Abstract
Cytolytic proteins and peptide toxins are classical virulence factors of several bacterial pathogens which disrupt epithelial barrier function, damage cells and activate or modulate host immune responses. Until now human pathogenic fungi were not known to possess such toxins. Here we identify the first fungal cytolytic peptide toxin in the opportunistic pathogen Candida albicans. This secreted toxin directly damages epithelial membranes, triggers a danger response signaling pathway and activates epithelial immunity. Toxin-mediated membrane permeabilization is enhanced by a positively charged C-terminus and triggers an inward current concomitant with calcium influx. C. albicans strains lacking this toxin do not activate or damage epithelial cells and are avirulent in animal models of mucosal infection. We propose the name ‘Candidalysin’ for this cytolytic peptide toxin; a newly identified, critical molecular determinant of epithelial damage and host recognition of the clinically important fungus, C. albicans.
Introduction
The ability of mucosal surfaces to discriminate between commensal and pathogenic microbes is essential to human health. The fungus Candida albicans is normally a benign member of the human microbiota but is also responsible for millions of mucosal infections each year in immunocompromised hosts, often with severe morbidity1. A defining feature of C. albicans pathogenesis is the transition from yeast to invasive filamentous hyphae2. Hyphae damage mucosal epithelia and induce activation of the activating protein-1 (AP-1) transcription factor c-Fos (via p38-MAPK) and the MAPK phosphatase MKP1 (via ERK1/2-MAPK), which trigger pro-inflammatory cytokine responses3–7. These signaling events constitute a ‘danger response’ against invasive hyphae, thus serving as a sensor of pathogenic C. albicans invasion8–14. However, it is unclear how C. albicans hyphae induce epithelial inflammatory responses and cell damage during mucosal infections. Here we identify and characterize Candidalysin, the first cytolytic peptide toxin isolated from any human fungal pathogen, as the hyphal factor critical for epithelial immune activation and C. albicans mucosal infection.
Ece1p is critical for epithelial activation and damage
Despite the well-known association between filamentation and virulence, the molecular mechanism underlying hypha-driven epithelial activation and mucosal damage has remained obscure. To elucidate this mechanism, we screened a panel of C. albicans gene deletion mutants that targeted key processes, pathways and proteins known or predicted to be associated with the yeast-hyphal transition and pathogenicity (62 strains). Only hypha-producing strains induced MKP1 phosphorylation (p-MKP1), c-Fos, cytokines (IL-1α, IL-6, G-CSF) and damage in oral epithelial cells (Extended Data Table 1). However, one C. albicans mutant (ece1Δ/Δ)15 formed normal hyphae but was incapable of inducing these epithelial danger responses. C. albicans ECE1 (extent of cell elongation) is highly expressed by hyphae during epithelial infection (Extended Data Fig. 1a, b) and is predicted to encode a secreted protein16. To probe its function we generated a panel of C. albicans ECE1mutants (Extended Data Table 2). The ece1Δ/Δ strain formed normal hyphae on (Extended Data Fig. 1c), and adhered to and invaded human epithelial cells similarly to wild type C. albicans (Extended Data Fig. 1d, e). Indeed, ece1Δ/Δ was capable of extensive epithelial invasion, penetrating through multiple epithelial cells (Extended Data Fig. 1f). Despite this, invasive ece1Δ/Δ hyphae did not damage epithelia or induce p-MKP1/c-Fos mediated danger responses or cytokine secretion (Fig. 1a–d). Thus, Ece1p is critical for epithelial damage and innate recognition of C. albicans hyphae in vitro.
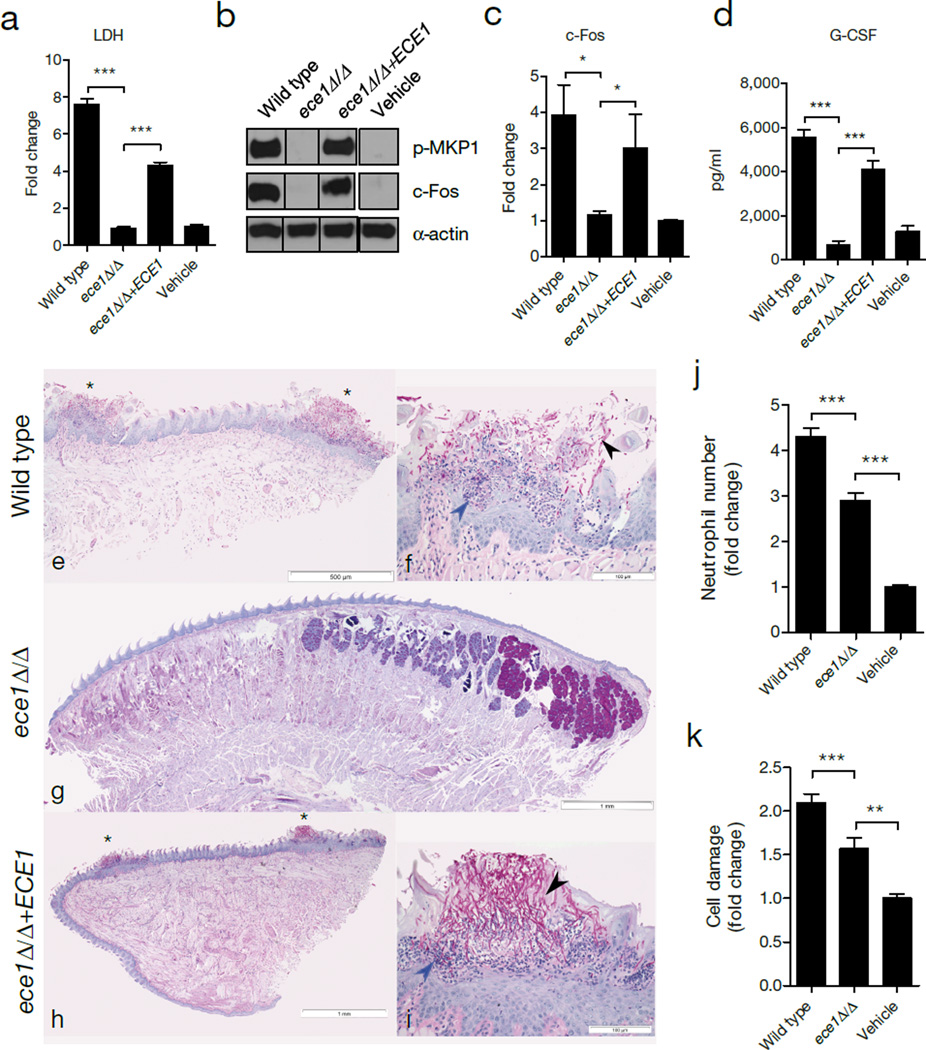
Figure 1. ECE1 is required for epithelial activation and C. albicans infection.
TR146 cells were infected with the indicated C. albicans strains. (a) LDH release 24 h post-infection (p.i.) (MOI = 0.1). (b) Induction of p-MKP-1 and c-Fos at 2 h p.i. (MOI = 10). (c) c-Fos DNA binding at 3 h p.i. (MOI = 10). (d) G-CSF production at 24 h p.i. (MOI = 0.01). (e-i) PAS-stained tongues from mice subjected to OPC 2 d p.i. (e, g, h) Whole-mount (x25) and (f, i) high-power (x200) views of PAS-stained tongues of mice infected with C. albicans wild type (e, f), ece1Δ/Δ (g) and ece1Δ/Δ+ECE1 (h, i). Invading hyphae (black arrow) and inflammatory cells (blue arrow) are indicated. (j) Quantification of neutrophils in zebrafish swimbladder following infection with wild type C. albicans (n (number of fish) = 47), ece1Δ/Δ (n = 53) or PBS (n = 40). (k) Quantification of damaged cells in zebrafish swimbladder after infection with C. albicans wild type (n = 73), ece1Δ/Δ (n = 59) or vehicle (n = 63). Data are representative (b, e-i) or the mean (a, c-d, j-k) of three biological replicates. Error bars ± SEM. Data were analyzed by one-way ANOVA (a, d), paired T test (c) or Kruskal-Wallis (j, k) and * = P < 0.05, ** = P < 0.01, *** = P < 0.001. For gel source data, see Supplementary Figure 1.
Ece1p is critical for mucosal pathogenesis
We next assessed the role of ECE1 in two in vivo models of C. albicans mucosal infection. In murine oropharyngeal candidiasis (OPC)17, mice infected with C. albicans wild type or ECE1 re-integrant (ece1Δ/Δ+ECE1) strains exhibited disease symptoms, including extensive hyphal invasion of the tongue epithelium, micro-abscesses of infiltrating neutrophils and tissue damage (Fig. 1e, f, h, i). In contrast, tongue tissue from ece1Δ/Δ-infected animals (n = 17/20) showed no invasive fungi and no inflammatory infiltrates or damage (Fig. 1g). We detected very low numbers of ece1Δ/Δ cells in only 3/20 mice (Extended Data Fig. 2a), which showed no evidence of local epithelial damage (not shown). Quantification of histology sections indicated that the percentage of epithelial surface infected was significantly greater with the wild type and ECE1 re-integrant strains (Extended DataFig. 2b). In a zebrafish swimbladder model of mucosal infection18,19, neutrophil recruitment and tissue damage were both significantly lower following ece1Δ/Δ infection as compared with the wild type strain (Fig. 1j, k, Extended DataFig. 2c, d). Therefore, C. albicans Ece1p is critical for mucosal pathogenesis and is an innate immune activator in vivo.
Ece1p encodes a cytolytic peptide toxin
Ece1p is an in vitro substrate for Kex2p, a Golgi-located protease that cleaves proteins after lysine-arginine (KR) motifs20. Ece1p contains seven KR-processing sites, suggesting it has the potential to produce eight secreted peptides from C. albicans20 (Extended Data Fig. 3a, b). Liquid chromatography – tandem mass spectrometry (LC-MS/MS) analysis confirmed that recombinant Kex2p (rKex2p) processes recombinant Ece1p (rEce1p) and that all eight peptides generated terminated in KR (and fragments thereof, showing that less efficient processing occurs also after a single K or R) (Supplementary information). The importance of Kex2p–mediated Ece1p processing was demonstrated using a kex2Δ/Δ null strain21, which was unable to damage oral epithelia or induce p-MKP1/c-Fos mediated danger responses or cytokine secretion (Extended Data Table 1). To determine which Ece1p peptide(s) were responsible for epithelial activation and damage, oral epithelial cells were incubated with peptides Ece1-I-VIII (1.5 – 70 µM). Only Ece1-III62–93 induced p-MKP1, c-Fos, cytokines and damage (Fig. 2a–c, Extended Data Fig. 3c–e). Notably, low Ece1-III62–93 concentrations (1.5 – 15 µM) were sufficient to induce c-Fos DNA binding (Fig. 2d), G-CSF and GM-CSF (Fig. 2c, Extended Data Fig. 3c), while high Ece1-III62–93 concentrations (70 µM) were required to induce damage (Fig. 2e) and the damage-associated cytokines IL-1α and IL-6, respectively (Extended Data Fig. 3d, e). Ece1-III62–93 could also directly lyse multiple human epithelial cell types and induce hemolysis of red blood cells, a classical test for cytotoxin activity (not shown). Neither the N-terminal hydrophobic region (Ece1-III62–85) nor the C-terminal hydrophilic region (Ece1-III86–93) induced p-MKP1, c-Fos, cytokines or damage of epithelial cells, either individually or in combination (Extended Data Fig. 3f–h), demonstrating that the peptide containing both regions is required for activity. Therefore, Ece1-III62–93 is the active region of Ece1p, acting as an epithelial immune activator and a cytolytic agent.
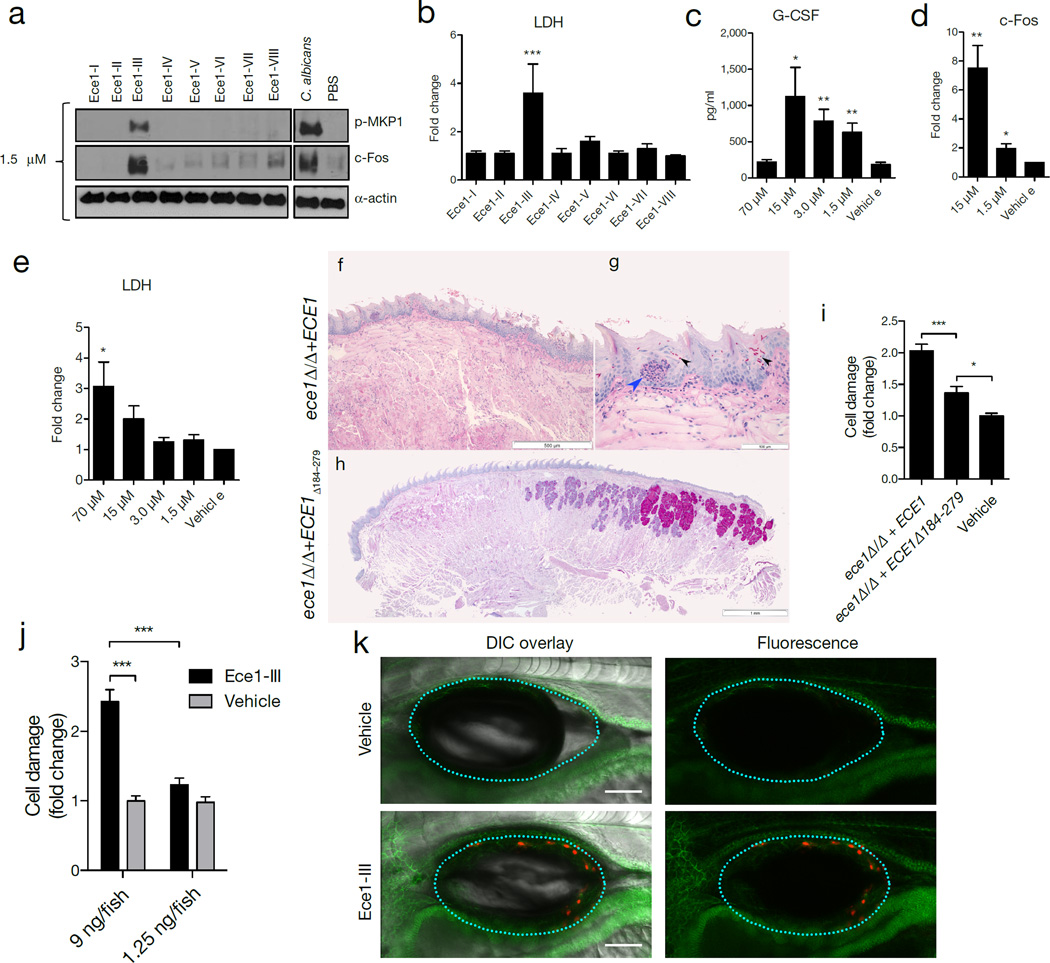
Figure 2. Ece1-III62–93 is the active region of Ece1p and is required for TR146 cell activation and mucosal C. albicans infection.
