Key Points
Dynamic intron retention programs exist in the murine megakaryocyte and erythroid and human erythroid lineages.
Intron retention inversely correlates with expression levels of a large set of transcripts.
Abstract
Intron retention (IR) is a form of alternative splicing that can affect mRNA levels through nonsense-mediated decay or by nuclear mRNA detention. A complex, dynamic IR pattern has been described in maturing mammalian granulocytes, but it is unknown whether IR occurs broadly in other hematopoietic lineages. We globally assessed IR in primary maturing mammalian erythroid and megakaryocyte (MK) lineages as well as their common progenitor cells (MEPs). Both lineages exhibit an extensive differential IR program involving hundreds of introns and genes with an overwhelming loss of IR in erythroid cells and MKs compared with MEPs. Moreover, complex IR patterns were seen throughout murine erythroid maturation. Similarly complex patterns were observed in human erythroid differentiation, but not involving the murine orthologous introns or genes. Despite the common origin of erythroid cells and MKs, and overlapping gene expression patterns, the MK IR program is entirely distinct from that of the erythroid lineage with regards to introns, genes, and affected gene ontologies. Importantly, our results suggest that IR serves to broadly regulate mRNA levels. These findings highlight the importance of this understudied form of alternative splicing in gene regulation and provide a useful resource for studies on gene expression in the MK and erythroid lineages.
Introduction
Alternative splicing is critical to mammalian transcription because it diversifies the proteome without a corresponding increase in genome complexity. In a given transcript, 5′ and 3′ splice sites can vary, exons can be spliced out, pairs of exons can be mutually exclusive, and introns can be retained.1 Intron retention (IR) is the least studied form of alternative splicing but is emerging as an important regulatory mechanism in several cell types.2-7 IR can result in a truncated protein, insert an amino acid sequence, or detain a transcript in the nucleus to prevent translation. Importantly, however, it often promotes mRNA destabilization via nonsense-mediated decay (NMD) in the cytoplasm or other mechanisms in the nucleus.2-5,8-10 The NMD machinery degrades IR mRNAs because of the presence of premature termination codons (PTCs) introduced by the introns.
Recent evidence argues that IR-induced mRNA modulation is not uncommon and is biologically important.2,3,5 Of note, even in the case of non-IR alternative splicing, exon splice variations can result in PTCs that activate NMD.11-13 In fact, it is estimated that one third of genes have an NMD-targeted splice variant.14,15 Thus, alternative splicing, including IR, can affect mRNA quality and quantity. In addition, understanding how alternative splicing and NMD together influence mRNA levels is important because their misregulation is linked to disease. It is estimated that one tenth to one third of known disease-causing mutations alter splicing16,17 and one third of hereditary diseases result from PTCs.18-20 Further, widespread IR was recently reported in cancer.21,22
Intricate hematopoietic gene expression and alternative splicing programs exist in the related megakaryocyte (MK) and erythrocyte lineages,23-28 and splicing-coupled NMD is observed during human erythropoiesis.11 However, if IR occurs in the MK and erythroid lineages, whether it is distinct between these lineages, and how it is modulated during maturation is unknown. Here, we used 3 RNA-seq data sets, the first comprising a trio of murine MK-erythrocyte precursors (MEPs, the common precursor giving rise to both lineages), MKs, and erythroid cells. The second and third represent terminal erythroid maturation stages derived from murine bone marrow and in vitro differentiated human cord blood cells, respectively. Using these data sets and the bioinformatics tool Intron Retention Finder (IRFinder), we uncovered a complex differential IR program that is distinct among these lineages and highly dynamic during cellular maturation.
Methods
We assessed IR using an improved version of the bioinformatics tool Intron Retention Finder (IRFinder, R.M. and W.R., manuscript in preparation). See the supplemental material, available on the Blood Web site, for details. For each intron, IRFinder estimates the abundance of transcripts retaining or not retaining it. The fraction of transcripts retaining that intron is then calculated by taking the ratio of transcripts retaining it to the sum of transcripts retaining or not retaining it. This is the “IR ratio” and ranges from 0 to 1. Partial IR resulting from splicing inside the intron is a distinct process and was not examined.29
To find introns differentially retained between 2 samples, we first identified those whose range of IR ratios in replicates of 1 sample does not overlap with the range of IR ratios in replicates of the other sample. For introns meeting this criterion, IR ratios were calculated in pooled replicates of each sample. P values indicating whether these IR ratios are different were calculated.3,30 Introns with a Benjamini-Hochberg adjusted P < .05 and IR ratio fold-change ≥2 were considered differentially retained.
