Abstract

Lantibiotics are a group of ribosomally synthesized and post-translationally modified peptides (RiPPs) exhibiting antimicrobial activity. They are characterized by the presence of the thioether-containing bisamino acids lanthionine and methyllanthionine. Here, we report a two-component lantibiotic from Bacillus cereus SJ1 with unusual structural features that we named bicereucin. Unlike all previous two-component lantibiotics, only one of the two peptides of bicereucin contains a lanthionine. The second peptide lacks any cysteines but contains several d-amino acids. These are installed by the dehydrogenase BsjJB, the activity of which was successfully reconstituted in vitro. The proteolytic removal of the leader peptide was also performed in vitro. Bicereucin displayed synergistic antimicrobial activities against Gram-positive strains including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci as well as hemolytic activity. To illustrate the utility of the enzymes, an analog of the d-amino acid containing opioid dermorphin was successfully produced in E. coli by employing the dehydratase BsjM and the dehydrogenase NpnJA.
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a major class of natural products, as revealed by the genome sequencing efforts of the past decade.1 One of the best-studied RiPP classes are the lanthipeptides, which are characterized by cross-linked thioether structures named lanthionine (Lan) and methyl-lanthionine (MeLan) (Figure 1a). They are formed by dehydration of Ser and Thr residues, followed by intramolecular Michael-type addition of Cys thiols to the resulting dehydroalanine (Dha) or dehydrobutyrine (Dhb) residues (Figure 1a).2−5 Many lanthipeptides exhibit potent antimicrobial activity and are named lantibiotics.6 Recently, we reported that several putative RiPP precusor peptides from Nostoc punctiforme PCC 73102 lack Cys residues,7 which are essential for either Lan or MeLan formation. This observation was intriguing since they are encoded in a gene cluster that contains the class II lanthipeptide synthetase NpnM. In addition, a zinc-dependent dehydrogenase NpnJA was encoded in the gene cluster. NpnJA was shown to hydrogenate Dha formed by NpnM to form d-alanine.8d-amino acids have advantageous properties as they stabilize peptide structures and reduce proteolytic susceptibility.9
Figure 1.
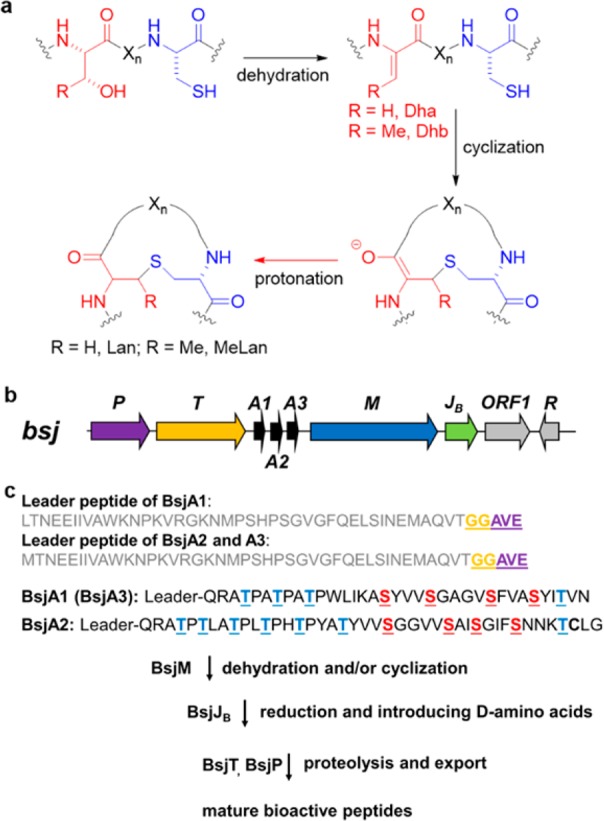
(a) General biosynthetic route toward Lan and MeLan. (b) The biosynthetic gene cluster for bicereucin. (c) Post-translational maturation of bicereucin. Proteolytic cleavage sites are highlighted in yellow and purple.
Thus, far, only a limited number of RiPPs containing d-amino acids have been reported,10−14 which prompted us to investigate other lanthipeptide gene clusters that encode putative precursor peptides lacking Cys residues.