(a) Induction of p-MKP-1 and c-Fos 2 h post-stimulation (p.s.) with Ece1 peptides at 1.5 µM. (b) LDH release 24 h p.s. with 70 µM Ece1 peptides. (c) Induction of G-CSF 24 h p.s. with Ece1-III62–93 (d) c-Fos DNA binding induction 3 h p.s. with sub-lytic concentrations of Ece1-III62–93 (e) LDH release 24 h p.s. with Ece1-III62–93 (f-h) PAS stained tongue sections from mice subjected to OPC, 2 d p.i. with (f, g) C. albicans ece1Δ/Δ+ECE1 (x25 and x200) or (h) ece1Δ/Δ+ECE1Δ184–279. Invading hyphae (black arrows) and infiltrating inflammatory cells (blue arrow) are shown. (i) Damaged cells in a zebrafish swimbladder 24 h p.i. with C. albicans ece1Δ/Δ+ECE1 (n (number of fish) = 44), ece1Δ/Δ+ECE1Δ184–279 (n = 58) or vehicle (n = 58). (j) Damaged cells in zebrafish swimbladders after stimulation with 9 ng (n = 51) or 1.25 ng (n = 56) Ece1-III62–93, or vehicle (40% DMSO, n = 54 and 5% DMSO, n = 55). (k) Co-localization of adherens junctions (α-catenin-citrine) with Ece1-III62–93-damaged cells (Sytox Orange-positive cells) in a zebrafish swimbladder. Data are representative (a, f-h, k) or mean (b-e, i-j) of three biological replicates (a-d) or ten mice or fish (f-h, k). Error bars show ± SEM. Data were analyzed by one-way ANOVA (b, c, e) paired T test (d) or Kruskal-Wallis (i, j). * = P < 0.05, ** = P < 0.01, *** = P < 0.001 (compared with vehicle control unless otherwise indicated). For gel source data, see Supplementary Figure 1.
To confirm that Ece1-III62–93 drives epithelial activation and fungal pathogenicity, we generated a C. albicans strain lacking only the Ece1-III62–93 region (ece1Δ/Δ+ECE1Δ184–279). LC-MS/MS analysis showed that the modified protein in this strain is stable, secreted and processed into each of the predicted peptide fragments, with the exception of the deleted peptide toxin (Supplementary information). Like ece1Δ/Δ, ece1Δ/Δ+ECE1Δ184–279 efficiently formed invasive hyphae (not shown). However, ece1Δ/Δ+ECE1Δ184–279 was unable to induce p-MKP1, c-Fos DNA binding, cytokines, or damage epithelia (Extended Fig. 3i–l). In murine OPC, unlike the ece1Δ/Δ+ECE1 complemented strain, ece1Δ/Δ+ECE1Δ184–279-infected mice demonstrated absent (n = 4/10) or low (n = 6/10) fungal burdens, with no evidence of inflammatory infiltrates or local epithelial damage (Fig. 2f–h, Extended Data Fig. 4a and 4b) Likewise, ece1Δ/Δ+ECE1Δ184–279 did not induce full damage in the zebrafish swimbladder model (Fig. 2i, Extended Data Fig. 4c). In contrast, injection of lytic doses of Ece1-III62–93 into the swimbladder induced epithelial damage (Fig. 2j, k). Thus, Ece1-III62–93 is both necessary and sufficient for epithelial immune activation, damage and mucosal infection in vivo.
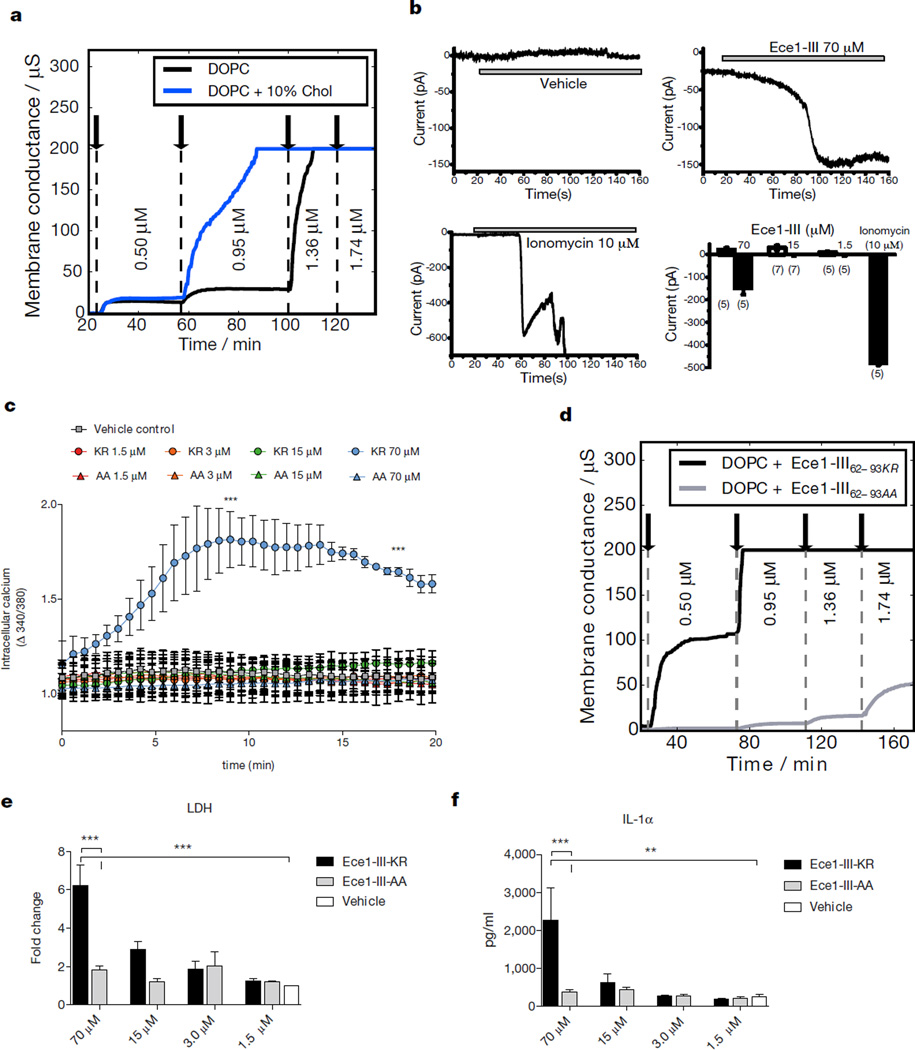
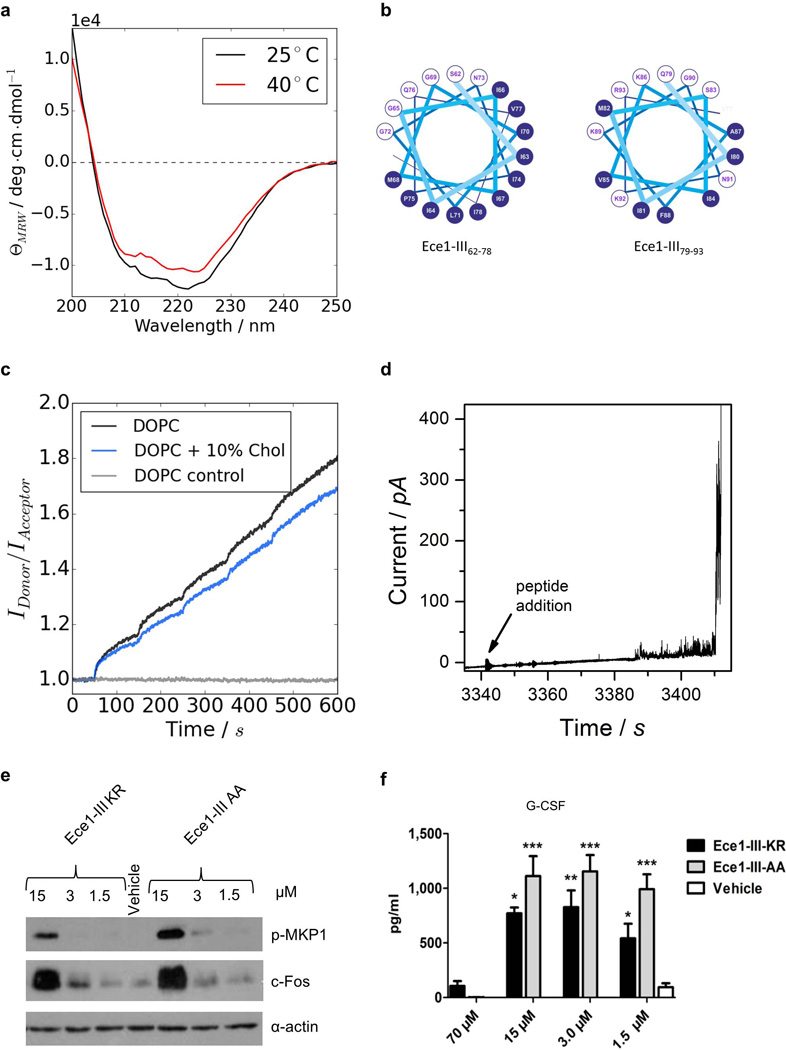
The amphipathic properties of Ece1-III62–93 (SIIGIIMGILGNIPQVIQIIMSIVKAFKGNKR) coupled with the α-helical structure of the N-terminal hydrophobic region (Extended Data Fig. 5a, b) indicated that this fungal peptide may act similarly to cationic antimicrobial peptides and peptide toxins such as melittin22 (honey bee), magainin 223 (African clawed frog) and alamethicin24 (Trichoderma viride). Cytolytic peptide toxins have not previously been found in human pathogenic fungi but bacterial cytolytic toxins are known to induce lesions after binding to target cell membranes25,26. To investigate the importance of lipid composition for Ece1-III62–93-mediated cytolysis, we used Förster resonance energy transfer (FRET) and electrical impedance spectroscopy to analyze the interactions of Ece1-III62–93 with model membranes comprised of lipid bilayers of dioleoylphosphatidylcholine (DOPC) with or without cholesterol. While Ece1-III62–93 was able to efficiently intercalate into DOPC membranes (Extended Data Fig. 5c), Ece1-III62–93 permeabilization was enhanced in the presence of cholesterol (Fig. 3a,). Ece1-III62–93-induced lesions were heterogeneous and transient (Extended Data Fig. 5d), indicating that the peptide may damage target membranes through a ‘carpet-like’ mechanism27. Patch-clamp analysis of epithelial cells demonstrated that lesion formation by Ece1-III62–93 is rapid and causes an inward current (Fig. 3b), associated with calcium influx (Fig. 3c). Similar phenomena occur with bacterial cytolytic toxins, which are known to trigger cell activation25,26,28.
Figure 3. Ece1-III62–93 functions as a cytolytic peptide toxin.
(a) Kinetic changes in conductance of tethered lipid membranes after exposure to different concentrations of Ece1-III62–93. (b) Evoked inward current at a membrane potential of −60 mV in TR146 cells post-addition of Ece1-III62–93 or ionomycin (positive control); individual (representative) and cumulative changes (bar chart - number of cells analyzed below each bar) shown. (c) Intracellular calcium level kinetics in TR146 cells post-stimulation (p.s.) with Ece1-III62–93 wild type (Ece1-III62–93KR) or Ece1-III62–93 AA C-terminal substitution (Ece1-III62–93AA). (d) Kinetic changes in conductance of tethered DOPC membranes after exposure to different concentrations of Ece1-III62–93KR and Ece1-III62–93AA. (e) LDH release and (f) Secretion of IL-1α from TR146 cells 24 h p.s. with Ece1-III62–93KR or Ece1-III62–93AA. Data shown are representative (a, d) or mean (b-c, e-f) of three biological replicates. Error bars show ± SEM. Data were analyzed by one-way ANOVA (c, e and f). ** = P < 0.01, *** = P < 0.001.
We postulated that the positively-charged C-terminal KR residues of Ece1-III62–93 might be critical for interacting with negatively-charged components of host membranes to promote lesion formation. Substitution of the KR motif to AA (alanine-alanine; Ece1-III62–93AA) did not affect membrane intercalation (not shown) but significantly reduced the peptide’s ability to permeabilize membranes, damage epithelial cells and induce calcium influx (Fig. 3c–e). Thus, the positive C-terminus of Ece1-III62–93 is critical for lesion formation and damage induction in epithelial membranes. Notably, Ece1-III62–93AA still induced p-MKP1, c-Fos and the non-damage associated cytokine G-CSF (Extended Data Fig. 5e, f) but not the damage-associated cytokine IL-1α (Fig. 3f), suggesting that Ece1-III62–93AA can be recognized by epithelial immunity without damaging cells. This finding is important as it means that epithelial cells are not only responding to damage but have evolved to specifically recognize the peptide.
