Abstract
DNA deaminases underpin pathways in antibody diversification (AID) and anti-viral immunity (APOBEC3s). Here we show how a high-throughput bacterial papillation assay can be used to screen for AID mutants with increased catalytic activity. The upmutations focus on a small number of residues, some highlighting regions likely implicated in AID’s substrate interaction. Notably, many of the upmutations bring the sequence of AID closer to that of APOBEC3s. AID upmutants can yield increased antibody diversification, raising the possibility that modification of AID’s specific activity might be used to regulate antibody diversification in vivo. However, upmutation of AID also led to increased frequency of chromosomal translocations suggesting that AID’s specific activity may have been limited by the risk of genomic instability.
INTRODUCTION
The immune system is unique in mammals in its use of active genomic mutation for a programmed physiological purpose. Within the adaptive immune system, the functionally rearranged immunoglobulin genes in activated B lymphocytes are targeted for deamination at cytosine residues by activation-induced deaminase (AID). Deamination within the immunoglobulin V gene triggers antibody gene diversification by somatic hypermutation and gene conversion, processes which underpin antibody affinity maturation. Deamination in the vicinity of the immunoglobulin switch (S) regions triggers the shift from the expression of IgM to that of one of the downstream isotypes (IgG/A/E)1,2. Programmed DNA deamination is also used within the innate immune system where deamination of retroviral replication intermediates by members of the APOBEC3 family (which show sequence homology to AID) is associated with pathways of retroviral restriction3,4.
Although such purposeful DNA deamination is beneficial to the host since it underpins major pathways in immunity, off-target action by the deaminases is likely to be harmful and lead to genomic instability and predisposition to cancer. Indeed many of the chromosomal translocations and point mutations associated with proto-oncogene activation in tumors of the B cell lineage can be ascribed to events triggered by the action of AID5. Thus the function of AID is likely regulated so as to achieve a balance between immunity and predisposition to cancer. Indeed, overexpression of AID in cell lines as well as in transgenic animals does indeed give rise to increased genomic instability and tumor incidence, possibly reflecting that increased AID abundance results in an increased likelihood of it accessing inappropriate targets6–8.
Here, in order to discover whether it is possible to enhance antibody diversification by increasing the specific activity of AID, we describe a high throughput bacterial (Escherichia coli) papillation assay which can be used to screen for AID mutants that exhibit increased activity. We use these mutants to show that the rate of antibody gene diversification is indeed limited by the specific activity of AID but show that whilst the rate of antibody diversification can be enhanced using AID upmutants, this leads to increased genomic instability. Intriguingly, a relatively small number of specific amino acid substitutions are repeatedly identified when selecting AID upmutants (several of which are close to the catalytic site) with many of these upmutations bringing the sequence of AID closer to that of its APOBEC3 relatives. The results suggest that AID, which functions in the nucleus, may have experienced more stringent limitations to the upward evolution of its specific activity than its APOBEC3 relatives, which largely function in the cytosol.
RESULTS
Mutator screen for DNA deaminases
Escherichia coli cells which harbour a mis-sense mutation within lacZ give rise to white colonies on MacConkey-lactose plates: within such white colonies, a small number of red microcolonies can often be discerned (papilli, typically 0-2 per colony) which reflect spontaneously-arising Lac+ revertants. Bacterial mutator clones which exhibit an elevated frequency of spontaneous mutation can be identified by virtue of an increased number of papilli.
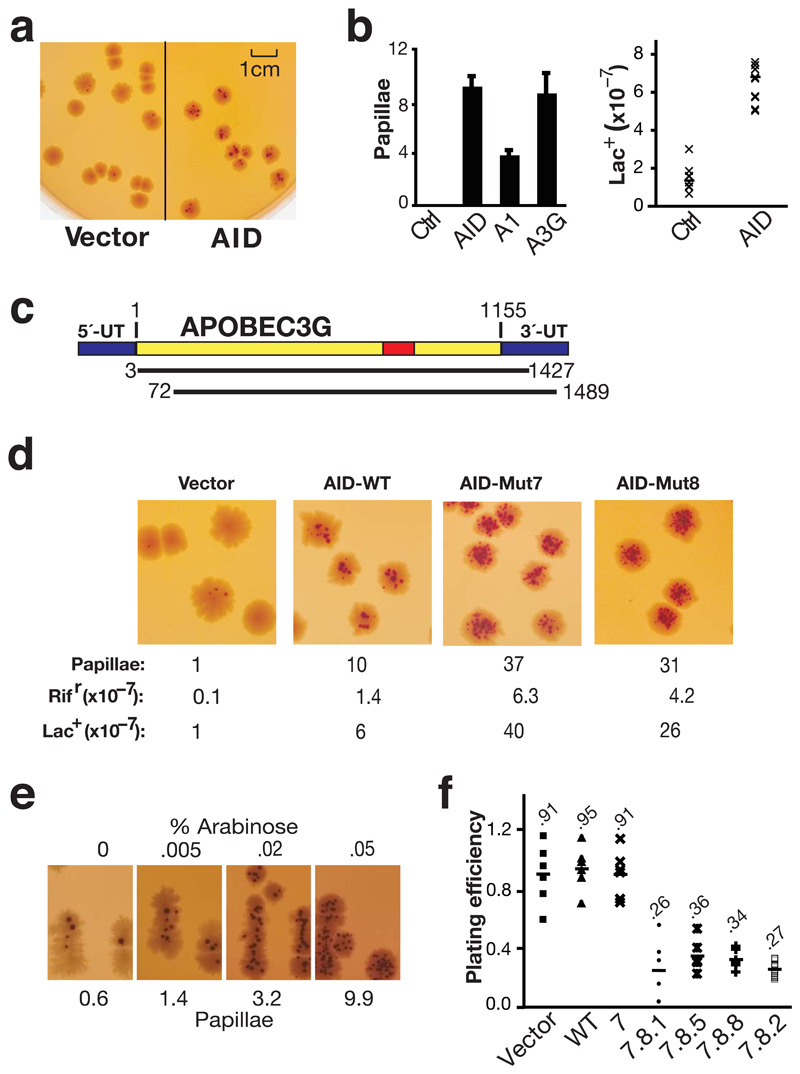
Although papillation assays have been used to screen for E. coli mutants that are defective in some aspect of DNA repair9,10, we wondered if the assay could be adapted to screen for active mutators. Strain CC102 carries a mis-sense mutation in codon 461 of lacZ with glutamate being substituted by glycine owing to a A:T to G:C transition mutation11. If expression of AID in CC102 were to increase the rate of cytosine deamination at codon 461, this might be expected to increase the frequency of Lac+ revertants. This is indeed the case. AID-expressing transformants of CC102 give an increased frequency of papillation on MacConkey-lactose plates (Fig. 1). The number of papilli per colony as assayed after 6 days of incubation increases from 0-2 per colony to 8-10: this correlates with a more than threefold increase in the frequency of Lac+ revertants in overnight cultures as judged on minimal-lactose plates. Sequence analysis of six such Lac+ revertants confirmed that they had indeed arisen through reversion at codon 461. The AID-related deaminases APOBEC1 and APOBEC3G are also able to trigger increased papillation when expressed in CC102 (Fig. 1b).
Figure 1.