Using genome mining (see SI) we identified one such gene cluster in Bacillus cereus SJ1, which encodes three precursor peptides BsjA1-3 (Figure 1b). Like most RiPPs, these peptides appear to have an N-terminal leader peptide, which would be removed during maturation, and a C-terminal core peptide where the post-translational modifications would take place. The leader peptide termini were predicted by the presence of a double-Gly motif (Figure 1b).15 BsjA1 and BsjA3 share near-identical sequences and have Ser/Thr-rich core peptides that lack Cys. BsjA2 has a nearly identical leader peptide but a different core peptide sequence and contains a sole Cys. The gene cluster also encodes a putative class II lanthipeptide synthetase BsjM, a peptidase-containing ATP-binding cassette transporter BsjT, a subtilisin-like serine protease BsjP, and a LanJB class dehydrogenase8 BsjJB (Table S2). The latter is a putative NAD(P)H- and flavin-dependent oxidoreductase that shows 27% protein sequence identity to CrnJ, another LanJB class flavin reductase involved in carnolysin biosynthesis. CrnJ was recently shown to convert Dha and Dhb to d-Ala and d-2-aminobutyrate (d-Abu) in vivo.13
BsjA1 or BsjA2 were individually co-expressed with BsjM in E. coli BL21 (DE3) as N-terminally His6-tagged peptides. After metal affinity purification, analysis by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) showed that BsjM-modified His6-BsjA1 exhibited a mass corresponding to eight dehydrations, whereas BsjM-modified His6-BsjA2 exhibited a mass corresponding to 11 dehydrations (Figures S1a, S2a). Next, BsjA1 and BsjA2 were individually co-expressed with BsjM and BsjJB in E. coli. After purification, the peptides were analyzed by MALDI-TOF MS demonstrating that His6-BsjA1 exhibited a mass consistent with eight dehydrations and four reductions, whereas His6-BsjA2 exhibited a mass corresponding to 11 dehydrations plus four reductions (Figures S1b, S2b). These results demonstrated that BsjJB reduced a subset of the Dha and/or Dhb residues to Ala and/or Abu, respectively, with four dehydro amino acids in BsjA1 and seven dehydro amino acids in BsjA2 escaping reduction (for positions, see below).
To determine the biological activity of the resulting products, proteolytic removal of the leader peptide was investigated. Since the biosynthetic gene cluster encodes a LanT-like peptidase-containing ATP-binding cassette transporter (BsjT) as well as a subtilisin-like serine protease (BsjP), the removal of the leader peptide likely takes place in two steps as observed for some two-component class II lanthipeptides such as lichenicidin and cytolysin.16,17 In these systems, LanT transporters containing a Cys protease domain made up of the N-terminal ∼150 amino acids cleave at the C-terminus of a conserved GG, GA, or GS motif.15 Then, a LanP protein performs a final cleavage extra-cellularly to release the mature peptides. Indeed, BsjP contains a secretion signal peptide.
We expressed the N-terminal 150 residues of BsjT (BsjT150) with an N-terminal His6-tag in E. coli BL21 (DE3) and purified the protein by metal affinity chromatography. Incubation of His6-BsjT150 with BsjM- and BsjJB-modified BsjA1 and BsjA2 in vitro using previously reported conditions18 and subsequent MALDI-TOF MS analysis showed that BsjT150 cleaved after the “GG” motif for both peptides (Figure S3a). BsjP was expressed with an N-terminal maltose-binding protein (MBP) tag in E. coli BL21 (DE3) and purified by affinity chromatography (see SI). After incubation of the BsjT150-cleaved BsjA1 or BsjA2 with MBP-BsjP, the N-terminal AVE sequence was removed for both peptides yielding the mature modified core peptides that we named Bsjα and Bsjβ (Figures S3b, S4, and S5). Knowing that BsjP cleaved after Glu and realizing that both Bsjα and Bsjβ do not contain any Glu residues, commercially available Glu-C endoprotease was employed for one-step removal of leader peptide in all subsequent experiments.
