Abstract
Several programmed lytic and necrotic-like cell death mechanisms have now been uncovered, including the recently described receptor interacting protein kinase-3 (RIPK3)-mixed lineage kinase domain-like (MLKL)-dependent necroptosis pathway. Genetic experiments have shown that programmed necrosis, including necroptosis, can play a pivotal role in regulating host-resistance against microbial infections. Alternatively, excess or unwarranted necroptosis may be pathological in autoimmune and autoinflammatory diseases. This review highlights the recent advances in our understanding of the post-translational control of RIPK3-MLKL necroptotic signaling. We discuss the critical function of phosphorylation in the execution of necroptosis, and highlight the emerging regulatory roles for several ubiquitin ligases and deubiquitinating enzymes. Finally, based on current evidence, we discuss the potential mechanisms by which the essential, and possibly terminal, necroptotic effector, MLKL, triggers the disruption of cellular membranes to cause cell lysis.
Keywords: necroptotic cell death, necroptosis, RIPK3, MLKL, mixed lineage kinase domain-like, receptor interacting protein kinase-3
Introduction
RIPK3 and MLKL are essential for necroptotic cell death
Necroptosis is a caspase-independent programmed cell death pathway 1. Necroptotic cell lysis and the release of intracellular pro-inflammatory molecules is dependent on phosphorylation of the pseudokinase, MLKL, by the protein kinase, RIPK3. Phosphorylation of MLKL leads to its activation, and subsequent cell death by mechanisms that are currently a matter of debate, although a weight of evidence suggests this involves disruption of cellular membranes, including the plasma membrane. Following tumor necrosis factor (TNF) receptor 1 (TNFR1) ligation, the protein kinase, RIPK1, activates RIPK3 presumably by promoting RIPK3 autophosphorylation, which in turn leads to MLKL activation. However, it is now apparent that RIPK3 can be activated independently of RIPK1 in many circumstances, and that RIPK1 can act to repress RIPK3 activation, both in vitro and in vivo 2– 8. Consequently, our current thinking is that RIPK3 and MLKL are the core machinery essential for all necroptotic cell death responses.
RIPK3 is a driver of MLKL dependent and independent inflammatory disease
Studies using RIPK3-deficient mice have implicated pathological RIPK3 signaling, and potentially necroptosis, in many inflammatory diseases, such as atherosclerosis, kidney ischemia reperfusion injury, liver injury, myocardial infarction, and multiple sclerosis (reviewed in 9). However, it has recently been posited that necroptosis may also act in an anti-inflammatory capacity, as cell death abrogates TNF- or toll-like receptor- (TLR) induced transcription of pro-inflammatory cytokines and the ensuing inflammatory response 10. Recent research has also revealed that RIPK3 has several non-necroptotic signaling capabilities, both in vitro and in vivo (reviewed in 11). These include the ability to activate caspase-8 dependent apoptosis 12– 14, trigger interleukin-1β (IL-1β)-dependent inflammation through caspase-8 and/or the Nod-like receptor 3 (NLRP3) inflammasome 15– 22, and regulate the transcription of cytokines 23, 24. Hence, the use of MLKL-deficient mice is required to validate necroptosis as a bona fide drug target in many inflammatory disease models where RIPK3 has been implicated. In this regard, murine genetic studies have started to document how unrestrained MLKL-dependent necroptotic signaling can result in embryonic lethality 3 and cause liver inflammation 25. In addition, the development of phospho-specific MLKL antibodies as markers of activated MLKL have shown that necroptosis is likely to occur in diseases such as toxic epidermal necrolysis 26, 27, drug-induced liver injury 28, and pathogen infection 21. Cancer cell lines have also been observed to suppress RIPK3 expression 29, which in some circumstances has been attributed to DNA methylation 30. As such, chemically induced hypomethylation can restore RIPK3 expression and promote RIPK3-MLKL-induced necroptosis in response to chemotherapeutic treatments. A greater understanding of the mechanisms of necroptosis signaling, and when it occurs, is therefore likely to yield new therapeutic opportunities in a number of different disease states.
Necroptosis is activated by a number of different receptors
Several signaling receptors have been documented to activate RIPK3-MLKL dependent necroptosis, including death receptors (i.e., TNFR1), TLRs, the DNA receptor DAI (DNA-dependent activator of interferon [IFN]-regulatory factors or ZBP1/DML-1), and the T-cell antigen receptor. Type I IFN and IFNγ-induced transcriptional responses have also been proposed to cause necroptosis, or to enhance TLR3/4 and TNFR1 necroptosis 31– 33. While protein kinase R (PKR) was suggested to directly trigger formation of the RIPK1-RIPK3 necrosome downstream of IFNγ signaling 33, PKR is not required for type I IFN killing 32, and hence the underlying mechanism for IFN-induced necroptosis requires further study. By comparison, necroptotic signaling caused by TNFR1 ligation is better defined (reviewed in 34). In most cases, TNF binding to TNFR1 triggers the formation of a cell surface complex, complex I, that induces the transcription of pro-survival genes and inflammatory cytokines. Mechanistically, the death domain (DD) of TNFR1 interacts with the DD of TNFR1-associated death domain (TRADD) (and potentially the DD of RIPK1) to nucleate the formation of a large multimeric TRADD-RIPK1-TRAF2- inhibitor of apoptosis (IAP) ubiquitylation platform 35– 38. For example, RIPK1 binding to this complex and its modification with ubiquitin chains by IAP proteins parallels IAP-dependent recruitment of the linear ubiquitin chain assembly complex (LUBAC). Ubiquitylated RIPK1, and LUBAC modification of NEMO (nuclear factor kappa-light-chain-enhancer of activated B cells [NFκB] essential modifier), subsequently activate canonical NFκB signaling. In the absence of optimal RIPK1 ubiquitylation (i.e., when IAPs are lost), RIPK1 dissociates into the cytosol to form a secondary death-inducing complex that can activate caspase-8 (complex II) to cause apoptosis. Caspase-8 represses necroptotic signaling, and hence, when caspase-8 activity is low, RIPK1 can bind RIPK3 to form the necrosome, activate MLKL, and induce necroptotic killing.
Physiological triggers of necroptosis
Because under normal cell culture conditions necroptosis is not induced by death receptor or TLR ligation, experimentally, necroptosis is usually studied by deleting or inhibiting key negative regulators of necroptotic signaling, such as caspase-8 or IAP proteins (see below). Physiological settings that trigger necroptosis have been less well defined, although situations where caspase-8 is down-regulated, such as following cutaneous wounding 39– 41, or IAP protein depletion, such as during TNF-like weak inducer of apoptosis (TWEAK)-FGF-inducible molecule 14 (FN14) TNF superfamily signaling 42, 43, clearly have the potential to promote a necroptotic response. Along these lines of evidence, biopsies from children suffering from inflammatory bowel disease display decreased caspase-8 expression and elevated RIPK3 and MLKL levels, and may indicate ongoing necroptosis 44. More direct experiments have been performed to suggest that bacterial and viral molecules can act to induce or inhibit necroptosis at multiple levels, including direct RIPK1/RIPK3 targeting, or downstream of MLKL phosphorylation 21, 45– 50.