Ece1-III62–92K is a secreted cytolytic peptide toxin
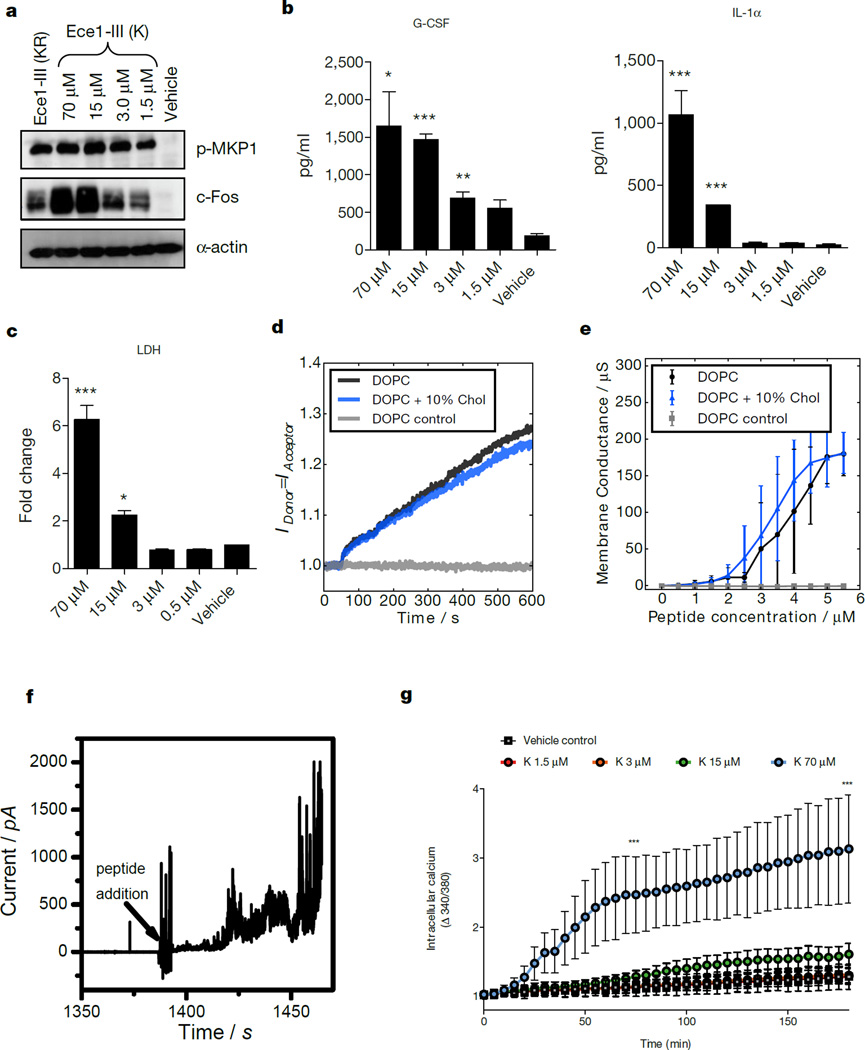
To demonstrate that Ece1-III is generated during epithelial infection, we performed LC-MS/MS analysis on the secretome from wild-type C. albicans hyphae grown in the presence and absence of epithelial cells (Supplementary information). Notably, Ece1-III was the only peptide detected in the presence of epithelial cells, indicating that the fungus secretes this toxin during mucosal infection. However, the predominant form of secreted Ece1-III terminated in a K residue (SIIGIIMGILGNIPQVIQIIMSIVKAFKGNK; Ece1-III62–92K) and not KR (SIIGIIMGILGNIPQVIQIIMSIVKAFKGNKR; Ece1-III62–93KR) (Extended Data Table 3). In fungi, it is known that following Kex2p processing, many proteins are subsequently cleaved by Kex1p29 (also in the Golgi), removing the C-terminal R. LC-MS/MS analysis on the hyphal secretome of a kex1Δ/Δ mutant demonstrated that the predominant peptide secreted terminates in KR (not K) (Supplementary information). Therefore, Ece1p is also subject to ordered Kex2p/Kex1p processing. Accordingly, we confirmed that Ece1-III62–92K functioned similarly to Ece1-III62–93KR with respect to epithelial cell activation. Specifically, Ece1-III62–92K is also α-helical (not shown) and induces c-Fos, p-MKP1, cytokines (IL-1α, G-CSF), damage (LDH), membrane intercalation and permeabilization, and calcium influx (Fig 4a–g). Thus, the dominant peptide secreted from C. albicans hyphae during mucosal infection is Ece1-III62–92K, which acts as a cytolytic peptide toxin that activates epithelial cells.
Figure 4. Ece1-III62–92K functions as a cytolytic peptide toxin that activates and damages epithelial cells.
(a) Induction of p-MKP-1 and c-Fos 2 h post-stimulation (p.s.), and (b) secretion of G-CSF and IL-1-α 24 h p.s., and (c) LDH release 24 h p.s. of TR146 cells with Ece1-III62–92K. (d) Förster resonance energy transfer (FRET) showing intercalation of Ece1-III62–92K (10 µM) into lipid liposomes. (e) Average peptide concentration-dependent changes in conductance of tethered lipid membranes. (f) Ece1-III62–92K (4 µM) induced permeabilization of planar lipid membranes showing heterogeneous and transient lesions leading to membrane rupture. (g) Intracellular calcium level kinetics in TR146 cells p.s. with Ece1-III62–92K. Data shown are representative (a, d, f) or mean (b-c, e, g) of three biological replicates. Error bars show ± SEM. Data are analyzed by one-way ANOVA (b, c). * = P < 0.05, ** = P < 0.01, *** = P < 0.001 (compared with vehicle control). For gel source data, see Supplementary Figure 1.
Based on these data, we propose a model of C. albicans mucosal infection whereby invasive hyphae secrete Ece1-62–92K into a membrane-bound ‘invasion pocket’30,31, facilitating peptide accumulation (Extended Data Fig 6). During early stages of infection, sub-lytic concentrations of Ece1-III62–92K induce epithelial immunity by activating the ‘danger response’ pathway (p-MKP1/c-Fos), alerting the host to the transition from colonizing yeast to invasive, toxin-producing hyphae. As infection progresses, Ece1-III62–92K levels accumulate and elicit direct tissue damage. Mechanistically, we propose that the asymmetric distribution of charge along the α-helix of Ece1-III62–92K facilitates correct peptide orientation relative to the host membrane, enabling intercalation, permeabilization and calcium influx. In conclusion, our data identifies C. albicans Ece1-III62–92K as the first cytolytic peptide toxin in a human fungal pathogen and reveals the molecular mechanisms of epithelial damage and host recognition of this clinically important fungus. We propose the name ‘Candidalysin’ for this newly discovered fungal toxin.
METHODS
Cell lines, reagents and Candida strains
Experiments were carried out using the TR146 buccal epithelial squamous cell carcinoma line32 obtained from the European Collection of Authenticated Cell Cultures (ECACC) and grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Cells were routinely tested for mycoplasma contamination using mycoplasma-specific primers and were found to be negative. Prior to stimulation, confluent TR146 cells were serum-starved overnight, and all experiments were carried out in serum-free DMEM. C. albicans wild type strains included the autotrophic strain BWP17+CIp3033 and the parental strain SC531434. Other C. albicans strains used and their sources are listed in Extended Data Tables 1 and 2. C. albicans cultures were grown in YPD medium (1% yeast extract, 2% peptone, 2% dextrose) at 30°C overnight. Cultures were washed in sterile PBS and adjusted to the required cell density. Antibodies to phospho-MKP1 and c-Fos were from Cell Signalling Technologies (New England Biolabs UK), mouse anti-human α-actin was from Millipore (UK), and goat anti-mouse and anti-rabbit horseradish peroxidase (HRP)-conjugated antibodies were from Jackson Immunologicals Ltd (Stratech Scientific, UK). Ece1p peptides were synthesized commercially (Proteogenix (France) or Peptide Synthetics (UK).
Generation of C. albicans ECE1 mutant strains
ECE1 deletion was performed as previously described35. Deletion cassettes were generated by PCR36. Primers ECE1-FG and ECE1-RG were used to amplify pFA-HIS1 and pFA-ARG4 -based markers. C. albicans BWP1737, was sequentially transformed38 with the ECE1-HIS1 and ECE1-ARG4 deletion cassettes and then transformed with CIp1039, yielding the ece1Δ/Δ deletion strain. For complementation, the ECE1 gene plus upstream and downstream intergenic regions were amplified with primers ECE1-RecF3k and ECE1-RecR and cloned into plasmid CIp10 at MluI and SalI sites. This plasmid was transformed into the uridine auxotrophic ece1Δ/Δ strain, yielding the ece1Δ/Δ+ECE1 complemented strain. For generation of the ece1Δ/Δ+ECE1Δ184–279 strain, the CIp10-ECE1 was amplified with primers Pep3-F1 and Pep3-R1, digested with ClaI and re-ligated, yielding the CIp10+ECE1Δ184–279 plasmid. This plasmid was transformed into the uridine auxotrophic ece1Δ/Δ strain, yielding the ece1Δ/Δ+ECE1Δ184–279 strain. All integrations were confirmed by PCR/sequencing and at least two independent isogenic transformants were created to confirm results. KEX1 deletion was performed exactly as the ECE1 deletion but using primers KEX1-FG and KEX1-RG for creating the deletion cassette. Fluorescent strains of ece1Δ/Δ and BWP17 were constructed as previously described40. Briefly, the ece1Δ/Δ and BWP17 strains were transformed with the pENO1-dTom-NATr plasmid. Primers used to clone and construct the ECE1 genes and intragenic regions are listed in Extended Data Table 4. Strains are listed in Extended Data Table 2.
Construction of C. albicans ECE1 promoter-GFP strain
ECE1 promoter (primers 5′ECE1prom-NarI / 3′ECE1prom-XhoI) and terminator (5′ECE1term-SacII / 5′ECE1term-SacI) were amplified and cloned into pADH1-GFP. Resulting pSK-pECE1-GFP was verified by sequencing. C. albicans SC5314 was transformed with the pECE1-GFP transformation cassette38. Resistance to nourseothricin was used as selective marker and correct integration of GFP into the ECE1 locus was verified by PCR. Primers for cloning and validation are listed in Extended Data Table 4. Strains are listed in Extended Data Table 2.
RNA isolation and real-time PCR analysis
C. albicans cells grown on TR146 epithelial cells were collected into RNA pure (PeqLab), centrifuged and the pellet resuspended in 400 µl AE buffer (50 mM Na-acetate pH 5.3, 10 mM EDTA, 1% SDS). Samples were vortexed (30 s), and an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) was added and incubated for 5 min (65°C) before subjected to 2x freeze-thawing. Lysates were clarified by centrifugation and the RNA precipitated with isopropyl alcohol/0.3 M sodium acetate by incubating for 1 h at −20°C. Precipitated pellets were washed (2× 1 ml 70% ice-cold ethanol), resuspended in DEPC-treated water and stored at −80°C. RNA integrity and concentration was confirmed using a Bioanalyzer (Agilent). RNA (500 ng) was treated with DNase (Epicenter) and cDNA synthesized using Reverse Transcriptase Superscript III (Invitrogen). cDNA samples were used for qPCR with EVAgreen mix (Bio&Sell). Primers (ACT1-F and ACT1-R for actin, ECE1-F and ECE1-R for ECE1 - Extended Data Table 4) were used at a final concentration of 500 nM. qPCR amplifications were performed using a Biorad CFX96 thermocycler. Data was evaluated using Bio-Rad CFX Manager 3.1 (Bio-Rad) with ACT1 as the reference gene and t0 as the control sample.
Western blotting
TR146 cells were lysed using a modified RIPA lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing protease (Sigma-Aldrich) and phosphatase (Perbio Science) inhibitors41, left on ice (30 min) and then clarified (10 min) in a refrigerated microfuge. Lysate total protein content was determined using the BCA protein quantitation kit (Perbio Science). 20 µg of total protein was separated on 12% SDS-PAGE gels before transfer to nitrocellulose membranes (GE Healthcare). After probing with primary (1:1000) and secondary (1:10,000) antibodies, membranes were developed using Immobilon chemiluminescent substrate (Millipore) and exposed to X-Ray film (Fuji film). Human α-actin was used as a loading control.
Transcription factor DNA binding assay
DNA binding activity of transcription factors was assessed using the TransAM transcription factor ELISA system (Active Motif) as previously described41,42. Serum-starved TR146 epithelial cells were treated for 3 h before being differentially lysed to recover nuclear proteins using a nuclear protein extraction kit (Active Motif) according to the manufacturer’s protocol. Protein concentration was determined (BCA protein quantitation kit (Perbio Science)) and 5 µg of nuclear extract was assayed in the TransAM system according to the manufacturer’s protocol. Data was expressed as fold-change in A450nm relative to resting cells.
Cytokine determination
Cytokine levels in cell culture supernatants were determined using the Performance magnetic Fluorokine MAP cytokine multiplex kit (Bio-techne) and a Bioplex 200 machine. The data were analyzed using Bioplex Manager 6.1 software to determine analyte concentrations.
Cell damage assay
Following incubation, culture supernatant was collected and assayed for lactate dehydrogenase (LDH) activity using the Cytox 96 Non-Radioactive Cytotoxicity Assay kit (Promega) according to the manufacturer’s instructions. Recombinant porcine LDH (Sigma-Aldrich) was used to generate a standard curve.
Epithelial adhesion assay
Quantification of C. albicans adherence to TR146 epithelial cells was performed as described previously43. Briefly, TR146 cells were grown to confluence on glass coverslips for 48 h in tissue culture plates in DMEM medium. C. albicans yeast cells (2 × 105) were added into 1 ml serum-free DMEM, incubated for 60 min (37°C/5% CO2) and non-adherent C. albicans cells removed by aspiration. Following washing (3× 1 ml PBS), cells were fixed with 4% paraformaldehyde (Roth) and adherent C. albicans cells stained with Calcofluor White and quantified using fluorescence microscopy. The number of adherent cells was determined by counting 100 high power fields of 200 µm × 200 µm size. Exact total cell numbers were calculated based on the quantified areas and the total size of the cover slip.
Epithelial invasion assay
C. albicans invasion of epithelial cells was determined as described previously43. Briefly, TR146 epithelial cells were grown to confluence on glass coverslips for 48 h and then infected with C. albicans yeast cells (1×105), for 3 h in a humidified incubator (37°C/5% CO2). Following washing (3x PBS), the cells were fixed with 4% paraformaldehyde. All surface adherent fungal cells were stained for 1 h with a rabbit anti-Candida antibody and subsequently with a goat anti-rabbit-Alexa Fluor 488 antibody. After rinsing with PBS, epithelial cells were permeabilized (0.1% Triton X-100 in PBS for 15 min) and fungal cells (invading and non-invading) were stained with Calcofluor White. Following rinsing with water, coverslips were visualized using fluorescence microscopy. The percentage of invading C. albicans cells was determined by dividing the number of (partially) internalized cells by the total number of adherent cells. At least 100 fungal cells were counted on each coverslip.
Imaging of C. albicans growth and invasion of epithelial cells
TR146 cells (105/ml) seeded on glass coverslips in DMEM/10% FBS were infected with C. albicans (2.5 × 104 cfu/ml) in DMEM and incubated for 6 h (37°C/5% CO2). Cells were washed with PBS, fixed overnight (4°C in 4% paraformaldehyde) and stained with Concanavalin A-Alexa Fluor 647 in PBS (10 µg/ml) for 45 min at room temperature in the dark with gentle shaking (70 rpm) to stain the fungal cell wall. Epithelial cells were permeabilised with 0.1% Triton X-100 for 15 min at 37°C in the dark, then washed and stained with 10 µg/ml Calcofluor White (0.1 M Tris-HCl pH 9.5) for 20 min at room temperature in the dark with gentle shaking. Cells were rinsed in water and mounted on slides with 6 µl of ProLong Gold anti-fade reagent, before air drying for 2 h in the dark. Fluorescence microscopy was performed on a Zeiss Axio Observer Z1 microscope, and 5 phase images were taken per picture.