Results
Dramatic IR changes during MEP differentiation
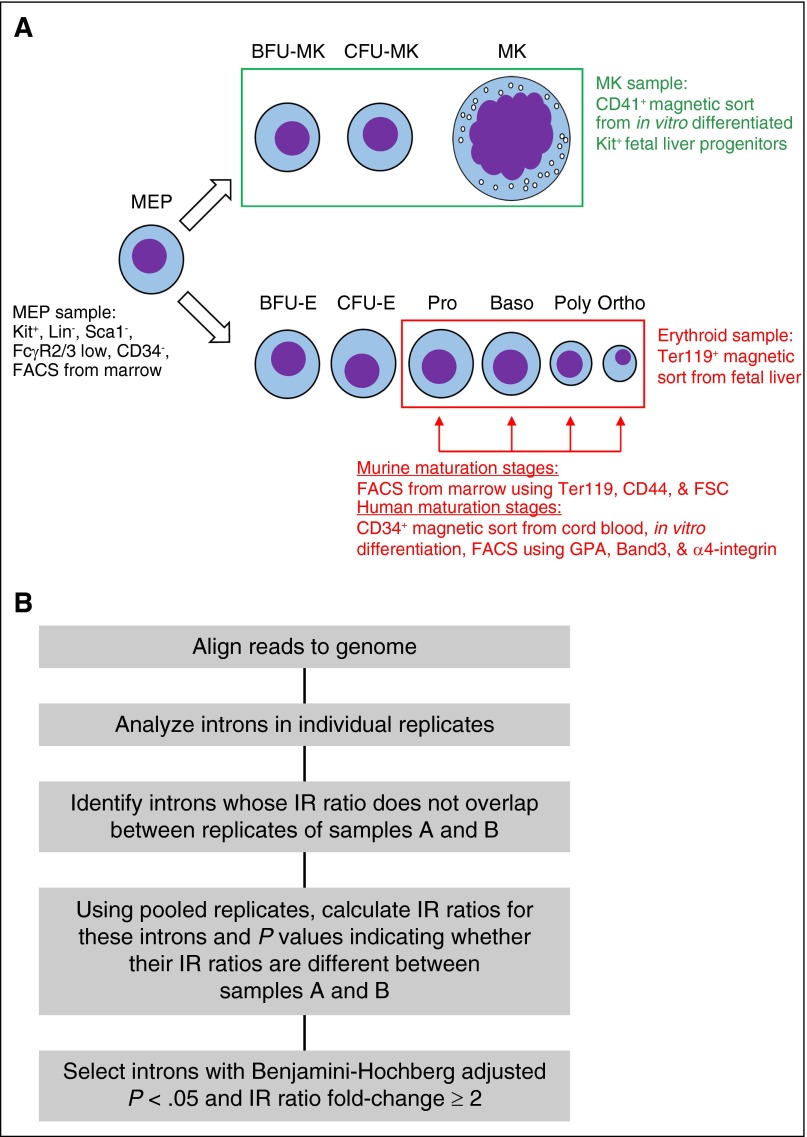
We investigated IR in murine MK and erythrocyte lineages using existing, directional, paired-end RNA-seq data sets.23 These data sets derive from primary tissues, including bone marrow MEPs, fetal liver Ter119+ erythroid populations, and CD41+ MKs differentiated in vitro from fetal liver progenitors (Figure 1A). These samples allow an initial global assessment of IR changes between MEPs and each differentiated lineage and between both lineages. Using IRFinder, we assessed IR in these cell types and identified differential retention by independently comparing MEPs to each lineage (Figure 1B).
Figure 1.
Experimental strategy. (A) Purification strategy for samples used to generate these data sets. Baso, basophilic erythroblast; BFU-E, burst-forming unit-erythroid; BFU-MK, burst-forming unit-megakaryocyte; CFU-E, colony-forming unit-erythroid; CFU-MK, colony-forming unit-megakaryocyte; MEP, megakaryocyte-erythrocyte precursor; MK, megakaryocyte; Ortho, orthochromatic erythroblast; Poly, polychromatic erythroblast; Pro, proerythroblast. Purification strategies for MEPs, the MK lineage in bulk, the erythroid lineage in bulk, and individual erythroid stages are indicated. (B) Strategy to identify differentially retained introns between pairs of samples using IRFinder.
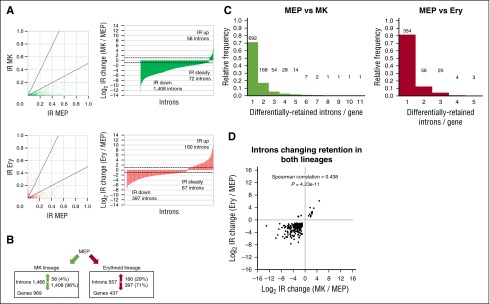
We analyzed approximately 200 000 introns in each sample and excluded those with extremely low splice-junction read counts (supplemental Methods). Most introns with measurable IR ratios have little to no retention, a minority has high levels of retention, and globally, MEPs have the most and MKs have the least IR (supplemental Figure 1A). We identified 1466 introns in 969 genes that change retention greater than or equal to twofold between MEPs and MKs, with 58 (4%) increasing and a remarkable 1408 (96%) decreasing their retention in MKs compared with MEPs (Figures 2A, upper panels, and 2B; supplemental Figure 2A and supplemental Table 1). Similarly, we identified 557 introns in 437 genes that change retention greater than or equal to twofold, with 160 (29%) increasing and 397 (71%) decreasing their retention in erythroid cells compared with MEPs (Figures 2A, lower panels, and 2B; supplemental Figure 2A and supplemental Table 2). It is worth noting that first, within either lineage, the majority of differential IR genes only possess one such intron (Figure 2C). Second, in genes bearing multiple differentially retained introns, retention almost exclusively changes in the same direction (not shown). Third, we observed that retained introns are shorter and have a higher GC content than nonretained introns (supplemental Figure 1B-D), consistent with previous reports.2-4 Finally, because the MKs were differentiated in vitro, we note that some MK IR events could possibly result from the culturing. In summary, a subset of the retained introns described before is differentially retained between samples and tends to have lower retention levels in differentiated cells relative to MEPs.