Papillation screen for active mutators. (a) Empty vector or AID-transformed CC102 was plated on MacConkey-lactose agar and grown at 37 °C for 4 days. (b) Frequency of reversion to Lac+ as determined by the average number papillae per colony on MacConkey-lactose agar (left panel; histogram showing the mean and s.d. of three experiments)) or by growth on minimal lactose plates (right panel: fluctuation analysis on six clones each) in CC102 transformants expressing vector control (Ctrl), AID, APOBEC1 (A1) or APOBEC3G (A3G). (c) Depiction of the two APOBEC3G cDNAs obtained by screening a human spleen cDNA library (50,000 colonies) for papillation on CC102. The wild type full length APOBEC3G mRNA is shown at the top and the structures of the two cDNAs below. The red shows the catalytic active site. The blue shows the untranslated region (UT). Nucleotide residues are numbered relative to the start of the open reading frame (+1). (d) Comparison of papillation (and mutation frequencies on lac and rifampicin) by wild type AID and by upmutants Mut7 (K10E/E156G) and 8 (K34E/K160E). (e) Papillation by AID Mut1.1 expressed from plasmid pBAD30 as a function of the concentration (% w/v) of L-arabinose (inducer). (f) Bacterial titers in cultures grown to saturation in LB/Amp under conditions of IPTG induction relative to the titers obtained from cultures grown in the absence of induction is presented for CC102 transformants expressing different AID upmutants.
To assess the robustness of the assay, we asked whether it could be used to isolate active mutators from a total splenic cDNA library. We introduced a human spleen cDNA library into CC102 and fifty thousand colonies screened for enhanced papillation. Thirty-six possible candidates were identified, which were retested by streaking on MacConkey lactose plates. Only two colonies were confirmed as giving increased papillation. Sequence analysis revealed that they carried distinct cDNAs derived from APOBEC3G (Fig. 1c), demonstrating that the assay can indeed be used as a high throughput screen for active mutators. The likely reason that the screen yielded APOBEC3G rather than AID relates to the fact that the cDNA library derives from unstimulated human spleen (containing both B and T cells, where APOBEC3G is well expressed) whereas AID expression is largely restricted to activated B cells.
Selection for AID upmutators
To see whether we could isolate variants of human AID exhibiting increased mutator activity in E. coli, a human AID cDNA was subjected to PCR-mutagenesis, cloned into plasmid pTrc99a and transformed into E. coli CC102. The diversity of the library of AID mutants thereby generated was assessed by sequencing 48 randomly selected clones. These clones each carried three to five nucleotide substitutions per Kb with no two clones carrying an identical mutation. Screening of a total of sixty thousand colonies from four independent PCR-mutagenesis experiments yielded 13 AID transformants which exhibited increased papillation on MacConkey-lactose plates (Figs. 1d and 2). The AID-containing plasmids from nine of these mutants were then tested following retransfection into E. coli strain KL16 for the frequency with which they yielded rifampicin-resistant colonies: all nine exhibited an increased frequency of mutation at the rpoB locus.
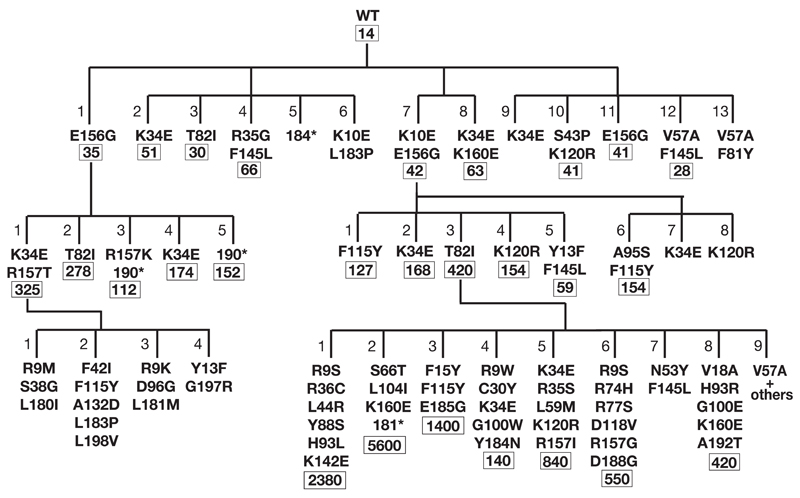
Figure 2.
Dynasty of AID upmutants selected by papillation screen. Upmutants obtained in three successive rounds of mutagenesis with mutants obtained from individual PCR-mutagenesis experiments grouped as families of siblings. The additional amino acid substitutions introduced in each round of mutagenesis are indicated with the numbers below giving the mean frequency of mutation to Rifr relative to vector. * indicates a C-terminal truncation caused by introduction of a premature stop codon at the indicated codon. Individual mutants are numbered according to their dynastic origin: thus, for example, Mut7 (K10E/E156G) is the parent of Mut7.1 (K10E/E156G/F115Y).
The AID cDNAs from two of the first-generation upmutants (Mut1 and Mut7) were then themselves subjected to PCR mutagenesis and second-generation mutants exhibiting enhanced papillation were obtained (Fig. 2). In fact the high papillation exhibited by these second generation mutants made it difficult to visually discern any additional increases in papillation. This meant that we needed to modify the assay in order to screen for further enhancement of mutator activity in a third round of mutation/selection. To this end, AID Mut1.1 and 7.3 cDNAs were moved to an arabinose-inducible expression vector such that the number of papilli obtained in CC102 transformants could be regulated by varying the concentration of arabinose in the medium (Fig. 1e). Screening for papillation under low (0.02%) arabinose, we obtained a third generation of AID upmutants, some of which gave a mutation frequency that was nearly 400-times greater than the wild type AID as judged by the frequency of mutation to rifampicin resistance (Fig. 2).
Several of the third generation mutants appeared to exhibit toxicity in E. coli as judged by smaller colony size when grown under inducing conditions; this was accompanied by a reduced viable cell count in bacterial cultures grown to saturation (Fig. 1f). This toxicity might have caused some highly papillating mutants to give anomalously low frequencies of mutation to Rifr (e.g. Mut7.3.4; Fig. 2) with AID expression possibly being downregulated during overnight culture.
Hotspots for upmutation of human AID
Apart from the premature stop codon mutations identified in three of the AID upmutants (Mut5, 1.3 and 1.5), analysis of the sequences of the various AID upmutants revealed a striking preference for certain amino acid substitutions. Thus, for example, the K34E, T82I and E156G substitutions (each of which is sufficient on its own to increase AID activity) have actually been selected in independent experiments. These mutations were not found amongst sequences of 48 random (i.e. unselected) clones from the PCR-generated libraries, where a wide spectrum of mutations was observed without indications of any major hotspots of the mutagenesis procedure itself. Thus, the repeat identification of a small number of amino acid substitutions suggests that there are a limited number of single amino acid substitutions in AID that yield increased papillation.