Scanning Electron Microscopy
For scanning electron microscopy (SEM) analysis, TR146 cells were grown to confluence on Transwell inserts (Greiner) and serum starved overnight in serum-free DMEM. After 5 h of C. albicans incubation on epithelial cells at an MOI of 0.01, cell media was removed and samples were fixed overnight at 4°C with 2.5% (v/v) glutaraldehyde in 0.05 M HEPES buffer (pH 7.2) and post-fixed in 1% (w/v) osmium tetroxide for 1 h at room temperature. After washing, samples were dehydrated through a graded ethanol series before being critical point dried (Polaron E3000, Quorum Technologies Ltd). Dried samples were mounted using carbon double side sticky discs (TAAB) on aluminium pins (TAAB) and gold coated in an Emitech K550X sputter coater (Quorum Technologies Ltd). Samples were examined and images recorded using a FEI Quanta 200 field emission scanning electron microscope operated at 3.5 kV in high vacuum mode.
Zebrafish swimbladder mucosal infection model
Zebrafish infections were performed in accordance with NIH guidelines under Institutional Animal Care and Use Committee (IACUC) protocol A2009-11-01 at the University of Maine. To determine sample size, a power calculation was done for all experiments based on 2-tails T-test in order to detect a minimum effect size of 0.8, with an alpha error probability of 0.05 and a power (1 – beta error probability) of 0.95. This gave a minimum number of 42 fish for each group. The fish selected for the experiments were randomly assigned to the different groups by picking them from a pool without bias and the groups were injected in different orders. No blinding was used to read the results. Ten to twenty zebrafish per group per experiment were maintained at 33°C in E3 + PTU and used as previously described40. Briefly, 4 day post-fertilization (dpf) larvae were treated with 20 µg/ml dexamethasone dissolved in 0.1% DMSO 1 h prior to infection and thereafter. For tissue damage and neutrophil recruitment, individual AB or mpo:GFP fish (respectively) were injected into the swimbladder with 4 nl of PBS with/without 25–40 C. albicans yeast cells of ece1Δ/Δ-dTomato, ece1Δ/Δ+ECE1+dTomato, ece1Δ/Δ+ECE1Δ184–279+dTomato or BWP17-dTomato. For tissue damage, 1 nl of Sytox green (0.05 mM in 1% DMSO) was injected at 20 h post-infection into the swimbladder and fish were imaged by confocal microscopy at 24 h post-infection. For neutrophil recruitment, fish were imaged at 24 h post-injection. For synthetic peptide damage, AB or α-catenin:citrine44 fish were injected with 2 nl of peptide (9 ng or 1.25 ng per fish) or vehicle (40% DMSO or 5% DMSO) + SytoxGreen (0.05 mM in 1% DMSO) or SytoxOrange (0.5 mM in 10% DMSO) and the fish imaged by confocal microscopy 4 h later. Numbers of neutrophils and damaged cells observed were counted and tabulated for each fish.
Zebrafish swimbladder fluorescence microscopy
Live zebrafish imaging was carried out as previously described40. Briefly, fish were anesthetized in Tris-buffered Tricaine (200 µg/ml, Western Chemicals) and further immobilized in a solution of 0.4% low-melting-point agarose (LMA, Lonza) in E3 + Tricaine in a 96-well plate glass-bottom imaging dish (Greiner Bio-On). Confocal imaging was carried out using an Olympus IX-81 inverted microscope with an FV-1000 laser scanning confocal system (Olympus). Images were collected and processed using Fluoview (Olympus) and Photoshop (Adobe Systems Inc.). Panels are either a single slice for the differential interference contrast channel (DIC) with maximum projection overlays of fluorescence image channels (red-green), or maximum projection overlays of fluorescence channels. The number of slices for each maximum projection is specified in the legend of individual figures.
Murine oropharyngeal candidiasis model
Murine infections were performed under UK Home Office Project Licence PPL 70/7598 in dedicated animal facilities at King’s College London. No statistical method was used to pre-determine sample size. No method of randomization was used to allocate animals to experimental groups. Mice in the same cage were part of the same treatment. The investigators were not blinded during outcome assessment. A previously described murine model of oropharyngeal candidiasis using female Balb/c mice45 was modified for investigating early infection events. Briefly, mice were treated sub-cutaneously with 3 mg/mouse (in 200 µl PBS with 0.5% Tween 80) of cortisone acetate on days −1 and 1 post-infection. On day 0, mice were sedated for ∼75 min with an intra-peritoneal injection of 110 mg/kg ketamine and 8 mg/kg xylazine, and a swab soaked in a 107 cfu/ml C. albicans yeast culture in sterile saline was placed sub-lingually for 75 min. After 2 days, mice were sacrificed, the tongue excised and divided longitudinally in half. One half was weighed, homogenized and cultured to derive quantitative Candida counts. The other half was processed for histopathology and immunohistochemistry.
Immunohistochemistry of murine tissue
C. albicans infected murine tongues were fixed in 10% (v/v) formal-saline before being embedded and processed in paraffin wax using standard protocols. For each tongue, 5 µm sections were prepared using a Leica RM2055 microtome and silane coated slides. Sections were dewaxed using xylene, before C. albicans and infiltrating inflammatory cells were visualized by staining using Periodic Acid-Schiff (PAS) stain and counterstaining with haematoxylin. Sections were then examined by light microscopy. Histological quantification of infection was undertaken by measuring the area of infected epithelium and expressed as a percentage relative to the entire epithelial area.
Whole cell patch clamp
TR146 epithelial cells were grown in 35 mm petri dishes (Nunc) for 48 h before recordings at low cell density (10–30% confluence). Cells were superfused with a modified Krebs solution (120 mM NaCl, 3 mM KCl, 2.5 mM CaCl2, 1.2 mM MgCl2, 22.6 mM NaHCO2, 11.1 mM glucose, 5 mM HEPES pH 7.4). Isolated cells were recorded at room temperature (21–23°C) in whole cell mode using microelectrodes (5–7 MΩ) containing 90 mM potassium acetate, 20 mM KCl, 40 mM HEPES, 3 mM EGTA, 3 mM MgCl2, 1 mM CaCl2 (free Ca2+ 40 nM), pH 7.4. Cells were voltage clamped at −60 mV using an Axopatch 200A amplifier (Axon Instruments) and current/voltage curves were generated by 1 s steps between −100 to + 50 mV. Treatments were applied to the superfusate to produce the final required concentration, with vehicle controls similarly applied. Data was recorded using Clampex software (PClamp 6, Axon Instrument) and analyzed with Clampfit 10.
Calcium flux
TR146 cells were grown in a 96-well plate overnight until confluent. The medium was removed and 50 µl of a Fura-2 solution (5 µl Fura-2 (Life Technologies)(2.5 mM in 50% Pluronic F-127 (Life technologies):50% DMSO), 5 µl probenecid (Sigma) in 5 ml saline solution (NaCl (140 mM), KCl (5 mM), MgCl2 (1 mM), CaCl2 (2 mM), Glucose (10 mM) and HEPES (10 mM), adjusted to pH 7.4)) was added and the plate incubated for 1 h at 37°C/5% CO2. The Fura-2 solution was replaced with 50 µl saline solution and baseline fluorescence readings (excitation 340 nm/emission 520nm) taken for 10 min using a FlexStation 3 (Molecular Devices). Ece1 peptides were added at different concentrations and readings immediately taken for up to 3 h. The data was analyzed using Softmax Pro software to determine calcium present in the cell cytosol and expressed as the ratio between excitation and emission spectra.
Impedance spectroscopy of tethered bilayer lipid membranes (tBLMs)
tBLMs with 10% tethering lipids and 90% spacer lipids (T10 slides) were formed using the solvent exchange technique46,47 according to the manufacturer’s instructions (SDx Tethered Membranes Pty Ltd, Sydney, Australia). Briefly, 8 µl of 3 mM lipid solutions in ethanol were added, incubated for 2 min and then 93.4 µl buffer (100 mM KCl, 5 mM HEPES, pH 7.0) was added. After rinsing 3x with 100 µl buffer the conductance and capacitance of the membranes were measured for 20 min before injection of Ece1 peptides at different concentrations. All experiments were performed at room temperature. Signals were measured using the tethaPod (SDx Tethered Membranes Pty Ltd, Sydney, Australia).
FRET intercalation experiments
Intercalation of Ece1 peptides into phospholipid liposomes was determined by FRET spectroscopy applied as a probe-dilution assay48. Phospholipids mixed with each 1% (mol/mol) of the donor dye NBD-phosphatidylethanolamine (NBD-PE) and of the acceptor dye rhodamine-PE, were dissolved in chloroform, dried, solubilized in 1 ml buffer (100 mM KCl, 5 mM HEPES, pH 7.0) by vortexing, sonicated with a titan tip (30 W, Branson sonifier, cell disruptor B15), and subjected to three cycles of heating to 60°C and cooling down to 4°C, each for 30 min. Lipid samples were stored at 4°C for at least 12 h before use. Ece1 peptide was added to liposomes and intercalation was monitored as the increase of the quotient between the donor fluorescence intensity ID at 531 nm and the acceptor intensity IA at 593 nm (FRET signal) independent of time.
Circular Dichroism spectroscopy
CD measurements were performed using a Jasco J-720 spectropolarimeter (Japan Spectroscopic Co., Japan), calibrated as described previously49. CD spectra represent the average of four scans obtained by collecting data at 1 nm intervals with a bandwidth of 2 nm. The measurements were performed in 100 mM KCl, 5 mM HEPES, pH 7.0 at 25°C and 40°C in a 1.0 mm quartz cuvette. The Ece1-III concentration was 15 µM.
Planar lipid bilayers
Planar lipid bilayers were prepared using the Montal-Mueller technique50 as described previously51. All measurements were performed in 5 mM HEPES, 100 mM KCl, pH 7.0 (specific electrical conductivity 17.2 mS/cm) at 37°C.
Hyphal secretome preparation for LC-MS/MS analysis
Candida strains were cultured for 18 h in hyphae inducing conditions (YNB medium containing 2% sucrose, 75 mM MOPSO buffer pH 7.2, 5 mM N-acetyl-D-glucosamine, 37°C). Hyphal supernatants were collected by filtering through a 0.2 µm PES filter, and peptides were enriched by Solid Phase Extraction (SPE) using first C4 and subsequently C18 columns on the C4 flowthrough. After drying in a vacuum centrifuge, samples were resolubilised in loading solution (0.2% formic acid in 71:27:2 ACN/H2O/DMSO (v/v/v)) and filtered through a 10 kDa MWCO filter. The filtrate was transferred into HPLC vials and injected into the LC-MS/MS system. LC-MS/MS analysis was carried out on an Ultimate 3000 nano RSLC system coupled to a QExactive Plus mass spectrometer (ThermoFisher Scientific). Peptide separation was performed based on a direct injection setup without peptide trapping using an Accucore C4 column as stationary phase and a column oven temperature of 50°C. The binary mobile phase consisting of A) 0.2% (v/v) formic acid in 95:5 H2O/DMSO (v/v) and B) 0.2% (v/v) formic acid in 85:10:5 ACN/H2O/DMSO (v/v/v) was applied for a 60 min gradient elution: 0–1.5 min at 60% B, 35–45 min at 96% B, 45.1–60 min at 60% B. The Nanospray Flex Ion Source (ThermoFisher Scientific) provided with a stainless steel emitter was used to generate positively charged ions at 2.2 kV spray voltage. Precursor ions were measured in full scan mode within a mass range of m/z 300–1600 at a resolution of 70k FWHM using a maximum injection time of 120 ms and an automatic gain control target of 1e6. For data-dependent acquisition, up to 10 most abundant precursor ions per scan cycle with an assigned charge state of z = 2–6 were selected in the quadrupole for further fragmentation using an isolation width of m/z 2.0. Fragment ions were generated in the HCD cell at a normalised collision energy of 30 V using nitrogen gas. Dynamic exclusion of precursor ions was set to 20 s. Fragment ions were monitored at a resolution of 17.5k (FWHM) using a maximum injection time of 120 ms and an AGC target of 2e5.
Protein database search
Thermo raw files were processed by the Proteome Discoverer (PD) software v1.4.0.288 (Thermo). Tandem mass spectra were searched against the Candida Genome Database (http://www.candidagenome.org/download/sequence/C_albicans_SC5314/Assembly22/current/C_albicans_SC5314_A22_current_orf_trans_all.fasta.gz;status: 2015/05/03) using the Sequest HT search algorithm. Mass spectra were searched for both unspecific cleavages (no enzyme) and tryptic peptides with up to 4 missed cleavages. The precursor mass tolerance was set to 10 ppm and the fragment mass tolerance to 0.02 Da. Target Decoy PSM Validator node and a reverse decoy database was used for (qvalue) validation of the peptide spectral matches (PSMs) using a strict target false discovery (FDR) rate of < 1%. Furthermore, we used the Score versus Charge State function of the Sequest engine to filter out insignificant peptide hits (xcorr of 2.0 for z=2, 2.25 for z=3, 2.5 for z=4, 2.75 for z=5, 3.0 for z=6). At least two unique peptides per protein were required for positive protein hits.
Statistics
TransAM and patch clamp data were analyzed using a paired t-test whilst cytokines, LDH and calcium influx data were analyzed using one-way ANOVA with all compared groups passing an equal variance test. Murine in vivo data was analyzed using the Mann-Whitney test. Zebrafish data was analyzed using the Kruskal-Wallis test with Dunn’s multiple comparison correction. In all cases, P < 0.05 was taken to be significant.
Extended Data
Extended Data Figure 1. C. albicans ECE1 expression and phenotypic effects of ECE1 gene deletion.