Figure 2.
IR dynamics between MEPs and MKs or MEPs and erythroid cells. (A) IR changes between MEPs and MKs (upper panels) or erythroid cells (lower panels). Scatterplots depict IR ratios between pairs of samples for introns with significantly (Ben-Hoch, P < .05) different IR ratios. Bar graphs depict log2 IR ratio fold-changes. Number of introns with IR ratios increasing greater than or equal to twofold, decreasing greater than or equal to twofold, or changing less than twofold are indicated. Dashed bars indicate twofold changes. Empty regions along bar graph x-axes are introns with IR = 0 in 1 sample, their fold-changes cannot be plotted on a log-transformed scale. (B) Differential IR among both lineages. Panels indicate introns with greater than or equal to twofold IR ratio changes. (C) Histograms of introns per gene with greater than or equal to twofold IR ratio changes. Absolute frequencies are indicated above bars. (D) Introns with greater than or equal to twofold IR ratio fold-changes in both lineages. Scatterplot depicts log2 IR ratio fold-changes between MEPs and MKs (x-axis) and MEPs and erythroid cells (y-axis). Introns with IR ratio = 0 in one sample cannot have their fold-changes plotted on a log-transformed scale and are omitted.
In a previous study, IR increases during granulocyte maturation correlated with expression decreases of a subset of splicing factors,3 suggesting splice factor downregulation as a cause. We examined splicing and NMD factor gene expression and found that at the mRNA level, the majority of factors show little change when comparing MEPs with MKs or erythroid cells (supplemental Figures 3A-B and 4A). The dynamic IR patterns thus might not result from broad changes in the splicing and NMD machinery. However, we cannot rule out a critical role for the few factors with changing mRNA levels, or changes in the splice or NMD machinery’s protein or activity levels.
Comparing differential IR between the MK and erythrocyte lineages
IR dramatically declines during MEP differentiation toward either lineage. We therefore asked whether there are commonalities among the dynamically retained introns. Two-hundred thirty introns in 198 genes change retention in both lineages (Figure 2D; Table 1), with nearly all introns changing retention in the same direction in each lineage. The majority of these shared IR events decline during differentiation. We additionally noted 258 genes with differential retention in both lineages, including those with different introns changing retention in each lineage. Finally, we assessed lineage-specific IR changes and found decreases in the majority of MK-specific IR events, whereas erythroid-specific IR has a more even mix of increasing and decreasing IR. The differentially retained introns in these lineages thus include a subset that changes IR ratios in both lineages, mostly decreasing, and additional subsets that are MK or erythroid-specific.
Table 1.
IR distribution among MK and erythroid lineages
| Introns and genes with IR ratios changing ≥twofold | |
|---|---|
| MK lineage-specific | |
| Introns with IR ratios changing | 1236 |
| Introns with IR ratios increasing | 47 (4%) |
| Introns with IR ratios decreasing | 1189 (96%) |
| Genes with IR ratios changing | 711 |
| Erythroid lineage-specific | |
| Introns with IR ratios changing | 327 |
| Introns with IR ratios increasing | 145 (44%) |
| Introns with IR ratios decreasing | 182 (56%) |
| Genes with IR ratios changing | 179 |
| Common to both lineages | |
| Introns with IR ratios changing | 230 |
| Introns with IR ratios increasing in both lineages | 11 |
| Introns with IR ratios decreasing in both lineages | 215 |
| Introns with IR changing in different directions | 4 |
| Genes with same intron changing in both lineages | 198 |
| Genes with same or different intron changing | 258 |
Enrichment of GO terms and KEGG pathways among differential IR genes during MEP differentiation
We searched the differential IR genes for enriched Gene Ontology (GO) terms and KEGG pathways using a background of all genes with measured IR ratios (supplemental Tables 5-12). In the MEP-Ery comparison, genes with decreasing IR are enriched for terms regarding ribosomes, translation, splicing, and adenosine triphosphate biosynthesis; genes with increasing IR are enriched for terms regarding mitosis, chromatin, and the spindle; and total genes with differential IR are enriched for all these terms. In contrast, in the MEP-MK comparison, where most genes have decreasing IR, differential IR genes are enriched for terms including actin, immunity, ncRNA, endocytosis, and the proteasome. Genes with the same intron changing retention in both lineages are enriched for translation and ncRNA processing terms. Finally, the lists of terms are fairly similar between total and lineage-specific differential IR genes within each lineage. The differential IR programs thus are enriched for functional terms that differ between lineages.
IR during murine terminal erythroid maturation
During erythropoiesis, cells undergo dramatic changes including diminishing cell size, chromatin condensation, and erythroid-specific gene upregulation.31,32 Complex transcription and splicing programs drive this process.11,25,27,28 We therefore determined whether an IR program exists during terminal maturation. We used nondirectional, single-end RNA-seq data sets from primary bone marrow–derived murine proerythroblasts and basophilic, polychromatic, and orthochromatic erythroblasts25 (Figure 1A). Corresponding human populations (including 2 basophilic stages) were differentiated in vitro from cord blood progenitors.25 These data sets were thus derived from pure stage-matched populations and allow comparisons of both species.