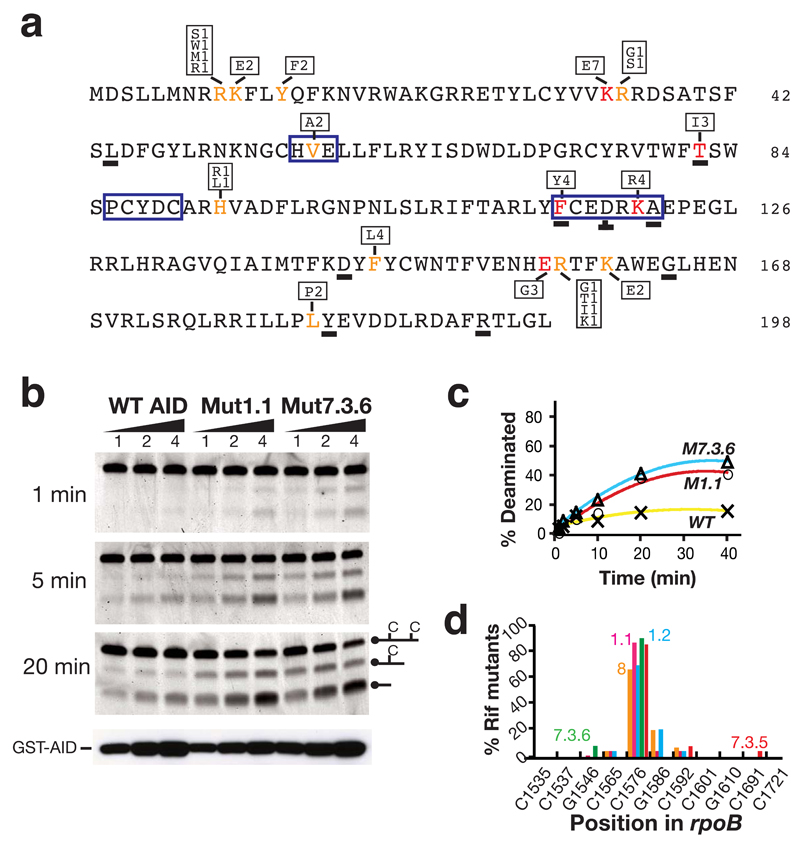
Although in some cases (especially in the third generation) the multiplicity of mutations introduced in a single round prevents unambiguous identification of those mutations responsible for the increased papillation, in many cases the relevant upmutation can be definitively identified because it constitutes the sole difference between a pair of differently-papillating AID sequences or (somewhat less definitively) because it was independently obtained in multiple PCRs. The locations of such upmutations are depicted in Fig. 3a, where it is seen that whereas some are located around the zinc-coordination motif in the vicinity of the likely catalytic site (V57A; T82I), others are in a region equivalent to a portion of APOBEC3s that have previously been suggested to be involved in polynucleotide binding (F115Y; K120R)12–14, several are clustered in regions whose function is unknown.
Figure 3.
Nature of the AID upmutations. (a) Primary sequence of human AID with upmutations identified in color. Mutations at residues in red constitute the sole difference between at least one pair of AID sequences resulting in > 2-fold increase in mutation frequencies at rpoB. Mutations at residues in orange have been identified in multiple independent upmutants but in the presence of other substitutions. The box above or below the highlighted residues shows the identity of the substitution mutations and the frequency with which each substitution was detected in the total of nine independent libraries. Underlined letters identify residues where the corresponding position in fugu AID also appears to be a site of selected upmutation, as judged by the fact that it is either the sole mutation or in multiple fugu upmutants. The zinc-coordination motifs (HVE, PCYDC) and regions of suggested polynucleotide contact (FCEDRKA)11–13, are highlighted by a blue box. (b) Deaminase activity of semi-purified GST-AID fusion proteins (100, 200, 400 ng) was analysed on an oligonucleotide substrate at indicated time points. Protein abundance was monitored by Western blot analysis using anti-AID antibody. (c) Quantification of extent of deamination over time. (d) The target specificity of the upmutants as judged by the spectrum of rpoB mutations in Rifr colonies. Transition mutations at any one of 11 C:G pairs within rpoB can give rise to Rifr: the distribution of such mutations amongst Rifr colonies is shown for AID upmutants Mut8 (orange), 1.1 (pink), 1.2 (blue), 7.3.5 (red) and 7.3.6 (green).
Upmutations increase AID’s specific activity
We were interested in ascertaining whether the mutations in AID that increased its bacterial mutator activity acted by increasing the yield of soluble protein or by increasing its specific activity. Initial screening of the sonic extracts of a large number of upmutants did not reveal any which exhibited a substantial increase in the fractional yield of soluble protein as judged by Western blot analysis. However, when GST-fusion proteins were generated from human upmutants Mut1.1 and 7.3.6, a clearly increase in specific activity was evident as judged by in vitro deamination assays performed on a single-stranded oligonucleotide substrate (Fig. 3b, c). The increased activity appears to reflect an increase in VMAX rather than any major change in KM. Thus, by performing the assay over a range of substrate concentrations, we estimate the KM of wild type GST-AID fusion protein as well as that of upmutants Mut1.1 and Mut7.3 for the oligonucleotide substrate all to be in the range 80 – 100 nM (values some fivefold greater than that determined by Larijani et al.15 for wild type GST-AID on a hotspot bubble substrate); however, the VMAX of Mut1.1 and Mut7.3 were calculated to be 0.4 and 0.3 fmol min-1ng-1 as compared to 0.07 fmol min-1ng-1 for the wild type fusion protein (Supplementary Fig. 1 online). Interestingly, although striking, the increase in VMAX displayed by the mutant proteins (4 to 6 fold) is notably less than the increase in the frequency of mutation to rifampicin resistance which they trigger in bacteria (20 to 30 fold). It is possible that this apparent discrepancy could reflect that the high frequency of DNA deamination catalysed by the AID upmutants might overwhelm the normal bacterial DNA repair pathways.
This increased specific activity does not appear to have been accompanied by any gross change in the target specificity since analysis of the rpoB mutations obtained using several human AID mutants did not reveal any major difference in mutation spectrum (Fig. 3d).