(a) Relative expression (vs t = 0) of ECE1 in C. albicans wild type over time after addition of yeast cells to TR146 epithelial cells as measured by RT-qPCR. (b) Imaging confirmation of ECE1 expression over time within C. albicans wild type. C. albicans cells expressing GFP under the control of the ECE1 5′ intragenic region, containing the ECE1 promoter, were grown on TR146 epithelial cells and stained with calcofluor white (CFW, post-permeabilization) to show cell wall chitin and Alexa-Fluor-647-labelled concanavalin A (ConA, pre-permeabilization) to show carbohydrates. A composite image showing CFW, ConA, GFP and the brightfield (BF) image is shown. (c) Scanning electron micrographs (top panels, 5 h) and light microscopy (bottom panels, 24 h) showing no gross abnormalities in hypha formation between C. albicans wild type (BWP17+CIp30), ECE1-deletion (ece1Δ/Δ) and ECE1 re-integrant (ece1Δ/Δ+ECE1) strains after infection of TR146 epithelial cells. (d) No difference in adhesion of C. albicans wild type, ece1Δ/Δ and ece1Δ/Δ+ECE1 strains to TR146 epithelial cells after 60 min. (e) No difference in invasion of C. albicans wild type, ece1Δ/Δ and ece1Δ/Δ+ECE1 strains into TR146 epithelial cells after 3 h. (f) Fluorescence staining of C. albicans wild type and ece1Δ/Δ hyphae invading through TR146 epithelial cells. Fungal cells are stained with calcofluor white (CFW, post-permeabilization) and Alexa-Fluor-647-labelled concanavalin A (ConA, pre-permeabilization) to show cell wall chitin and carbohydrates, respectively, and to distinguish between invading hyphae (only stained after permeabilization) and non-invading hyphae (stained both pre- and post-permeabilization). Levels of chitin and β-glucan are comparable in both strains. White arrows indicate invasion into epithelial cells. Data shown are representative (b, c, f) or the mean (a, d, e) of three biological replicates. Error bars show ± SEM.
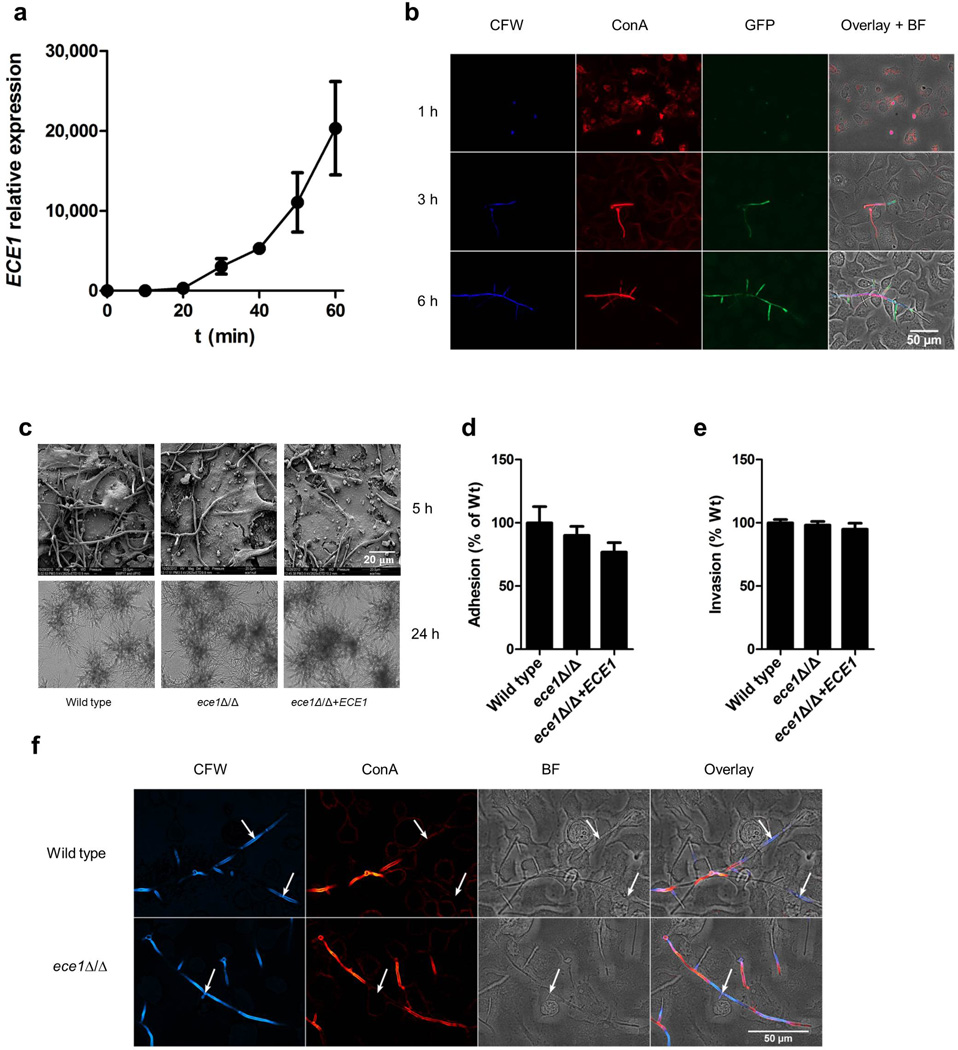
Extended Data Figure 2. C. albicans Ece1p is critical for mucosal virulence in vivo.
(a) Fungal burdens recovered from the tongues of mice infected with C. albicans wild type (BWP17+CIp30) (n (number of mice) = 13), ECE1-deletion (ece1Δ/Δ) (n = 20) and ECE1 re-integrant (ece1Δ/Δ+ECE1) (n = 24) strains after 2 day oropharyngeal infection. (b) Average percentage of the entire tongue epithelium area infected in different groups of mice infected with the different C. albicans strains. (c) Confocal imaging of 4 day post-fertilization (dpf) mpo-gfp transgenic zebrafish swimbladders infected with C. albicans wild type (BWP17+CIp30+dTomato), ECE1-deletion (ece1Δ/Δ+dTomato) and ECE1 re-integrant (ece1Δ/Δ+ECE1+dTomato) strains for 24 h. C. albicans cells appear red whilst neutrophils appear green. Red dots outline the swimbladder. Images are composites of maximum projections in the red and green channels (25 slices each, approximately 100 µm depth) with (left) or without (right) a single slice in the DIC channel overlay. Scale bars represent 100 µm. (d) Confocal imaging of 4 dpf zebrafish swimbladders infected with C. albicans wild type (BWP17+CIp30+dTomato), ECE1-deletion (ece1Δ/Δ+dTomato) and ECE1 re-integrant (ece1Δ/Δ+ECE1+dTomato) strains for 24 h stained with the fluorescent exclusion dye Sytox Green. C. albicans cells appear red and damaged epithelial cells appear green. White dots outline the pronephros and red dots outline the swimbladder. Images are composites of maximum projections in the red and green channels (25 slices each, approximately 100 µm depth) with (left) or without (right) a single slice in the DIC channel overlay. High magnification images of the white boxes are shown. Scale bars (bottom right) represent 100 µm (low magnification) and 30 µm (high magnification). Data shown are the mean (a, b) or representative (c, d) of at least three biological replicates. Error bars show ± SEM. Data were analyzed by Mann-Whitney test. *** = P < 0.001.
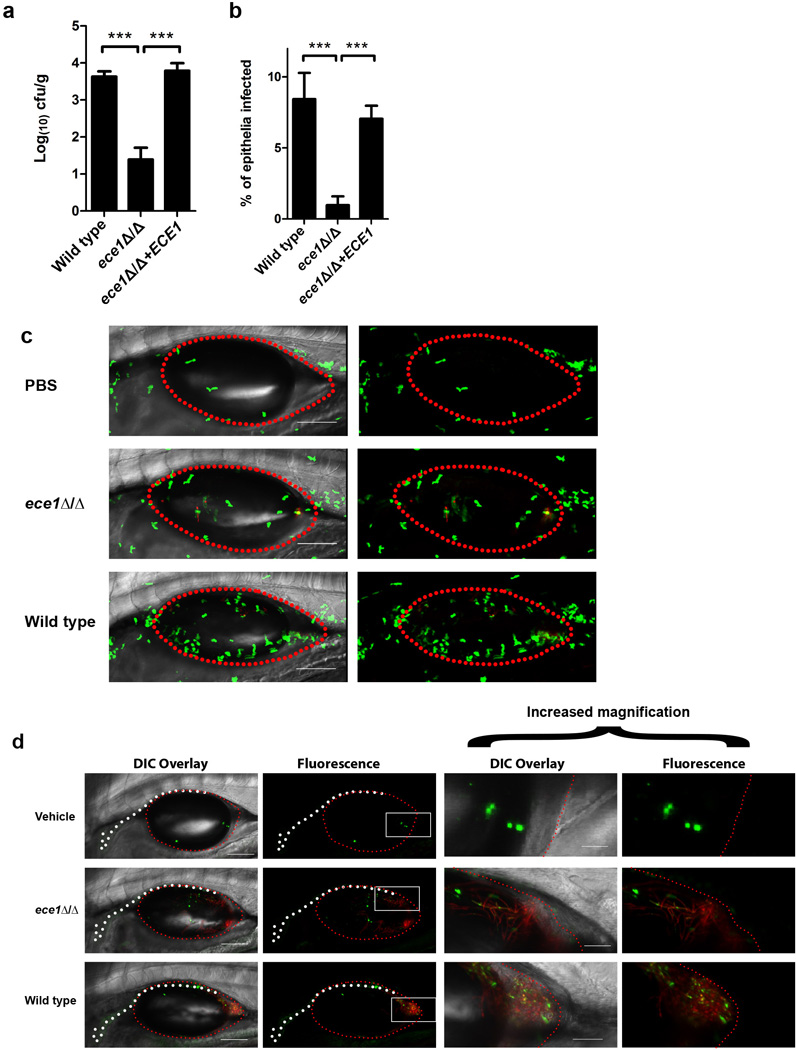
Extended Data Figure 3. Ece1-III62–93 is the active region of Ece1p.
(a) Amino acid sequence of Ece1p and a schematic of the protein, indicating the signal peptide (SP), lysine-arginine motifs (KR) at the C-terminus of each peptide, and the processed peptides (Ece1-I-VIII) produced by Kex2p cleavage. (b) Amino acid sequences of the processed peptides (Ece1-I-VIII) produced by Kex2p cleavage. Induction of (c) GM-CSF, (d) IL-1α and (e) IL-6 secreted after stimulation of TR146 epithelial cells for 24 h with varying concentrations of Ece1-III62–93 (70 µM - 1.5 µM). (f) Phosphorylation of MKP-1 and c-Fos production after 2 h treatment of TR146 epithelial cells with 15 µM of Ece1-III62–85 (hydrophobic region), Ece1-III86–93 (hydrophillic region), Ece1-III62–85 and Ece1-III86–93 together, or Ece1-III62–93 alone. (g) Induction of G-CSF secretion after 24 h treatment of TR146 epithelial cells with 15 µM of Ece1-III62–85, Ece1-III86–93, Ece1-III62–85 and Ece1-III86–93 together, or Ece1-III62–93 alone. (h) Fold change induction of LDH release after 24 h treatment of TR146 epithelial cells with 70 µM of Ece1-III62–85, Ece1-III86–93, Ece1-III62–85 and Ece1-III86–93 together, or Ece1-III62–93 alone. (i) Induction of p-MKP-1 and c-Fos 2 h post-infection (p.i.) with the indicated C. albicans strains (MOI = 10). (j) c-Fos DNA binding induction 3 h p.i. with indicated C. albicans strains (MOI = 10). (k) G-CSF secretion and (l) LDH release 24 h p.i. with indicated C. albicans strains (MOI = 0.01). Data shown are representative (f, i) or the mean (c-e, g-h, j-l) of three biological replicates. Error bars show ± SEM. Data were analyzed by one-way ANOVA (c-e, g-h,k-l) or T test (j). * = P < 0.05, ** = P < 0.01, *** = P < 0.001 (compared with vehicle control). For gel source data, see Supplementary Figure 1.
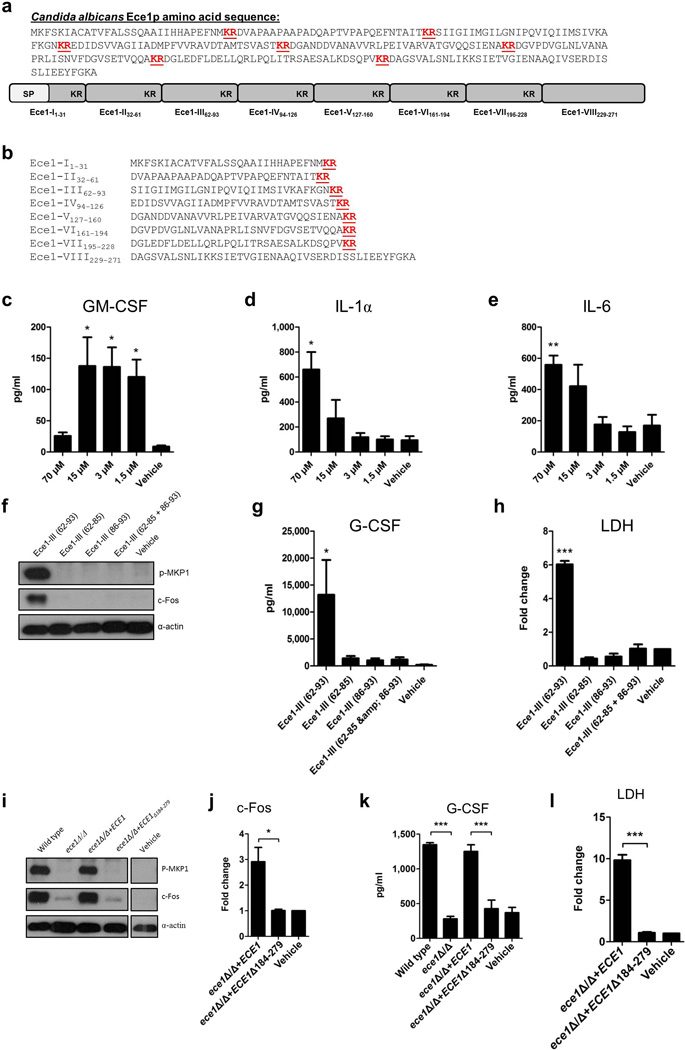
Extended Data Figure 4. Ece1-III62–93 is required for C. albicans mucosal infection.
(a) Fungal burdens recovered from the tongues of mice infected with C. albicans wild type (BWP17+CIp30) (n (number of mice) = 13), ECE1-deletion (ece1Δ/Δ) (n = 20), ECE1 re-integrant (ece1Δ/Δ+ECE1) (n = 24) and Ece1-III62–93 deletion (ece1Δ/Δ+ECE1Δ184–279) (n = 10) strains after 2 day oropharyngeal infection. (b) Average percentage of the entire tongue epithelium area infected in different groups of mice infected with the different C. albicans strains. (c) Confocal imaging of 4 dpf zebrafish swimbladders infected with C. albicans Ece1-III62–93 deletion (ece1Δ/Δ+ECE1Δ184–279+dTomato) and ECE1 re-integrant (ece1Δ/Δ+ECE1+dTomato) strains for 24 h stained with the fluorescent exclusion dye Sytox Green. C. albicans cells appear red and damaged cells appear green. White dots outline the pronephros and red dots outline the swimbladder. Images are composites of maximum projections in the red and green channels (25 slices each, approximately 100 µm depth) with (left) or without (right) a single slice in the DIC channel overlay. Scale bars (bottom right) represent 100 µm. Data shown are the mean (a-b) or representative (c) of at least three biological replicates. Error bars show ± SEM. Data were analyzed by Mann-Whitney test. ** = P < 0.01, *** = P < 0.001.
