Abstract
PURA is the leading candidate gene responsible for the developmental phenotype in the 5q31.3 microdeletion syndrome. De novo mutations in PURA were recently reported in 15 individuals with developmental features similar to the 5q31.3 microdeletion syndrome. Here we describe six unrelated children who were identified by clinical whole-exome sequencing (WES) to have novel de novo variants in PURA with a similar phenotype of hypotonia and developmental delay and frequently associated with seizures. The protein Purα (encoded by PURA) is involved in neuronal proliferation, dendrite maturation, and the transport of mRNA to translation sites during neuronal development. Mutations in PURA may alter normal brain development and impair neuronal function, leading to developmental delay and the seizures observed in patients with mutations in PURA.
Keywords: central hypotonia, generalized clonic seizures, generalized tonic seizures, severe global developmental delay
INTRODUCTION
Whole-exome sequencing (WES) provides a comprehensive strategy to identify pathogenic genetic variants in patients with developmental abnormalities (Veltman and Brunner 2012; Yang et al. 2013, 2014). The 5q31.3 microdeletion syndrome is associated with hypotonia, feeding difficulties, severe developmental delay, and epilepsy. Genomic 5q31.3 deletions range in size from 101 kb to 5 Mb and include the purine-rich element binding protein A (PURA), a candidate gene for the developmental manifestations (Shimojima et al. 2011; Hosoki et al. 2012; Brown et al. 2013). PURA is essential for normal brain development, synapse formation and proliferation of neurons, oligodendrocytes, and astrocytes in the central nervous system (Khalili et al. 2003; Johnson et al. 2006; Hokkanen et al. 2012). Mutations in PURA have previously been associated with moderate to severe developmental delay, learning disability, hypotonia, neonatal respiratory issues, feeding difficulties, and seizures or “seizure-like” movements in 15 patients (Hunt et al. 2014; Lalani et al. 2014). Here we describe six unrelated patients with de novo mutations in PURA identified through WES associated with the consistent phenotype of hypotonia and developmental delay and frequently associated with seizures.
RESULTS
Clinical Presentation
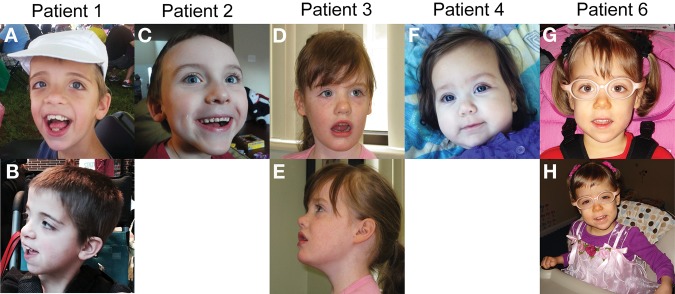
Six unrelated patients all share similar clinical features of hypotonia and developmental delay (Table 1). The children range in age from 6 mo to 15 yr, and none of the children are functionally verbal. There is no history of regression in any of the children. Patient 1 had poor suck, bradycardia, and apnea during the neonatal period. “Seizure-like” activity was observed in two patients. One had “seizure-like” activity (twitching, stiffening, staring spells, collapsing) with a normal EEG, and a second developed myoclonic movements and possible gelastic seizures at 3 yr of age. One child has a history of infantile spasms. Vision has been variably and minimally impaired in five of the children. All have normal hearing. One child also has mild osteopenia and a history of fractures. There is no evidence of other significant medical problems or birth defects. Four of the children are dysmorphic with varying features (Fig. 1). Two of the patients have only minor features such as epicanthal folds or highly arched palate, while two other patients have more significant findings such as dolichocephaly, hypertelorism, broad forehead, and persistent fetal pads. Height and weight are within the normal range. Head circumferences ranged from less than the 10th to the 97th percentile. Brain MRI demonstrates delayed myelination and nonspecific enlargement of the subarachnoid spaces, cortical sulci, and ventricular system in one patient, periventricular white matter changes reflective of a stroke in the second patient, periventricular leukomalacia in the third, and mild corpus callosum volume loss in the fourth. Three of the patients have constipation and one also has gastroesophageal reflux disease. All children had clinical WES as part of a trio analysis wherein the proband and both parents were sequenced. There was no known history of consanguinity in any of the families.