Upmutations in fugu AID are in analogous positions
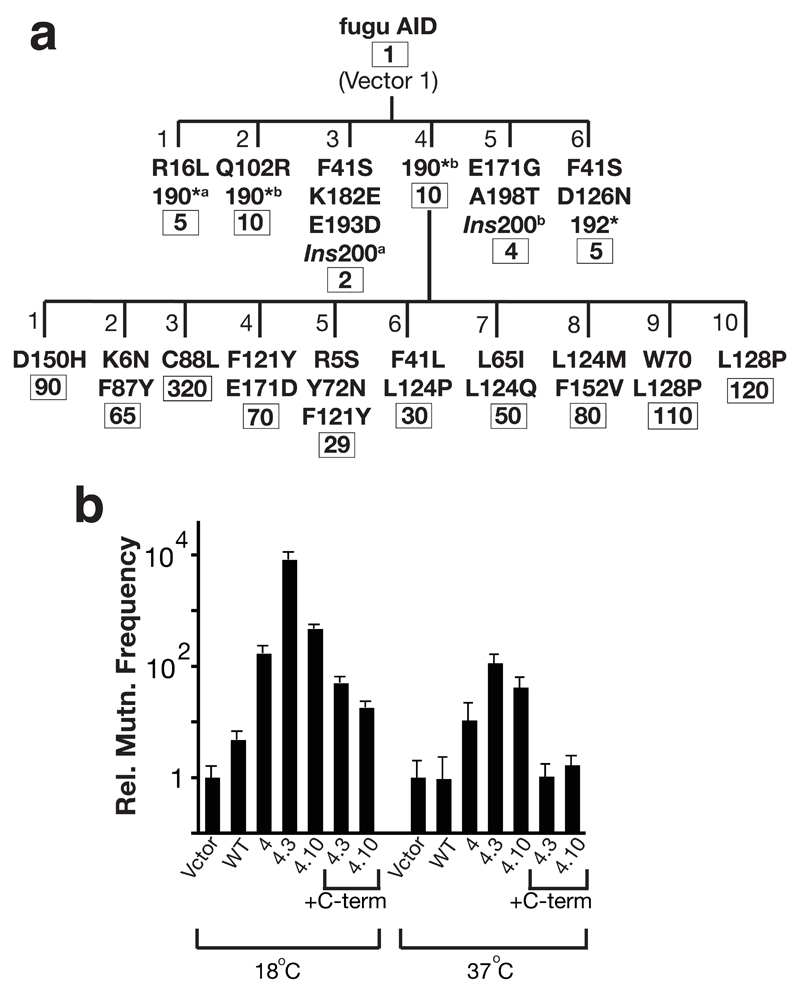
We were interested in extending this study to an evolutionarily distant AID to see if analogous mutations would underpin increased mutator activity. We have previously shown that AID from pufferfish (which live at around 26 °C) exhibits little bacterial mutator activity when assayed at 37 °C whereas mutator activity can be detected at 18 °C16. We therefore asked whether the papillation assay could be used to select mutants of fugu AID that gave robust papillation at 37 °C. Such mutants could be isolated but, as shown in Fig. 4, all the first generation mutants isolated harbored C-terminal truncation mutations, with the six mutants obtained harboring 5 distinct truncation mutations. A variety of amino acid substitutions, however, could then lead to enhanced papillation in second generation mutants (Fig. 4a), with several of these occurring at positions analogous to the upmutations identified in human AID (Figs. 3a, 4a and Supplementary Fig. 2 online). Thus, the mutation (C88L) responsible for the increased activity of fugu AID Mut1.3 occurs at the equivalent position to the T82I mutation in human AID (Supplementary Fig. 2 online). Similarly, residues Phe121, Leu124 and Leu128 in fugu AID (each of which is a target for mutation in either two or three fugu upmutants) are all located in a stretch of fugu AID corresponding to 115-121 in human AID where upmutations were also obtained.
Figure 4.
Upmutants of fugu AID. (a) Dynasty of upmutants of fugu AID selected at 37 °C. * indicates a C-terminal truncation caused by introduction of a premature stop codon; *a and *b indicate different single nucleotide substitutions at codon 190 causing the premature stop codon; Ins200a and Ins200b indicate different single nucleotide insertion mutations at codon 200 which cause the C-terminal region to be read out-of-frame. Numbers below gives the mean frequency of mutation to Rifr relative to vector. (b) The frequency of mutation to Rifr relative to vector-only transformants at either 18 °C or 37 °C is shown for E. coli K16 transformed with plasmids encoding wild type or mutated fugu AID as indicated. Derivatives of Mut4.3 and 4.10 were constructed in which the nonsense mutation at 190 had been reverted, thereby yielding a wild-type C-terminus (+C-term).
Although C-terminal truncations were detected amongst the panel of human AID upmutants and such truncations have previously been shown to give higher mutator activity in E. coli (refs 17 and 18 and our unpublished data), it was nevertheless striking that all the first generation mutants of fugu AID selected at 37 °C carried truncations at the C-terminus. This led us to wonder whether these C-terminal mutations underpinned increased thermal stability and that the amino acid substitutions giving rise to increased papillation in the second generation fugu upmutants might not have been discernible at 37 °C in the absence of a C-terminal truncation mutation. This does indeed appear a likely explanation. The C88L and L128P substitutions both give increased frequency of mutation to Rifr as assayed at 18 °C in the presence or absence of a C-terminal truncation. However, when assayed at 37 °C, these amino acid substitutions do not give any discernible increase in mutation frequency in the absence of the C-terminal truncation (Fig. 4b).
AID upmutation increases antibody diversification
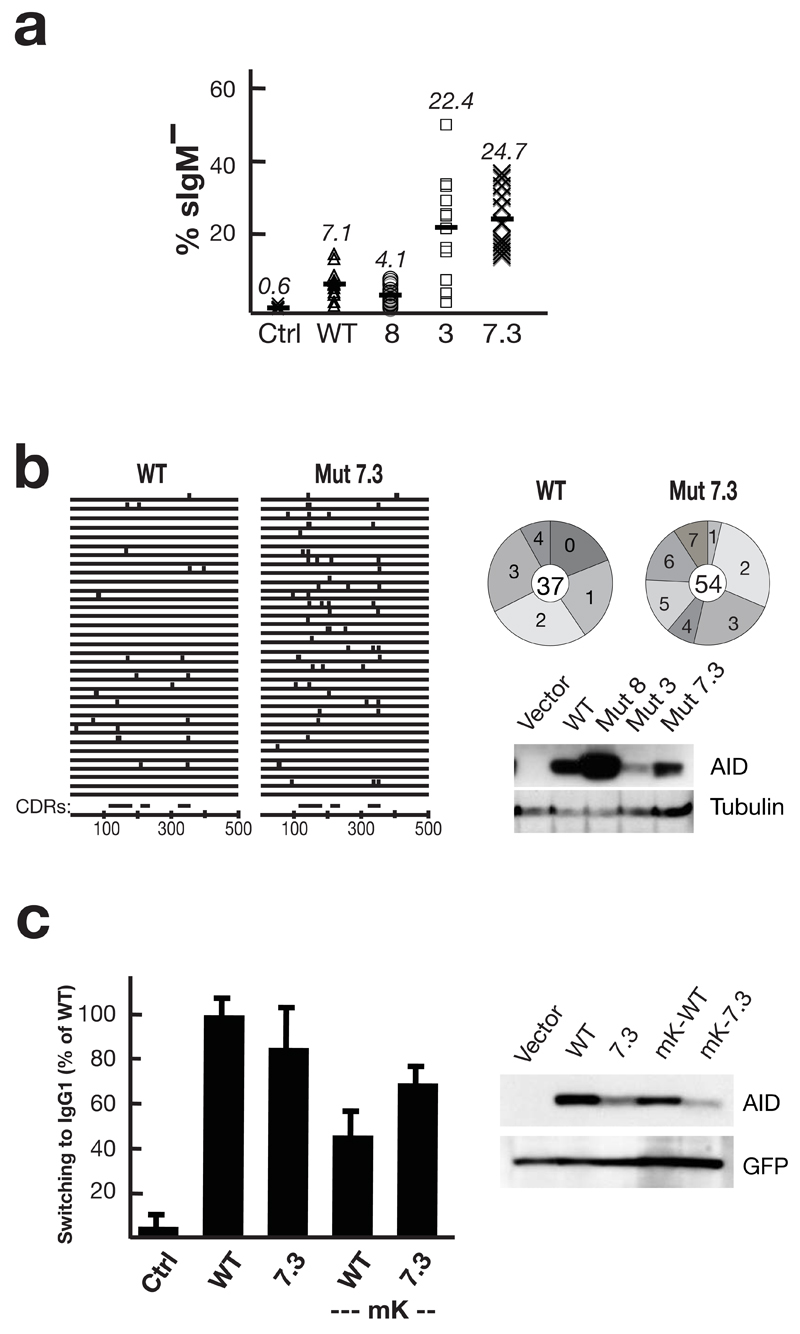
Although several studies have revealed that the efficacy of class switching is limited by the abundance of AID polypeptide19–25, it is unknown whether the efficacy of antibody gene diversification is also limited by the specific activity of AID. We were therefore interested in ascertaining whether AID upmutants exhibiting increased specific activity would also yield increased antibody gene diversification. Mutants 3 (T82I), 8 (K34E, K160E) and 7.3 (K10E, E156G, T82I) were expressed in an AID-deficient chicken DT40 B cell line in which somatic mutation of the IgV can be inferred from the frequency of generation of sIgM-loss variants26. Both Mut3 and Mut7.3 appeared to give substantially enhanced somatic mutation as judged by this sIgM-loss assay. Furthermore, sequence analysis revealed that after one month of clonal expansion, cells expressing these mutant AIDs did indeed carry a higher mutation load in the IgVλ gene than did control cells expressing the wild type enzyme (Fig. 5). Not only did a higher proportion of sequences carry mutations but those that did carry mutations also carried a higher mutation load. This effect is particularly marked when account is taken of the fact that the mutant AID is expressed at lower abundance than its wild type counterpart in these transfectants. In contrast, mutant 8 did not give enhanced somatic mutation indicating that the K34E and/or K160E substitutions are likely to diminish aspects of AID’s function in B cells. Interestingly, Mut8 polypeptide is found at much higher abundance in the DT40 transfectants than are the Mut3 or 7.3 polypeptides. This is consistent with observations that we have made in other work (ref 27 and our unpublished data) that AID mutants which exhibit compromised activity in antibody diversification/genomic mutation in DT40 cells tend to be expressed at higher abundance without any evident alteration in intracellular localisation. We suspect that the explanation for these differences in expression levels is that, in cell transfectants, there is selection against cells expressing high levels of AID proteins which are active in chromosomal mutation.