Extended Data Figure 5. Ece1-III62–93 is a cytolytic α-helical peptide.
(a) Circular dichroism spectra showing the α-helical conformation of Ece1-III62–93 in buffer (100 mM KCl, 5 mM HEPES, pH 7). Increasing the temperature from 25°C to 40°C did not affect the stability of the α-helical structure. (b) Diagram to illustrate the amphipathic nature of Ece1-III62–93 (residues 62–78, left panel; residues 79–93, right panel). Residues with hydrophobic or polar/charged side chains are displayed with a blue and white background, respectively. Modified from output generated in PEPWHEEL (http://emboss.bioinformatics.nl/cgi-bin/emboss/pepwheel). (c) Förster resonance energy transfer (FRET) experiments show the intercalation of Ece1-III62–93 into lipid liposomes (10 µM) composed of DOPC in the absence or presence of cholesterol. Peptide titration of Ece1-III62–93 to liposomes showed slightly enhanced intercalation for pure DOPC. (d) Ece1-III62–93 induced the permeabilization of planar lipid membranes composed of DOPC. The graph shows heterogeneous and transient lesions leading finally to a rupture of the membrane. Ece1-III62–93 concentration was 0.125 µM. (e) Induction of p-MKP-1 and c-Fos 2 h in TR146 cells post stimulation (p.s.) with Ece1-III62–93KR or Ece1-III62–93AA(f) Secretion of G-CSF from TR146 cells 24 h p.s. with Ece1-III62–93KR or Ece1-III62–93AA. Data shown are representative (a-e) or mean (f) of at least three biological replicates. Error bars show ± SEM. Data were analyzed by one-way ANOVA (f). * = P < 0.05, ** = P < 0.01, *** = P < 0.001 (compared with vehicle control). For gel source data, see Supplementary Figure 1.
Extended Data Figure 6. Schematic of the role of Ece1-III in C. albicans infection of epithelial cells.
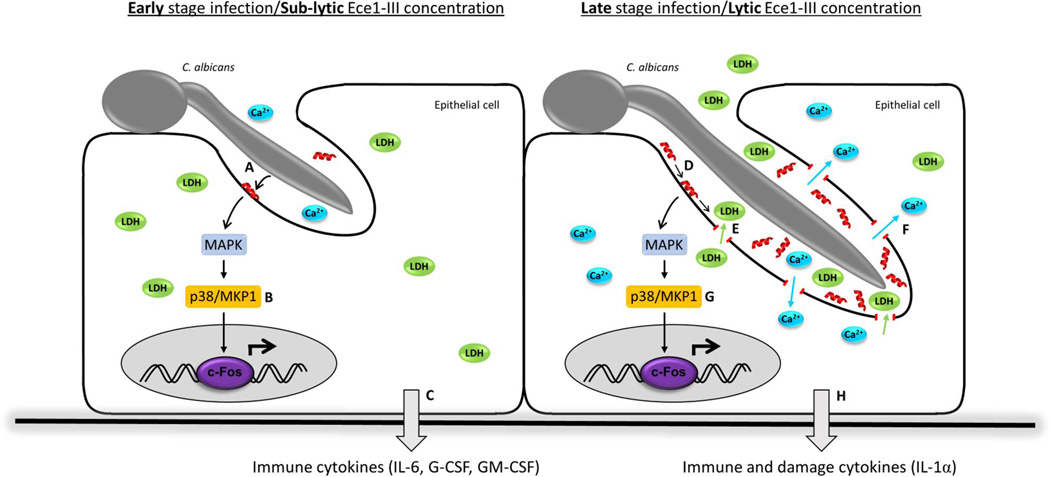
During early stage infection of the mucosal surface by C. albicans, Ece1-III (red α-helix) is secreted into the invasion pocket created by the invading hypha (a). Sub-lytic concentrations of Ece1-III trigger epithelial signal transduction through MAPK, p38/MKP-1 and c-Fos (b) resulting in the production of immune regulatory cytokines (c). As the severity of the infection increases, Ece1-III accumulates (d) and once lytic concentrations are reached, causes membrane damage and the release of lactate dehydrogenase from the host epithelium (e), concomitant with calcium influx (f). Epithelial signal transduction is maintained (g) and additionally induces the release of damage associated cytokines, such as IL-1α (h). Ece1-III may also have activity on the epithelial surface outside of the invasion pocket and on neighboring cells not in contact with hyphae if Ece1-III is produced in sufficient concentrations.
Extended Data Table 1.
C. albicans strains used in this study.
| Strain name | Strain/Gene Function | Strain Reference |
Morphology* | Phospho MKP1† |
c-Fos† | Cytokines‡ | Damage§ | Phenotype Reference |
|---|---|---|---|---|---|---|---|---|
| Controls | ||||||||
| SC5314 | Wild type | [52] | Hyphae | Yes | Yes | Yes | Yes | This study |
| BWP17 & CIp30 | Parental strain | [53] | Hyphae | Yes | Yes | Yes | Yes | This study |
| CAI-4 & CIp10 | Parental strain | [54] | Hyphae | Yes | Yes | Yes | Yes | This study |
| CAF2-1 | Parental strain | [54] | Hyphae | Yes | Yes | Yes | Yes | This study |
| DAY286 | Parental strain | [55] | Hyphae | Yes | Yes | Yes | Yes | This study |
| Yeast-locked | ||||||||
| efg1Δ/Δ | Transcription factor | [56] | Yeast | No | No | No | No | This study |
| efg1/cph1Δ/Δ | Transcription factor/ Transcription factor | [57] | Yeast | No | No | No | No | [58]/This study |
| eed1Δ/Δ | RNA polymerase II regulator | [59] | Yeast | No | No | No | No | [58]/This study |
| flo8Δ/Δ | Transcription factor | [60] | Yeast | No | No | No | No | This study |
| tpk1Δ/Δ | cAMP-dependent protein kinase | [61] | Yeast | No | No | No | No | This study |
| tpk2Δ/Δ | cAMP-dependent protein kinase | [62] | Yeast | No | No | No | No | This study |
| vps11Δ/Δ | Protein trafficking | [63] | Yeast | No | No | No | No | This study |
| cap1Δ/Δ | Transcription factor | [64] | Yeast | No | No | Yes | No | This study |
| och1Δ/Δ | Alpha-1,6-mannosyltransferase | [65] | Yeast | No | No | No | No | [66] |
| kex2Δ/Δ | Processing enzyme | [67] | Yeast | No | No | No | No | This study |
| Hypha-producing | ||||||||
| nrg1Δ/Δ | Transcriptional corepressor | [68] | Hyphae | Yes | Yes | Yes | Yes | [58]/This study |
| cph1Δ/Δ | Transcription factor | [69] | Hyphae | Yes | Yes | Yes | Yes | This study |
| cph2Δ/Δ | Transcription factor | [70] | Hyphae | Yes | Yes | Yes | Yes | This study |
| efh1Δ/Δ | Transcription factor | [71] | Hyphae | Yes | Yes | Yes | Yes | This study |
| czf1Δ/Δ | Transcription factor | [72] | Hyphae | Yes | Yes | Yes | Yes | This study |
| rfg1Δ/Δ | Transcriptional repressor | [73] | Hyphae | Yes | Yes | Yes | Yes | This study |
| hog1Δ/Δ | MAP kinase | [74] | Hyphae | Yes | Yes | Yes | Yes | This study |
| sun42Δ/Δ | Adhesin-like protein | [75] | Hyphae | Yes | Yes | Yes | Yes | This study |
| pga29Δ/Δ | GPI-anchored yeast-associated protein | [76] | Hyphae | Yes | Yes | Yes | Yes | This study |
| phr2Δ/Δ | Glycosidase | [77] | Hyphae | Yes | Yes | Yes | Yes | This study |
| pga36Δ/Δ | GPI-anchored protein | [78] | Hyphae | Yes | Yes | Yes | Yes | This study |
| ece1Δ/Δ | Hypha-associated protein | [79]║ | Hyphae | No | No | No | No | This study |
| mkc1Δ/Δ | MAP kinase | [80] | Hyphae | Yes | Yes | Yes | Yes | This study |
| bud2Δ/Δ | GTPase activating protein | [81] | Hyphae | Yes | Yes | Yes | Yes | This study |
| pra1Δ/Δ | Zinc binding protein | [82] | Hyphae | Yes | Yes | Yes | Yes | This study |
| utr2/crh11/crh12Δ/Δ |
Putative
wall glycosidase/transglycosylase |
[83] | Hyphae | Yes | Yes | Yes | Yes | This study |
| wap1Δ/Δ | Surface antigen on hyphae/buds | [84] | Hyphae | Yes | Yes | Yes | Yes | This study |
| sod5Δ/Δ | Superoxide dismutase | [85] | Hyphae | Yes | Yes | Yes | Yes | This study |
| hwp1Δ/Δ | Adhesin | [86] | Hyphae | Yes | Yes | Yes | Yes | This study |
| rbt1Δ/Δ | Putative GPI-modified cell wall protein | [84] | Hyphae | Yes | Yes | Yes | Yes | This study |
| rbt5Δ/Δ | Heme binding | [84] | Hyphae | Yes | Yes | Yes | Yes | This study |
| hyr1Δ/Δ | GPI-anchored hyphal cell wall protein | [87] | Hyphae | Yes | Yes | Yes | Yes | This study |
| mp65Δ/Δ | Cell surface mannoprotein | [88] | Hyphae | Yes | Yes | yes | Yes | This study |
| cek1Δ/Δ | ERK-family protein kinase | [89] | Hyphae | Yes | Yes | Yes | Yes | This study |
| sap2Δ/Δ | Secreted aspartyl protease | [90] | Hyphae | Yes | Yes | Yes | Yes | This study |
| sap7Δ/Δ | Secreted aspartyl protease | [91] | Hyphae | Yes | Yes | Yes | Yes | This study |
| sap9/sap10Δ/Δ | Secreted aspartyl proteases | [92] | Hyphae | Yes | Yes | Yes | Yes | This study |
| als1Δ/Δ | Agglutinin-like sequence protein | [93] | Hyphae | Yes | Yes | Yes | Yes | [94] |
| als2Δ/ PMAL-ALS2 | Agglutinin-like sequence protein | [95] | Hyphae | Yes | Yes | Yes | Yes | [94] |
| als3Δ/Δ | Adhesin | [93] | Hyphae | Yes | Yes | Partial¶ | Partial¶ | [94] |
| als4Δ/Δ | Agglutinin-like sequence protein | [95] | Hyphae | Yes | Yes | Yes | Yes | [94] |
| als5Δ/Δ | Agglutinin-like sequence protein | [96] | Hyphae | Yes | Yes | Yes | Yes | [94] |
| als6Δ/Δ | Agglutinin-like sequence protein | [96] | Hyphae | Yes | Yes | Yes | Yes | [94] |
| als7Δ/Δ | Agglutinin-like sequence protein | [96] | Hyphae | Yes | Yes | Yes | Yes | [94] |
| als9Δ/Δ | Agglutinin-like sequence protein | [97] | Hyphae | Yes | Yes | Yes | Yes | [94] |
| pmt1Δ/Δ | Mannosyltransferase | [98] | Hyphae | Partial¶ | Partial¶ | Partial¶ | Partial¶ | [66] |
| pmr1Δ/Δ | Secretory pathway ATPase | [99] | Hyphae | Partial¶ | Partial¶ | Partial¶ | Partial¶ | [66] |
| mnn4Δ/Δ | Regulator of mannosylphosphorylation | [100] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| mnn9Δ/Δ | Putative mannosyltransferase | [101] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| mnt1/mnt2Δ/Δ | Mannosyltransferases | [102] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| chs2/chs3Δ/Δ | Chitin synthase/ Chitin synthase | [103] | Hyphae | Yes | Yes | Yes | Yes | This study |
| mit1Δ/Δ | Mannose:Inositolphosphoceramide mannose transferase |
[104] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| bmt1Δ/Δ | Beta-mannosyltransferase | [105] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| bmt2Δ/Δ | Putative beta-mannosyltransferase | [105] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| bmt3Δ/Δ | Beta-mannosyltransferase | [105] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| bmt4Δ/Δ | Beta-mannosyltransferase | [105] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| bmt5Δ/Δ | Putative beta-mannosyltransferase | [106] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| bmt6Δ/Δ | Beta-mannosyltransferase | [106] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| gsc1Δ/GSC1 | Beta-1,3-glucan synthase
catalytic subunit |
[107] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| gsl1Δ/Δ | Beta-1,3-glucan synthase subunit | [107] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| gsl2Δ/Δ | Beta-1,3-glucan synthase subunit | [107] | Hyphae | Yes | Yes | Yes | Yes | [66] |
| kre6Δ/KRE6 | Beta-1,6-glucan synthase subunit | [108] | Hyphae | Yes | Yes | Yes | Yes | This study |
Morphology recorded 2 h post-infection on TR146 buccal epithelial cell monolayers; hyphae includes pseudohyphae.
Data based on Western blotting.
Cytokines includes IL-1α, IL-6 and G-CSF.
Damage measured by LDH assay.
New ece1Δ/Δ also created in this study (See Extended Data Table 2). Original mutant (in red) produced by [27] using the URA-blaster protocol [3]. A set of ece1 mutants, including partial deletion of ECE1 and a revertant, was produced in this study in the same genetic background using strain BWP17 to avoid a URA3 effect based on genomic location [109,110].
Partial activation is due to lack of adhesion.
Extended Data Table 2.