Table 1.
Clinical features of patients with mutations in PURA
| Patient | Age | Sex | Mutation | Hypotonia | Head circumference | Age at sitting | Age at walking | Verbal skills | Vision | Brian MRI | EEG | Seizure |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 8 yr | M | c.563T>C | Y | 97th %ile | 1 yr | 3 yr | One word approximations | Normal | Slightly delayed myelination | Normal | “Seizure-like” activity |
| p.Ile188Thr | ||||||||||||
| 2 | 4 yr | M | c.768dupC | Y | Unknown | 1 yr | 4 yr with a walker | Nonverbal | Optic nerve pallor, esotropia | White matter changes | Normal | None |
| p.Ile257Hisfs*37 | ||||||||||||
| 3 | 10 yr | F | c.1A>T | Y | 50th %ile | 1 yr | 4 yr | Nonverbal | Strabismus, esotropia | Normal | Unknown | Myoclonic, possible gelatic seizures |
| p.Met1? | ||||||||||||
| 4 | 6 mo | F | c.697_699delTTC | Y | 5–10th %ile | N/A | N/A | N/A | Cortical visual impairment | Periventricular leukomalacia | Normal | None |
| p.Phe233del | ||||||||||||
| 5 | 15 yr | F | c.4_8delGCGGA | Y | 25–50th %ile | Unknown | 4 yr | Nonverbal | Exotropia | Mild corpus callosum volume loss | Abnormal | Infantile spasms |
| p.A1a2Profs*197 | ||||||||||||
| 6 | 5 yr | F | 302_310delCTCTCTCCA | Y | 33rd %ile | 5 yr | Not walking | Nonverbal | Myopia and strabismus | Normal | Normal | Spasticity in foot and lower extremities |
| p.Thre101_Ser103del | ||||||||||||
| Hunt et al. #1 | 4 yr | F | c.726_727delGT | Y | 9–25th %ile | 2 yr | Nonambulatory | Nonverbal | Nystagmus with preserved optokinetic reflex, dysconjugate gaze | Delayed myelination | Normal | “Seizure-like” episodes |
| p.Phe243Tyrfs*50 | ||||||||||||
| Hunt et al. #2 | 14 yr | F | c.847delG | N | <0.4th %ile | 1 yr | 2 yr | Sentences; limited vocabulary | Normal | Normal | N/A | None |
| p.Glu283Argfs*45 | ||||||||||||
| Hunt et al. #3 | 12 yr | F | c.616A>T | Y | 75–91st %ile | 1 yr | 1 yr | Short phrases, repetitive | Normal | Normal | Abnormal | “Seizure-like” episodes |
| p.Ile206Phe | ||||||||||||
| Hunt et al. #4 | 6 yr | F | c.697_699delTTC | Y | 75th %ile | Cannot sit unassisted | Nonambulatory | Nonverbal | Early cortical visual impairment, eye movements dysconjugate | Delayed myelination | Abnormal | Infantile spasms, progressed to tonic, focal dyscognitive seizures |
| p.Phe233del | ||||||||||||
| Lalani et al. #1 | 6 mo | M | c.812_814delTCT | Y | 86th %ile | Cannot sit unassisted | N/A | N/A | Normal | Normal | Abnormal | Infantile myoclonus, partial onset with secondary clonic generalization |
| p.Phe271del | ||||||||||||
| Lalani et al. #2 | 7 mo | M | c.307_308delTC | Y | 20th %ile | Cannot sit unassisted | N/A | N/A | Intermittent exotrophia | Dysmorphic corpus callosum | N/A | None |
| p.Ser103Hisfs*97 | ||||||||||||
| Lalani et al. #3 | 10 mo | M | c.556C>T | Y | 73rd %ile | Cannot sit unassisted | N/A | N/A | Normal | Normal | Normal | “Seizure-like” episodes in the neonatal period, myoclonic jerks |
| p.Gln186* | ||||||||||||
| Lalani et al. #4 | 1 yr | F | c.289A>G | Y | 65th %ile | 10 mo | Nonambulatory | Nonverbal | Strabismus | Normal | N/A | Myoclonic jerks, exaggerated startle |
| p.Lys97Glu | ||||||||||||