Figure 5.
Enhanced antibody diversification by AID upmutants. (a) Somatic mutation of the IgV was assayed by monitoring surface IgM-loss (% sIgM–) in AID–/– φV–/– sIgM+ DT40 cells that had been stably transfected with constructs co-expressing the indicated AID mutants together with GFP (mean of 12 independent clonal transfectants). (b) The distribution of mutations in 34 IgVλ sequences obtained by PCR amplification from unsorted WT/Mut7.3-transfected DT40 one month after transfection is shown on the left. The corresponding positions of the complementarity determining regions (CDRs) are shown below. On the right, pie charts depict the number of mutations in PCR amplified IgVλ sequences that had been obtained from the same transfectants but after sorting for sIgM-loss: the number in the middle of the pie chart indicates the total number of sequences analysed. Below the pie charts, Western blots show AID abundance in the DT40 cell extracts with tubulin as loading control. The IgVλ sequences are provided in Supplementary Fig. 3 online. (c) Histogram showing switching to IgG1 in LPS+IL4 cultures of AID-deficient B cells that have been transduced with GFP-(WT/Mut7.3) (mean and s.d. of four experiments). ‘mK’ indicates where transduction was performed using vectors with a mutated Kozak sequence, so as to reduce the extent of AID overexpression. A Western blot shows AID abundance in the B cells extracts 3 days after retroviral transduction; the blot was reprobed with anti-GFP antibodies as a control. Representative flow cytometry plots for (a) and (c) are shown in Supplementary Fig. 4 online.
In order to assay the activity of the mutant AID in class switch recombination, we used an assay based on retroviral transduction of the mutant enzymes into AID-deficient mouse B cells. We were concerned that this assay involves considerable over expression of AID such that the amount of protein might saturate the switching assay. We therefore performed the assay using both the conventional pMX-Ig virus as well as a variant in which the transduced AID is expressed at lower levels through mutation of the Kozak sequence21. As seen in Fig. 5, AID Mut7.3 was more effective in promoting CSR than the wild type counterpart although expressed at lower levels. However, upmutation of AID did not always yield increased class switching. Thus, for example, Mut7.3.2, though a potent mutator, gave no detectable switching in transduced B cells – presumably owing to its C-terminal truncation (data not shown).
AID upmutation increases chromosomal translocations
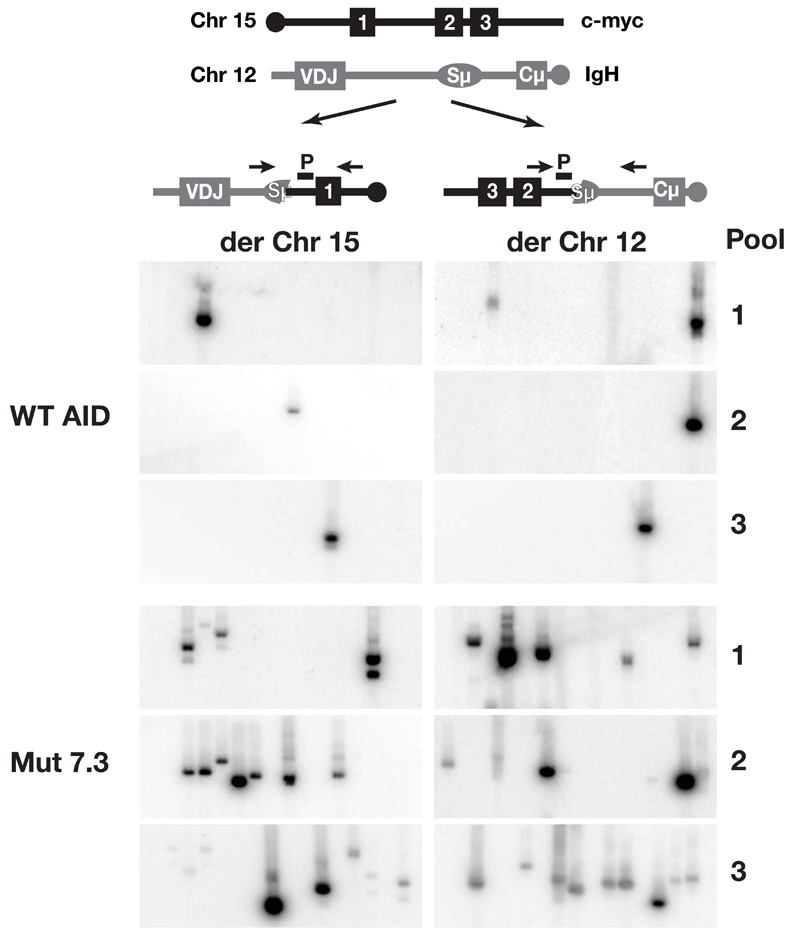
To ascertain whether the AID upmutants also gave increased chromosomal translocations, we exploited a PCR-based assay28 which can be used to detect c-myc/IgH translocations in B cells from AID deficient mice which have been retrovirally transduced for AID expression and cultured for 1-2 days in vitro7. Wild type AID gave rise to an estimated frequency of one c-myc/IgH translocation per 2 × 105 cells. In contrast, Mut7.3 gave a substantially higher frequency of one translocation per 5 × 104 cells (Fig. 6) with the increase in translocation frequency being similar to the increase in Mut7.3’s specific deaminase activity.
Figure 6.
Increased chromosomal translocations by AID upmutants. (a) Scheme of reciprocal translocation between c-myc and IgH locus indicating the primers used for PCR amplification (arrows) and the probes (P) used for Southern blot hybridization. (b) Chromosome 15 and 12 derived translocations (der Chr 15, der Chr 12) were amplified by PCR on three independent pools of genomic DNA from 2 × 105 cell-equivalents of AID-deficient B cells retrovirally transduced with pMx-GFP constructs directing the expression of either wild type or Mut7.3 AID.