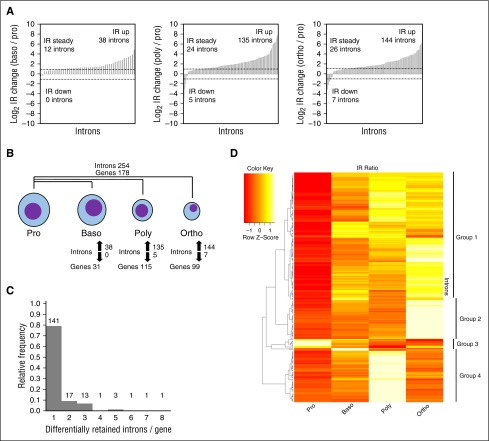
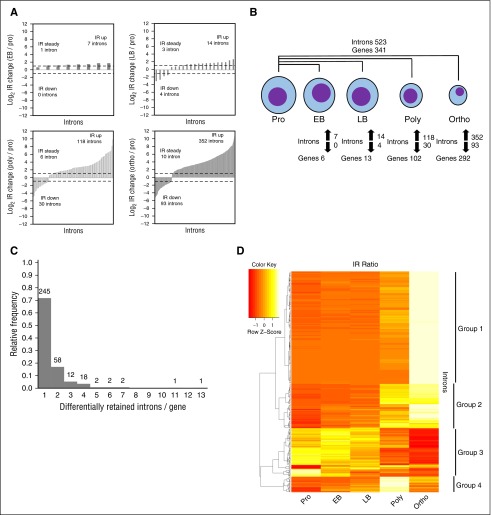
We examined the murine lineage by independently comparing proerythroblasts to each subsequent stage. Among all 3 comparisons, we identified 254 differentially retained introns in 178 genes, the majority of genes containing one such intron (Figure 3A-C; supplemental Figure 2B and supplemental Table 3). In surprising contrast to the MEP/erythroid comparison, IR ratios almost exclusively increase during maturation with a jump in the number of introns at the polychromatic stage. The IR signature thus changes considerably during maturation. The IR patterns across the stages are complex (Figure 3D), with several intron groups discernible. In the largest, IR ratios are generally higher after the proerythroblast stage. In 2 smaller groups, IR ratios peak strongly in either polychromatic or orthochromatic erythroblasts. The smallest group has IR ratios descending during maturation.
Figure 3.
Differential IR at multiple stages during murine erythroid maturation. (A) Bar graphs depicting IR ratio fold-changes between pairs of samples. Number of introns with IR ratios increasing greater than or equal to twofold, decreasing greater than or equal to twofold, or changing less than twofold are indicated. Dashed bars indicate twofold changes. (B) Number of introns and genes with IR ratios changing greater than or equal to twofold between proerythroblasts and each downstream stage. Total introns and genes among all pairwise comparisons is shown above. (C) Histograms of introns per gene with greater than or equal to twofold IR ratio changes. Absolute frequencies are indicated above bars. (D) Heat map of IR ratios across stages. Rows represent introns whose IR ratio changes greater than or equal to twofold between proerythroblasts and at least 1 downstream stage. Columns represent stages. Colors indicate Z-scores of IR ratios across each row. Rows were ordered using the Pearson distance metric and complete clustering method. Clusters were identified visually and using the dendrogram and are indicated to the right.
Some splicing and NMD factors have decreasing mRNA levels during maturation (supplemental Figures 3C-D and 4B). Smg8, a factor essential for human NMD,33 also has moderately lower transcript levels during the final stage. The extent to which these mRNA changes influence IR is unclear and again, unseen changes in these factors’ protein and activity levels may shape the IR program. However, global changes in the splicing or NMD machineries seem insufficient to account for the selectivity and complex dynamics of IR, favoring the existence of cell type and/or stage-specific mechanisms.
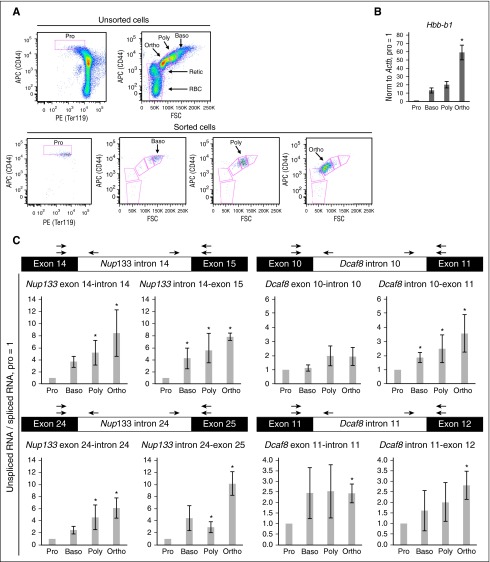
To validate IR changes, we independently purified the same murine erythroid maturational stages used for the RNA-seq data sets (Figures 1A and 4A) and performed quantitative reverse-transcription polymerase chain reaction. Rising β-major (Hbb-b1) mRNA levels confirmed increasing maturity (Figure 4B). We selected 4 introns with predicted retention increases during maturation. The ratios of intron-exon junctions (IR transcript) to exon-exon junctions (spliced transcript) were compared between proerythroblasts and downstream stages and increased during maturation (Figure 4C).
Figure 4.