| Lalani et al. #5 | 4 yr | F | c.299T>C | Y | 66th %ile | 1 yr | Nonambulatory | Nonverbal | Nystagmus, Brown syndrome | Mild myelin maturation delay | N/A | Single generalized tonic clonic seizure at 5 mo |
| p.Leu100Pro | ||||||||||||
| Lalani et al. #6 | 2 yr | F | c.363C>G | Y | 91st %ile | 1 yr | Non-ambulatory | Nonverbal | Normal | Absent septum pellucidum | Abnormal | Myoclonic jerks |
| p.Tyr121* | ||||||||||||
| Lalani et al. #7 | 2 yr | F | c.783C>G | Y | 76th %ile | 1 yr | Non-ambulatory | Nonverbal | Left eye estropia | Normal | Abnormal | Seizure-like episode in the neonatal period, myoclonic jerks |
| p.Tyr261* | ||||||||||||
| Lalani et al. #8 | 5 yr | F | c.470T>A | Y | 85th %ile | Cannot sit unassisted | Nonambulatory | Nonverbal | Nystagmus, strabismus | Normal | N/A | None |
| p.Mct157Lys | ||||||||||||
| Lalani et al. #9 | 12 yr | M | c.265G>C | Y | >97th %ile | Cannot sit unassisted | Nonambulatory | Nonverbal | Nystagmus | Hypomyelination | Abnormal | Myoclonic, generalized tonic and atonic seizures |
| p.Ala89Pro | ||||||||||||
| Lalani et al. #10 | 12 yr | F | c.263_265delTCG | Y | 50th %ile | 2 yr | 3 yr | Nonverbal | Strabismus, myopia | Normal | Abnormal | Lennox-Gastaut syndrome, intractable generalized epilepsy |
| p.Ile88_Ala89delins Thr | ||||||||||||
| Lalani et al. #11 | 15 yr | F | c.596G>C | Y | N/A | Unknown | Nonambulatory | Nonverbal | Nystagmus, extremely farsighted | N/A | Abnormal | Lennox-Gastaut syndrome, myoclonic drops, tonic clonic, startle seizures |
| p.Arg199Pro |
MRI, magnetic resonance imaging; EEG, electroencephalogram.
Figure 1.
Photographs of patients. (A,B) Patient 1. (C) Patient 2. (D,E) Patient 3. (F) Patient 4. (G,H) Patient 6. Patients 1 and 2 exhibit dolichocephaly and Patients 1–5 all have a broad forehead. Note hypertelorism and highly arched palate in Patient 1 and epicanthal folds in Patient 4.
Exome sequencing produced an average of ∼13 Gb of sequence per sample (Table 2). The mean coverage of captured regions was ∼150× per sample with >98% covered with at least 10× coverage, an average of 92% of base call quality of Q30 or greater, and an overall average mean quality score of >Q36. Filtering of common SNPs (>10% frequency present in 1000 Genomes database) resulted in ∼4500 variants per proband sample. In total, 232 genes (257 unique sequence changes) of interest were identified across the six families when considering all possible modes of inheritance. Evaluation of these 232 genes eliminated 231 genes lacking clinical overlap with the patients’ phenotype, leaving one gene.
Table 2.
Sequencing results
| Patient | 10× cov. (%) | Mean cov. | Yield (Gb) | Q30 | MeanQ | Filtered vars | PURA mean CDS cov. | Var. total fam. cov. | Samples | Mean per-sample var. cov. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 98.57 | 122 | 14.6 | 88 | 34 | 4973 | 105 | 491 | 3 | 164 |
| 2 | 98.79 | 166 | 12.5 | 94 | 36 | 4057 | 128 | 721 | 3 | 240 |
| 3 | 98.61 | 165 | 11.6 | 95 | 37 | 4475 | 143 | 63 | 3 | 21 |
| 4 | 98.71 | 201 | 14.6 | 88 | 35 | 4613 | 149 | 811 | 3 | 270 |
| 5 | 98.45 | 155 | 11.1 | 93 | 36 | 4504 | 125 | 41 | 3 | 14 |
| 6 | 97.22 | 122 | 12.3 | 91 | 36 | 5445 | 70 | 244 | 3 | 81 |
| Mean | 98.39 | 155 | 13 | 92 | 36 | 4678 | 120 | 395 | 3 | 132 |
cov., coverage; CDS, coding sequence; var., variance; fam, family.