Upmutations bring AID sequence closer to APOBEC3s
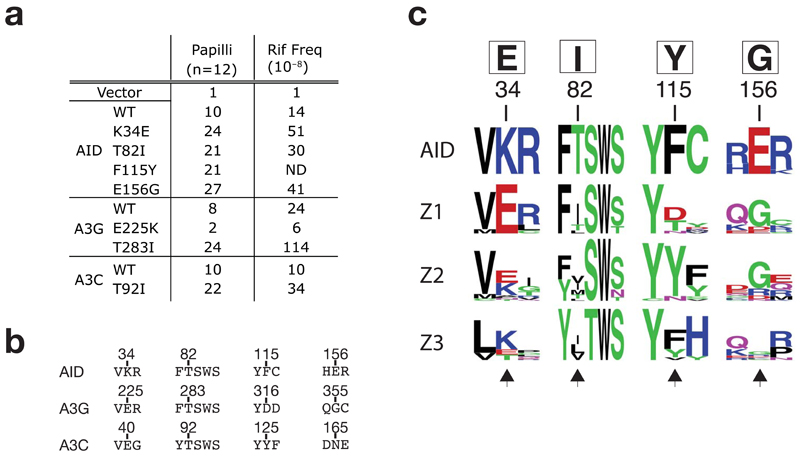
The analysis of the human AID upmutants identifies a collection of single amino acid substitutions each of which can be demonstrated to be individually sufficient to increase the enzyme’s mutator activity (Fig. 7a). Based on the sequence alignment of AID and APOBEC3s, we thought it should now be possible to design specific amino acid substitutions in individual APOBEC3 family members that could be predicted to similarly up- or down-regulate their activity. Thus by comparing the sequence of AID to that of APOBEC3G (Fig. 7b), we predicted that mutation of the APOBEC3G Glu225 residue ‘back’ to lysine should diminish its mutator activity. This is indeed the case (Fig. 7a). Similarly, we successfully predicted that - analogously to what we found with AID - the mutator activity of both APOBEC3C and APOBEC3G should also be increased by substituting the threonine residue preceding their SWS motif (Thr283 in APOBEC3G; Thr92 in APOBEC3C) by an isoleucine (Fig. 7a).
Figure 7.
Comparison of AID with APOBEC3s. (a) Mutator activity of mutants of AID, APOBEC3G and APOBEC3C carrying the indicated single amino acid substitutions as monitored in the papillation and rifampicin-resistance assays. (b) Sequence alignments of selected regions of AID, APOBEC3G and 3C. Number above the sequences indicate the positions of the residues in the parental sequence. (c) Web LOGO alignment (http://weblogo.berkeley.edu/) depicting amino acid conservation surrounding the major sites of upmutation of AID and the homologous regions in the Z1, Z2 and Z3 domains of mammalian APOBEC3s (cow, sheep, pig, dog, peccary, horse, cat, dog, mouse, rat, human and macaque: sequence accession numbers are provided in supplementary Table 1). Any sequence with over 90% amino acid identity to any other sequence was discarded from generation of the LOGO profiles. The AID upmutations identified in this work are shown in the box above the numbered residues. Black arrows at the bottom of the alignment highlight the homologous residues in the APOBEC3s.
A striking feature of the AID upmutations emerges when the sequence of AID is aligned to those of the family of APOBEC3 deaminases (Fig. 7c). The APOBEC3 family is presumed to have been derived from a duplication of AID and subsequently underwent rapid evolution to yield multiple APOBEC3 copies in higher animals: their zinc-coordination domains can be classified by sequence homology into one of three subgroupings (Z1, Z2 and Z3)16,29. Alignment of AID sequences with those of the APOBEC3s reveals that most of the frequently selected upmutations in human AID serve to bring the sequence of AID closer to that of its APOBEC3 relatives (Fig. 7c). In fact, whereas the AID upmutation at Phe115 substitutes the amino acid preferred at the corresponding position in APOBEC3 Z2 domains (Tyr), the upmutations at Lys34, Thr82 and Glu156 all substitute to the preferred amino acid at the corresponding position in the APOBEC3 Z1 domains. Interestingly, it is these Z1 domains which LaRue et al (2008)30 found to be the most catalytically active of APOBEC3 domains. Thus, it appears that whereas the deamination of activity of AID can be artificially increased by specific upmutations, such upmutations may have been counterselected during the evolution of AID but not during the evolution of most APOBEC3s.
DISCUSSION
In this work, we have shown how it is possible to use a bacterial papillation assay to screen for active mutators and also obtain AID mutants with increased catalytic activity, with some of these AID mutants giving more efficacious antibody diversification albeit at the expense of increased chromosomal translocations.
Recent studies have revealed that reducing the abundance of AID polypeptide reduces the efficiency of class-switch recombination or that increasing its abundance increases the risk of chromosomal translocations19–25. However, the finding that switching and translocations can both be increased by increasing the specific activity of AID (whilst actually diminishing its abundance) was a surprise to us. The results strongly suggest that the targeting of AID to the immunoglobulin locus will not always lead to deamination events and raise the possibility that post-translational modification of AID (e.g. phosphorylation) could be used to regulate antibody diversification by modulating AID’s specific activity not just by controlling its interactions with partners31,32. This will be an important topic for further work.
It will be interesting to ascertain whether the higher frequencies of somatic mutation that can be achieved with the AID upmutants could also lead to more efficient antibody mutation in vivo. The antibody hypermutation rate in vivo has been estimated to be in the order of 10-3 mutations per base pair per generation, with theoretical studies suggesting that this frequency might be optimal: a lower frequency might generate beneficial mutations too rarely whereas a higher mutation rate might cause an overload of deleterious mutations33–38. The construction of transgenic mouse lines expressing some of the AID upmutants described in this work might now allow one to address the issue of whether antibody maturation in vivo could be accelerated by increasing the hypermutation rate, provided that such mice do not exhibit too severe an increase in cancer predisposition. Furthermore, if such upmutants do allow more efficient antibody maturation in vivo, the mouse lines might find utility in monoclonal antibody generation. Alternatively, the upmutants might be well be used to allow more efficient antibody maturation through in vitro in approaches that exploit hypermutating cell-lines39–41.
Although a large number of independent AID upmutants were selected in this work, it is notable that increased AID activity was repeatedly obtained through a relatively small cohort of individual amino acid substitutions. Several of the upmutations were obtained in equivalent positions in the human and fugu AID sequences. Some of the favored sites (Val57 and Thr82 in human AID; Cys88 in fugu AID) lie within the Zn-coordination motif and are presumably located at or very close to the active site. Several of the others (Phe115 and Lys120 in human AID; Phe121, Leu124 and Leu128 in fugu AID) lie within a short stretch of AID that is equivalent to a region of APOBEC3 that has been speculated might be implicated in DNA binding12–14. The AID upmutants give good support to the idea that this region is indeed implicated in AID’s interaction with DNA. The remaining hotspots of human AID upmutation are largely clustered in the regions 9-13, 34-35 and 156-160. We note that Bransteitter et al.42 also observed that a double mutation at 35/36 affected AID’s catalytic activity. The mechanism by why these changes affect AID’s actvity is, however, less readily explicable although more insight will presumably arise once the three-dimensional structure of AID has been elucidated.