In this review we summarize recent advances in identifying and understanding positive and negative regulators of necroptotic signaling ( Table 1), focusing on findings with strong genetic evidence. We begin by a brief discussion on the best studied necroptotic components; RIPK1, RIPK3 and MLKL.
Table 1. Post-translational modification of RIPK1, RIPK3 and MLKL.
| Protein | Target
residue |
PTM | Writer | Eraser | Impact of PTM | References |
|---|---|---|---|---|---|---|
| Mouse RIPK1 | S89 | Putative
phosphorylation |
Putative pS89; suppresses
necroptosis in Jurkat cells |
113 | ||
| Mouse/Human
RIPK1 |
K377 and
others |
K63-, K48-, K11- linked
ubiquitylation |
cIAP1/
cIAP2 |
A20 | Ubiquitination prevents death
signalling |
103, 114– 118 |
| Mouse RIPK3 | S204 | Phosphorylation | pS204 promotes necroptosis | 113 | ||
| Human RIPK3 | S199 | Phosphorylation | RIPK3 | pS199 promotes necroptosis | 119 | |
| Mouse RIPK3 | T231/S232 | Phosphorylation | Ppm1b | pT231/pS232 promotes
MLKL interaction and necroptosis |
101, 102, 113 | |
| Human RIPK3 | S227 | Phosphorylation | pS227 promotes MLKL
interaction and necroptosis |
57 | ||
| Mouse RIPK3 | K5 | Ubiquitylation | A20 | K5Ub promotes necrosome
assembly and necroptosis |
15, 96 | |
| Mouse RIPK3 | C119 | S-nitrosylation | Promotes necroptosis | 120 | ||
| Mouse MLKL | S345, S347,
T349 |
Phosphorylation | RIPK3 | pS345 and pS347 promote
necroptosis |
60, 64, 65 | |
| Human MLKL | T357, S358 | Phosphorylation | RIPK3 | pT357/pS358 promote
necroptosis |
57 | |
| Mouse MLKL | S124 | Phosphorylation | Nil. Counterpart of human
S125 |
65, 121 | ||
| Mouse MLKL | S158 | Phosphorylation | pS158 likely suppresses
necroptosis |
65 | ||
| Mouse MLKL | S228 | Phosphorylation | RIPK3 | pS228 relieves possible
RIPK3-mediated suppression of necroptosis |
65 | |
| Mouse MLKL | Ubiquitylation | Unknown. Correlates with
necroptosis |
15 |
The core necroptotic machinery
RIPK3
The defining feature that triggers RIPK3 serine/threonine kinase activity and its phosphorylation of MLKL is RIPK3 oligomerization via RIP homotypic interaction motif (RHIM) containing proteins. For example, RHIM containing signaling components of TNFR1 (RIPK1), TLR3 and TLR4 (TRIF) and DAI itself can all form RHIM-RHIM interactions with RIPK3 to trigger necroptotic death. Similarly, the artificial dimerization/oligomerization of RIPK3 directly suffices to cause its activation and recruitment of MLKL in the absence of any upstream signal 4, 14, 51. The importance of RHIM-RHIM interactions for RIPK3 signaling is highlighted by several viral RHIM-containing proteins, such as cytomegalovirus vIRA and herpes simplex virus ICP-6 and ICP-10, which bind RIPK1 and RIPK3 and can regulate necroptosis to alter host anti-viral responses 46– 49.
The interaction of RIPK1 and RIPK3 RHIMs induces the formation of a large amyloid-like necrosome signaling platform 52. Recent work has suggested that RIPK1-RIPK3 heterodimers do not suffice to trigger necroptosis, and instead RIPK3 homo-oligomer formation within the necrosome complex is essential for RIPK3 activation, intramolecular RIPK3 phosphorylation, and MLKL recruitment 51. This model fits with recent genetic studies showing that the deletion of RIPK1 in vivo can trigger RIPK3-MLKL induced necroptosis 3, as the loss of RIPK1 may enhance the propensity for RIPK3 oligomerization 4.
Activated RIPK3 phosphorylates MLKL, thereby allowing MLKL association with, and disruption of, phospholipid membranes 28, 53– 55. RIPK3-deficient mice display no overt phenotype, and hence targeting RIPK3 kinase activity with small-molecule inhibitors represents one strategy for the treatment of necroptotic-driven diseases 13, 56. It is noteworthy, however, that RIPK3 kinase inhibition 13, or a D161N mutation in the RIPK3 kinase domain in vivo 12, can drive lethal caspase-8-dependent apoptosis by triggering RIPK1 recruitment to RIPK3, and RIPK1-mediated activation of caspase-8. Interestingly, RIPK3 kinase activity per se is not essential for mammalian viability, as an independently generated kinase-dead (K51A) RIPK3 mouse is viable and fertile 13. Hence, the development of RIPK3 kinase inhibitors that avoid lethal RIPK3 conformational changes are required. Alternatively, MLKL inhibitors 54, 57 may be a more attractive strategy for specifically targeting necroptosis, as these would avoid complications that may result from altering non-necroptotic RIPK3 signaling capabilities.
MLKL
MLKL is an essential necroptosis effector that operates downstream of RIPK3 57– 60. Unlike RIPK3, which is a catalytically active, conventional protein kinase, MLKL belongs to a group of related proteins that are enzymatically-dead, termed pseudokinases 61, 62. In addition to its C-terminal pseudokinase domain, MLKL contains an N-terminal four-helix bundle (4HB) domain, which is now known to mediate MLKL’s killer function 53– 55, 63. Our current understanding of MLKL activation is that RIPK3 phosphorylates the activation loop in MLKL’s pseudokinase domain 57, 59, 60, 64, 65, which induces a conformational change that relieves the suppressive function of the pseudokinase domain, allowing the unleashing of the executioner N-terminal 4HB domain 54. Several different models for how the MLKL 4HB domain can induce cell death have been proposed, ranging from a direct membrane permeabilization or pore-forming capacity 28, 55, 66, to the implication of downstream effectors 53, 63 (summarized in Figure 1). Although details are still emerging, recombinant MLKL was found to bind to various lipids in “fat Western” lipid arrays 28, 55, which was further reflected in a capacity to permeabilize liposomes in vitro 28, 55, 66. Curiously, MLKL most readily permeabilized liposomes containing 15% cardiolipin, a lipid believed to be confined to the mitochondrial inner membrane. The necessity of mitochondria for necroptosis has been challenged 67, making the preference for cardiolipin difficult to reconcile. At the molecular level, the mode of 4HB domain engagement of lipids remains of outstanding interest. While initial reports suggested this interaction might be mediated by positively-charged residues in the human MLKL 4HB domain 55, these residues are poorly conserved among MLKL orthologs and indeed, curiously, mutation of the acidic counterparts in the mouse 4HB domain similarly compromised the capacity of the domain to induce cell death 54. It is therefore possible that, rather than a direct interaction with negatively-charged phospholipids, these charged residues in the MLKL 4HB domain are important to the MLKL homo- and/or hetero-oligomerisation that is necessary for necroptosis.