C. albicans mutant strains constructed and used in this study.
| Strain description | Strain name | Genotype |
|---|---|---|
| BWP17+Clp30 | M1477 |
ura3: :λim m434/ura3:
:λ
imm434 iro1::λimm434/iro1::λimm434 his1 ::hisG/his1 ::hisG arg4: :hisG/arg4: :hisG RPS1/rps1::(URA3-HIS1-ARG4) |
| ece1 Δ/Δ | M2057 |
ura3: :λim m434/ura3:
:λ imm434 iro1 ::Aimm434/iro1 ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG ece1::HIS1/ece1::ARG4 RPS1/rps1::URA3 |
| ece1Δ/Δ+ECE1 | M2059 |
ura3: :λim m434/ura3:
:λimm434 iro1 ::λimm434/iro1 ::λimm434 his1::hisG/his1: :hisG arg4: :hisG/arg4: :hisG ece1 ::HIS1/ece1 ::ARG4 RPS1/rps1 ::(URA3-ECE1) |
| ece1Δ/Δ+ECElΔ184_279 | M2174 |
ura3: :λim m434/ura3:
:λimm434 iro1 :λimm434/iro1 ::λimm434 his1::hisG/his1::hisG arg4: :hisG/arg4: :hisG ece1::HIS1/ece1::ARG4 RPS1/rps1 ::(URA3-ECE1Δ184–279) |
| Kex1Δ/Δ | M2258 |
ura3: :λim m434/uro3:
:λimm434 iro1 ::λimm434/iro1 ::λimm434 his1::hisG/his1: :hisG arg4::hisG/arg4: :hisG kex1::HIS1/kex1::ARG4 RPS1/rps1::URA3 |
| SC5314+pECE1-GFP (ECE1 promoter-GFP) | CA58 | ECE1/ece1::GFP-SAT1 |
| BWP17+Clp30+pENO1-dTom (ENO1 promoter-dTom) | RWC83 |
ura3::λimm434/ura3::λimm434 iro1 ::λimm434/iro1 ::λimm434 his1 ::hisG/his1 ::hisG arg4: :hisG/arg4: :hisG RPS1rps1 ::(URA3-HIS1-ARG4) EN01/eno1::dTom-SAT1 |
| ece1Δ/Δ+pENO1-dTom (ENO1 promoter-dTom) | RWC84 |
ura3: :λim m434/ura3:
:λimm434 iro1 ::λimm434/iro1 ::λimm434 his1::hisG/his1: :hisG arg4: :hisG/arg4: :hisG ece1 ::HIS1/ece1 ::ARG4 RPS1/rps1::URA3 EN01/enol::dTom-SAT1 |
| ece1Δ/Δ+ECE1+ dTomato | RWC85 |
ura3: :λim m434/ura3:
:λimm434 iro1: :λimm 43 4/iro1: :λim m434 his1::hisG/his1::hisG arg4: :hisG/arg4: :hisG ece1 ::HIS1/ece1 ::ARG4 RPS1/rps1::(URA3-ECE1) EN01/eno1::dTomato-NAT |
| ece1Δ/Δ+ECE1Δ184_279 + dTomato | RWC86 |
ura3::λimm434/ura3::λimm434 iro1 ::λimm434/iro1 ::λimm434 his1::hisG/his1: :hisG arg4: :hisG/arg4: :hisG ece1 ::HIS1/ece1 ::ARG4 RPS1/rps1 ::(URA3-ECE1Δ184–279) EN01/eno1::dTomato-NAT |
Extended Data Table 3.
LC-MS/MS analysis of C. albicans Ece1-III
| Ece1-III sequence | |||||
|---|---|---|---|---|---|
| Wild Type | ece1Δ/Δ+ECE1 | TR146+Wild type | rEce1p+rKex2p | kex1Δ/Δ | |
| SIIGIIMGILGNIPQVIQIIMSIVKAFKGNK | n/d§ | ||||
| SIIGIIMGILGNIPQVIQIIMSIVKAFKGNKR | |||||
The number of peptide spectrum matches. Data for ece1Δ/Δ and ece1Δ/Δ+ECE1Δ184–279 are not included as no Ece1-III peptides were detected in either strain, as expected.
% of SIIGIIMGILGNIPQVIQIIMSIVKAFKGNK or SIIGIIMGILGNIPQVIQIIMSIVKAFKGNKR detected amongst all Ece1-III peptides found by LC-MS/MS.
% of SIIGIIMGILGNIPQVIQIIMSIVKAFKGNK or SIIGIIMGILGNIPQVIQIIMSIVKAFKGNKR detected amongst all Ecelp peptides found by LC-MS/MS.
n/d; not detected.
Extended Data Table 4.
Oligonucleotide primers used in this study.
| Primer name | Application | Sequence (5′-3′) | Description |
|---|---|---|---|
| ECE1-FG | PCR | atcaaataacccacctatttcaaaattgttttatttttgtttatctctacaaca aacaactttcctttattttactaccaactattttccattcgttaaagaagcttc gtacgctgcaggtc |
Construction of ECE1 deletion construct |
| ECE1-RG | PCR | cacaaaaaacaacaattaaaaaaatcagttacagcaaaagtgtcacaag acttatggaataaaagattaagcttgtggaaaacaaatttttatctgctgag cattctgatatcatcgatgaattcgag |
Construction of ECE1 deletion construct |
| ECE1-RecF3k | PCR | gcacgcgtctaaagtggagtaacaac | Construction of ECE1
complementation plasmid |
| ECE1-RecR | PCR | ggtcgaccccagacgttggttgc | Construction of ECE1
complementation plasmid |
| ECE1-F1 | PCR | ggcttctcataaatgaagggctcag | Confirmation of ECE1 deletion |
| ECE1-R1 | PCR | gccgaatcaatcttgtcgtgccac | Confirmation of ECE1 deletion |
| KEX1-FG | PCR | tatctttttttgttttatttacccatccttcatatctttacaacccttgatacctt acctaaacaacacacatctatctttaatcaatcaaacacaaatcaattgaa gcttcgtacgctgcaggtc |
Construction of KEX1 deletion construct |
| KEX1-RG | PCR | tcacaatctagattattgtaggttgtatagacaaaaaataaaaatcaaact attattcgttatataaatctacaagatctctaatctccactgtaccgaaaaat tctgatatcatcgatgaattcgag |
Construction of KEX1 deletion construct |
| KEX1-F1 | PCR | ggaagcccataagaaattgga | Confirmation of KEX1 deletion |
| KEX1-R1 | PCR | aggaagctgtggtggtagtg | Confirmation of KEX1 deletion |
| HIS-F2 | PCR | ggacgaattgaagaaagctggtgcaaccg | Confirmation of ECE1/KEX1 deletion |
| HIS-R2 | PCR | caacgaaatggcctcccctaccacag | Confirmation of ECE1/KEX1 deletion |
| ARG-F2 | PCR | ggatatgttggctactgatttag | Confirmation of ECE1/KEX1 deletion |
| ARG-R2 | PCR | aatggatcagtggcaccggtg | Confirmation of ECE1/KEX1 deletion |
| ECE1-Flnt1 | PCR | ctaacgtttttgatggcgtcctgg | Confirmation of plasmid integration |
| URAF2 | PCR | ggagttggattagatgataaaggtgatgg | Confirmation of plasmid integration |
| RPF-1 | PCR | gagcagtgtacacacacacatcttg | Confirmation of plasmid integration |
| RPF-2 | PCR | cgccaaagagtttcccctattatc | Confirmation of plasmid integration |
| Pep3-F1 | PCR | gaagatatcgattctgttgttgctgg | Excision of Ece1-llI62–93 from ECE1 |
| Pep3-R1 | PCR | cagaatcgatatcttctcttttggtaatagcagtattgaattcttg | Excision of Ece1-llI62–93 from ECE1 |
| 5′ECE1prom- Nar1 |
PCR | gatcggcgcctccagccactattttgtacctgt | Amplification of ECE1
promoter region for ECE1 promoter-GFP construct |
| 3′ECE1prom- Xhol |
PCR | tcagctcgagtttaacgaatggaaaatagttggtag | Amplification of ECE1
promoter region for ECE1 promoter-GFP construct |
| 5′ECE1term- Sacll |
PCR | gatcccgcggcagcagataaaaatttgttttccacaag | Amplification of ECE1
terminator region for ECE1 promoter-GFP construct |
| 5′ECE1term- Sacl |
PCR | tcaggagctccgttaagaatatgaatgacagttggtc | Amplification of ECE1
terminator region for ECE1 promoter-GFP construct |
| G1-ECE1 | PCR | ctcgctgattagagttcaagagt | Confirmation of ECE1-GFP
plasmid integration (5′ end) |
| GFP veri rev | PCR | tgatctgggtatctcgcaaagcat | Confirmation of ECE1-GFP
plasmid integration (5′ end) |
| G4-ECE1 | PCR | tggaagattcacttgagttggaac | Confirmation of ECE1-GFP
plasmid integration (3′ end) |
| X3-SAT1 | PCR | gtgaagtgtgaagggggag | Confirmation of ECE1-GFP
plasmid integration (3′ end) |
| pENO1FW | PCR | tccttggctggca ctga a ctcg | Confirmation of pENO1-dTom
plasmid integration |
| dTom REV | PCR | aaggtctaccttcaccttcacc | Confirmation of pENO1-dTom
plasmid integration |
| ACT1-F | qPCR | tcagaccagctgatttaggtttg | Quantification of actin cDNA |
| ACT1-R | qPCR | gtgaacaatggatggaccag | Quantification of actin cDNA |
| ECE1-F | qPCR | atcgaaaatgccaagagag | Quantification of ECE1 cDNA |
| ECE1-R | qPCR | agcattttcaataccgacag | Quantification of ECE1 cDNA |
Supplementary Material
Acknowledgments
We thank Sarah Gaffen, Bruce Klein, Christian Hertweck, Abigail Tucker, Jeremy Green and Stephen Challacombe for comments on the manuscript. For experimental assistance we thank Stuart Bevan and David Andersson (calcium assays), Deepa Nayar (histology), Durdana Rahman and Mukesh Mistry (murine model), Mark Nilan (zebrafish model), Sabrina Groth (FRET spectroscopy), Nadine Gebauer (Impedance experiments), Daniela Schulz (kex1Δ/Δ strain) and our colleagues for supplying fungal mutant strains. This work was supported by grants from the Medical Research Council (MR/J008303/1, MR/M011372/1), Biotechnology & Biological Sciences Research Council (BB/J015261/1), FP7-PEOPLE-2013-Initial Training Network (606786) to JRN; Wellcome Trust Strategic Award for Medical Mycology and Fungal Immunology (097377/Z/11/Z) to JRN and DW; Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (102549/Z/13/Z) to DW; Deutsche Forschungsgemeinschaft CRC/TR124 FungiNet Project C1 and Z2, Deutsche Forschungsgemeinschaft SPP 1580 (Hu 528/17-1) and CSCC, German Federal Ministry of Education and Health [BMBF] 01EO1002 to BHu; Cluster of Excellence ‘Inflammation at interfaces’ and Deutsche Forschungsgemeinschaft SPP1580 project GU 568/5-1 to TG; National Institutes of Health (R15AI094406) and the Burroughs Wellcome Fund to RTW.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions DLM, JPR, SXT, MR, CM, MB, SII, NK performed signaling, transcription factor, calcium and cytokine assays, and murine work; DW, SH, SM, TMF, BHe, LK AH, OB and OKu created fungal strains and performed fluorescent microscopy, adhesion, invasion, gene expression and damage assays; RLG and RTW performed zebrafish experiments; JW and TG performed biophysical analysis with artificial membranes; JR performed whole patch clamp analysis; GV performed electron microscopy; ST performed histological analysis; SM, TL, TK and OKn performed LC-MS analyses; JRN, BHu, DLM, JPR and DW wrote the paper; JRN, BHu and EC supervised the project.
The authors declare no competing financial interests.