A wholly unexpected finding of this work was that many of the favoured amino acid substitutions selected in the AID upmutants (e.g at positions 34, 82, 115 and 156) had, in a sense, already occurred during natural evolution of APOBEC3 family members. So why had they occurred during the evolution of APOBEC3s but not in AID? In the case of K34E substitution, it is likely that this mutation was not selected during AID evolution since, whilst increasing AID’s deaminase activity, the mutation destroys AID’s in vivo antibody diversifying activity (possibly by precluding a necessary interaction with a partner). The same argument, however, does not however apply to the T82I and E156G mutations, since AID upmutants harboring these substitutions do increased antibody diversification in B cells.
Although it is possible that increased efficacy of antibody diversification might not benefit the host organism, we believe as discussed above that a more likely explanation is that the risk of genomic instability has limited the upward evolution of AID’s specific activity. It is possible that such restrictions may have been less stringent in limiting the specific activity of most APOBEC3 deaminases. Thus, the best characterised of the APOBEC3 family (APOBEC3G) is largely restricted to, and indeed retained in the cytoplasm where it can become incorporated into viral capsids43–45. In contrast, AID shuttles into the nucleus in order to carry out its physiological mutator function. So the risk of genomic stability may have placed different constraints on the mutator activity of AID and some of the APOBEC3s. Consistent with this speculation, whilst enforced expression of AID in transgenic mice has been shown to lead to cancer6, we have observed no increased tumorigenesis in transgenic mice expressing high levels of APOBEC3C or APOBEC3G (C. Thomas and M.S.Neuberger, Cambridge, UK unpublished data). Nevertheless, limiting deaminase specific activity is unlikely to be the sole mechanism for minimising the risk of ectopic mutation. APOBEC3A despite being the most active deaminase of the APOBEC3 family is nevertheless found in the nucleus46; it will be interesting to find out whether APOBEC3A overexpression is tumorigenic or whether other mechanisms act to limit its off-target activity.
Finally, we note that whilst we have shown in this work that a bacterial papillation assay can be used as a high throughput screen for AID upmutants, we can readily envisage that the assay could be extended to screen for upmutants of other members of the AID/APOBEC family or for deaminase mutants with altered target specificity. Furthermore, it is also possible that a papillation screen could be used to identify as yet uncharacterised active mutators: one could envisage that such active mutators might play a role in, for example, the initiation of variant surface glycoprotein variation in trypanosomes or in repeat-induced point mutation in fungi.
METHODS
Bacterial mutation
For papillation assays, we transformed AID/APOBEC cDNAs in plasmid pTrc9947 into Escherichia coli K12 strain CC102 [araΔ(lac proB)XIII carrying F lacI– Z– proAB+ episome in which the lacZ carries a GAG->GGG missense mutation at codon 461 (ref 11) and plated on MacConkey-lactose agar (BD Biosciences) supplemented with ampicillin (100 μg ml−1) and isopropyl β-D-1-thiogalactopyranoside (IPTG; 1 mM). Plates were incubated at 37 °C for 3–6 days with papilli becoming visible after 3 days and their numbers increasing up till day 7. For analysis of arabinose-inducible expression, AID was expressed in plasmid pBAD30 (ref. 48).
The frequency of reversion of CC102 [pTrc99-AID/APOBEC] transformants to Lac+ was determined by plating cultures grown overnight to saturation in LB medium supplemented with ampicillin (100 μg ml−1) and IPTG (1 mM) on M9+0.2% (w/v) lactose agar whereas mutation to rifampicin resistance (50 μg ml−1) was assessed following transformation into E. coli strain KL16 (Hfr (PO-45) relA1 spoT1 thi-1). Mutation frequencies were measured by determining the median number of colony-forming cells that survived selection per 107 viable cells plated with each median determined from 12 independent cultures. The identity of mutations was determined by sequencing PCR-amplified relevant sections of lacZ and rpoB (lacZ: (5’-AGAATTCCTGAAGTTCAGATGT and 5’-GGAATTCGAAACCGCCAAGAC; rpoB: 5'-TTGGCGAAATGGCGGAAAACC-3' and 5'-CACCGACGGATACCACCTGCTG-3').
PCR Mutagenesis
The first and second generation human AID mutant libraries were generated by error-prone PCR using Taq polymerase (2.5 U; Bioline) on 1 ng of template DNA with 1 µM forward and reverse primers (5’-ATGGAATTCATGGACAGCCTCTTG; 5’-CTGAAGCTTTCAAAGTCCCAAAGTA), 250 µM-dNTPs, 10 mM-MgCl2 in Taq buffer at 94 °C (2 min), followed by 30 cycles of 94 °C (30 s), 65 °C (30 s) and, 72 °C (1 min). The fugu AID and third generation human AID mutant libraries were generated using Genemorph II Random Mutagenesis Kit (Stratagene) on 0.1 ng DNA template according to the manufacturer’s instructions.
Antibody diversification
For assaying class-switching, surface IgG1 expression was analysed by flow cytometry in B cells that had been purified from AID–/– mice and cultured in the presence of LPS+IL4 (48 h) following a 24 h-infection with AID-encoding retroviruses as previously described49. To facilitate a diminution in the extent of AID overexpression in the transduced B cells, a retroviral vector with a mutated Kozak sequence was used as described by McBride et al21. AID abundance in extracts prepared by heating cells (106) in 50 µl of reducing SDS-sample buffer was monitored following SDS-PAGE by Western blot analysis using rabbit anti-AID antiserum (Abcam); GFP was detected using HRP conjugated goat anti-GFP antiserum (Abcam).
AID-induced somatic mutation was monitored by measuring the frequency of sIgM-loss variants in AID–/– φV–/– sIgM+ DT40 cells26 transfected with AID-encoding vectors based on pExpressPuro2 (gift from J-M Buerstedde, Munich, Germany). For each construct, the percentage of sIgM– cells was monitored in 12–24 independent transfectants that had been expanded under selection (0.25 μg ml−1 puromycin) for 3 weeks prior to flow cytometry. Mutations in the IgVλ region were characterised by sequencing genomic DNA that was PCR-amplified from either 100,000 unsorted or from (GFP+, sIgM–)-sorted cell equivalents50.
Chromosomal translocations
B cells from AID-deficient were transduced with AID-expressing retrovirus and cultured in medium containing LPS (20 μg ml−1) and IL4 (50 ng ml−1) as described for the class-switching assays, seeding 8 × 105 cell per ml in 6-well plates. Genomic DNA from 2 × 105 cells that had been prepared using DirectPCR (Viatech) from sorted GFP+ cells 36 h after transduction was subjected to two rounds of nested PCR with Expand Long Template PCR system (Roche) followed by Southern blotting to amplify and detect both der12 c-myc/Igμ and der15 c-myc/Igμ translocations and the specific products as described by Ramiro et al7.