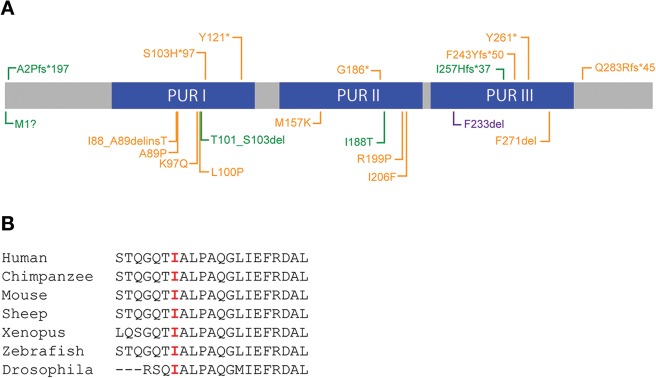
Novel de novo heterozygous variants in PURA were detected by WES and confirmed by Sanger sequencing in six affected children (Table 1 and Fig. 2). The PURA p.Ile188Thr (c.563T>C) mutation in Patient 1 represents a nonconservative amino acid substitution of a hydrophobic residue replaced by a polar residue. Amino acid isoleucine 188 is highly conserved throughout evolution (Fig. 2B). The c.768dupC (p.Ile257Hisfs*37) mutation in Patient 2 produces a frameshift starting with codon isoleucine 257 that changes this amino acid to a histidine residue and creates a premature termination codon that deletes the last 66 amino acids after the insertion of 36 novel amino acids. This mutation is predicted to cause a loss of normal protein function through protein truncation. In Patient 3, the de novo c.1A>T (p.Met1?) mutation alters the initiator methionine codon (Met1). It is not known whether the loss of Met1 disrupts all protein translation from that allele or whether an abnormal protein is produced using an alternate Met start codon. The c.697_699delTTC (p.Phe233del) mutation in Patient 4 is an in-frame deletion, previously reported in a patient by Hunt et al. (2014). The c.4_8delGCGGA (p.Ala2Profs*197) mutation identified in Patient 5 causes a frameshift starting with codon alanine 2, changes this amino acid to a proline residue and creates a premature termination codon and replaces the last 321 residues with 196 different ones. This mutation is predicted to cause a loss of normal protein function through protein truncation. Patient 6 has a c.302_310delCTCTCTCCA (p.Thr101_Ser103del) in-frame deletion of three amino acids in the highly conserved Pur I domain. None of the de novo variants identified in these six patients were detected in the Database of Single Nucleotide Polymorphisms (dbSNP; http://www.ncbi.nlm.nih.gov/SNP/), 1000 Genomes (1000G; http://www.1000genomes.org/), Exome Aggregation Consortium (ExAc; http://exac.broadinstitute.org), or Exome Variant Server (ESP; http://evs.gs.washington.edu/EVS/). All six mutations are located in highly conserved regions and all except for the mutation of Patient 3 are predicted to be deleterious by SIFT (http://sift.jcvi.org/), CADD (http://cadd.gs.washington.edu/), and MutationTaster (http://www.mutationtaster.org/). The de novo c.1A>T (p.Met1?) mutation in Patient 3 is presumed deleterious by CADD and Mutation Taster.
Figure 2.
Mutations in PURA. (A) Diagram of Purα with mutations identified in our patients in green. Previously identified mutations are in orange (Hunt et al. 2014; Lalani et al. 2014). Missense and in-frame deletion mutations are shown below the protein diagram and gene disrupting truncating mutations are shown above. (B) Sequence alignment of Ile188 in PUR II region illustrating conservation of amino acids at amino acid 188.
