Abstract
Metabolomics, which is defined as the comprehensive analysis of metabolites in a biological specimen, is an emerging technology that holds promise to inform the practice of precision medicine. Historically, small numbers of metabolites have been used to diagnose complex metabolic diseases as well as monogenic disorders such as inborn errors of metabolism. Current metabolomic technologies go well beyond the scope of standard clinical chemistry techniques and are capable of precise analyses of hundreds to thousands of metabolites. Consequently, metabolomics affords detailed characterization of metabolic phenotypes and can enable precision medicine at a number of levels, including the characterization of metabolic derangements that underlie disease, discovery of new therapeutic targets, and discovery of biomarkers that may be used to either diagnose disease or monitor activity of therapeutics.
INTRODUCTION
The goal of precision medicine is to design disease prevention and clinical care strategies taking into account individual variability in environment, lifestyle, genetics, and molecular phenotype. The application of clinical genomics in cancer to inform selection of therapies and predict outcomes has been the vanguard of the field. Using a microscope as an analogy, genomic tools constitute a powerfully informative objective lens through which to examine individual variability, but it does not provide a view to other biomolecules, such as metabolites, which also define molecular phenotypes. Ideally, a molecular microscope for precision medicine would be equipped with additional objectives to examine biochemistry more broadly. There are decades of precedence for using analyses of small numbers of metabolites to diagnose disease and impact clinical care—for example, the development of blood glucose test strips in the 1950s to test for diabetes (Clarke and Foster 2012) or measuring phenylalanine in newborns to screen for phenylketonuria (Guthrie and Susi 1963). Indeed, measurable changes in metabolite levels occur in response to both complex disease and monogenic disorders, and in contrast to the genome, these changes can exhibit tissue specificity and temporal dynamics. Metabolomics is an emerging field and is broadly defined as the comprehensive measurement of all metabolites and low-molecular-weight molecules in a biological specimen. Because metabolomics affords profiling of much larger numbers of metabolites than are presently covered in standard clinical laboratory techniques, and hence comprehensive coverage of biological processes and metabolic pathways, it holds promise to serve as an essential objective lens in the molecular microscope for precision medicine.
METABOLOMICS IS AN EMERGING AND EVOLVING TECHNOLOGY
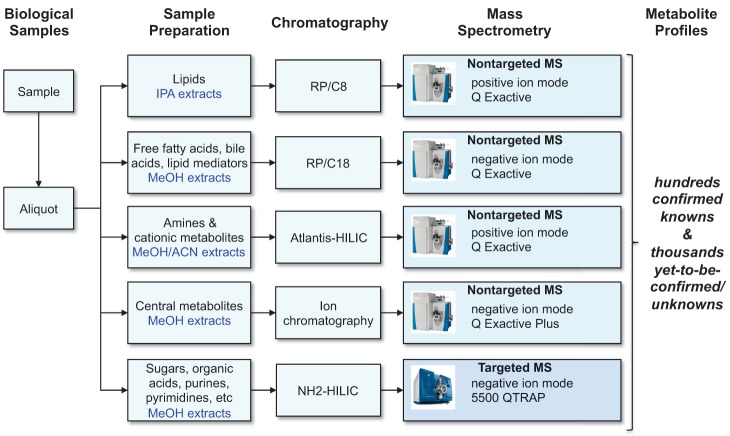
In practice, metabolomics presents a significant analytical challenge because, unlike genomic and proteomic methods, it aims to measure molecules that have disparate physical properties (e.g., ranging in polarity from very water soluble organic acids to very nonpolar lipids [Kuehnbaum and Britz-McKibbin 2013]). Accordingly, comprehensive metabolomic technology platforms typically take the strategy of dividing the metabolome into subsets of metabolites—often based on compound polarity, common functional groups, or structural similarity—and devise specific sample preparation and analytical procedures optimized for each, as illustrated in Figure 1. The metabolome is therefore measured as a patchwork of results from different analytical methods. As an emerging field that has been enabled, at least in part, by steady development of analytical instrumentation with new capabilities each year, the methods used in metabolomics continue to evolve and improve (van der Greef et al. 2013). However, a consequence of metabolomics laboratories using multiple procedures that are potentially subject to frequent refinement is that individual laboratories tend to have unique methods and there are comparatively few standard operating procedures commonly adopted across laboratories. Although this diversity of technologies is linked to innovation in the field, it lends itself to potential challenges when comparing data between laboratories because of issues like differences in precision of measurement for select classes of metabolites or nonoverlapping metabolite coverage. In addition, the degree of certainty in metabolite identification can vary among methods, ranging from metabolite identities rigorously confirmed using authentic reference standards to putative identifications made using reference databases to signals that remain as “unknowns.” The need for standardization in metabolomics has been appreciated by its practitioners and has given rise to a number of initiatives toward realizing this aim, such as the Metabolomics Standards Initiative to develop guidelines for data reporting (Sansone et al. 2007, Sumner et al. 2007); ring tests to evaluate the capabilities of different metabolomics methods and laboratories to obtain similar results (Martin et al. 2015); and open-access repositories for metabolomics results and related metadata such as the MetaboLights database in Europe (http://www.ebi.ac.uk/metabolights) (Haug et al. 2013) and the Metabolomics Workbench (http://www.metabolomicsworkbench.org) funded by the National Institutes of Health Common Fund in the United States.