Assaying deaminase activity
GST-AID fusion proteins were purified from pOPTG-AID transformants of E. coli strain Rosetta (DE3) pLysS (pOPTG vector a gift from O. Perisic, Cambridge, UK). Cells were grown at 37 °C in 2 × TY containing 100 μg ml−1 ampicillin and 100 µM ZnCl2 until the culture reached an absorbance of 0.8 at 600 nM when it was induced with 1 mM IPTG for 16 h at 18 °C and the pelleted cells then lysed by a 30 min incubation on ice in lysis buffer (20 mM-Tris pH 7.4, 100 mM-NaCl, 0.1% (w/v) Triton X-100, 5 mM-DTT, 4 μg ml−1 RNase A and Roche complete EDTA-free protease inhibitor cocktail) followed by sonication. Cell lysates were clarified by centrifugation (95,000 g; 1 h) and GST-AID was purified from these lysates by absorption onto glutathione-Sepharose (Amersham Pharmacia) at 4 °C for 5 h and elution following extensive washing with lysis buffer supplemented with 50 mM reduced glutathione lacking Triton X-100. Eluted samples were stored at 4 ºC for up to one week.
Deaminase activity of semi-purified GST-AID (100−400 ng) was assayed at 37 °C in 10 µl of reaction buffer (8 mM-Tris, pH 8.0, 8 mM-KCl, 10 mM-NaCl, 2.5 mM-EDTA, 0.2 mM-dithiothreitol, 5 μg RNase A and 0.4 units uracil-DNA glycosylase d(NEB)) with 0.5 pmol oligodeoxyribonucleotide (fluorescein-5'-ATATGAATAGAATAGAGGGGTGAGCTGGGGTGAGCTGGGGTGAG-3'-biotin). Reactions were terminated at indicated times by addition of an equal volume of loading dye (formamide, 0.5 mM EDTA) and heating at 98 °C for 3 min. The resultant cleaved oligonucleotides were subjected electrophoresis in 10% PAGE-urea gels and fluorescence detected with a Typhoon Phosphoimager (Molecular Dynamics). The extent of deamination was determined from the scanned images, expressing the pixel volume of the cleaved product bands (following background subtraction) as a percentage of the combined pixel volume of product and residual substrate bands.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Jeffrey Miller (Molecular Biology Institute and Department of Biology, University of California) for kindly providing E. coli strain CC102 and recommendations regarding plating, to Silvestro G. Conticello (Istituto Toscano Tumori, Italy) for helpful suggestions and the James Baird and Frank Elmore funds for support to MW.
Footnotes
AUTHOR CONTRIBUTIONS
M.W. and Z.Y performed the papillation assay and selection of human AID upmutants. M.W performed all other experiments. C.R. designed and assisted with the class switching assay and translocation assay. M.S.N. designed the overall research and wrote the manuscript.
References
- 1.Alt FW, Honjo T, editors. Adv Immunol. Vol. 94. Elsevier; Amsterdam: 2007. AID for Immunoglobulin Diversity. [Google Scholar]
- 2.Di Noia JM, Neuberger MS. Molecular Mechanisms of Antibody Somatic Hypermutation. Anu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 3.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 4.Haché G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006;8:148–157. [PubMed] [Google Scholar]
- 5.de Yebenes VG, Ramiro AR. Activation-induced deaminase: light and dark sides. Trends Mol Med. 2006;12:432–439. doi: 10.1016/j.molmed.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki IM, et al. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc–Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: A locus in Escherichia coli that generates G·C→T·A transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz SM, Létourneau S, Cupples CG. Isolation and characterization of an Escherichia coli strain with a high frequency of C-to-T mutations at 5-methylcytosines. J Bacteriol. 1993;175:4985–4989. doi: 10.1128/jb.175.16.4985-4989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conticello SG, Langlois MA, Neuberger MS. Insights into DNA deaminases. Nat Struct Mol Biol. 2007;14:7–9. doi: 10.1038/nsmb0107-7. [DOI] [PubMed] [Google Scholar]
- 13.Chen KM, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 14.Holden LG, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larijani M, et al. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol Cell Biol. 2007;27:20–30. doi: 10.1128/MCB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 17.Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 18.Ta VT, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 19.de Yébenes VG, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBride KM, et al. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauklin S, Sernandez IV, Bachmann G, Ramiro AR, Petersen-Mahrt SK. Estrogen directly activates AID transcription and function. J Exp Med. 2009;206:99–111. doi: 10.1084/jem.20080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sernández IV, de Yébenes VG, Dorsett Y, Ramiro AR. Haploinsufficiency of Activation-Induced Deaminase for Antibody Diversification and Chromosome Translocations both In Vitro and In Vivo. PLoS ONE. 2008;3:e3927. doi: 10.1371/journal.pone.0003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takizawa M, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1945–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arakawa H, Saribasak H, Buerstedde JM. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2004;2:E179. doi: 10.1371/journal.pbio.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conticello SG, et al. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell. 2008;31:474–484. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Janz S, Müller J, Shaughnessy J, Potter M. Detection of recombinations between c-myc and immunoglobulin switch alpha in murine plasma cell tumors and preneoplastic lesions by polymerase chain reaction. Proc Natl Acad Sci U S A. 1993;90:7361–7365. doi: 10.1073/pnas.90.15.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaRue RS, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaRue RS, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 32.Basu U, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 33.Clarke SH, et al. Inter- and intraclonal diversity in the antibody response to influenza hemagglutinin. J Exp Med. 1985;161:687–704. doi: 10.1084/jem.161.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berek C, Milstein C. Mutation drift and repertoire shift in the maturation of the immune response. Immunol Rev. 1987;96:23–41. doi: 10.1111/j.1600-065x.1987.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 35.Celada F, Seiden PE. Affinity maturation and hypermutation in a simulation of the humoral immune response. Eur J Immunol. 1996;26:1350–1358. doi: 10.1002/eji.1830260626. [DOI] [PubMed] [Google Scholar]
- 36.Kepler TB, Perelson AS. Somatic hypermutation in B cells: an optimal control treatment. J Theor Biol. 1993;164:37–64. doi: 10.1006/jtbi.1993.1139. [DOI] [PubMed] [Google Scholar]
- 37.Kepler TB, Perelson AS. Modeling and optimization of populations subject to time-dependent mutation. Proc Natl Acad Sci U S A. 1995;92:8219–8223. doi: 10.1073/pnas.92.18.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinstein SH, Louzoun Y, Shlomchik MJ. Estimating hypermutation rates from clonal tree data. J Immunol. 2003;171:4639–4649. doi: 10.4049/jimmunol.171.9.4639. [DOI] [PubMed] [Google Scholar]
- 39.Arakawa H, et al. Protein evolution by hypermutation and selection in the B cell line DT40. Nucleic Acids Res. 2008;36:e1. doi: 10.1093/nar/gkm616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cumbers SJ, et al. Generation and iterative affinity maturation of antibodies in vitro using hypermutating B-cell lines. Nat Biotechnol. 2002;20:1129–1134. doi: 10.1038/nbt752. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–167459. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem. 2004;279:51612–51621. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- 43.Bennett RP, et al. APOBEC-1 and AID are nucleo-cytoplasmic trafficking proteins but APOBEC3G cannot traffic. Biochem Biophys Res Commun. 2006;350:214–219. doi: 10.1016/j.bbrc.2006.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett RP, Presnyak V, Wedekind JE, Smith HC. Nuclear Exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J Biol Chem. 2008;283:7320–7327. doi: 10.1074/jbc.M708567200. [DOI] [PubMed] [Google Scholar]
- 45.Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J Virol. 2008;82:9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 47.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 48.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Noia JM, et al. Dependence of antibody gene diversification on uracil excision. J Exp Med. 2007;204:3209–3219. doi: 10.1084/jem.20071768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.