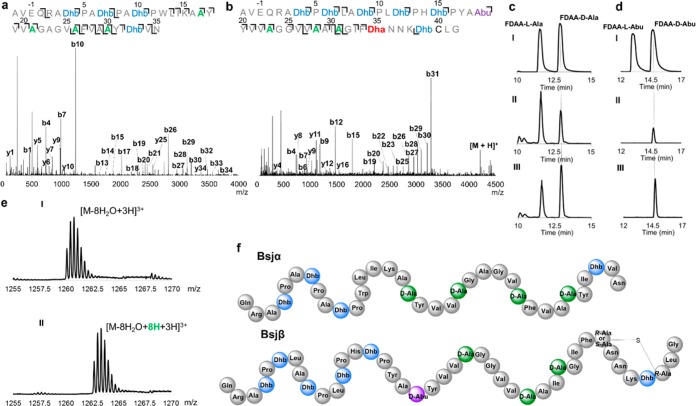
Purified AVE-Bsjα and AVE-Bsjβ were subjected to ESI quadrupole TOF tandem MS analysis (ESI-Q-TOF MS-MS). The results illustrated that Dha residues at positions 17, 21, 26, and 30 were reduced to Ala in Bsjα and that Dha residues at positions 23, 28, and 31 were reduced in Bsjβ (Figure 2a,b). In addition, Dhb19 was reduced to Abu in Bsjβ (Figure 2b). This observation shows that BsjJB, like its ortholog CrnJB in carnolysin biosynthesis,13 can reduce both Dha and Dhb, unlike the LanJA enzymes characterized to date, including NpnJA, which only reduce Dha. The residues that escape reduction are either flanked by Pro or are near the N- and C-termini of the core peptide (Figure 2a,b), suggesting positional and/or sequence dependence of BsjJB. We also incubated purified His6-BsjJB with BsjM-modified His6-BsjA1 in the presence of NADH and FMN. Following digestion by BsjT150, LC-ESI-Q-TOF MS analysis showed a mass increase of 8 Da (Figure 2e), consistent with the co-expression data (Figure S4).
Figure 2.
Structural analysis of Bsjα and Bsjβ. MS-MS fragmentation pattern of (a) AVE-Bsjα and (b) AVE-Bsjβ. (c) Stereochemistry of the introduced Ala using Marfey’s method: (I) Extracted ion chromatogram (EIC) of [M + H] = 342.1 ± 0.2 Da corresponding to FDAA-l- and d-Ala standards. (II) EIC of hydrolysate of AVE-Bsjα derivatized with FDAA. (III) EIC of hydrolysate of AVE-Bsjα derivatized with FDAA coinjected with d-Ala derivatized with FDAA. (d) Stereochemistry of Abu: (I) EIC of [M + H] = 356.1 ± 0.2 Da corresponding to FDAA-Abu standards. (II) EIC of hydrolysate of AVE-Bsjβ derivatized with FDAA. (III) EIC of hydrolysate of AVE-Bsjβ derivatized with FDAA coinjected with authentic d-Abu derivatized with FDAA. (e) ESI-Q-TOF MS of in vitro assay of BsjJB with BsjM-modified His6-BsjA1 followed by BsjT-150 digestion: (I) incubation with heat-denatured His-BsjJB as negative control. (II) Assay with His-BsjJB. (f) Structures of Bsjα and Bsjβ assigned in this work.
To determine the stereochemistry of reduction by BsjJB, AVE-Bsjα and AVE-Bsjβ were hydrolyzed in 6 M HCl, and the resulting amino acids were derivatized using Marfey’s reagent, 1-fluoro-2,4-dinitrophenyl-5-l-alanine amide (FDAA).19 Quantitative liquid chromatography (LC)-MS analysis and comparison with authentic l- and d-Ala or l- and d-Abu derivatized under same conditions revealed that in the Bsjα hydrolysate the FDAA-l- and d-Ala stereoisomers were present in a 1.75:1 ratio. This ratio is close to the calculated ratio of 7:4 if all four Dha were converted to d-Ala (Figure 2c; AVE-Bsjα contains seven gene-encoded l-Ala residues). Therefore, for Bsjα, the four newly introduced Ala residues exhibit the d-configuration (Figure 2f). In the hydrolysate of AVE-Bsjβ, the detected FDAA-l- and d-Ala isomers were observed in a 1.65:1 ratio, which is very close to the calculated ratio of 5:3 if all 3 Dha were converted to d-Ala (Figure S6, AVE-Bsjβ contains five gene encoded l-Ala residues). Hence, for Bsjβ the three newly introduced Ala residues also exhibit the d-configuration. A peak at the same retention time of FDAA-d-Abu standard was also observed in the Bsjβ hydrolysate, indicating that the introduced Abu also exhibits the d-configuration (Figure 2d).