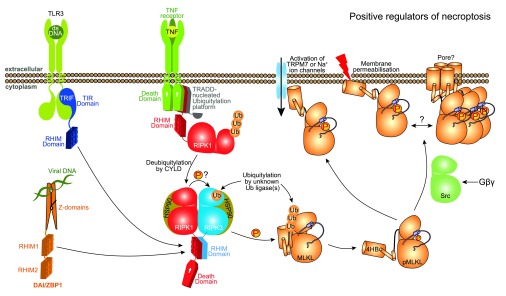
Figure 1. Positive regulators of necroptosis.
The core necroptosis machinery, comprising RIPK3 and MLKL, are activated following RIPK3 interaction with RIPK1, TRIF or DAI via their RHIMs. CYLD-mediated deubiquitylation of RIPK1 is necessary for its participation in cell death pathways, while ubiquitylation of RIPK3 (at Lys5) and MLKL by as-yet-unidentified E3 ligases may promote necroptosis 15, 97. HSP90 is known to augment the necroptotic functions of RIPK1 and RIPK3. MLKL is believed to induce cell death via a membrane-directed process 54, perhaps by directly permeabilizing membranes 28, 55, with some debate over whether channel activation might be involved 28, 53, 63, or, as one report suggests, there may be a role for Src in promoting MLKL-mediated death downstream of Gβγ. Abbreviations: 4HB, four-helix bundle; CYLD, cylindromatosis; DAI, DNA-dependent activator of interferon [IFN]-regulatory factors; HSP90, heat shock protein 90; IAP, inhibitor of apoptosis proteins; MLKL, mixed lineage kinase domain-like; RHIM, RIP homotypic interaction motif; RIPK, receptor interacting protein kinase; TLR, toll-like receptor; TNF, tumour necrosis factor; TNFR1, TNF receptor 1; TRADD, TNFR1-associated death domain; TRAF2, TNF receptor associated factor 2; TRPM7, transient receptor potential cation channel, subfamily M, member 7.
Another outstanding question is whether the MLKL 4HB domain forms a transmembrane pore or merely somehow compromises the integrity of the membrane. In concert with a greater understanding of which membrane within a cell is the target of MLKL action, knowledge of the means by which MLKL breaches membranes is essential for a comprehensive understanding of its action. Even though the membrane compromising activity of recombinant MLKL in liposome assays is highly suggestive of MLKL acting as a lone gunman, several lines of evidence have implicated other proteins as either co-effectors that augment or negate the activities of RIPK3 or MLKL, or as effectors that function downstream of MLKL activation ( Figure 1 and Figure 2). Of note, the induction of necroptosis is associated with the ubiquitylation of RIPK3 and MLKL 15, although the function of these modifications requires further study. We have summarized important necroptotic regulators that promote ( Figure 1) or negate ( Figure 2) MLKL-mediated death, and elaborate on current knowledge of their activities below.
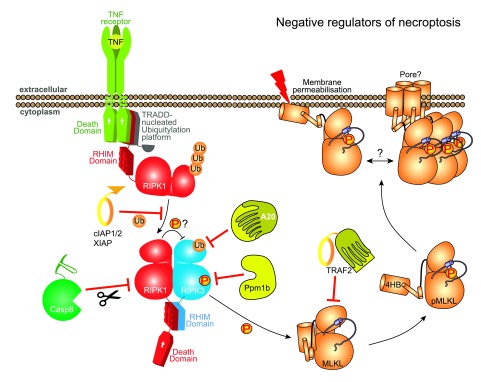
Figure 2. Negative regulators of necroptosis.
Negative regulation at the levels of RIPK1, RIPK3 and MLKL have been reported to attenuate necroptotic signalling. Not only is necroptosis by definition a caspase-independent process, but caspase-8 negates cell death, potentially by proteolytically-cleaving CYLD, RIPK1 and RIPK3. Reversible tuning of the pathway is accomplished by introduction or removal of post-translational modifications, including: ubiquitylation of RIPK1 by IAPs; deubiquitylation of RIPK3 on Lys5 by A20; and dephosphorylation of mouse RIPK3 at S231/T232 by Ppm1b. Whether an analogous phosphatase exists to dephosphorylate activated MLKL and inhibit necroptosis is not clear. However, other regulators of MLKL have been proposed, such as TRAF2. It is currently unknown what factors might govern phospho-MLKL assembly into higher order oligomers on the membrane, although a role for additional factors in mediating death is suggested by the lag between MLKL membrane translocation/oligomerisation and cell death 65. Abbreviations: 4HB, four-helix bundle; CYLD, cylindromatosis; IAP, inhibitor of apoptosis proteins; MLKL, mixed lineage kinase domain-like; RHIM, RIP homotypic interaction motif; RIPK, receptor interacting protein kinase; TNF, tumour necrosis factor; TRADD, TNFR1-associated death domain; TRAF2, TNF receptor associated factor 2.
RIPK1
An important role for RIPK1 and its kinase activity in death receptor-induced necroptosis was documented long before RIPK3 was identified as the essential necroptotic RIPK1 binding partner 68. The development of the RIPK1 kinase inhibitor necrostatin-1 69, 70 has been widely used to demonstrate the therapeutic potential of targeting RIPK1 kinase activity and necroptosis in disease models. The plausibility of this strategy was recently validated when, in contrast to the embryonic lethality of RIPK1-deficient mice caused by unrestrained apoptotic and necroptotic signaling 2, 3, 5, RIPK1 kinase-dead knockin mice, containing K45A 2 or D138N 12, 71 alleles, were shown to be viable and fertile. Hence, while RIPK1 kinase activity is often a requisite for necroptotic killing, it is not critical for mammalian survival. Instead the kinase-independent scaffolding role of RIPK1 is vital in preventing lethal RIPK3-MLKL and caspase-8 signaling.
Mechanistically, RIPK1 phosphorylation of RIPK3 is an attractive hypothesis for why RIPK1 kinase function is required for TNF-induced necroptosis. However, RIPK1 mediated phosphorylation of RIPK3 has not been formally reported to date, and the key target(s) of RIPK1 kinase activity remain unclear. Recently it has been suggested that the inhibition of RIPK1 kinase activity enhances RIPK1’s anti-necroptotic function, and consistent with this, depletion of total RIPK1 in several cell types was shown to sensitize to TNF and TLR-induced necroptotic killing 7. Why RIPK1 loss sensitizes some cell lines to TNF-induced necroptosis, while in others it confers protection, may be due to the expression levels of RIPK3 in different cells or tissues, dictating its ability to efficiently engage TNFR1 or TLR complexes and form RIPK3 oligomers. Consistent with this notion, the expression levels of RIPK3 have been shown to largely determine whether TNF can engage RIPK3 killing in the absence of RIPK1 72. Unlike the direct recruitment of RIPK3 to the RHIM containing receptor DAI 46, or the TLR adaptor protein TRIF 56, 73, the way RIPK3 is recruited to TNF receptor complexes in RIPK1 deficient cells is unknown.
Positive regulators of necroptosis
CYLD
CYLD (Cylindromatosis) is a deubiquitinating enzyme that removes K63-linked and linear ubiquitin chains from its target proteins, which include RIPK1 and TRAFs. This deubiquitinase activity of CYLD is linked with increased TNF-induced death signaling, including RIPK1-RIPK3 necrosome formation and necroptotic cell death 74– 76. Mechanistically, CYLD has been proposed to deubiquitylate RIPK1 during necrosome complex formation to facilitate RIPK1/3 kinase activity 76. In this context, it is interesting to note that the LUBAC component, HOIL-1 interacting protein (HOIP), can ubiquitylate RIPK1 within the necrosome 77. Although HOIP loss did not impact necroptotic killing, the functional consequences of necroptotic-associated RIPK1, RIPK3 and MLKL ubiquitylation warrant further investigation.