Experimental validation of differential IR. (A) Fluorescence-activated cell sorting profiles of unsorted and sorted murine erythroid marrow cells. Target antigens and fluorophores are indicated along axes. (B) Bar graph depicts quantitative reverse-transcription polymerase chain reaction of β-major (Hbb-b1) mRNA levels normalized to β-actin mRNA levels. (C) Genes and introns are indicated above, samples are indicated below, and the ratio of intron-exon signals (intron retained) to exon-exon signals (intron spliced out) are indicated to the left of each graph. Graphs depict mean and standard error of the mean of 4 to 6 biological replicates. Asterisks indicate significance (Wilcoxon rank-sum test, P < .05). Arrows indicate quantitative polymerase chain reaction primer locations.
IR during human terminal erythroid maturation
We next examined the human erythroid lineage. Among all 4 comparisons, we identified 523 differentially retained introns in 341 genes, the majority of genes containing one such intron (Figure 5A-C; supplemental Figure 2C and supplemental Table 4). Like the murine program, IR patterns across stages are complex (Figure 5D). Approximately half of the introns have retention peaking in orthochromatic erythroblasts. Two smaller intron groups have retention peaking in polychromatic erythroblasts or the last 2 stages, and a fourth group decreases retention during maturation. Unlike murine erythropoiesis, however, the fractions of introns with retention peaking in the final stage or dropping during late stages are larger. The orthochromatic erythroblast IR peak is striking. We noticed a general decrease in the expression of splicing and NMD factors during the later stages (supplemental Figures 3E-F and 4C), which might account for some IR increases, but would fail to explain the simultaneous IR decreases. A complex dynamic IR program thus exists during terminal human erythropoiesis, with similarities to the murine program.
Figure 5.
Differential IR at multiple stages during human erythroid maturation. (A) Bar graphs depicting IR ratio fold-changes between pairs of samples. Number of introns with IR ratios increasing greater than or equal to twofold, decreasing greater than or equal to twofold, or changing less than twofold are indicated. Dashed bars indicate twofold changes. (B) Number of introns and genes with IR ratios changing greater than or equal to twofold between proerythroblasts and each downstream stage. Total introns and genes among all pairwise comparisons is shown above. (C) Histograms of introns per gene with greater than or equal to twofold IR ratio changes. Absolute frequencies are indicated above bars. (D) Heat map of IR ratios across stages. Rows represent introns whose IR ratio changes greater than or equal to twofold between proerythroblasts and at least 1 downstream stage. Columns represent stages. Colors indicate Z-scores of IR ratios across each row. Rows were ordered using the Pearson distance metric and complete clustering method. Clusters were identified visually and using the dendrogram and are indicated to the right. EB, early basophilic erythroblast; LB, late basophilic erythroblast.
Enrichment of GO terms and KEGG pathways among differential IR genes during terminal erythroid maturation
During murine terminal erythropoiesis, GO terms related to mitosis and the KEGG pathway porphyrin and chlorophyll metabolism (which includes heme metabolism genes) are enriched (supplemental Table 13). In humans, enriched GO terms include heme biosynthesis, iron homeostasis, splicing, post-transcriptional gene regulation, translation, and glycosylation (supplemental Table 14). We noted spectrin-associated cytoskeleton genes (dematin, ankyrin, and α/β spectrin). In addition, genes with IR spiking in orthochromatic erythroblasts are enriched for splicing and transcription terms (supplemental Table 15). These results are notable because erythropoiesis involves producing optimal heme levels, metabolizing iron, controlling the number of cell divisions, globally changing mRNA levels, mass-producing erythroid proteins, and remodeling the cell membrane and cytoskeletal scaffold.31,32 In summary, dynamic IR genes are enriched for biologically important GO terms and KEGG pathways during murine and human erythroid maturation.
IR conservation during erythroid maturation stages
To ask whether total IR is conserved between murine and human erythroid differentiation, we identified introns with orthologs in both species (UCSC Liftover) and IR measurements in equivalent stages. We compared each murine stage with its human counterpart with murine basophilic erythroblasts compared with early or late human basophilic erythroblasts. IR ratios showed a weak positive correlation in every stage (supplemental Table 16). Importantly, in each stage-matched comparison, 30% to 40% of murine-retained introns are retained in humans (supplemental Table 17). Likewise, 40% to 50% of human-retained introns are retained in mice. This overlap is significant (hypergeometric test P < 7E−212, 2-sided Fisher exact test P < 2.2E−16).
We similarly asked whether the murine and human differential IR programs overlap. Among all introns with orthologs and IR measurements in both species, 0.27% (84) and 0.65% (204) are differentially retained in mice and humans, respectively. Importantly, only 6 are differentially retained in both species (supplemental Figure 5A). This overlap is significant (hypergeometric test P = 1.37E−6, 2-sided Fisher exact test P = 1.94E−5) but miniscule. Of the overall murine differential IR program’s 178 genes, nearly all have a human ortholog but only 18% (32) are in the human differential IR program (supplemental Figure 5B). Thus, total IR, but not differential IR, is conserved between murine and human terminal erythropoiesis.
NMD inhibition did not upregulate retention of differentially retained introns
IR can result in mRNA nuclear sequestration or cytoplasmic degradation by PTC-triggered NMD.2-5,8,9 NMD operates in the erythroid lineage11,34 and its factors are expressed in our samples (supplemental Figure 4). More than 99% of the differentially retained introns potentially introduce a PTC and more than 90% are predicted to trigger NMD (supplemental Tables 18-21). To ask whether NMD degrades our differential IR transcripts, we purified adult murine bone marrow proerythroblasts and polychromatic erythroblasts and inhibited NMD with cycloheximide (CHX, 100 μM for Pro, 100 μM or 500 μM for Poly; supplemental Methods). Proerythroblasts received only 1 concentration because of low cell numbers. We assessed retention in the same 4 introns examined in Figure 4C, again checking IR at the 5′ and 3′ intron-exon junctions. These four have NMD predicted and increase retention during differentiation, whereas their transcript levels trend downward (not shown).