DISCUSSION
Six unrelated patients ranging in age from 6 mo to 15 yr with common clinical features of hypotonia and developmental delay were all found to have heterozygous de novo predicted deleterious rare or novel variants in PURA, identified through WES (Table 1). PURA is the leading candidate gene responsible for the developmental phenotype in the 5q31.3 microdeletion syndrome (Brown et al. 2013; Hunt et al. 2014; Lalani et al. 2014). Two papers recently reported de novo mutations in PURA in 15 individuals with developmental features equivalent to the 5q31.3 microdeletion syndrome (Hunt et al. 2014; Lalani et al. 2014). A separate large-scale WES study implicated 12 novel genes enriched for damaging de novo mutations with evidence for a role in developmental disorders (The Deciphering Developmental Disorders Study 2015). The analysis of the exomes of 1133 children with severe, undiagnosed developmental disorders and their parents identified PURA in three of the patients. We report six additional unrelated patients with de novo mutations in PURA to provide further evidence that PURA is largely, if not solely, responsible for the developmental delay, hypotonia, and seizures observed in the 5q31.3 microdeletion syndrome. Our additional patients expand the number and location of mutations in PURA and the associated phenotypes. Four of our patients had variants in the PUR domains, which are highly conserved throughout evolution (Fig. 2).
PURA is located on 5q31.2 and is encoded by a single exon that encodes a highly conserved multifunctional protein, Purα. PURA is expressed ubiquitously, including the brain, muscle, heart, and blood. Purα is a member of the Pur family of nucleic acid binding proteins which consist of a glycine-rich flexible amino terminus, a central core region and a potential carboxy-terminal protein binding region. All human Pur proteins have three highly conserved sequence-specific repeats, Pur repeats I–III, of 64–80 amino acids that are the hallmark of the Pur proteins. Purα has helix-unwinding capability (Khalili et al. 2003) and has been shown to bind specific sequences of ssDNA, dsRNA, and ssRNA with preference for GGN-repeats (Gallia et al. 2000; Graebsch et al. 2009; Johnson et al. 2013) to regulate a variety of cellular processes including DNA replication, gene transcription, RNA transport, and mRNA translation (White et al. 2009; Johnson et al. 2013).
Studies on mice have shown that Purα is involved in neuronal proliferation, dendrite maturation, and the transport of mRNA to translation sites in hippocampal neurons (Khalili et al. 2003; Kanai et al. 2004; Johnson et al. 2006; Hokkanen et al. 2012). Two independently generated knockout mouse models demonstrate that mice appear normal at birth but develop neurologic features, including ataxic gait, hind limb weakness, and continuous and increasingly severe tremors (Khalili et al. 2003; Hokkanen et al. 2012). The PURA knockout mice eat and sleep normally but do not gain weight. Throughout postnatal development, these mice exhibit mislamination of the cerebellum and cerebrum and low numbers of Purkinje cells in the hippocampus and cerebellum associated with uncoordinated movements, tremors, and lethargy with death by 21–25 d. Heterozygous PURA+/− mice appeared normal but exhibited neurologic and myeloid defects that were intermediate in severity compared with PURA−/− mice. Both PURA−/− and PURA+/− mice had myeloid defects with reduced splenic monocyte development; however, the effect on PURA+/− mice was not as severe as the knockout mice due to PURA haploinsufficiency. Heterozygous PURA+/− mice also had occasional spontaneous seizures upon handling (Khalili et al. 2003; Hokkanen et al. 2012).
In the central nervous system, Purα has been detected in large neuronal mRNA-containing complexes with the fragile X mental retardation protein (FMRP), encoded by FMR1, an RNA-binding protein that is required for normal neural development. Johnson et al. (2006, 2013) showed that Purα is specifically expressed in the dendrites of hippocampal neurons in rats, and colocalizes with FMRP at dendritic junctional translation sites. Purα also binds to CGG repeats in FMR1, which is silenced in individuals with fragile X syndrome (FXS) and is overexpressed in patients with fragile X–associated tremor/ataxia syndrome (FXTAS). It is possible that impaired binding of Purα to CGG repeats in FMR1 may play a role in disease progression, resulting in defective neural and brain development in both settings.
Purα is also known to regulate expression of the myelin proteolipid protein Plp1, which is the predominant structural component of myelin sheaths in the central nervous system (Dobretsova et al. 2008). This interaction may be related to the delayed and decreased myelination observed in the brain MRIs. Mutations in PLP1 are associated with Pelizaeus–Merzbacher disease (PMD; MIM#312080) (Torii et al. 2014). We hypothesize that abnormal or decreased binding of Purα to its targets may be responsible for defective brain development and function and related to the seizures seen in patients with mutations in PURA. Additional studies on patients with PURA mutations are necessary to better understand the correlation between genotype and phenotype and further investigation of the molecular mechanism of PURA during brain development and function.