Following TNF stimulation, aberrant CYLD necroptotic activity is held in check by caspase-8 mediated CYLD processing and inactivation 74. Consistent with this, colitis induced by deletion of the essential caspase-8 adaptor protein Fas-associated protein with death domain (FADD) in intestinal epithelial cells was ameliorated by loss of either RIPK3 or CYLD activity 78. CYLD is thereby important for necroptotic signaling in some situations in vitro and in vivo. In contrast, however, unlike RIPK3-deletion, the in vivo inactivation of CYLD delays, but does not prevent, the inflammation caused by unrestrained necroptotic killing of FADD deficient keratinocytes 79. Hence, although CYLD tunes necroptotic activity, it is not essential for it to occur in all cell types.
HSP90
The Cdc37-heat shock protein 90 (HSP90) co-chaperone system has been widely implicated as a regulator of protein kinase “clients”. Conventionally, HSP90 interaction is thought to enhance protein stability, often through protection from proteasomal degradation, as is believed to be the case with RIPK1 80– 85. RIPK3 has long been recognized as an HSP90 client protein 86, 87, although only recently has a less passive regulatory role been suggested 88. Inhibition of HSP90 or genetic knockdown of the kinase-interaction adaptor, Cdc37, inhibited RIPK3’s capacity to phosphorylate MLKL to induce necroptosis, but only conferred modest effects on RIPK3 abundance 88. The precise underlying mechanism remains a matter of outstanding interest.
Channels
MLKL was implicated as a regulator of plasma membrane ion channels in two independent studies: one suggested a role for TRPM7 (transient receptor potential cation channel, subfamily M, member 7) in necroptotic killing 53, and another more broadly for Na + ion channel signaling 63. Subsequent studies using ion-free media, however, have challenged whether ion channels play an obligate role in mediating necroptosis 28, but instead suggest they contribute to varying extents in different cultured cell lines under some circumstances.
Gβγ-Src signaling
A genetic screen for additional necroptosis effectors identified the transmembrane G-proteins, Gβ and Gγ, as instigators of an alternative pathway that operates in parallel with TNF-induced necroptosis 89. Perturbation of Gβγ signaling disrupted MLKL oligomerization and translocation to membranes, a finding attributed to perturbed activation of the protein kinase, Src. The mechanistic underpinnings and universality of this pathway have not yet been fully elucidated, but are illustrative of the many possible signaling effectors that could potentially intersect with and tune the necroptosis signaling pathway. This idea is supported by the recent identification and functional characterization of MLKL phosphorylation sites outside the best-understood phosphorylation sites within the MLKL pseudokinase domain activation loop 65. This study is illustrative of a broader potential role of post-translational modifications in modulating the activities of RIPK3 and MLKL.
Negative regulators of necroptosis
Caspase-8
Caspase-8-deficient embryonic lethality, or inflammatory disease resulting from tissue or cell type specific caspase-8 deletion, is rescued by RIPK3 or MLKL loss (reviewed in 9), demonstrating that caspase-8 is an essential repressor of potentially lethal necroptotic activity. Notably, this pro-survival function for caspase-8 does not require caspase-8 processing 90, but appears to be mediated by the catalytic activity of caspase-8/FLICE-inhibitory protein (cFLIP) heterodimers 91. The critical caspase-8 targets required to repress necroptosis remain unclear, although caspase-8 cleavage of key necroptotic inducers, including RIPK1 92, RIPK3 93, and CYLD 74, have been described and are likely to be important.
IAP proteins
Mammalian IAP proteins are ubiquitin E3 ligases and include cIAP1, cIAP2 and XIAP (reviewed in 94). The cIAPs target RIPK1 for ubiquitylation to propagate TNF-induced NFκB activation and pro-survival responses, and also to prevent RIPK1 from associating with, and activating, caspase-8. XIAP, on the other hand, is a direct inhibitor of apoptotic caspases, although its deletion in mice does not result in an overt phenotype. In contrast, the co-deletion of cIAP1 and cIAP2 causes embryonic lethality, which is rescued in part by loss of RIPK1, RIPK3 or TNFR1 95. Strikingly, while cIAP1/2 loss in combination with caspase inhibition is sufficient to trigger necroptosis in several cell types, in bone marrow derived macrophages and dendritic cells XIAP is more important for limiting TNF- and TLR-induced killing 15, 19. For example, XIAP loss alone can confer some sensitivity to TNF- and TLR-induced necroptosis and apoptosis, which is greatly enhanced by cIAP1/2 co-deletion 15. Lipopolysaccharide (LPS) treatment of wildtype macrophages triggers IAP-dependent RIPK3 ubiquitylation 15. Because LPS stimulation alone does not perturb macrophage viability, this may indicate that, akin to RIPK1 ubiquitylation, RIPK3 ubiquitylation under these conditions regulates transcriptional and/or pro-survival responses. However, LPS-induced necroptotic signaling caused by the loss of IAPs and caspase inhibition is also associated with increased RIPK3 (and MLKL) ubiquitylation 15, which in this case may play a pro-necroptotic role 96. At this stage the identity of the key XIAP substrate required to limit RIPK3-MLKL signaling remains unclear. It is possible that other ubiquitin E3 ligases also negatively regulate necroptosis signaling. Consistent with this, it has been suggested that the ubiquitin ligase MKRN1 (Makorin RING finger protein-1) represses RIPK1-RIPK3 complex formation, although how it does so is incompletely understood 97.
TRAF2
TNF receptor associated factor 2 (TRAF2) binds to cIAP1/2 via a cIAP interaction motif (CIM) and is required for cIAP1/2 recruitment into TNF receptor complexes 98. Hence, as one might predict, the loss of TRAF2 may sensitize cells to death receptor mediated necroptotic killing by preventing cIAP1/2 recruitment and ubiquitylation of RIPK1 99, 100. Remarkably, however, TRAF2 was recently reported to also exert anti-necroptotic activity independently of cIAP1/2 by binding directly to MLKL to limit the association of MLKL with RIPK3 100. The steady state association of TRAF2 with MLKL was diminished upon TNF-induced necroptosis induction, and this correlated with CYLD dependent deubiquitylation of TRAF2. It will be interesting to define the TRAF2 interacting residues of MLKL, and whether their mutation impacts MLKL binding to RIPK3 and cellular necroptotic activity.
Ppm1b
Ppm1b was recently identified as a phosphatase that dephosphorylates mouse RIPK3 at T231/S232 101, two sites at which phosphorylation is known to enhance RIPK3 catalytic activity 102. While deficiency in Ppm1b led to a modest elevation in cell death in the absence of stimulation (5–10% vs 2–5% cell death, depending on cell line studied), the enhancement of cell death observed upon TNF-stimulation of L929 fibrosarcoma cells and TNF+pan-caspase inhibitor (zVAD-fmk) treatment of mouse embryonic fibroblasts was profound 103. This is the first illustration that necroptotic signaling can be tuned by dephosphorylation of RIPK3. It remains of outstanding interest whether there are other phosphatases that modulate the activity of RIPK3 and whether dephosphorylation of MLKL might serve as a mechanism to disarm its pro-necroptotic activity.
