ, Guilhot F, et al. IRIS (International Randomized Interferon vs STI571) Study Group. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS study. Blood. 2008;111(8):4022–4028. doi: 10.1182/blood-2007-10-116475. [DOI] [PubMed] [Google Scholar]
- 44.Peng B, Hayes M, Resta D, et al. Pharmacokinetics and pharmacodynamics of imatinib in a phase I trial with chronic myeloid leukemia patients. J Clin Oncol. 2004;22(5):935–942. doi: 10.1200/JCO.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Mishra N, Raina D, Saxena S, Kufe D. Abrogation of the cell death response to oxidative stress by the c-Abl tyrosine kinase inhibitor STI571. Mol Pharmacol. 2003;63(2):276–282. doi: 10.1124/mol.63.2.276. [DOI] [PubMed] [Google Scholar]
- 46.Burton EA, Plattner R, Pendergast AM. Abl tyrosine kinases are required for infection by Shigella flexneri. EMBO J. 2003;22(20):5471–5479. doi: 10.1093/emboj/cdg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Campobasso N, Biju MP, et al. Discovery and characterization of a cell-permeable, small-molecule c-Abl kinase activator that binds to the myristoyl binding site. Chem Biol. 2011;18(2):177–186. doi: 10.1016/j.chembiol.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 48.Guo XY, Fontana J, Kufe D, Deisseroth A. Antagonistic effects of ABL and BCRABL proteins on proliferation and the response to genotoxic stress in normal and leukemic myeloid cells. Leuk Lymphoma. 1998;30(3-4):225–235. doi: 10.3109/10428199809057536. [DOI] [PubMed] [Google Scholar]
- 49.Wetzler M, Talpaz M, Van Etten RA, Hirsh-Ginsberg C, Beran M, Kurzrock R. Subcellular localization of Bcr, Abl, and Bcr-Abl proteins in normal and leukemic cells and correlation of expression with myeloid differentiation. J Clin Invest. 1993;92(4):1925–1939. doi: 10.1172/JCI116786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slupianek A, Gurdek E, Koptyra M, et al. BLM helicase is activated in BCR/ABL leukemia cells to modulate responses to cisplatin. Oncogene. 2005;24(24):3914–3922. doi: 10.1038/sj.onc.1208545. [DOI] [PubMed] [Google Scholar]
- 51.Slupianek A, Nowicki MO, Koptyra M, Skorski T. BCR/ABL modifies the kinetics and fidelity of DNA double-strand breaks repair in hematopoietic cells. DNA Repair (Amst). 2006;5(2):243-250. [DOI] [PMC free article] [PubMed]
- 52.Slupianek A, Schmutte C, Tombline G, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8(4):795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 53.Canitrot Y, Falinski R, Louat T, et al. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood. 2003;102(7):2632–2637. doi: 10.1182/blood-2002-10-3207. [DOI] [PubMed] [Google Scholar]
- 54.Cramer K, Nieborowska-Skorska M, Koptyra M, et al. BCR/ABL and other kinases from chronic myeloproliferative disorders stimulate single-strand annealing, an unfaithful DNA double-strand break repair. Cancer Res. 2008;68(17):6884–6888. doi: 10.1158/0008-5472.CAN-08-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolanin K, Magalska A, Kusio-Kobialka M, et al. Expression of oncogenic kinase Bcr-Abl impairs mitotic checkpoint and promotes aberrant divisions and resistance to microtubule-targeting agents. Mol Cancer Ther. 2010;9(5):1328–1338. doi: 10.1158/1535-7163.MCT-09-0936. [DOI] [PubMed] [Google Scholar]
- 56.Chen X, Zhang J, Lee J, et al. A kinase-independent function of c-Abl in promoting proteolytic destruction of damaged DNA binding proteins. Mol Cell. 2006;22(4):489–499. doi: 10.1016/j.molcel.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Kharbanda S, Pandey P, Jin S, et al. Functional interaction between DNA-PK and c-Abl in response to DNA damage. Nature. 1997;386(6626):732–735. doi: 10.1038/386732a0. [DOI] [PubMed] [Google Scholar]
- 58.Yuan Z-M, Huang Y, Ishiko T, et al. Regulation of Rad51 function by c-Abl in response to DNA damage. J Biol Chem. 1998;273(7):3799–3802. doi: 10.1074/jbc.273.7.3799. [DOI] [PubMed] [Google Scholar]
- 59.Chen G, Yuan SS, Liu W, et al. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J Biol Chem. 1999;274(18):12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 60.Yuan SS, Chang HL, Lee EY. Ionizing radiation-induced Rad51 nuclear focus formation is cell cycle-regulated and defective in both ATM(-/-) and c-Abl(-/-) cells. Mutat Res. 2003;525(1-2):85–92. doi: 10.1016/s0027-5107(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 61.Shangary S, Brown KD, Adamson AW, et al. Regulation of DNA-dependent protein kinase activity by ionizing radiation-activated abl kinase is an ATM-dependent process. J Biol Chem. 2000;275(39):30163–30168. doi: 10.1074/jbc.M004302200. [DOI] [PubMed] [Google Scholar]