METHODS
Whole-Exome Sequencing
Genomic DNA was extracted from whole blood from 1098 affected children with developmental delay and their parents. Exome sequencing was performed on exon targets isolated by capture using the Agilent SureSelect Human All Exon V4 (50-Mb) kit (Agilent Technologies). One microgram of DNA from blood specimen was sheared into 350–400-bp fragments, which were then repaired, ligated to adaptors, and purified for subsequent PCR amplification. Amplified products were then captured by biotinylated RNA library baits in solution following the manufacturer's instructions. Bound DNA was isolated with streptavidin-coated beads and reamplified. The final isolated products were sequenced using the Illumina HiSeq 2000 or 2500 sequencing system with 100-bp paired-end reads (Illumina). DNA sequence was mapped to the published human genome build UCSC hg19/GRCh37 reference sequence using BWA with the latest internally validated version at the time of sequencing, progressing from BWA v0.5.8 through BWA-Mem v0.7.8 (Li and Durbin 2009; Li 2012). Targeted coding exons and splice junctions of known protein-coding RefSeq genes were assessed for average depth of coverage with a minimum depth of 10× required for inclusion in downstream analysis. Local realignment around insertion-deletion sites was performed using the Genome Analysis Toolkit v1.6 (DePristo et al. 2011). Variant calls were generated simultaneously on all sequenced family members using SAMtools v0.1.18 (Li et al. 2009). All coding exons and surrounding intron/exon boundaries were analyzed. Automated filtering removed common sequence changes (defined as >10% frequency present in 1000 Genomes database). The targeted coding exons and splice junctions of the known protein-coding RefSeq genes were assessed for the average depth of coverage and data quality threshold values. Whole-exome sequence data for all sequenced family members were analyzed using GeneDx's XomeAnalyzer (a variant annotation, filtering, and viewing interface for WES data), which includes nucleotide and amino acid annotations, population frequencies (NHLBI Exome Variant Server and 1000 Genomes databases), in silico prediction tools, amino acid conservation scores, and mutation references. Variants were filtered based on inheritance patterns, gene lists of interest, phenotype and population frequencies, as appropriate. Resources including the Human Gene Mutation Database (HGMD), 1000 Genomes database, NHLBI Exome Variant Server, OMIM, PubMed, and ClinVar were used to evaluate genes and detected sequence changes of interest (Table 3). Additional searches were performed using specific gene lists related to ID. Identified sequence changes of interest were confirmed in all members of the trio by conventional di-deoxy DNA sequence analysis using an ABI3730 (Life Technologies) and standard protocols with a new DNA preparation.
Table 3.
Variants identified from whole-exome sequencing of six families
| Filtering results | Manual review | Resulting genes of interest | |
|---|---|---|---|
| Homozygous (# seq changes) | 80 (83) | 0 (0) | 0 (0) |
| Compound heterozygous (# seq changes) | 15 (32) | 5 (10) | 0 (0) |
| De novo (# seq changes) | 127 (132) | 1 (1) | 1 (1) |
| X-linked genes (# seq changes) | 10 (10) | 1 (1) | 0 (0) |
| Total genes (# seq changes) | 232 (257) | 7 (12) | 1 (1) |
ADDITIONAL INFORMATION
Ethics Statement
The study was approved by the Institutional Review Board of Columbia University and written consent was obtained for collecting blood samples and sequencing from all study participants.
Data Deposition and Access
Whole-exome sequencing data is not publicly available because patient consent could not be obtained. The PURA variants found in this study have been deposited in ClinVar under accession numbers SCV000223979, SCV000223981, SCV000223986, SCV000223987, SCV000223989, and SCV000223991.
Acknowledgments
We thank the families for their generous contribution.