Figure 1.
Illustration of the liquid chromatography–mass spectrometry (LC-MS)-based metabolomics platform used at the Broad Institute of MIT and Harvard. Our laboratory has developed a comprehensive metabolomics platform that uses targeted and nontargeted LC-MS methods to measure lipids, metabolites of intermediate polarity such as free fatty acids and bile acids, and polar metabolites. IPA, isopropanol; RP, reversed phase; HILIC, hydrophilic interaction liquid chromatography.
As the field matures over time, it is conceivable that there might be convergence among laboratories on particular standard operating procedures, but in the meantime we must evaluate methods and the results they generate on a case-by-case basis. Necessary information that can aid this evaluation is comprehensive descriptions of methods, including details such as instrument settings and the means by which each metabolite was identified; data from repeated measures of representative samples, such as pooled samples, to enable determination of coefficients of variation for each metabolite measured by a particular method; and access to data sets submitted either as electronic supplements or to metabolomics data repositories such as those described above.
DEFINING THE METABOLOME AND IDENTIFICATION OF NEW DISEASE INDICATORS
While we remain in the midst of metabolomics technology refinement, we are still learning about what actually constitutes the human metabolome. A recent estimate of the entire complement of small molecules expected to be found in the human body exceeds 19,000 (Wishart et al. 2013). This number includes not only metabolites directly linked to endogenous enzymatic activities encoded by the human genome, but also those derived from food, medications, the microbiota that inhabit the body, and the environment. Our dependence on diet as a source for nine of the 20 amino acids for which there are codons in the human genome but no endogenous biosynthetic route is an example that highlights why it is important to account for “exogenous” metabolites in our study of the metabolome. Although broadening the scope of analyses to measure more metabolites increases the difficulty level, it is clear that comprehensive metabolomics heralds exciting new opportunities for discovery. Metabolomics is an objective lens to view the complex nature of how physiology is linked to external events and conditions, as well as measure its response to perturbations such as those associated with disease.
Metabolites have been described as proximal reporters of disease because their abundances in biological specimens are often directly related to pathogenic mechanisms (Gerszten and Wang 2008) and this concept is routinely demonstrated in clinical chemistry laboratory results. Historically, often a decade or more could pass between the initial discovery of a disease marker, its validation in human trials, and its routine implementation as a clinical test. To realize the potential of precision medicine, we need to accelerate the discovery of specific markers of disease and drug pharmacodynamics, as well as metabolite profiles associated with external environment and their associations with disease risk. Current metabolomics technologies can enable more rapid discovery and validation of metabolic indicators of disease. Techniques used in metabolomics, such as liquid chromatography–mass spectrometry (LC-MS), can routinely measure tens to hundreds of metabolites with excellent precision and are suitable for discovery studies in human cohorts. Confidence comes from experience with recent applications to find early metabolic indicators of disease in longitudinal cohorts years before symptoms are clinically apparent—for example, in pancreatic cancer (Mayers et al. 2014), type 2 diabetes (Rhee et al. 2011, Wang et al. 2011; 2013), memory impairment (Mapstone et al. 2014), and many other conditions. Metabolomics studies have also inspired work revealing novel insights into relationships between diet and disease, such as observations linking elevated branched chain amino acids and obesity to insulin resistance (Newgard et al. 2009). Bringing the microbiome into the mix, metabolite profiling studies by Koeth et al. (2013) recently showed that elevated plasma levels of trimethylamine-N-oxide are at the nexus of a relationship among diets abundant in red meat, the composition of the gut microbiome, and risk for cardiac events. Following the discovery of metabolic indicators of disease, it is relatively straightforward to create specific assays for metabolites of interest to enable follow-up studies, mainly because the discovery method can serve as a starting point and different technology may not be required. For this reason, the lag time between marker discovery and deployment of specific assay for clinical use may be brief. Finally, as proximal reporters of disease, metabolic indicators are well suited to serve as efficacy markers during drug development, which will be further enabled as more markers are discovered.
PROBING METABOLISM IN VIVO USING TRACERS
Specific aspects of metabolism that are centrally important to disease can be probed directly and dynamically in vivo using labeled tracer substrates. Tissues that are characterized by significant glucose metabolism, such as malignant tumors and brain tissue, can be imaged radiologically using 18F-fluorodeoxyglucose-positron emission tomography (18F-FDG-PET). An important application of this technique is diagnosing and staging malignancies, as well as measuring responses to therapies, but the tool extends to other pathologies such as inflammation (Hess et al. 2014). Nonradioactive, stable isotope-labeled tracers are also powerful metabolic probes, offering good safety profiles for in vivo dosing and the opportunity to measure the metabolism of just about any biological molecule that can be synthesized. Using techniques such as 13C hyperpolarization magnetic resonance, metabolism can be monitored in vivo (Brindle 2015). Following administration of stable isotope-labeled substrates, it can also be informative to collect biological fluids or tissues, via biopsy or surgical resection, and apply analytical methods to samples ex vivo to finely trace metabolism of the substrate. For example, Sellers et al. (2015) recently reported results from a 13C6-glucose tracer study in human subjects with non-small-cell lung cancer (NSCLC) that revealed enhanced pyruvate carboxylase pathway activity, as read out by increased levels of the downstream metabolite 13C3-aspartate, occurred in cancer bearing lung tissue. Additional experiments confirmed that pyruvate carboxylase activity was essential for NSCLC proliferation and tumorigenesis. This effort therefore serves as a valuable example of how metabolic tracer studies may reveal potentially novel therapeutic targets in precision medicine.