- 62.Amarante-Mendes GP, Naekyung Kim C, Liu L, et al. Bcr-Abl exerts its antiapoptotic effect against diverse apoptotic stimuli through blockage of mitochondrial release of cytochrome C and activation of caspase-3. Blood. 1998;91(5):1700–1705. [PubMed] [Google Scholar]
- 63.Kumar S, Bharti A, Mishra NC, et al. Targeting of the c-Abl tyrosine kinase to mitochondria in the necrotic cell death response to oxidative stress. J Biol Chem. 2001;276(20):17281–17285. doi: 10.1074/jbc.M101414200. [DOI] [PubMed] [Google Scholar]
- 64.Kharbanda S, Yuan ZM, Weichselbaum R, Kufe D. Determination of cell fate by c-Abl activation in the response to DNA damage. Oncogene. 1998;17(25):3309–3318. doi: 10.1038/sj.onc.1202571. [DOI] [PubMed] [Google Scholar]
- 65.Kurzrock R, Kantarjian HM, Druker BJ, Talpaz M. Philadelphia chromosome-positive leukemias: from basic mechanisms to molecular therapeutics. Ann Intern Med. 2003;138(10):819–830. doi: 10.7326/0003-4819-138-10-200305200-00010. [DOI] [PubMed] [Google Scholar]
- 66.Petzer AL, Eaves CJ, Barnett MJ, Eaves AC. Selective expansion of primitive normal hematopoietic cells in cytokine-supplemented cultures of purified cells from patients with chronic myeloid leukemia. Blood. 1997;90(1):64–69. [PubMed] [Google Scholar]
- 67.Voss J, Posern G, Hannemann JR, et al. The leukaemic oncoproteins Bcr-Abl and Tel-Abl (ETV6/Abl) have altered substrate preferences and activate similar intracellular signalling pathways. Oncogene. 2000;19(13):1684–1690. doi: 10.1038/sj.onc.1203467. [DOI] [PubMed] [Google Scholar]
- 68.Wu JJ, Phan H, Lam KS. Comparison of the intrinsic kinase activity and substrate specificity of c-Abl and Bcr-Abl. Bioorg Med Chem Lett. 1998;8(17):2279–2284. doi: 10.1016/s0960-894x(98)00413-2. [DOI] [PubMed] [Google Scholar]
- 69.Guo XY, Balague C, Wang T, et al. The presence of the Rb c-box peptide in the cytoplasm inhibits p210bcr-abl transforming function. Oncogene. 1999;18(8):1589–1595. doi: 10.1038/sj.onc.1202479. [DOI] [PubMed] [Google Scholar]
- 70.McWhirter JR, Wang JY. An actin-binding function contributes to transformation by the Bcr-Abl oncoprotein of Philadelphia chromosome-positive human leukemias. EMBO J. 1993;12(4):1533–1546. doi: 10.1002/j.1460-2075.1993.tb05797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Asimakopoulos FA, Shteper PJ, Krichevsky S, et al. ABL1 methylation is a distinct molecular event associated with clonal evolution of chronic myeloid leukemia. Blood. 1999;94(7):2452–2460. [PubMed] [Google Scholar]
- 72.Diamond J, Goldman JM, Melo JV. BCR-ABL, ABL-BCR, BCR, and ABL genes are all expressed in individual granulocyte-macrophage colony-forming unit colonies derived from blood of patients with chronic myeloid leukemia. Blood. 1995;85(8):2171–2175. [PubMed] [Google Scholar]
- 73.Gupta M, Milani L, Hermansson M, et al. Expression of BCR-ABL1 oncogene relative to ABL1 gene changes overtime in chronic myeloid leukemia. Biochem Biophys Res Commun. 2008;366(3):848–851. doi: 10.1016/j.bbrc.2007.12.029. [DOI] [PubMed] [Google Scholar]
- 74.Taipale M, Krykbaeva I, Whitesell L, et al. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotechnol. 2013;31(7):630–637. doi: 10.1038/nbt.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grover P, Shi H, Baumgartner M, Camacho CJ, Smithgall TE. Fluorescence polarization screening assays for small molecule allosteric modulators of ABL kinase function. PLoS One. 2015;10(7):e0133590. doi: 10.1371/journal.pone.0133590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong X, Cao P, Washio Y, et al. Structure-guided optimization of small molecule c-Abl activators. J Comput Aided Mol Des. 2014;28(2):75–87. doi: 10.1007/s10822-014-9731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Branford S, Yeung DT, Parker WT, et al. Prognosis for patients with CML and >10% BCR-ABL1 after 3 months of imatinib depends on the rate of BCR-ABL1 decline. Blood. 2014;124(4):511–518. doi: 10.1182/blood-2014-03-566323. [DOI] [PubMed] [Google Scholar]
- 78.Fielding AK, Zakout GA. Treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia. Curr Hematol Malig Rep. 2013;8(2):98–108. doi: 10.1007/s11899-013-0155-4. [DOI] [PubMed] [Google Scholar]