To determine whether the Cys residue of Bsjβ was involved in a Lan or MeLan, the modified peptide was treated with iodoacetamide (IAA) to test for the presence of a free thiol.20 Subsequent analysis by MALDI-TOF MS showed only a trace amount of an acetamidomethyl adduct indicating that Cys40 is mostly cyclized (Figure S7). To determine which Dha/Dhb reacted with Cys40, Bsjβ was hydrolyzed in acid, and the resulting amino acids were derivatized and analyzed by gas chromatography (GC) MS using a column containing a chiral stationary phase.21,22 Interestingly, a Lan residue was observed as a mixture of dl- and ll-configurations (Figure S8). When combined with the information from the tandem MS analysis, Dha35 is the only possible residue that could have reacted with Cys40. The deduced structures of Bsjα and Bsjβ are shown in Figure 2f. We hypothesize that the reason for the low stereoselectivity of cyclization by BsjM is the absence of the catalytic residues that are usually conserved in LanM cyclization domains,23,24 including the His acid that stereoselectively protonates the enolate in other LanMs (Figure S9).25
We were unable to elicit production of bicereucin from B. cereus SJ1 to confirm these structures. In lieu of such confirmation, we investigated their antimicrobial activity. Agar diffusion growth inhibition assays demonstrated that bicereucin is a two-component system with synergistic activity against a range of Gram-positive bacteria (Figures 3a and S10, Tables S3, S4). The optimal molar ratios of Bsjα and Bsjβ as well as the minimal inhibitory concentrations (MICs) were determined by liquid growth inhibition assays, resulting in low micromolar to submicromolar MICs (Table S5). The resulting isobologram demonstrated that the optimal molar ratio between Bsjα and Bsjβ is 2:1 (Figure 3b). Since modification of BsjA1 and BsjA3 both result in Bsjα, the optimal ratio is the same as the stoichiometry of the genes (bsjA1 + bsjA3:bsjA2 = 2:1). The MICs of the combination of Bsjα and Bsjβ (2:1) was about 50–100 lower than their independent MICs. We also examined the order of binding for cell growth inhibitory activity by sequential addition of the peptides as reported previously.26−28 The results suggest that Bsjα binds a target on the outer surface of the bacterial cell and that the resulting complex is required for Bsjβ to perform its synergistic effect (Figure S11). Interestingly, bicereucin also exhibits weak synergistic hemolytic activity against rabbit red blood cells (Figure S12), but no antifungal activity was observed. Synergistic antimicrobial and hemolytic activity is reminiscent of the enterococcal cytolysin.29 To investigate the importance of the Lan in Bsjβ, we generated a Bsjβ-C40A variant (Figure S13) and performed antimicrobial activity assays independently or combined with Bsjα. In both cases, a dramatic drop in inhibitory activity was observed (Figure S14, Table S6), illustrating the importance of the Lan in Bsjβ for activity. Dha35 and Dhb39 were not reduced by BsjJB in this BsjA mutant illustrating that they do not escape reduction because of the formation of the Lan ring.
Figure 3.

Antimicrobial activity assays of bicereucin against Bacillus subtilis ATCC6633. (a) Agar diffusion growth assay: (I) blank, (II) Bsjα, (III) Bsjβ, (IV) Bsjα combined with Bsjβ; spotted total peptide amount: 1 nmol. (b) Isobologram demonstrating the optimal inhibitory ratio of Bsjα and Bsjβ.
The clean biosynthetic transformations and the synergistic activity and stoichiometry strongly suggest that the structures in Figure 2f represent native bicereucin. The structure of Bsjα is noteworthy since it is not a lanthipeptide. We searched the databases for other gene clusters encoding a lanthipeptide synthetase, a LanJ, and precursor peptide(s) lacking cysteine (Tables S7, S8). These clusters are potentially encoding the biosynthesis of non-cross-linked d-amino acid-containing peptides, which would be a new RiPP class.