A20
A20 is a deubiquitinating enzyme that is rapidly induced following TNF- and TLR-stimulation as part of a negative feedback loop to restrict NFκB activation and inflammatory cytokine production. Hence, its mutation or loss in mice and humans results in pronounced inflammatory disease. A20 has been proposed to restrict TNF-induced NFκB by removing K63-linked ubiquitin chains from RIPK1, and at the same time can target RIPK1 for proteasomal degradation through the addition of K48-linked ubiquitin chains 103. However, it also exerts its anti-inflammatory properties by targeting a number of TNF receptor and TLR signaling components 104. Recently it was found that A20 deletion in T-cells led to RIPK3 dependent T-cell killing, and that the perinatal lethality of A20 deficient mice was significantly rescued by RIPK3 co-deletion 96. Mechanistically, A20 loss promoted RIPK1-RIPK3 necrosome formation as a consequence of RIPK3 Lys5 ubiquitylation, while A20 expression and catalytic activity correlated with decreased RIPK3 ubiquitination.
Open questions surrounding MLKL function
Because MLKL was only recently identified as an effector of the necroptosis cell death pathway, much remains to be garnered about how it functions in this pathway, whether it has additional “moonlighting” functions, and how it might contribute to other death signaling pathways. Should moonlighting functions exist for MLKL, they are likely very subtle, since genetic deletion of MLKL in mice does not noticeably impact viability, fertility or development 58, 60. The recent observation that, upon phosphorylation, MLKL translocates to the nucleus 105 suggests either a second (as-yet-unknown) function for MLKL in the nucleus or that the necroptosis signaling pathway relies on a transition via the nucleus to other membranes. This remains a matter of ongoing interest. The subcellular destination for MLKL that leads to cell death has also been a matter of debate: does MLKL-mediated death rely on translocation to mitochondrial or plasma membranes or another subcellular organelle? Initially, MLKL was implicated as being upstream of the mitochondrial phosphatase, phosphoglycerate mutase family member 5 (PGAM5), whose activation was thought to drive cell death through mitochondrial fragmentation via Dynamin-related protein 1 (Drp1) 106. However, several studies indicated that both PGAM5 and Drp1 are dispensable for necroptosis 60, 107– 109 and, as a result, the ubiquity of their involvement in the pathway has been questioned. Indeed, depletion of mitochondria by mitophagy did not prevent necroptotic death 67, suggesting that mitochondria are dispensable for necroptosis, at least in a subset of commonly studied laboratory cell lines. Nonetheless, reactive oxygen species (ROS), which emanate from mitochondria, have been widely implicated in necroptotic cell death 59, 110– 112. It remains to be determined whether ROS arise as a consequence of cell death or whether their generation is a driving force or augments necroptotic death and/or contributes to inflammatory responses. As described above, it is of enormous interest to understand the precise mechanism by which MLKL kills cells and whether other factors are involved downstream of MLKL phosphorylation and whether MLKL function can be modulated by ubiquitylation, as suggested by a recent study. Moreover, these modifications may govern whether MLKL can participate in other intersecting signaling pathways, such as inflammatory signaling in the absence of cell death 18 or cell death by pyroptosis.
Abbreviations
4HB, four-helix bundle; CYLD, cylindromatosis; DAI, DNA-dependent activator of IFN-regulatory factors; DD, death domain; Drp1, Dynamin-related protein 1; FADD, Fas-associated protein with death domain; HOIP, HOIL-1 interacting protein; HSP90, heat shock protein 90; IAP, inhibitor of apoptosis proteins; IFN, interferon; LPS, lipopolysaccharide; LUBAC, linear ubiquitin chain assembly complex; MLKL, mixed lineage kinase domain-like; NFκB, nuclear factor kappa-light-chain-enhancer of activated B cells; PGAM5, phosphoglycerate mutase family member 5; PKR, protein kinase R; RHIM, RIP homotypic interaction motif; RIPK, receptor interacting protein kinase; ROS, reactive oxygen species; TLR, toll-like receptor; TNF, tumour necrosis factor; TNFR1, TNF receptor 1; TRADD, TNFR1-associated death domain; TRAF2, TNF receptor associated factor 2.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Jiahuai Han, School of Life Sciences, Xiamen University, Xiamen, China
Peter Vandenabeele, Unit of Molecular Signaling and Cell Death, VIB Inflammation Research Center - UGent, Gent, 9052, Belgium
Martin Leverkus, Universitätsklinikum der RWTH Aachen, Aachen, Germany
Pascal Meier, Division of Breast Cancer Research, The Institute of Cancer Research, London, SW3 6JB, UK
Funding Statement
The authors are grateful to the National Health and Medical Research Council of Australia for support via a fellowship to JEV (1052598), project grants (1057905, 1067289, 1051210) and the IRIISS scheme (9000220), and the Australian Research Council for fellowship support to JMM (FT100100100). Additional support was provided by Victorian State Government Operational Infrastructure Support Scheme.
We confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 4 approved]
References
- 1. Vandenabeele P, Galluzzi L, Vanden Berghe T, et al. : Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11(10):700–14. 10.1038/nrm2970 [DOI] [PubMed] [Google Scholar]
- 2. Kaiser WJ, Daley-Bauer LP, Thapa RJ, et al. : RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proc Natl Acad Sci U S A. 2014;111(21):7753–8. 10.1073/pnas.1401857111 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 3. Rickard JA, O'Donnell JA, Evans JM, et al. : RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell. 2014;157(5):1175–88. 10.1016/j.cell.2014.04.019 [DOI] [PubMed] [Google Scholar]
- 4. Orozco S, Yatim N, Werner MR, et al. : RIPK1 both positively and negatively regulates RIPK3 oligomerization and necroptosis. Cell Death Differ. 2014;21(10):1511–21. 10.1038/cdd.2014.76 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Dillon CP, Weinlich R, Rodriguez DA, et al. : RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157(5):1189–202. 10.1016/j.cell.2014.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Roderick JE, Hermance N, Zelic M, et al. : Hematopoietic RIPK1 deficiency results in bone marrow failure caused by apoptosis and RIPK3-mediated necroptosis. Proc Natl Acad Sci U S A. 2014;111(40):14436–41. 10.1073/pnas.1409389111 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Kearney CJ, Cullen SP, Clancy D, et al. : RIPK1 can function as an inhibitor rather than an initiator of RIPK3-dependent necroptosis. FEBS J. 2014;281(21):4921–34. 10.1111/febs.13034 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Dannappel M, Vlantis K, Kumari S, et al. : RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature. 2014;513(7516):90–4. 10.1038/nature13608 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Silke J, Rickard JA, Gerlic M: The diverse role of RIP kinases in necroptosis and inflammation. Nat Immunol. 2015;16(7):689–97. 10.1038/ni.3206 [DOI] [PubMed] [Google Scholar]
- 10. Kearney CJ, Cullen SP, Tynan GA, et al. : Necroptosis suppresses inflammation via termination of TNF- or LPS-induced cytokine and chemokine production. Cell Death Differ. 2015;22(8):1313–27. 10.1038/cdd.2014.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khan N, Lawlor KE, Murphy JM, et al. : More to life than death: molecular determinants of necroptotic and non-necroptotic RIP3 kinase signaling. Curr Opin Immunol. 2014;26:76–89. 10.1016/j.coi.2013.10.017 [DOI] [PubMed] [Google Scholar]
- 12. Newton K, Dugger DL, Wickliffe KE, et al. : Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343(6177):1357–60. 10.1126/science.1249361 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Mandal P, Berger SB, Pillay S, et al. : RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol Cell. 2014;56(4):481–95. 10.1016/j.molcel.2014.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Cook WD, Moujalled DM, Ralph TJ, et al. : RIPK1- and RIPK3-induced cell death mode is determined by target availability. Cell Death Differ. 2014;21(10):1600–12. 10.1038/cdd.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lawlor KE, Khan N, Mildenhall A, et al. : RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat Commun. 2015;6: 6282. 10.1038/ncomms7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vince JE, Wong WW, Gentle I, et al. : Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity. 2012;36(2)215–27. 10.1016/j.immuni.2012.01.012 [DOI] [PubMed] [Google Scholar]
- 17. Duong BH, Onizawa M, Oses-Prieto JA, et al. : A20 restricts ubiquitination of pro-interleukin-1β protein complexes and suppresses NLRP3 inflammasome activity. Immunity. 2015;42(1):55–67. 10.1016/j.immuni.2014.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Kang TB, Yang SH, Toth B, et al. : Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 2013;38(1):27–40. 10.1016/j.immuni.2012.09.015 [DOI] [PubMed] [Google Scholar]
- 19. Yabal M, Müller N, Adler H, et al. : XIAP restricts TNF- and RIP3-dependent cell death and inflammasome activation. Cell Rep. 2014;7(6):1796–808. 10.1016/j.celrep.2014.05.008 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Moriwaki K, Bertin J, Gough PJ, et al. : A RIPK3-caspase 8 complex mediates atypical pro-IL-1β processing. J Immunol. 2015;194(4):1938–44. 10.4049/jimmunol.1402167 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Kitur K, Parker D, Nieto P, et al. : Toxin-induced necroptosis is a major mechanism of Staphylococcus aureus lung damage. PLoS Pathog. 2015;11(4):e1004820. 10.1371/journal.ppat.1004820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deutsch M, Graffeo CS, Rokosh R, et al. : Divergent effects of RIP1 or RIP3 blockade in murine models of acute liver injury. Cell Death Dis. 2015;6:e1759. 10.1038/cddis.2015.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moriwaki K, Balaji S, McQuade T, et al. : The necroptosis adaptor RIPK3 promotes injury-induced cytokine expression and tissue repair. Immunity. 2014;41(4):567–78. 10.1016/j.immuni.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Wong WW, Vince JE, Lalaoui N, et al. : cIAPs and XIAP regulate myelopoiesis through cytokine production in an RIPK1- and RIPK3-dependent manner. Blood. 2014;123(16):2562–72. 10.1182/blood-2013-06-510743 [DOI] [PubMed] [Google Scholar]
- 25. Rickard JA, Anderton H, Etemadi N, et al. : TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. Elife. 2014;3:e03464. 10.7554/eLife.03464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panayotova-Dimitrova D, Feoktistova M, Leverkus M: RIPping the Skin Apart: Necroptosis Signaling in Toxic Epidermal Necrolysis. J Invest Dermatol. 2015;135(8):1940–3. 10.1038/jid.2015.159 [DOI] [PubMed] [Google Scholar]
- 27. Kim SK, Kim WJ, Yoon JH, et al. : Upregulated RIP3 Expression Potentiates MLKL Phosphorylation-Mediated Programmed Necrosis in Toxic Epidermal Necrolysis. J Invest Dermatol. 2015;135(8):2021–30. 10.1038/jid.2015.90 [DOI] [PubMed] [Google Scholar]
- 28. Wang H, Sun L, Su L, et al. : Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell. 2014;54(1):133–46. 10.1016/j.molcel.2014.03.003 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Geserick P, Wang J, Schilling R, et al. : Absence of RIPK3 predicts necroptosis resistance in malignant melanoma. Cell Death Dis. 2015;6:e1884. 10.1038/cddis.2015.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koo GB, Morgan MJ, Lee DG, et al. : Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015;25(6):707–25. 10.1038/cr.2015.56 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Robinson N, McComb S, Mulligan R, et al. : Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol. 2012;13(10):954–62. 10.1038/ni.2397 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. McComb S, Cessford E, Alturki NA, et al. : Type-I interferon signaling through ISGF3 complex is required for sustained Rip3 activation and necroptosis in macrophages. Proc Natl Acad Sci U S A. 2014;111(31):E3206–13. 10.1073/pnas.1407068111 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Thapa RJ, Nogusa S, Chen P, et al. : Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc Natl Acad Sci U S A. 2013;110(33):E3109–18. 10.1073/pnas.1301218110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. de Almagro MC, Vucic D: Necroptosis: Pathway diversity and characteristics. Semin Cell Dev Biol. 2015;39:56–62. 10.1016/j.semcdb.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 35. Ermolaeva MA, Michallet MC, Papadopoulou N, et al. : Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol. 2008;9(9):1037–46. 10.1038/ni.1638 [DOI] [PubMed] [Google Scholar]
- 36. Pobezinskaya YL, Kim YS, Choksi S, et al. : The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol. 2008;9(9):1047–54. 10.1038/ni.1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsu H, Huang J, Shu HB, et al. : TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4(4):387–96. 10.1016/S1074-7613(00)80252-6 [DOI] [PubMed] [Google Scholar]
- 38. Hsu H, Shu HB, Pan MG, et al. : TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84(2):299–308. 10.1016/S0092-8674(00)80984-8 [DOI] [PubMed] [Google Scholar]
- 39. Lee DJ, Du F, Chen SW, et al. : Regulation and Function of the Caspase-1 in an Inflammatory Microenvironment. J Invest Dermatol. 2015;135(8):2012–20. 10.1038/jid.2015.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee P, Lee DJ, Chan C, et al. : Dynamic expression of epidermal caspase 8 simulates a wound healing response. Nature. 2009;458(7237):519–23. 10.1038/nature07687 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Vince JE: When Beauty Is Skin Deep: Regulation of the Wound Response by Caspase-8, RIPK3, and the Inflammasome. J Invest Dermatol. 2015;135(8):1936–9. 10.1038/jid.2015.185 [DOI] [PubMed] [Google Scholar]