References
- 1.Brown GD, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004404. 165rv113. [DOI] [PubMed] [Google Scholar]
- 2.Jacobsen ID, et al. Candida albicans dimorphism as a therapeutic target. Expert Rev Anti Infect Ther. 2012;10:85–93. doi: 10.1586/eri.11.152. [DOI] [PubMed] [Google Scholar]
- 3.Moyes DL, et al. A Biphasic Innate Immune MAPK Response Discriminates between the Yeast and Hyphal Forms of Candida albicans in Epithelial Cells. Cell Host Microbe. 2010;8:225–235. doi: 10.1016/j.chom.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moyes DL, et al. Candida albicans yeast and hyphae are discriminated by MAPK signaling in vaginal epithelial cells. PLoS ONE. 2011;6:e26580. doi: 10.1371/journal.pone.0026580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murciano C, et al. Candida albicans cell wall glycosylation may be indirectly required for activation of epithelial cell proinflammatory responses. Infect Immun. 2011;79:4902–4911. doi: 10.1128/IAI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyes DL, et al. Activation of MAPK/c-Fos induced responses in oral epithelial cells is specific to Candida albicans and Candida dubliniensis hyphae. Med Microbiol Immunol. 2012;201:93–101. doi: 10.1007/s00430-011-0209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murciano C, et al. Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PLoS ONE. 2012;7:e33362. doi: 10.1371/journal.pone.0033362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moyes DL, Naglik JR. Mucosal Immunity and Candida albicans Infection. Clin Dev Immunol. 2011;2011 doi: 10.1155/2011/346307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naglik JR, Moyes D. Epithelial Cell Innate Response to Candida albicans . Adv Dent Res. 2011;23:50–55. doi: 10.1177/0022034511399285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naglik JR, Moyes DL, Wachtler B, Hube B. Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 2011;13:963–976. doi: 10.1016/j.micinf.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebecker B, Naglik JR, Hube B, Jacobsen ID. Pathogenicity mechanisms and host response during oral Candida albicans infections. Expert Rev Anti Infect Ther. 2014;12:867–879. doi: 10.1586/14787210.2014.916210. [DOI] [PubMed] [Google Scholar]
- 12.Naglik JR. Candida Immunity. New Journal of Science. 2014;2014 Article ID 390241, 390227 pages. [Google Scholar]
- 13.Naglik JR, Richardson JP, Moyes DL. Candida albicans Pathogenicity and Epithelial Immunity. PLoS Pathog. 2014;10:e1004257. doi: 10.1371/journal.ppat.1004257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moyes DL, Richardson JP, Naglik JR. Candida albicans-epithelial interactions and pathogenicity mechanisms: scratching the surface. Virulence. 2015;6:338–346. doi: 10.1080/21505594.2015.1012981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birse CE, Irwin MY, Fonzi WA, Sypherd PS. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans . Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohm M, et al. A family of secreted pathogenesis-related proteins in Candida albicans . Mol Microbiol. 2013;87:132–151. doi: 10.1111/mmi.12087. [DOI] [PubMed] [Google Scholar]
- 17.Kamai Y, Kubota M, Hosokawa T, Fukuoka T, Filler SG. New model of oropharyngeal candidiasis in mice. Antimicrob Agents Chemother. 2001;45:3195–3197. doi: 10.1128/AAC.45.11.3195-3197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brothers KM, et al. NADPH Oxidase-Driven Phagocyte Recruitment Controls Candida albicans Filamentous Growth and Prevents Mortality. PLoS Pathog. 2013;9:e1003634. doi: 10.1371/journal.ppat.1003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gratacap RL, Rawls JF, Wheeler RT. Mucosal candidiasis elicits NF-kappaB activation, proinflammatory gene expression and localized neutrophilia in zebrafish. Dis Model Mech. 2013;6:1260–1270. doi: 10.1242/dmm.012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader O, Krauke Y, Hube B. Processing of predicted substrates of fungal Kex2 proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris . BMC Microbiol. 2008;8:116. doi: 10.1186/1471-2180-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newport G, Agabian N. KEX2 influences Candida albicans proteinase secretion and hyphal formation. J Biol Chem. 1997;272:28954–28961. doi: 10.1074/jbc.272.46.28954. [DOI] [PubMed] [Google Scholar]
- 22.Liu P, Huang X, Zhou R, Berne BJ. Observation of a dewetting transition in the collapse of the melittin tetramer. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 23.Bechinger B, Salnikov ES. The membrane interactions of antimicrobial peptides revealed by solid-state NMR spectroscopy. Chem Phys Lipids. 2012;165:282–301. doi: 10.1016/j.chemphyslip.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Pieta P, Mirza J, Lipkowski J. Direct visualization of the alamethicin pore formed in a planar phospholipid matrix. Proc Natl Acad Sci U S A. 2012;109:21223–21227. doi: 10.1073/pnas.1201559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bischofberger M, Iacovache I, van der Goot FG. Pathogenic pore-forming proteins: function and host response. Cell Host Microbe. 2012;12:266–275. doi: 10.1016/j.chom.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Los FC, Randis TM, Aroian RV, Ratner AJ. Role of pore-forming toxins in bacterial infectious diseases. Microbiol Mol Biol Rev. 2013;77:173–207. doi: 10.1128/MMBR.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oren Z, Shai Y. Selective lysis of bacteria but not mammalian cells by diastereomers of melittin: structure-function study. Biochem. 1997;36:1826–1835. doi: 10.1021/bi962507l. [DOI] [PubMed] [Google Scholar]
- 28.Walev I, et al. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proc Natl Acad Sci U S A. 2001;98:3185–3190. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt MJ, Breinig F. Yeast viral killer toxins: lethality and self-protection. Nat Rev Microbiol. 2006;4:212–221. doi: 10.1038/nrmicro1347. [DOI] [PubMed] [Google Scholar]
- 30.Zakikhany K, et al. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol. 2007;9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]
- 31.Wachtler B, et al. Candida albicans-epithelial interactions: dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS ONE. 2012;7:e36952. doi: 10.1371/journal.pone.0036952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rupniak HT, et al. Characteristics of four new human cell lines derived from squamous cell carcinomas of the head and neck. J Natl Cancer Inst. 1985;75:621–635. [PubMed] [Google Scholar]
- 33.Mayer FL, et al. The novel Candida albicans transporter Dur31 Is a multi-stage pathogenicity factor. PLoS pathog. 2012;8:e1002592. doi: 10.1371/journal.ppat.1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 35.Citiulo F, et al. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS pathog. 2012;8:e1002777. doi: 10.1371/journal.ppat.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gola S, Martin R, Walther A, Dunkler A, Wendland J. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast. 2003;20:1339–1347. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- 37.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walther A, Wendland J. An improved transformation protocol for the human fungal pathogen Candida albicans . Curr Genet. 2003;42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- 39.Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans . Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 40.Gratacap RL, Rawls JF, Wheeler RT. Mucosal candidiasis elicits NF-kappaB activation, proinflammatory gene expression and localized neutrophilia in zebrafish. Dis Model Mech. 2013;6:1260–1270. doi: 10.1242/dmm.012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyes DL, et al. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe. 2010;8:225–235. doi: 10.1016/j.chom.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moyes DL, et al. Candida albicans yeast and hyphae are discriminated by MAPK signaling in vaginal epithelial cells. PloS ONE. 2011;6:e26580. doi: 10.1371/journal.pone.0026580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wachtler B, Wilson D, Haedicke K, Dalle F, Hube B. From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PloS ONE. 2011;6:e17046. doi: 10.1371/journal.pone.0017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trinh le A, et al. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Gene Devel. 2011;25:2306–2320. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat Protoc. 2012;7:637–642. doi: 10.1038/nprot.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cranfield C, Carne S, Martinac B, Cornell B. The assembly and use of tethered bilayer lipid membranes (tBLMs) Methods Mol Biol. 2015;1232:45–53. doi: 10.1007/978-1-4939-1752-5_4. [DOI] [PubMed] [Google Scholar]
- 47.Cranfield CG, et al. Transient potential gradients and impedance measures of tethered bilayer lipid membranes: pore-forming peptide insertion and the effect of electroporation. Biophys J. 2014;106:182–189. doi: 10.1016/j.bpj.2013.11.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schromm AB, et al. Lipopolysaccharide-binding protein mediates CD14-independent intercalation of lipopolysaccharide into phospholipid membranes. FEBS Lett. 1996;399:267–271. doi: 10.1016/s0014-5793(96)01338-5. [DOI] [PubMed] [Google Scholar]
- 49.Chen GC, Yang JT. 2-Point Calibration of Circular Dichrometer with D-10-Camphorsulfonic Acid. Anal. Lett. 1977;10:1195–1207. [Google Scholar]
- 50.Montal M, Mueller P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc Natl Acad Sci U S A. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutsmann T, Heimburg T, Keyser U, Mahendran KR, Winterhalter M. Protein reconstitution into freestanding planar lipid membranes for electrophysiological characterization. Nat Protoc. 2015;10:188–198. doi: 10.1038/nprot.2015.003. [DOI] [PubMed] [Google Scholar]
Extended Data Table references
- 52.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3, E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 53.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans . Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans . Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braun BR, Johnson AD. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans . Genetics. 2000;155:57–67. doi: 10.1093/genetics/155.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo HJ, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 58.Moyes DL, et al. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe. 2010;8:225–235. doi: 10.1016/j.chom.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zakikhany K, et al. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol. 2007;9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]
- 60.Cao F, et al. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans . Mol Biol Cell. 2006;17:295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bockmuhl DP, Krishnamurthy S, Gerads M, Sonneborn A, Ernst JF. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans . Mol Microbiol. 2001;42:1243–1257. doi: 10.1046/j.1365-2958.2001.02688.x. [DOI] [PubMed] [Google Scholar]
- 62.Sonneborn A, et al. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans . Mol Microbiol. 2000;35:386–396. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- 63.Palmer GE, Cashmore A, Sturtevant J. Candida albicans VPS11 is required for vacuole biogenesis and germ tube formation. Eukaryot Cell. 2003;2:411–421. doi: 10.1128/EC.2.3.411-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zou H, Fang HM, Zhu Y, Wang Y. Candida albicans Cyr1, Cap1 and G-actin form a sensor/effector apparatus for activating cAMP synthesis in hyphal growth. Mol Microbiol. 2010;75:579–591. doi: 10.1111/j.1365-2958.2009.06980.x. [DOI] [PubMed] [Google Scholar]
- 65.Bates S, et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans . J Biol Chem. 2006;281:90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- 66.Murciano C, et al. Candida albicans cell wall glycosylation may be indirectly required for activation of epithelial cell proinflammatory responses. Infect Immun. 2011;79:4902–4911. doi: 10.1128/IAI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newport G, Agabian N. KEX2 influences Candida albicans proteinase secretion and hyphal formation. J Biol Chem. 1997;272:28954–28961. doi: 10.1074/jbc.272.46.28954. [DOI] [PubMed] [Google Scholar]
- 68.Murad AM, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans . EMBO J. 2001;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 70.Lane S, Zhou S, Pan T, Dai Q, Liu H. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1 . Mol Cell Biol. 2001;21:6418–6428. doi: 10.1128/MCB.21.19.6418-6428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White SJ, et al. Self-regulation of Candida albicans population size during GI colonization. PLoS pathog. 2007;3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown DH, Jr, Giusani AD, Chen X, Kumamoto CA. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol Microbiol. 1999;34:651–662. doi: 10.1046/j.1365-2958.1999.01619.x. [DOI] [PubMed] [Google Scholar]
- 73.Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans . Mol Cell Biol. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.San Jose C, Monge RA, Perez-Diaz R, Pla J, Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans . J Bacteriol. 1996;178:5850–5852. doi: 10.1128/jb.178.19.5850-5852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Firon A, et al. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans . Mol Microbiol. 2007;66:1256–1275. doi: 10.1111/j.1365-2958.2007.06011.x. [DOI] [PubMed] [Google Scholar]
- 76.de Boer AD, et al. The Candida albicans cell wall protein Rhd3/Pga29 is abundant in the yeast form and contributes to virulence. Yeast. 2010;27:611–624. doi: 10.1002/yea.1790. [DOI] [PubMed] [Google Scholar]
- 77.Muhlschlegel FA, Fonzi WA. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol Cell Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin R, et al. A core filamentation response network in Candida albicans is restricted to eight genes. PloS ONE. 2013;8:e58613. doi: 10.1371/journal.pone.0058613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Birse CE, Irwin MY, Fonzi WA, Sypherd PS. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans . Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Navarro-Garcia F, Sanchez M, Pla J, Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol Cell Biol. 1995;15:2197–2206. doi: 10.1128/mcb.15.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hausauer DL, Gerami-Nejad M, Kistler-Anderson C, Gale CA. Hyphal guidance and invasive growth in Candida albicans require the Ras-like GTPase Rsr1p and its GTPase-activating protein Bud2p. Eukaryot Cell. 2005;4:1273–1286. doi: 10.1128/EC.4.7.1273-1286.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sentandreu M, Elorza MV, Sentandreu R, Fonzi WA. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans . J Bacteriol. 1998;180:282–289. doi: 10.1128/jb.180.2.282-289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pardini G, et al. The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans . J Biol Chem. 2006;281:40399–40411. doi: 10.1074/jbc.M606361200. [DOI] [PubMed] [Google Scholar]
- 84.Braun BR, Head WS, Wang MX, Johnson AD. Identification and characterization of TUP1-regulated genes in Candida albicans . Genetics. 2000;156:31–44. doi: 10.1093/genetics/156.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fradin C, et al. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- 86.Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 87.Bailey DA, Feldmann PJ, Bovey M, Gow NA, Brown AJ. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sandini S, La Valle R, De Bernardis F, Macri C, Cassone A. The 65 kDa mannoprotein gene of Candida albicans encodes a putative beta-glucanase adhesin required for hyphal morphogenesis and experimental pathogenicity. Cell Microbiol. 2007;9:1223–1238. doi: 10.1111/j.1462-5822.2006.00862.x. [DOI] [PubMed] [Google Scholar]
- 89.Csank C, et al. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hube B, et al. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taylor BN, et al. Induction of SAP7 correlates with virulence in an intravenous infection model of candidiasis but not in a vaginal infection model in mice. Infect Immun. 2005;73:7061–7063. doi: 10.1128/IAI.73.10.7061-7063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schild L, et al. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot Cell. 2011;10:98–109. doi: 10.1128/EC.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao X, et al. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiol. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- 94.Murciano C, et al. Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PloS ONE. 2012;7:e33362. doi: 10.1371/journal.pone.0033362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao X, Oh SH, Yeater KM, Hoyer LL. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiol. 2005;151:1619–1630. doi: 10.1099/mic.0.27763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao X, Oh SH, Hoyer LL. Deletion of ALS5 ALS6 or ALS7 increases adhesion of Candida albicans to human vascular endothelial and buccal epithelial cells. Med Mycol. 2007;45:429–434. doi: 10.1080/13693780701377162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao X, Oh SH, Hoyer LL. Unequal contribution of ALS9 alleles to adhesion between Candida albicans and human vascular endothelial cells. Microbiol. 2007;153:2342–2350. doi: 10.1099/mic.0.2006/005017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Timpel C, Strahl-Bolsinger S, Ziegelbauer K, Ernst JF. Multiple functions of Pmt1p–mediated protein O-mannosylation in the fungal pathogen Candida albicans . J Biol Chem. 1998;273:20837–20846. doi: 10.1074/jbc.273.33.20837. [DOI] [PubMed] [Google Scholar]
- 99.Bates S, et al. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J Biol Chem. 2005;280:23408–23415. doi: 10.1074/jbc.M502162200. [DOI] [PubMed] [Google Scholar]
- 100.Hobson RP, et al. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J Biol Chem. 2004;279:39628–39635. doi: 10.1074/jbc.M405003200. [DOI] [PubMed] [Google Scholar]
- 101.Southard SB, Specht CA, Mishra C, Chen-Weiner J, Robbins PW. Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins. J Bacteriol. 1999;181:7439–7448. doi: 10.1128/jb.181.24.7439-7448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Munro CA, et al. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J Biol Chem. 2005;280:1051–1060. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mio T, et al. Role of three chitin synthase genes in the growth of Candida albicans . J Bacteriol. 1996;178:2416–2419. doi: 10.1128/jb.178.8.2416-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mille C, et al. Inactivation of CaMIT1 inhibits Candida albicans phospholipomannan beta-mannosylation, reduces virulence, and alters cell wall protein beta-mannosylation. J Biol Chem. 2004;279:47952–47960. doi: 10.1074/jbc.M405534200. [DOI] [PubMed] [Google Scholar]
- 105.Mille C, et al. Identification of a new family of genes involved in beta-1,2-mannosylation of glycans in Pichia pastoris and Candida albicans . J Biol Chem. 2008;283:9724–9736. doi: 10.1074/jbc.M708825200. [DOI] [PubMed] [Google Scholar]
- 106.Mille C, et al. Members 5 and 6 of the Candida albicans BMT family encode enzymes acting specifically on beta-mannosylation of the phospholipomannan cell-wall glycosphingolipid. Glycobiol. 2012;22:1332–1342. doi: 10.1093/glycob/cws097. [DOI] [PubMed] [Google Scholar]
- 107.Mio T, et al. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis. J Bacteriol. 1997;179:4096–4105. doi: 10.1128/jb.179.13.4096-4105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mio T, et al. Isolation of the Candida albicans homologs of Saccharomyces cerevisiae KRE6 and SKN1: expression and physiological function. J Bacteriol. 1997;179:2363–2372. doi: 10.1128/jb.179.7.2363-2372.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Staab JF, Sundstrom P. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 2003;11:69–73. doi: 10.1016/s0966-842x(02)00029-x. [DOI] [PubMed] [Google Scholar]
- 110.Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans . Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.