β-major (Hbb-b1) and IR levels changed as expected during maturation (supplemental Figure 6A,C, compare first and third bars). Genes Srsf7 and Srsf3 served as positive controls for NMD inhibition. Analyses of their human orthologs35 and UCSC genome tracks suggest they contain differentially spliced “poison exons,” exons introducing a PTC and triggering NMD. Despite sizeable quantitative polymerase chain reaction variation in some CHX-treated samples, 2 things were evident. First, for Srsf7 and Srsf3, CHX treatment dramatically increased the ratio of transcripts containing the “poison” exons to transcripts lacking them (supplemental Figure 6B). Second, surprisingly, CHX caused no general IR increase (supplemental Figure 6C). Although NMD thus functions during murine terminal erythropoiesis, we found no evidence that it degrades our differential IR transcripts.
IR regulates gene expression levels
Multiple studies have reported an inverse correlation between IR and mRNA levels,2,3,21 though not all.4 Although our CHX experiments did not show a role for NMD in IR mRNA destabilization, we did not look globally and mRNA degradation may occur via other mechanisms. We therefore determined globally whether IR and mRNA levels inversely correlate at the MEP to MK or erythroid transitions, and during the erythroid stages with the most pronounced IR changes, namely between human proerythroblasts and polychromatic or orthochromatic erythroblasts.
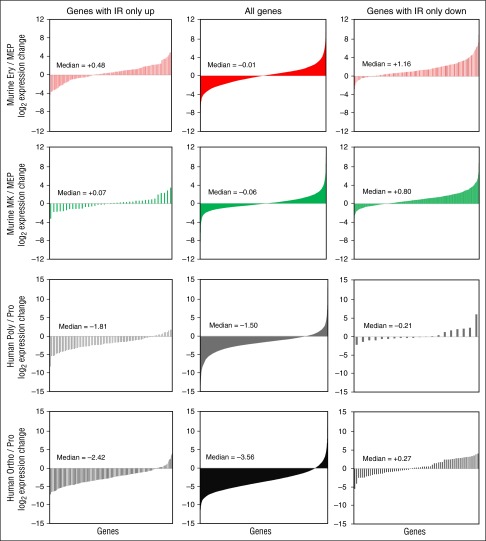
We grouped genes into those with IR increasing or decreasing greater than or equal to twofold, excluded those with mixed IR change directions, and compared expression changes between those genes and with all genes with IR measurements. All genes with IR measurements have a fairly even mix of up- and downregulation during murine MEP to MK or erythroid transitions (Figure 6, top 2 rows, center column). Genes with increasing IR have similar or only slightly different expression patterns (compare center and left columns). In contrast, decreasing IR has a strong impact on expression with a dramatically larger fraction of mRNAs upregulated (compare center and right columns).
Figure 6.
An inverse correlation between gene expression changes and IR changes. Graphs depict log2 expression changes between pairs of samples for all genes with measured IR ratios (center panels), genes with IR ratios only increasing greater than or equal to twofold (left panels), and genes with IR ratios only decreasing greater than or equal to twofold (right panels). Pairwise comparisons of samples are indicated to the left. Median log2 expression changes are indicated within each graph.
During human proerythroblast to polychromatic or orthochromatic erythroblast transitions, genes with IR measurements and genes with increasing IR tend to be downregulated (Figure 6, bottom 2 rows, center and left columns). Again, when considering genes with reduced IR, a shift occurs with a considerably larger fraction of genes increasing their expression (compare center and right columns). Median expression changes are highest for genes with decreasing IR (Wilcoxon rank-sum, P < .05). The inverse correlation between IR and expression is unaltered by raising the minimum IR fold-change from two- to tenfold (not shown). In addition, these trends are unchanged when parsing genes into those with only 1 or multiple differentially retained introns (supplemental Figure 7). We note that whether the multiple differential IR introns coexist or are in separate mRNA molecules is unknown. Thus, genes with decreasing IR tend to be more upregulated than either all genes with IR measurements or the subset with increasing IR.
The inverse correlation between IR and mRNA changes is not absolute, likely because of confounding effects from transcription changes and the fact that IR does not guarantee mRNA degradation. During murine terminal erythropoiesis, we observed almost no genes with decreasing IR, contrasting with human cells. Regardless, overall, our results add support to the hypothesis that IR serves to negatively regulate mRNA levels.
Discussion
Using an improved version of IRFinder (R.M. and W.R., manuscript in preparation), we discovered a program of differential IR in mammalian MEP, MK, and erythroid cells. MEPs have more IR compared with erythroid and MK lineages. In addition, throughout erythroid maturation, complex layers of IR are superimposed on the murine and human gene expression programs. Importantly, dynamic IR correlates inversely with transcript level changes, suggesting a major involvement of IR-coupled mRNA degradation in regulating gene expression. Our IR analysis thus adds further complexity to the mammalian MK and erythroid lineages, emphasizes that gene regulation must be considered post-transcriptionally, and presents a resource for those studying gene expression in these lineages.
Our IR analysis strategy incorporated features worth considering. IRFinder’s methods minimized the effects of small RNAs and local coverage spikes on IR measurements. The algorithm considered both canonical splicing (2 adjacent exons joined) and alternative splicing (1 adjacent and 1 distal exon joined). Finally, RNA-seq data sets were derived from polyA+ RNA, excluding nascent transcripts and free introns.
Gene expression studies typically focus on transcript synthesis. However, mRNA levels are influenced post-transcriptionally by mechanisms such as the regulation of mRNA turnover with or without miRNAs.36,37 Recent studies add alternative splicing and IR to this list of mechanisms.2,3,5,11,21 Our analyses suggest that the mammalian MK and erythroid lineage differential IR programs may play such a role. We observed that expression of genes with decreasing IR during differentiation trends upward relative to all genes with IR measurements. This is consistent with IR triggering mRNA destabilization. However, we observed a second gene set with IR increasing during differentiation, but with similar expression changes compared with all genes with IR measurements. The reason for this asymmetric correlation between IR changes and transcript level changes is unclear. One possibility is that the 2 transcript groups differ in sensitivity to mRNA destabilization pathways. Our analyses are confounded by transcriptional changes and the fact that IR does not guarantee mRNA destabilization. In addition, we note that because degraded IR transcripts cannot be detected, our IR level assessments may be underestimates. Surprisingly, 4 differentially retained introns we examined with regard to NMD regulation did not generally increase retention upon NMD inhibition. This clearly indicates that IR with a PTC does not guarantee NMD. Moreover, NMD does not account for all IR-related changes in transcript levels and IR mRNAs may be detained and degraded in the nucleus. This last point is consistent with reports that retained introns are NMD-resistant and enriched in the nuclei of human erythroid cells38 (discussed next). However, it is possible, if not probable, that NMD might play a role at other IR transcripts. Overall, our results strongly suggest a role for differential IR programs in regulating mRNA levels.
Using another algorithm, others recently assessed terminal human erythropoiesis IR38 (see supplemental Methods for a comparison). Similar to our findings, they observed increased retention of a set of introns in gene groups enriched for mRNA processing GO terms. Tested introns were NMD-resistant. Moreover, we noted some overlap between our introns with increasing retention during late erythropoiesis and one of their identified intron clusters (cluster C1) which also has increasing retention during late erythropoiesis. It should be noted that their strategy grouped introns into clusters based on patterns of IR values throughout differentiation, whereas we identified introns with statistically significant IR fold-changes between maturational stages. Thus, although discrepancies between the IR patterns identified by the 2 algorithms may result from mapping or methodologic differences, they may also result from differences in the organization and presentation of IR patterns. Overall, both approaches yielded similar conclusions regarding IR patterns in human erythroid maturation.
Downregulating or limiting transcript levels via IR-coupled degradation seems counterintuitive because it is conceptually more complex than silencing transcription and involves wasteful transcription cycles. There are several possibilities why this mechanism evolved. First, this gives cells an additional mechanism for adjusting transcript levels. Second, it allows cells to coordinately regulate genes lacking common miRNA target sequences or transcription or translation regulators. Third, this mechanism utilizes existing pathways, obviating the need to evolve or upregulate new proteins. Fourth, protein production, and thus transcription repressor production, might be at least as energetically expensive as mRNA production.39 Hence, altering splicing might be more energy efficient than producing repressor proteins. Finally, altering splicing might be kinetically favorable compared with transcribing and translating a repressor. Of course, this strategy of leaving transcription active and intercepting the mRNAs is not unprecedented, as exemplified by miRNA-mediated repression.
Although IR is typically associated with mRNA degradation, it can also influence protein production. IR transcripts for tissue factor and IL-1β are present in MKs and carry over into anucleate platelets.40,41 Introns are spliced out upon platelet activation and only then are the transcripts successfully translated. Our MK and orthochromatic erythroblast IR transcripts might similarly carry over into and affect protein production in anucleate platelets and reticulocytes, respectively. One corollary of this is that IR mRNAs would need to escape degradation. This might account for the general lack of transcript destabilization with increasing IR. Further investigations are needed to reveal whether these IR programs have this role.
Many introns are retained at low levels, prompting questions regarding their biological impact. In this regard, 3 points should be considered. First, as mentioned earlier, IR transcripts may be degraded and their true extent and contribution to mRNA levels may thus be higher than appears. Second, IR may affect cell biology via small but collective effects on many transcripts. Finally, IR might serve to subtly fine-tune gene expression.
It is unclear what triggers differential IR in these lineages, but many possible mechanisms exist. Splice factors can be regulated in terms of expression, protein abundance, transcript-specific recruitment, and RNA-binding and catalytic activity. Splice-site strength2,4,38 and intronic and exonic splice enhancers and silencers can also modulate splicing.42,43 In addition, signaling pathways, the transcription machinery, chromatin modifications, nucleosome occupancy, transcription elongation kinetics, and ncRNAs may influence splicing.1,2,44 Finally, degradation of intron-retaining transcripts can involve nuclear ribonucleases, the NMD pathway, which is linked to the translation machinery, and other factors, any of which might be targets for modulation. In our murine samples, there is no universal change in splice or NMD factor mRNAs, suggesting that IR patterns might not result from nonspecific global changes. Transcript levels change for a few factors and we cannot formally rule out their involvement in differential IR. In human erythroid cells, maturation is accompanied by more substantial downregulation of splice and NMD factor transcripts. Yet, because IR does not increase globally, one would have to invoke differential sensitivity of introns to alterations in the splicing or NMD pathways. In addition, reductions in these factors do not easily explain the IR decrease during maturation. Finally, unseen changes in the factors’ protein or activity levels may shape the IR programs. In sum, this work suggests that IR is a mode of mRNA regulation that functions in a gene-specific manner. Further studies of IR in a variety of organisms will shed light on how this intriguing mechanism evolved.
Acknowledgments
The authors thank Perry Evans, Joanna Balcerek, and Chris Hsiung for computational support, sample purification guidance, and heat map guidance, respectively.
This work was supported by Hematology Training grant T32-HL007439 (C.R.E.), National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grants RO1-DK054937 (G.A.B.) and R37-DK026263 (N.M.), the NSW Cancer Institute (W.R., J.J.-L.W.), National Health and Medical Research Council grants 1061906 and 1030830 (J.E.J.R.), the Cancer Council of NSW (J.E.J.R.), Cure the Future (J.E.J.R.), and an anonymous foundation (J.E.J.R.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.R.E. and G.A.B. designed experiments and wrote the manuscript; C.R.E. analyzed and interpreted data and performed experiments; R.M., W.R., J.J.-L.W., U.S., J.E.J.R., X.A., and N.M. analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerd A. Blobel, 316H Abramson Research Center, 3615 Civic Center Blvd, Philadelphia, PA 19104; e-mail: blobel@email.chop.edu.
References
- 1.Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14(3):153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 2.Braunschweig U, Barbosa-Morais NL, Pan Q, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24(11):1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong JJ, Ritchie W, Ebner OA, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154(3):583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 4.Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29(1):63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26(11):1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell TJ, Miyashiro KY, Sul JY, et al. Cytoplasmic BK(Ca) channel intron-containing mRNAs contribute to the intrinsic excitability of hippocampal neurons. Proc Natl Acad Sci USA. 2008;105(6):1901–1906. doi: 10.1073/pnas.0711796105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galante PAF, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10(5):757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13(11):700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829(6-7):612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Wong JJL, Au AYM, Ritchie W, Rasko JEJ. Intron retention in mRNA: No longer nonsense: Known and putative roles of intron retention in normal and disease biology. BioEssays. 2016;38(1):41–49. doi: 10.1002/bies.201500117. [DOI] [PubMed] [Google Scholar]
- 11.Pimentel H, Parra M, Gee S, et al. A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2014;42(6):4031–4042. doi: 10.1093/nar/gkt1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13(1):91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 13.Sureau A, Gattoni R, Dooghe Y, Stévenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20(7):1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weischenfeldt J, Waage J, Tian G, et al. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 2012;13(5):R35. doi: 10.1186/gb-2012-13-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIlwain DR, Pan Q, Reilly PT, et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci USA. 2010;107(27):12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci USA. 2011;108(27):11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewandowska MA. The missing puzzle piece: splicing mutations. Int J Clin Exp Pathol. 2013;6(12):2675–2682. [PMC free article] [PubMed] [Google Scholar]
- 18.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29(8):1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 19.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36(8):801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 20.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8(10):1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 21.Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7(1):45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung H, Lee D, Lee J, et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47(11):1242–1248. doi: 10.1038/ng.3414. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Kostadima M, Martens JH, et al. BRIDGE Consortium. Transcriptional diversity during lineage commitment of human blood progenitors. Science. 2014;345(6204):1251033. doi: 10.1126/science.1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paralkar VR, Mishra T, Luan J, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123(12):1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An X, Schulz VP, Li J, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123(22):3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Wang H, Chang KH, et al. Transcriptome dynamics during human erythroid differentiation and development. Genomics. 2013;102(5-6):431–441. doi: 10.1016/j.ygeno.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng AW, Shi J, Wong P, et al. Muscleblind-like 1 (Mbnl1) regulates pre-mRNA alternative splicing during terminal erythropoiesis. Blood. 2014;124(4):598–610. doi: 10.1182/blood-2013-12-542209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong P, Hattangadi SM, Cheng AW, Frampton GM, Young RA, Lodish HF. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood. 2011;118(16):e128–e138. doi: 10.1182/blood-2011-03-341404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amit M, Donyo M, Hollander D, et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Reports. 2012;1(5):543–556. doi: 10.1016/j.celrep.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 31.Dzierzak E, Philipsen S. Erythropoiesis: development and differentiation. Cold Spring Harb Perspect Med. 2013;3(4):a011601. doi: 10.1101/cshperspect.a011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61(8):800–830. doi: 10.1002/iub.226. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita A, Izumi N, Kashima I, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23(9):1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981;27(3 Pt 2):543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 35.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446(7138):926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 36.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13(4):246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 38.Pimentel H, Parra M, Gee SL, Mohandas N, Pachter L, Conboy JG. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016;44(2):838–851. doi: 10.1093/nar/gkv1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwanhäusser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 40.Denis MM, Tolley ND, Bunting M, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122(3):379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwertz H, Tolley ND, Foulks JM, et al. Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenicity of human platelets. J Exp Med. 2006;203(11):2433–2440. doi: 10.1084/jem.20061302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]