Author Contributions
A.J.T. analyzed the data, drafted, and critically reviewed the manuscript. R.B. analyzed the data and critically reviewed the manuscript. M.T.C. analyzed the data and critically reviewed the manuscript. K.A.-Y. provided the clinical data and critically reviewed the manuscript. P.A. provided the clinical data and critically reviewed the manuscript. A.L.W. provided the clinical data and critically reviewed the manuscript. F.K. provided the clinical data and critically reviewed the manuscript. B.H. provided the clinical data and critically reviewed the manuscript. T.M. provided the clinical data and critically reviewed the manuscript. M.N. provided the clinical data and critically reviewed the manuscript. K.R. generated and analyzed the data and critically reviewed the manuscript. J.J. analyzed the data and critically reviewed the manuscript. W.K.C. conceived of the study, analyzed the data, drafted and critically reviewed the manuscript.
Funding
This work was generously supported by a grant from the Simons Foundation.
Competing Interest Statement
R.B., K.R., and J.J. are employees of GeneDx. M.T.C. is an employee of BioReference Laboratories. W.K.C. is a consultant to BioReference Laboratories.
REFERENCES
- Brown N, Burgess T, Forbes R, McGillivray G, Kornberg A, Mandelstam S, Stark Z. 2013. 5q31.3 Microdeletion syndrome: clinical and molecular characterization of two further cases. Am J Med Genet A 161A: 2604–2608. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsova A, Johnson JW, Jones RC, Edmondson RD, Wight PA. 2008. Proteomic analysis of nuclear factors binding to an intronic enhancer in the myelin proteolipid protein gene. J Neurochem 105: 1979–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallia GL, Johnson EM, Khalili K. 2000. PURα: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res 28: 3197–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graebsch A, Roche S, Niessing D. 2009. X-ray structure of Pur-α reveals a Whirly-like fold and an unusual nucleic-acid binding surface. Proc Natl Acad Sci 106: 18521–18526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokkanen S, Feldmann HM, Ding H, Jung CK, Bojarski L, Renner-Muller I, Schuller U, Kretzschmar H, Wolf E, Herms J. 2012. Lack of Pur-α alters postnatal brain development and causes megalencephaly. Hum Mol Genet 21: 473–484. [DOI] [PubMed] [Google Scholar]
- Hosoki K, Ohta T, Natsume J, Imai S, Okumura A, Matsui T, Harada N, Bacino CA, Scaglia F, Jones JY, et al. 2012. Clinical phenotype and candidate genes for the 5q31.3 microdeletion syndrome. Am J Med Genet A 158A: 1891–1896. [DOI] [PubMed] [Google Scholar]
- Hunt D, Leventer RJ, Simons C, Taft R, Swoboda KJ, Gawne-Cain M; DDD study, Magee AC, Turnpenny PD, Baralle D. 2014. Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet 51: 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, Khalili K, Winckler B, Gordon J. 2006. Role of Pur α in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res 83: 929–943. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Daniel DC, Gordon J. 2013. The Pur protein family: genetic and structural features in development and disease. J Cell Physiol 228: 930–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43: 513–525. [DOI] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, et al. 2003. Purα is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol 23: 6857–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Zhang J, Schaaf CP, Brown CW, Magoulas P, Tsai AC, El-Gharbawy A, Wierenga KJ, Bartholomew D, Fong CT, et al. 2014. Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet 95: 579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2012. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28: 1838–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima K, Isidor B, Le Caignec C, Kondo A, Sakata S, Ohno K, Yamamoto T. 2011. A new microdeletion syndrome of 5q31.3 characterized by severe developmental delays, distinctive facial features, and delayed myelination. Am J Med Genet A 155A: 732–736. [DOI] [PubMed] [Google Scholar]
- The Deciphering Developmental Disorders Study. 2015. Large-scale discovery of novel genetic causes of developmental disorders. Nature 12: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii T, Miyamoto Y, Tamauchi J, Tanoue A. 2014. Pelizaeus–Merzbacher disease: cellular pathogenesis and pharmacologic therapy. Pediatr Int 56: 659–666. [DOI] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. 2012. De novo mutations in human genetic disease. Nat Rev Genet 13: 565–575. [DOI] [PubMed] [Google Scholar]
- White MK, Johnson EM, Khalili K. 2009. Multiple roles for PURα in cellular and viral regulation. Cell Cycle 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, et al. 2013. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med 369: 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, et al. 2014. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312: 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]