- 42. Vince JE, Chau D, Callus B, et al. : TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1-TRAF2 complex to sensitize tumor cells to TNFalpha. J Cell Biol. 2008;182(1):171–84. 10.1083/jcb.200801010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Varfolomeev E, Blankenship JW, Wayson SM, et al. : IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131(4):669–81. 10.1016/j.cell.2007.10.030 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Pierdomenico M, Negroni A, Stronati L, et al. : Necroptosis is active in children with inflammatory bowel disease and contributes to heighten intestinal inflammation. Am J Gastroenterol. 2014;109(2):279–87. 10.1038/ajg.2013.403 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Li S, Zhang L, Yao Q, et al. : Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature. 2013;501(7466):242–6. 10.1038/nature12436 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Upton JW, Kaiser WJ, Mocarski ES: DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe. 2012;11(3):290–7. 10.1016/j.chom.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Upton JW, Kaiser WJ, Mocarski ES: Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe. 2010;7(4):302–13. 10.1016/j.chom.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo H, Omoto S, Harris PA, et al. : Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe. 2015;17(2):243–51. 10.1016/j.chom.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang Z, Wu SQ, Liang Y, et al. : RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe. 2015;17(2):229–42. 10.1016/j.chom.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 50. Omoto S, Guo H, Talekar GR, et al. : Suppression of RIP3-dependent necroptosis by human cytomegalovirus. J Biol Chem. 2015;290(18):11635–48. 10.1074/jbc.M115.646042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu XN, Yang ZH, Wang XK, et al. : Distinct roles of RIP1-RIP3 hetero- and RIP3-RIP3 homo-interaction in mediating necroptosis. Cell Death Differ. 2014;21(11):1709–20. 10.1038/cdd.2014.77 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Li J, McQuade T, Siemer AB, et al. : The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150(2):339–50. 10.1016/j.cell.2012.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Cai Z, Jitkaew S, Zhao J, et al. : Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol. 2014;16(1):55–65. 10.1038/ncb2883 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Hildebrand JM, Tanzer MC, Lucet IS, et al. : Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc Natl Acad Sci U S A. 2014;111(42):15072–7. 10.1073/pnas.1408987111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dondelinger Y, Declercq W, Montessuit S, et al. : MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 2014;7(4):971–81. 10.1016/j.celrep.2014.04.026 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Kaiser WJ, Sridharan H, Huang C, et al. : Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J Biol Chem. 2013;288(43):31268–79. 10.1074/jbc.M113.462341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun L, Wang H, Wang Z, et al. : Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148(1–2):213–27. 10.1016/j.cell.2011.11.031 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Wu J, Huang Z, Ren J, et al. : Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res. 2013;23(8):994–1006. 10.1038/cr.2013.91 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Zhao J, Jitkaew S, Cai Z, et al. : Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci U S A. 2012;109(14):5322–7. 10.1073/pnas.1200012109 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Murphy JM, Czabotar PE, Hildebrand JM, et al. : The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39(3):443–53. 10.1016/j.immuni.2013.06.018 [DOI] [PubMed] [Google Scholar]
- 61. Murphy JM, Zhang Q, Young SN, et al. : A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J. 2014;457(2):323–34. 10.1042/BJ20131174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Manning G, Whyte DB, Martinez R, et al. : The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Chen X, Li W, Ren J, et al. : Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24(1):105–21. 10.1038/cr.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Rodriguez DA, Weinlich R, Brown S, et al. : Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 2015. 10.1038/cdd.2015.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tanzer MC, Tripaydonis A, Webb AI, et al. : Necroptosis signalling is tuned by phosphorylation of MLKL residues outside the pseudokinase domain activation loop. Biochem J. 2015;471(2):255–65. 10.1042/BJ20150678 [DOI] [PubMed] [Google Scholar]
- 66. Su L, Quade B, Wang H, et al. : A plug release mechanism for membrane permeation by MLKL. Structure. 2014;22(10):1489–500. 10.1016/j.str.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Tait SW, Oberst A, Quarato G, et al. : Widespread mitochondrial depletion via mitophagy does not compromise necroptosis. Cell Rep. 2013;5(4):878–85. 10.1016/j.celrep.2013.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Holler N, Zaru R, Micheau O, et al. : Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000;1(6):489–95. 10.1038/82732 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Degterev A, Hitomi J, Germscheid M, et al. : Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4(5):313–21. 10.1038/nchembio.83 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 70. Degterev A, Huang Z, Boyce M, et al. : Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1(2):112–9. 10.1038/nchembio711 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 71. Polykratis A, Hermance N, Zelic M, et al. : Cutting edge: RIPK1 Kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J Immunol. 2014;193(4):1539–43. 10.4049/jimmunol.1400590 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Moujalled DM, Cook WD, Okamoto T, et al. : TNF can activate RIPK3 and cause programmed necrosis in the absence of RIPK1. Cell Death Dis. 2013;4:e465. 10.1038/cddis.2012.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. He S, Liang Y, Shao F, et al. : Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc Natl Acad Sci U S A. 2011;108(50):20054–9. 10.1073/pnas.1116302108 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. O'Donnell MA, Perez-Jimenez E, Oberst A, et al. : Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol. 2011;13(12):1437–42. 10.1038/ncb2362 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Hitomi J, Christofferson DE, Ng A, et al. : Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135(7):1311–23. 10.1016/j.cell.2008.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Moquin DM, McQuade T, Chan FK: CYLD deubiquitinates RIP1 in the TNFα-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS One. 2013;8(10):e76841. 10.1371/journal.pone.0076841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. de Almagro MC, Goncharov T, Newton K, et al. : Cellular IAP proteins and LUBAC differentially regulate necrosome-associated RIP1 ubiquitination. Cell Death Dis. 2015;6:e1800. 10.1038/cddis.2015.158 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Welz PS, Wullaert A, Vlantis K, et al. : FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477(7364):330–4. 10.1038/nature10273 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Bonnet MC, Preukschat D, Welz PS, et al. : The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity. 2011;35(4):572–82. 10.1016/j.immuni.2011.08.014 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Chen WW, Yu H, Fan HB, et al. : RIP1 mediates the protection of geldanamycin on neuronal injury induced by oxygen-glucose deprivation combined with zVAD in primary cortical neurons. J Neurochem. 2012;120(1):70–7. 10.1111/j.1471-4159.2011.07526.x [DOI] [PubMed] [Google Scholar]
- 81. Fearns C, Pan Q, Mathison JC, et al. : Triad3A regulates ubiquitination and proteasomal degradation of RIP1 following disruption of Hsp90 binding. J Biol Chem. 2006;281(45):34592–600. 10.1074/jbc.M604019200 [DOI] [PubMed] [Google Scholar]