CLOSING THOUGHTS
It is still early with respect to the application of metabolomics in precision medicine and its impact will be ultimately be demonstrated by improvements in care for individuals. A developing story that demonstrates potential for metabolomics began with the characterization of cells expressing a mutant isoform of cytosolic isocitrate dehydrogenase (IDH1), R132H, that had been found to occur at high frequency in gliomas and glioblastomas. Dang et al. (2009) investigated the metabolic consequences of this mutation by applying metabolomics to extracts from cells transfected with either wild-type IDH1 or R132H mutant IDH1 and observed significantly higher levels (>20-fold) of 2-hydroxyglutarate (2HG), a known component of the metabolome but of unknown physiologic importance. Additional experiments showed >10-fold higher concentrations of 2HG in human malignant glioma tissues bearing IDH1 mutations relative to those with wild-type IDH1 (Dang et al. 2009) and, subsequently, it was found that other mutant isoforms of IDH associated with cancer also produce 2HG (Gross et al. 2010). Presently, 2HG is being measured as a pharmacodynamic marker in clinical trials designed to test inhibitors of mutant IDH (Dimitrov et al. 2015). As metabolomics finds wider application in lines of investigation similar to this example, more stories will surely emerge. It will be exciting to both share results from and follow these stories in the pages of this new journal.
REFERENCES
- Brindle KM. 2015. Imaging metabolism with hyperpolarized (13)C-labeled cell substrates. J Am Chem Soc 137: 6418–6427. [DOI] [PubMed] [Google Scholar]
- Clarke SF, Foster JR. 2012. A history of blood glucose meters and their role in self-monitoring of diabetes mellitus. Br J Biomed Sci 69: 83–93. [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov L, Hong CS, Yang C, Zhuang Z, Heiss JD. 2015. New developments in the pathogenesis and therapeutic targeting of the IDH1 mutation in glioma. Int J Med Sci 12: 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerszten RE, Wang TJ. 2008. The search for new cardiovascular biomarkers. Nature 451: 949–952. [DOI] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. 2010. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 207: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie R, Susi A. 1963. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 32: 338–343. [PubMed] [Google Scholar]
- Haug K, Salek RM, Conesa P, Hastings J, de Matos P, Rijnbeek M, Mahendraker T, Williams M, Neumann S, Rocca-Serra P, et al. 2013. MetaboLights—an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res 41: D781–D786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S, Blomberg BA, Zhu HJ, Hoilund-Carlsen PF, Alavi A. 2014. The pivotal role of FDG-PET/CT in modern medicine. Acad Radiol 21: 232–249. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. 2013. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19: 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehnbaum NL, Britz-McKibbin P. 2013. New advances in separation science for metabolomics: resolving chemical diversity in a post-genomic era. Chem Rev 113: 2437–2468. [DOI] [PubMed] [Google Scholar]
- Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, Hall WJ, Fisher SG, Peterson DR, Haley JM, et al. 2014. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med 20: 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JC, Maillot M, Mazerolles G, Verdu A, Lyan B, Migne C, Defoort C, Canlet C, Junot C, Guillou C, et al. 2015. Can we trust untargeted metabolomics? Results of the metabo-ring initiative, a large-scale, multi-instrument inter-laboratory study. Metabolomics 11: 807–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, Yuan C, Bao Y, Townsend MK, Tworoger SS, et al. 2014. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med 20: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. 2009. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9: 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O'Donnell CJ, et al. 2011. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest 121: 1402–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone SA, Fan T, Goodacre R, Griffin JL, Hardy NW, Kaddurah-Daouk R, Kristal BS, Lindon J, Mendes P, Morrison N, et al. 2007. The metabolomics standards initiative. Nat Biotechnol 25: 846–848. [DOI] [PubMed] [Google Scholar]
- Sellers K, Fox MP, Bousamra M II, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, et al. 2015. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest 125: 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, et al. 2007. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Greef J, van Wietmarschen H, van Ommen B, Verheij E. 2013. Looking back into the future: 30 years of metabolomics at TNO. Mass Spectrom Rev 32: 399–415. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. 2011. Metabolite profiles and the risk of developing diabetes. Nat Med 17: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, Ghorbani A, O'Sullivan J, Cheng S, Rhee EP, et al. 2013. 2-aminoadipic acid is a biomarker for diabetes risk. J Clin Invest 123: 4309–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. 2013. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res 41: D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]