Introduction of d-amino acids into ribosomally synthesized peptides is highly valuable since such residues reduce susceptibility to proteolysis.30 A limited number of enzymes have been reported that post-translationally introduce d- stereocenters into peptides,31−33 but they have not yet been used for synthetic purposes. We investigated the utility of LanJ enzymes to prepare d-amino acid containing natural products. Dermorphin is a d-Ala-containing heptapeptide(H-Tyr-d-Ala-Phe-Gly-Tyr-Pro-Ser-NH2) isolated from the skin of South American frogs.34 Dermorphin is a potent natural agonist of μ opioid receptors.34−36 We envisioned making dermorphin via a synthetic biology approach by appending the BsjA leader peptide to a peptide sequence based on dermorphin but replacing d-Ala with Ser. We termed this precursor peptide BsjA-leader-dermorphin. We cloned the gene for this peptide into multicloning site (MCS) 1 of pRSF-Duet-1 and bsjM into MCS2 and co-expressed the proteins with BsjJB. MALDI-TOF MS showed the desired dehydration at Ser2, but no reduction was observed. This result suggested that Dha2 may be too close to the leader peptide, since BsjJB does not reduce the dehydroamino acids near the leader peptide in its cognate substrates. We therefore replaced BsjJB with NpnJA, which previously showed high substrate tolerance.8 After co-expression and purification, one dehydration and one reduction were observed. Trypsin was used to remove the leader peptide, and high-resolution ESI-Q-TOF MS-MS and Marfey analysis confirmed that Ser2 was converted into d-Ala (Figures S15, S16). Thus, E. coli produced an analog of dermorphin that only lacks the C-terminal amide, which can be introduced by one-step enzymatic transformation.37,38
In summary, we describe a novel two-component lantibiotic bicereucin in which one of the peptides contains a single Lan and the second peptide is devoid of any cross-links. Bicereucin is the first example of a nonlanthipeptide that is important for the bioactivity of a lanthipeptide. Bsjα and Bsjβ contain multiple d-amino acids, and Bsjβ is, to our knowledge, only the second example of a RiPP containing d-Abu after carnolysin.13 We illustrate the potential utility of LanJ enzymes for synthetic biology by presenting the formal biosynthesis of the d-Ala containing natural opioid dermorphin.
Acknowledgments
This work was supported by the US National Institutes of Health (R01 GM 058822 to W.A.v.d.D.). We thank Dr. Weixin Tang and Dr. Xiao Yang for initial bioinformatic searches.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b02513.
Procedures and supporting figures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Arnison P. G.; et al. Nat. Prod. Rep. 2013, 30, 108. 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey J. M.; van der Donk W. A. Annu. Rev. Microbiol. 2007, 61, 477. 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Chatterjee C.; Paul M.; Xie L.; van der Donk W. A. Chem. Rev. 2005, 105, 633. 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- Bierbaum G.; Sahl H. G. Curr. Pharm. Biotechnol. 2009, 10, 2. 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- Cotter P. D.; Hill C.; Ross R. P. Curr. Protein Pept. Sci. 2005, 6, 61. 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- Montalban-Lopez M.; Zhou L.; Buivydas A.; van Heel A. J.; Kuipers O. P. Expert Opin. Drug Discovery 2012, 7, 695. 10.1517/17460441.2012.693476. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Yang X.; Wang H.; van der Donk W. A. ACS Chem. Biol. 2014, 9, 2686. 10.1021/cb500622c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; van der Donk W. A. J. Am. Chem. Soc. 2015, 137, 12426. 10.1021/jacs.5b05207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rodriguez S.; Martinez-Gomez A. I.; Rodriguez-Vico F.; Clemente-Jimenez J. M.; Las Heras-Vazquez F. J. Chem. Biodiversity 2010, 7, 1531. 10.1002/cbdv.200900245. [DOI] [PubMed] [Google Scholar]
- Skaugen M.; Nissen-Meyer J.; Jung G.; Stevanovic S.; Sletten K.; Inger C.; Abildgaard M.; Nes I. F. J. Biol. Chem. 1994, 269, 27183. [PubMed] [Google Scholar]
- Martin N. I.; Sprules T.; Carpenter M. R.; Cotter P. D.; Hill C.; Ross R. P.; Vederas J. C. Biochemistry 2004, 43, 3049. 10.1021/bi0362065. [DOI] [PubMed] [Google Scholar]
- Cotter P. D.; O’Connor P. M.; Draper L. A.; Lawton E. M.; Deegan L. H.; Hill C.; Ross R. P. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 18584. 10.1073/pnas.0509371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohans C. T.; Li J. L.; Vederas J. C. J. Am. Chem. Soc. 2014, 136, 13150. 10.1021/ja5070813. [DOI] [PubMed] [Google Scholar]
- Freeman M. F.; Gurgui C.; Helf M. J.; Morinaka B. I.; Uria A. R.; Oldham N. J.; Sahl H. G.; Matsunaga S.; Piel J. Science 2012, 338, 387. 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- Håvarstein L. S.; Diep D. B.; Nes I. F. Mol. Microbiol. 1995, 16, 229. 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- Tang W. X.; Dong S. H.; Repka L. M.; He C.; Nair S. K.; van der Donk W. A. Chem. Sci. 2015, 6, 6270. 10.1039/C5SC02329G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano T.; Krawczyk J. M.; Mosker E.; Süssmuth R. D.; Mendo S. Chem. Biol. 2011, 18, 90. 10.1016/j.chembiol.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Furgerson Ihnken L. A.; Chatterjee C.; van der Donk W. A. Biochemistry 2008, 47, 7352. 10.1021/bi800278n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan R.; Bruckner H. Amino Acids 2004, 27, 231. 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- Gomi T.; Fujioka M. Biochemistry 1982, 21, 4171. 10.1021/bi00260a039. [DOI] [PubMed] [Google Scholar]
- Liu W.; Chan A. S.; Liu H.; Cochrane S. A.; Vederas J. C. J. Am. Chem. Soc. 2011, 133, 14216. 10.1021/ja206017p. [DOI] [PubMed] [Google Scholar]
- Tang W.; van der Donk W. A. Biochemistry 2012, 51, 4271. 10.1021/bi300255s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L.; van der Donk W. A. Curr. Opin. Chem. Biol. 2004, 8, 498. 10.1016/j.cbpa.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Paul M.; Patton G. C.; van der Donk W. A. Biochemistry 2007, 46, 6268. 10.1021/bi7000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; van der Donk W. A. ACS Chem. Biol. 2015, 10, 1234. 10.1021/acschembio.5b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan S. M.; O’Connor P. M.; Cotter P. D.; Ross R. P.; Hill C. Antimicrob. Agents Chemother. 2005, 49, 2606. 10.1128/AAC.49.7.2606-2611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N.; Oman T. J.; Andrew Wang T. S.; De Gonzalo C. V.; Walker S.; van der Donk W. A. J. Antibiot. 2014, 67, 133. 10.1038/ja.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oman T. J.; van der Donk W. A. ACS Chem. Biol. 2009, 4, 865. 10.1021/cb900194x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C. R.; Coburn P. S.; Gilmore M. S. Curr. Protein Pept. Sci. 2005, 6, 77. 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- Jungheim L. N.; Shepherd T. A.; Baxter A. J.; Burgess J.; Hatch S. D.; Lubbehusen P.; Wiskerchen M.; Muesing M. A. J. Med. Chem. 1996, 39, 96. 10.1021/jm950576c. [DOI] [PubMed] [Google Scholar]
- Heck S. D.; Faraci W. S.; Kelbaugh P. R.; Saccomano N. A.; Thadeio P. F.; Volkmann R. A. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 4036. 10.1073/pnas.93.9.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka B. I.; Vagstad A. L.; Helf M. J.; Gugger M.; Kegler C.; Freeman M. F.; Bode H. B.; Piel J. Angew. Chem., Int. Ed. 2014, 53, 8503. 10.1002/anie.201400478. [DOI] [PubMed] [Google Scholar]
- Murkin A. S.; Tanner M. E. J. Org. Chem. 2002, 67, 8389. 10.1021/jo0204653. [DOI] [PubMed] [Google Scholar]
- Melchiorri P.; Negri L. Gen. Pharmacol. 1996, 27, 1099. 10.1016/0306-3623(95)02149-3. [DOI] [PubMed] [Google Scholar]
- Kreil G. J. Biol. Chem. 1994, 269, 10967. [PubMed] [Google Scholar]
- Amiche M.; Delfour A.; Nicolas P. EXS 1998, 85, 57. 10.1007/978-3-0348-8837-0_4. [DOI] [PubMed] [Google Scholar]
- Nuijens T.; Piva E.; Kruijtzer J. A. W.; Rijkers D. T. S.; Liskamp R. M. J.; Quaedflieg P. J. L. M. Tetrahedron Lett. 2012, 53, 3777. 10.1016/j.tetlet.2012.05.039. [DOI] [Google Scholar]
- Boeriu C. G.; Frissen A. E.; Boer E.; van Kekem K.; van Zoelen D. J.; Eggen I. F. J. Mol. Catal. B: Enzym. 2010, 66, 33. 10.1016/j.molcatb.2010.03.010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.