- 82. Gentle IE, Wong WW, Evans JM, et al. : In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8. J Biol Chem. 2011;286(15):13282–91. 10.1074/jbc.M110.216226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lewis J, Devin A, Miller A, et al. : Disruption of hsp90 function results in degradation of the death domain kinase, receptor-interacting protein (RIP), and blockage of tumor necrosis factor-induced nuclear factor-kappaB activation. J Biol Chem. 2000;275(14):10519–26. 10.1074/jbc.275.14.10519 [DOI] [PubMed] [Google Scholar]
- 84. Pantano C, Shrivastava P, McElhinney B, et al. : Hydrogen peroxide signaling through tumor necrosis factor receptor 1 leads to selective activation of c-Jun N-terminal kinase. J Biol Chem. 2003;278(45):44091–6. 10.1074/jbc.M308487200 [DOI] [PubMed] [Google Scholar]
- 85. Vanden Berghe T, Kalai M, van Loo G, et al. : Disruption of HSP90 function reverts tumor necrosis factor-induced necrosis to apoptosis. J Biol Chem. 2003;278(8):5622–9. 10.1074/jbc.M208925200 [DOI] [PubMed] [Google Scholar]
- 86. Cho YS, Challa S, Moquin D, et al. : Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137(6):1112–23. 10.1016/j.cell.2009.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. Park SY, Shim SH, Cho YS: Distinctive roles of receptor-interacting protein kinases 1 and 3 in caspase-independent cell death of L929. Cell Biochem Funct. 2014;32(1):62–9. 10.1002/cbf.2972 [DOI] [PubMed] [Google Scholar]
- 88. Li D, Xu T, Cao Y, et al. : A cytosolic heat shock protein 90 and cochaperone CDC37 complex is required for RIP3 activation during necroptosis. Proc Natl Acad Sci U S A. 2015;112(16):5017–22. 10.1073/pnas.1505244112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li L, Chen W, Liang Y, et al. : The Gβγ-Src signaling pathway regulates TNF-induced necroptosis via control of necrosome translocation. Cell Res. 2014;24(4):417–32. 10.1038/cr.2014.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kang TB, Oh GS, Scandella E, et al. : Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice. J Immunol. 2008;181(4):2522–32. 10.4049/jimmunol.181.4.2522 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 91. Oberst A, Dillon CP, Weinlich R, et al. : Catalytic activity of the caspase-8-FLIP L complex inhibits RIPK3-dependent necrosis. Nature. 2011;471(7338):363–7. 10.1038/nature09852 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 92. Lin Y, Devin A, Rodriguez Y, et al. : Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13(19):2514–26. 10.1101/gad.13.19.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Feng S, Yang Y, Mei Y, et al. : Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal. 2007;19(10):2056–67. 10.1016/j.cellsig.2007.05.016 [DOI] [PubMed] [Google Scholar]
- 94. Silke J, Meier P: Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation. Cold Spring Harb Perspect Biol. 2013;5(2): pii: a008730. 10.1101/cshperspect.a008730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Moulin M, Anderton H, Voss AK, et al. : IAPs limit activation of RIP kinases by TNF receptor 1 during development. EMBO J. 2012;31(7):1679–91. 10.1038/emboj.2012.18 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Onizawa M, Oshima S, Schulze-Topphoff U, et al. : The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol. 2015;16(6):618–27. 10.1038/ni.3172 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 97. Lee E, Kim J, Ahn Y, et al. : Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat Commun. 2012;3:978. 10.1038/ncomms1981 [DOI] [PubMed] [Google Scholar]
- 98. Vince JE, Pantaki D, Feltham R, et al. : TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (TNF) to efficiently activate NF-{kappa}B and to prevent TNF-induced apoptosis. J Biol Chem. 2009;284(51):35906–15. 10.1074/jbc.M109.072256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Karl I, Jossberger-Werner M, Schmidt N, et al. : TRAF2 inhibits TRAIL- and CD95L-induced apoptosis and necroptosis. Cell Death Dis. 2014;5:e1444. 10.1038/cddis.2014.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Petersen SL, Chen TT, Lawrence DA, et al. : TRAF2 is a biologically important necroptosis suppressor. Cell Death Differ. 2015;22(11):1846–57. 10.1038/cdd.2015.35 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 101. Chen W, Wu J, Li L, et al. : Ppm1b negatively regulates necroptosis through dephosphorylating Rip3. Nat Cell Biol. 2015;17(4):434–44. 10.1038/ncb3120 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 102. Chen W, Zhou Z, Li L, et al. : Diverse sequence determinants control human and mouse receptor interacting protein 3 (RIP3) and mixed lineage kinase domain-like (MLKL) interaction in necroptotic signaling. J Biol Chem. 2013;288(23):16247–61. 10.1074/jbc.M112.435545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wertz IE, O'Rourke KM, Zhou H, et al. : De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–9. 10.1038/nature02794 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 104. Wertz I, Dixit V: A20--a bipartite ubiquitin editing enzyme with immunoregulatory potential. Adv Exp Med Biol. 2014;809:1–12. 10.1007/978-1-4939-0398-6_1 [DOI] [PubMed] [Google Scholar]
- 105. Yoon S, Bogdanov K, Kovalenko A, et al. : Necroptosis is preceded by nuclear translocation of the signaling proteins that induce it. Cell Death Differ. 2015. 10.1038/cdd.2015.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Z, Jiang H, Chen S, et al. : The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148(1–2):228–43. 10.1016/j.cell.2011.11.030 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 107. Moujalled DM, Cook WD, Murphy JM, et al. : Necroptosis induced by RIPK3 requires MLKL but not Drp1. Cell Death Dis. 2014;5(2):e1086. 10.1038/cddis.2014.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Remijsen Q, Goossens V, Grootjans S, et al. : Depletion of RIPK3 or MLKL blocks TNF-driven necroptosis and switches towards a delayed RIPK1 kinase-dependent apoptosis. Cell Death Dis. 2014;5(1):e1004. 10.1038/cddis.2013.531 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 109. Schenk B, Fulda S: Reactive oxygen species regulate Smac mimetic/TNFα-induced necroptotic signaling and cell death. Oncogene. 2015. 10.1038/onc.2015.35 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 110. Lin Y, Choksi S, Shen HM, et al. : Tumor necrosis factor-induced nonapoptotic cell death requires receptor-interacting protein-mediated cellular reactive oxygen species accumulation. J Biol Chem. 2004;279(11):10822–8. 10.1074/jbc.M313141200 [DOI] [PubMed] [Google Scholar]
- 111. Vanlangenakker N, Vanden Berghe T, Bogaert P, et al. : cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ. 2011;18(4):656–65. 10.1038/cdd.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 112. Marshall KD, Baines CP: Necroptosis: is there a role for mitochondria? Front Physiol. 2014;5:323. 10.3389/fphys.2014.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. McQuade T, Cho Y, Chan FK, et al. : Positive and negative phosphorylation regulates RIP1- and RIP3-induced programmed necrosis. Biochem J. 2013;456(3):409–15. 10.1042/BJ20130860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bertrand MJ, Milutinovic S, Dickson KM, et al. : cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30(6):689–700. 10.1016/j.molcel.2008.05.014 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 115. Mahoney DJ, Cheung HH, Mrad RL, et al. : Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105(33):11778–83. 10.1073/pnas.0711122105 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 116. O'Donnell MA, Legarda-Addison D, Skountzos P, et al. : Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17(5):418–24. 10.1016/j.cub.2007.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Varfolomeev E, Goncharov T, Fedorova AV, et al. : c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem. 2008;283(36):24295–9. 10.1074/jbc.C800128200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Dynek JN, Goncharov T, Dueber EC, et al. : c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29(24):4198–209. 10.1038/emboj.2010.300 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 119. He S, Wang L, Miao L, et al. : Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137(6):1100–11. 10.1016/j.cell.2009.05.021 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 120. Miao W, Qu Z, Shi K, et al. : RIP3 S-nitrosylation contributes to cerebral ischemic neuronal injury. Brain Res. 2015;1627:165–76. 10.1016/j.brainres.2015.08.020 [DOI] [PubMed] [Google Scholar]
- 121. Dephoure N, Zhou C, Villén J, et al. : A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105(31):10762–7. 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation