Abstract
During 3' end formation most pre-mRNAs undergo endonucleolytic cleavage and polyadenylation in the 3' untranslated region. Very little is known concerning the role that post-translational modifications play in the function and regulation of the factors required for 3' cleavage. Using the reconstituted pre-mRNA cleavage reaction, we find that non-specific dephosphorylation of HeLa cell nuclear extract leads to the loss of 3' cleavage activity. A variety of serine/threonine phosphatases inhibited cleavage activity, while a tyrosine phosphatase did not. When the three major cleavage factor activities—CPSF, CstF and CFm (containing CFIm and CFIIm)—were separated and dephosphorylated individually, only CFm was found to lose activity, indicating that the target of dephosphorylation resides within this fraction. In accordance with this result, only CFm was able to restore cleavage activity to HeLa nuclear extract whose 3' cleavage activity had been completely inactivated by dephosphorylation. We conclude that at least one subunit of either CFIm or CFIIm requires serine or threonine phosphorylation to function during 3' cleavage. Our data suggest that cleavage factor phosphorylation may serve as a regulatory on/off switch to control pre-mRNA 3' end formation.
Keywords: cleavage and polyadenylation, protein phosphorylation, pre-mRNA processing, alternative polyadenylation
Introduction
For the majority of mammalian pre-mRNAs, 3' end formation is a two-step process involving, first, the cleavage of the pre-mRNA in the 3' untranslated region (UTR) and, second, poly(A) tail addition to the upstream, message-bearing fragment (reviewed in refs. 1 and 2). Cleavage and polyadenylation are coupled to one another, but can be studied separately in vitro using systems reconstituted from HeLa cell nuclear extract3 or its partially purified factors.4–6 At least thirteen polypeptides, including four multiple subunit factors, CPSF, CstF, CFIm and CFIIm, are required for in vitro cleavage. The requirement for so many proteins may reflect the need to cut the pre-mRNA sequence specifically, and to coordinate 3' cleavage with other transcription related events. The combination of poly(A) site cis-acting elements in the 3'-UTR with the sequence preferences of the RNA-binding proteins found among the cleavage factors accounts for much of the cleavage site definition.7–9 However, actual cleavage site selection is known to vary in many pre-mRNAs, and this can have important consequences on the outcome of a gene’s expression.10 For example, many 3'-UTRs contain sequences that contribute to the stability and subsequent localization of the mRNA. Additionally, most microRNAs appear to downregulate translation through targeting mRNAs in their 3' UTR,11 which raises the possibility that alternative poly(A) site selection may influence microRNA function, since removal of a targeted sequence through alternative polyadenylation would permit a message to escape translation repression by this mechanism. In addition to alterations in the 3' UTR, alternative polyadenylation, often under the influence of alternative splicing, can produce changes in the amino acid sequence near the C-terminus of a protein, even leading to exclusion of small protein domains.10,12 It was recently estimated that 54% of human genes undergo some form of alternative polyadenylation.13 In most cases, the biochemical basis for choosing among the different poly(A) signals is not known.
In vitro, the mammalian 3' cleavage reaction requires the cleavage polyadenylation specificity factor (CPSF, five subunits), the cleavage stimulation factor (CstF, three subunits), cleavage factor I (CFIm, a heterodimer composed of a 25 kD subunit bound to one of three larger subunit of 59 kD, 68 kD or 72 kD), cleavage factor II (CFIIm, containing two verified subunits), the RNA polymerase II (Pol II) largest subunit C-terminal domain (CTD) and, for most substrates, poly(A) polymerase (PAP). The CFIm and CFIIm activities cofractionate on anion exchange columns such as DEAE and are, when not separated, referred to in this paper as CFm, similar to the CF designation of earlier reports.14 The mammalian cleavage factor I activity has been reconstituted from the 25 kD and 68 kD subunits,15 while cleavage factor II remains incompletely characterized.16 No snRNA or other RNA component is required for the transcription-independent basal cleavage reaction in vitro,9 though coordination with splicing involves at least U2 snRNA.17 Many of the mammalian cleavage factor subunits share sequence homology with proteins found in other eukaryotes, with those in yeast being the most frequently studied.2,18 In mammals, among the factors required for cleavage, only CPSF and PAP are necessary to begin the coupled polyadenylation of the upstream fragment, suggesting that 3' cleavage must be immediately followed by a large rearrangement of the precleavage complex, with most factors dissociating from the cleavage site. In yeast, it has been proposed that protein phosphorylation/dephosphorylation cycles within the large cleavage polyadenylation factor (CPF) complex, which contains many of the mammalian cleavage factor homologs, are part of the normal 3' end formation mechanism as it switches from cleavage to polyadenylation, and that this mechanism may work by altering the composition of, or arrangement within, CPF.19
Reversible phosphorylation is one of the most widespread post-translational control mechanisms found in eukaryotic cells.20 Among 3' cleavage factors, the phosphorylation of PAP and the C-terminal domain of Pol II largest subunit (CTD) is known to affect 3' processing. Hyperphosphorylation of PAP’s ser/thr-rich C-terminal domain during M phase represses PAP’s catalytic activity,21 whereas the Pol II CTD becomes highly phosphorylated during transcription.22 Within the 26 and 52 heptapeptide repeats found in the yeast and mammalian CTDs, respectively, low resolution analyses have shown the pattern of phosphorylation to be dynamic, with Ser-5 of the heptad repeats being most heavily phosphorylated near the promoter, and Ser-2 most heavily phosphorylated near the 3'-end.7,23 Likely as a function of its changing phosphorylation pattern throughout the transcription cycle, the CTD is believed to work as a staging area for the accumulation of a variety of proteins, enabling the CTD to concentrate them near the site of transcription, from which vantage point they presumably scan the nascent RNA for cis-acting sequences.24 However, in vitro, in the absence of transcription, the CTD, whether as part of the Pol II holoenzyme or simply fused to an affinity tag, stimulates 3' pre-mRNA cleavage independent of its phosphorylation status.25,26 Moreover, because PAP is not in all cases required for cleavage, it has not been suggested that the mammalian cleavage reaction is under the influence of protein phosphorylation control, though as stated above, this was recently suggested to be the case in yeast.
A long-known but still unexplained feature of the in vitro mammalian cleavage reaction is that a high concentration of creatine phosphate (or related phosphoamino acid) is required to bring about efficient cleavage.27 An investigation of this requirement led to the proposal that creatine phosphate may function as a phosphoprotein mimic, an idea which in turn led to the discovery that the Pol II CTD plays a direct role in the cleavage reaction.26 However, as pointed out above, the phosphorylation state of the CTD, at least as it is represented by its hyper-, hypo- and unphosphorylated forms, does not substantially affect the cleavage reaction, making it unlikely that creatine phosphate actually works as a phospho-CTD mimic. The work presented in this report began with the idea that creatine phosphate, by virtue of its structural similarity to a phosphorylated protein residue, may work instead as a competitive inhibitor of a protein phosphatase that normally acts to suppress cleavage under in vitro conditions. Were such a phosphatase to exist, the fortuitous inhibition of its activity over the years by inclusion of creatine phosphate in standard in vitro cleavage reactions might have masked a requirement for the phosphorylation of one of the highly purified core cleavage factors. This idea has led in the work presented here to the finding that the cleavage activity in HeLa cell nuclear extract, or in the cleavage factor activities fractionated from it, is lost upon treatment with ser/thr-phosphatases, implying the existence of a functionally phosphorylated cleavage factor. The susceptible component of nuclear extract is found in the CFm fraction, suggesting further that either CF Im or CF IIm contains a subunit whose phosphorylation allows cleavage to take place, but whose dephosphorylation prevents it. Thus, reversible phosphorylation among the basal cleavage factors may provide higher eukaryotes with an additional layer of gene expression control at the level of pre-mRNA processing.
Materials and Methods
Buffers
Buffer D contained 20% glycerol, 20 mM Na-HEPES, pH 7.9, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT and 50 mM ammonium sulfate. Buffer DMg, the CIP dialysis buffer, contained 6.25 mM MgCl2 in place of EDTA.
Calf intestinal alkaline phosphatase (CIP)
CIP was purchased from Promega (cat. no. M2825) and dialyzed 2 × 4 h at 4°C each time against 10,000 volumes of Buffer DMg, aliquotted and stored at −80°C or −20°C. Protein content was estimated before and after dialysis using the Bio-Rad Bradford protein assay with BSA (Roche, Molecular Biology grade) as the standard. The enzymatic activity was estimated before and after dialysis using p-nitrophenyl phosphate as the substrate28 and setting the predialysis activity equal to the vendor supplied value. On average, during dialysis the phosphatase activity declined from approximately 4.7 units/µg to 3.8 units/µg. A slight decline in phosphatase activity during long periods of storage was noticed when prepared in this way.
Nuclear extracts and fractionated cleavage factor preparations
HeLa cells were purchased as pellets from the National Cell Culture Center (Biovest International), and the nuclei were extracted according to Dignam et al.,29 with minor modifications,6 and dialyzed into Buffer D. Total protein content of nuclear extracts used in cleavage reactions ranged from 2.2 to 2.7 mg/ml, measured as described above. For fractionation, nuclear extract containing approximately 75 mg total protein was loaded onto a 19 ml hand-packed DEAE-Sepharose Fast Flow (GE Healthcare) column and eluted as previously described.25 Active fractions were identified either by in vitro polyadenylation (CPSF, fractions supplemented with recombinant bovine PAP II), or by in vitro cleavage (CFm) using small amounts of previously purified CPSF and CstF.25 CstF (along with PAP) was assumed to be in the flow-through.16 Active fractions were pooled, precipitated at 70% ammonium sulfate saturation, then resuspended in a minimum volume of Buffer D, and dialyzed 2 × 4 h at 4°C in Buffer D. Protein concentrations of extracts and fractionated cleavage factors were estimated as described above. Fractionated cleavage factor protein concentrations were as follows: CPSF (2.4 mg/ml, 1 µl used per 12.5 µl cleavage reaction), CstF (3.1 mg/ml, 0.5 µl used per 12.5 µl cleavage reaction), CFm (4.8 mg/ml, 2 µl used per 12.5 µl cleavage reaction).
RNA substrates
SV40 late pre-mRNA (233-nt) was transcribed in vitro by SP6 RNA polymerase (Promega) from thepG3SVL-A plasmid linearized at the Dra I site.6 The Ad2 L3 RNA (178-nt) was similarly made from the pG3L3-A plasmid linearized at the Bam H1 site.6 The transcripts were uniformly labeled by including [α-32P]-UTP (Perkin-Elmer Life Sci.), and 5' capped during transcription by including the 7-MeGpppG cap analog (NEB), gel-purified, eluted, precipitated and resuspended in 2 mM Tris-HCl, pH 7.
3' pre-mRNA cleavage reactions using nuclear extract ± CIP, general procedure
Nuclear extract (4.25–5.25 µl) was mixed with Buffer D and, as indicated in the figure legends, CIP or CIP dialysis buffer (indicated by dash), including 1 mM MgCl2, protease inhibitors or β-phosphoglycerol as indicated, to a total volume of 6.25 µl, and placed at 30°C for 30 min. EDTA was then added to a concentration of 2 mM above that of the total MgCl2. This mixture was returned to 30°C for 20 min to incapacitate the CIP prior to creatine phosphate addition, in order to rule out any possibility that CIP-mediated hydrolysis of creatine phosphate contributes to cleavage inhibition. This mixture was then iced, and the following components were added (final concentrations): 2'-dATP (2 mM), tRNA (0.1 µg/µl), DTT (1.85 mM), creatine phosphate (50 mM), RNAse inhibitor (Promega, 0.32 u/µl), polyvinyl alcohol (2.5 %), pre-mRNA (1–3 nM), 10% glycerol, 10 mM HEPES (pH 7.9), EDTA 2 mM above final MgCl2 concentration, phenylmethylsulfonyl fluoride (PMSF, 0.1 mM), ammonium sulfate (25 mM). The resulting 12.5 µl processing reaction was incubated at 30°C for 2 h and the isolated RNA was resolved on a 6% denaturing polyacrylamide gel and visualized and quantitated using a Molecular Dynamics Storm Phosphorimager.
PP1 (α-Isoform from Rabbit), YOP and lambda phosphatase reactions
Identical to CIP procedure except: 0.4 mM Mn2+ was used in place of 1 mM Mg for PP1 and λ-PPase, and the YOP reaction contained no divalent cation. These PPases (NEB) were not dialyzed, but were used with the supplied buffers. Mock reactions contained identical buffer components without enzyme. Manufacturer units are shown.
3' pre-mRNA cleavage reactions using cleavage factor activities, general procedure
Similar to the above nuclear extract cleavage procedure except: the fractionated cleavage factor preparations were mixed and supplemented with 0.5 µg BSA. When the factors were treated with CIP individually, EDTA was added to 2 mM above the MgCl2 concentration at the end of the CIP pretreatment of an individual factor (containing the factor, 1 mM MgCl2, BSA and CIP or CIP dialysis buffer for mock reactions), and the reaction was placed at 30°C for 20 additional min to inhibit CIP activity. An independent phosphatase assay28 employing p-nitrophenyl phosphate under simulated cleavage conditions was used to verify that CIP has less than 0.7% of its original activity when treated in this way (not shown). In Figure 4, lanes 10 and 20, the CIP was inhibited in this way before adding it to the mixture of all three factors, verifying that CIP is rendered inactive when treated in this way. The reaction was then iced and the other two (untreated) cleavage factor activities were added, followed by the pre-mRNA and other ingredients to initiate 3' processing, as described above for the nuclear extract cleavage reaction.
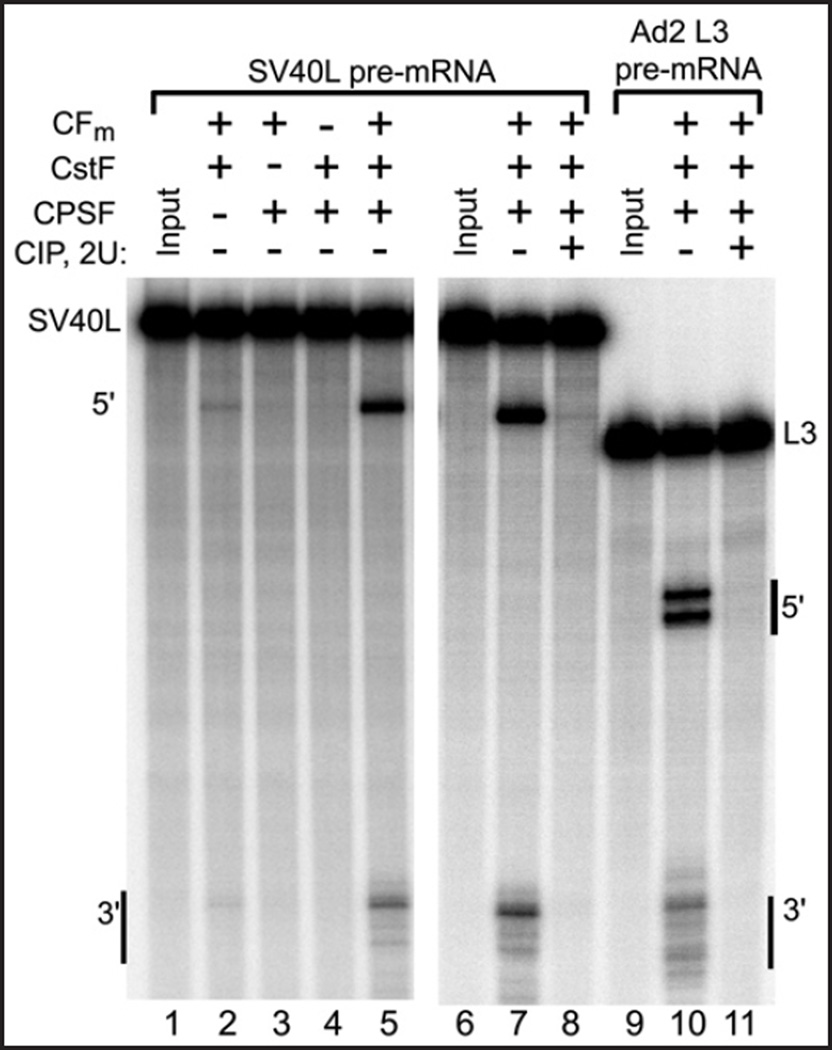
Figure 4.
Only the CFm fraction, which contains mammalian cleavage factors Im and IIm, loses its activity when dephosphorylated by CIP. The fractionated cleavage factors, as indicated above lanes, were pretreated with CIP individually, and the CIP enzymatic activity was then inactivated by addition of EDTA before the other two (untreated) cleavage factors were added and processing begun by the addition of the pre-mRNA. In lanes 2–3 and 12–13 all three factors were combined before dephosphorylation by CIP. *indicates control reactions where the CIP was inactivated by EDTA treatment prior to being added to the mixture of the three factors, demonstrating the efficacy of the CIP inactivation by EDTA.
Cleavage factor add-vack reactions
5.25 µl nuclear extract was treated with CIP at 1 mM MgCl2 in 5.5 µl at 30°C for 30 min, as described above. The reaction was iced and EDTA was added in a 0.5 µl volume to 2 mM above the MgCl2 concentration. The reaction was incubated at 30°C for an additional 20 min to inhibit CIP activity, and then iced again. The amount of a single fractionated cleavage factor activity normally used in a cleavage reaction was then added to the treated nuclear extract in a volume of 2 µl and the reaction was incubated at 30°C for 15 min to allow for protein complex subunit exchange, and once again iced. Pre-mRNA and other reaction components were finally added for a total of 12.5 µl, and processing was allowed to proceed as described above. In order to treat the added-back CFm fraction with CIP (or mock treat) prior to add-back, 4 µl of the CFm fraction was treated with CIP (1 U and 1 mM MgCl2 final at this stage, added in 0.5 µl) at 30°C for 30 min, and the subsequent EDTA required to inhibit the CIP activity was then added in 0.5 µl, followed by the usual 20 min 30°C incubation to stop all CIP activity. Half (i.e., 2.5 µl) of these mixtures (CIP or mock CIP) was added back to the CIP-inhibited nuclear extract and the mixture allowed to incubate at 30°C for 15 min. Finally, the pre-mRNA and other reaction components were added, and processing was allowed to proceed as described above.
Results
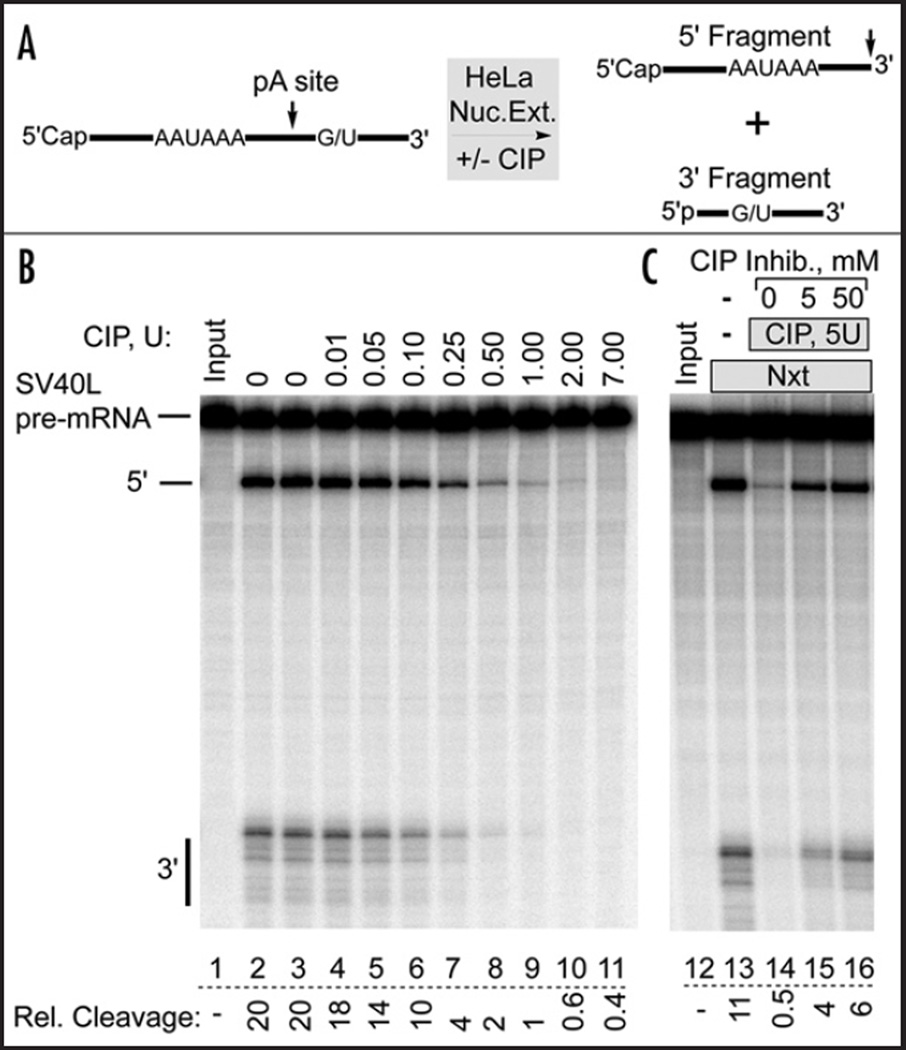
The hypothesis that creatine phosphate may stimulate in vitro 3' cleavage by inhibiting a negatively regulating phosphatase leads to a testable prediction: treatment of nuclear extract with an exogenous, non-specific phosphatase prior to adding creatine phosphate should prevent creatine phosphate from stimulating 3' cleavage, since the phosphatase would have already dephosphorylated the substrate of the endogenous phosphatase before creatine phosphate and the RNA substrate are added. In addition, the low ATP concentration found in nuclear extract,30,31 should not support rephosphorylation by kinases in the extract. If creatine phosphate were to work by another mechanism, for example by bringing about a conformational change in one of the cleavage factors,27 non-specific phosphatase treatment would not be expected to have an effect. Accordingly, calf intestinal alkaline phosphatase (CIP), an enzyme that hydrolyzes phosphate monoesters indiscriminately,32 was used to dephosphorylate the components of HeLa cell nuclear extract. This well-characterized enzyme hydrolyzes phosphate monoesters of protein alcohol residues, and is regularly used to dephosphorylate phosphoproteins.33,34 As shown in Figure 1B, CIP pretreatment of nuclear extract inhibited in a concentration dependent manner subsequent processing of the SV40 Late (SV40L) pre-mRNA substrate. This substrate does not require poly(A) polymerase (PAP) for processing,14 rendering unlikely an effect due to dephosphorylation of this phosphoprotein. As little as 0.01 units of CIP (lane 4, ~2.4 ng, or 2.9 nM) led to a detectable reduction in cleavage activity, with 0.1 units reducing cleavage to 50%. In all cases, 50 mM creatine phosphate was present during the subsequent processing reaction. Thus, CIP appears to be a potent inhibitor of HeLa cell nuclear extract 3' cleavage activity, possibly preventing creatine phosphate from stimulating cleavage.
Figure 1.
Dephosphorylation of HeLa cell nuclear extract inhibits 3' pre-mRNA cleavage activity. (A) Schematic diagram of the in vitro pre-mRNA cleavage reaction. (B) HeLa Cell nuclear extract was dephosphorylated by calf intestinal alkaline phosphatase (CIP) pretreatment. CIP activity was then inhibited with EDTA before adding the SV40L pre-mRNA to start the processing reaction. EDTA and 2’-dATP were included in all experiments to prevent polyadenylation. 6% denaturing polyacrylamide gel. Lanes 2 and 3: no CIP. Lane 2: no MgCl2. Relative cleavage is expressed as: [5' fragment/(5' fragment + Uncut)] × 100. (C) The CIP inhibitor β-phosphoglycerol was added at the indicated concentrations during pretreatment of the nuclear extract with CIP. In the absence of CIP, 50 mM inhibitor did not affect normal 3' cleavage (not shown).
We verified that 3' cleavage inhibition was indeed caused by CIP, and not some contaminating activity, by using a known CIP inhibitor, β-phosphoglycerol.35 Adding this compound to HeLa cell nuclear extract during the pretreatment reaction reduced the CIP-mediated inhibition of 3' cleavage activity in a concentration dependent manner (Fig. 1C, lanes 14–16). This result directly implicated the CIP phosphatase activity as the inhibitor of 3' cleavage. We also tested a variety of protease inhibitors to ensure that 3' cleavage inhibition was not due to contaminating protease activity, and also used the chromogenic protease substrate Azocoll to test the CIP preparation directly for protease contamination.36 No protease activity was detected, and protease inhibitors had no effect (Fig. S1). In addition, a 6 µg sample of the CIP preparation revealed only a single Coomassie-stained band, which at 1 µg per gel lane migrated at approximately 66 kD, close to the expected CIP monomer size of 65 kD37 (not shown). Thus, a variety of protease inhibitors did not alter the CIP effect, and the CIP preparation appears to be homogeneous and to have no detectable protease activity.
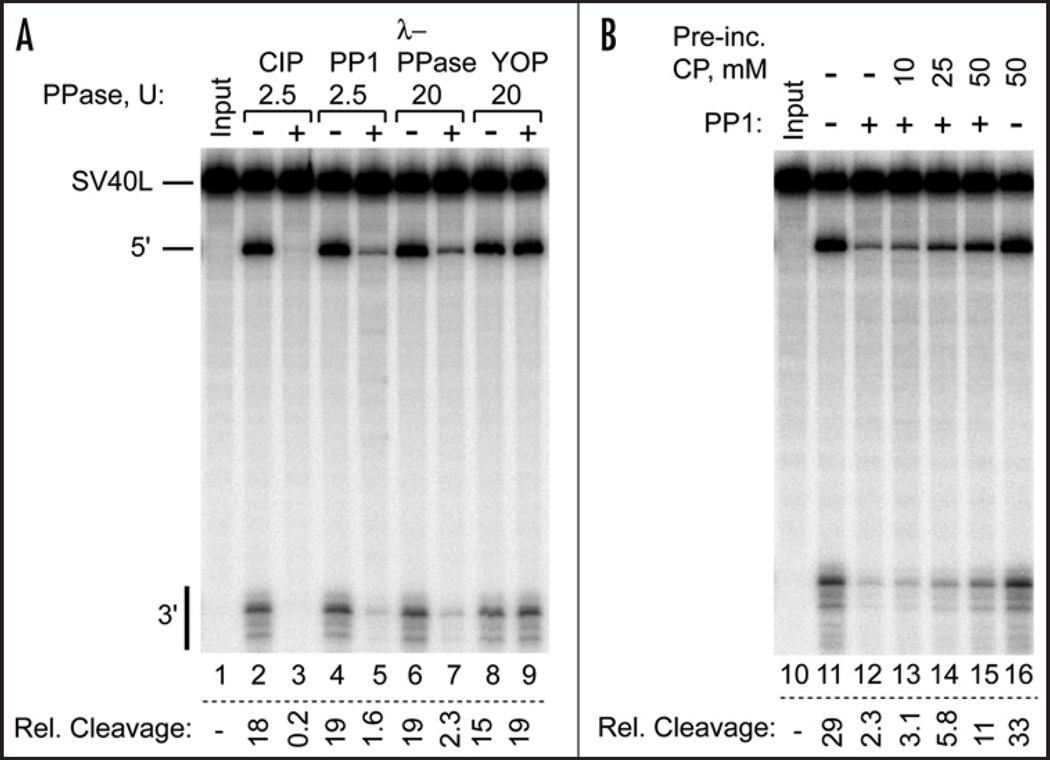
In addition to CIP we tested other commercially available protein phosphatases (Fig. 2). Protein phosphatase 1 (PP1) and lambda phosphatase are similar to CIP in that they are known to dephosphorylate phosphoserine (pS), pT and pY residues in vitro, whereas YOP protein tyrosine phosphatase only dephosphorylates pY residues.38 Of these phosphatases, YOP alone was unable to inhibit 3' cleavage activity in HeLa nuclear extract, suggesting that removal of a phosphate from serine or threonine residue(s) on one of the 3' cleavage factors is responsible for the loss of processing activity.
Figure 2.
Serine/threonine phosphatases inhibit 3' cleavage activity. (A) HeLa Cell nuclear extract was dephosphorylated by treatment with the indicated phosphatases (PPase) before use in processing the SV40L pre-mRNA substrate. Only YOP, a phosphotyrosine phosphatase, failed to inhibit 3' cleavage. (B) Creatine phosphate antagonizes PP1 inhibition of nuclear extract 3' cleavage activity. The indicated concentration of creatine phosphate was present during PP1 pretreatment of the nuclear extract prior to use in SV40L pre-mRNA processing. All reactions had 50 mM creatine phosphate during processing.
To explore the possibility that creatine phosphate stimulates in vitro 3' cleavage by fortuitously inhibiting a negatively regulating mammalian phosphatase, we included increasing amounts of creatine phosphate in a PP1 dephosphorylation reaction of nuclear extract. As shown in Figure 2B, creatine phosphate reduced the loss of subsequent 3' processing activity in a concentration dependent manner. This result demonstrated that at these concentrations creatine phosphate is a PP1 inhibitor. However, in a 3' processing reaction where no exogenous PP1 was added, the known PP1 inhibitors okadaic acid and Inhibitor-2 could not substitute for creatine phosphate by stimulating 3' cleavage (data not shown). Thus, although the phosphatase-inhibiting activity of creatine phosphate could be responsible for its ability to stimulate 3' processing in vitro, creatine phosphate does not appear to stimulate 3' cleavage by inhibiting PP1. More importantly, the creatine phosphate hypothesis led us to find that the dephosphorylation of nuclear extract inhibits 3' pre-mRNA cleavage.
As a first step towards identifying the component(s) of nuclear extract that are susceptible to phosphatase treatment, HeLa cell nuclear extract was fractionated into its three major component cleavage activities, CPSF, CstF and CFm (CFIm and CFIIm), by passing it through DEAE-sepharose and eluting with increasing salt concentration, as previously described25 (see Materials and Methods). Although these crude fractions contain many other nuclear proteins, Figure 3 (lanes 1–5) shows that all three activities have been successfully separated, since all three must be added to reconstitute efficient pre-mRNA cleavage. Western blotting confirmed the identity of the CPSF and CstF fractions (Fig. S2). The trace of processing in Figure 3, lane 2 is consistent with the low level of CPSF detected in the CstF and CFm fractions (Fig. S2).
Figure 3.
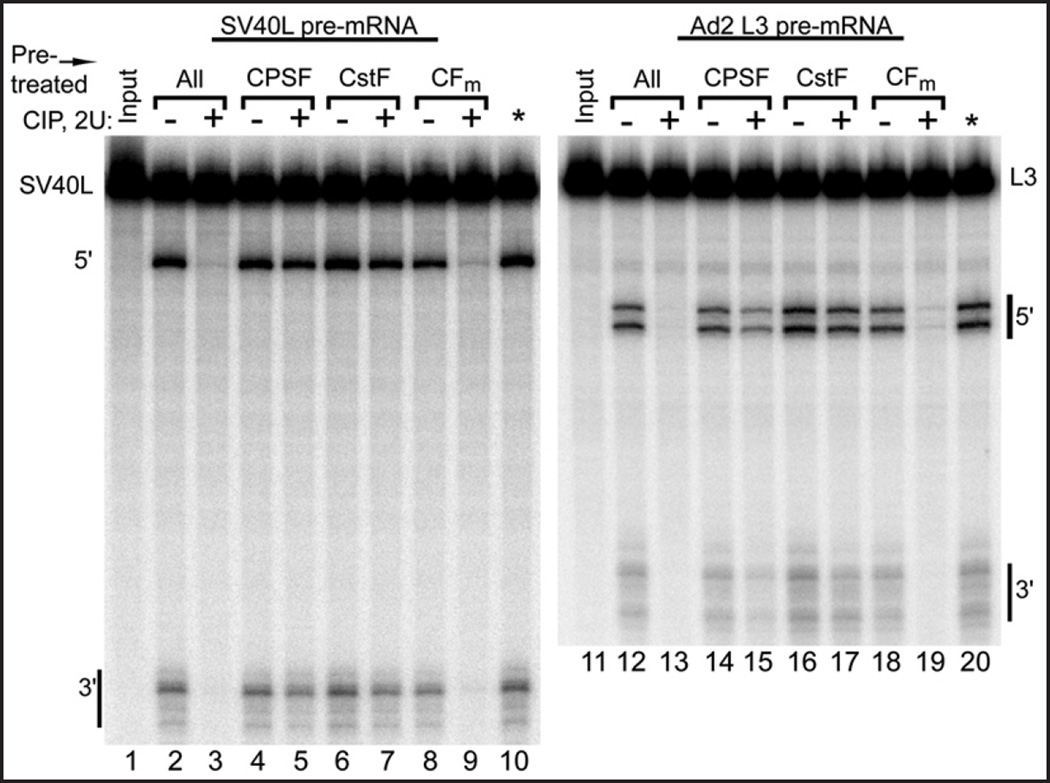
Processing of SV40L, a PAP-independent pre-mRNA substrate, and Ad2 L3, a PAP-dependent substrate, is inhibited by CIP pretreatment of DEAE-fractionated HeLa cleavage factors. Lanes 1–5 demonstrate DEAE-sepharose separation of the three main cleavage factor activities in nuclear extract: CPSF, CstF and CFm (containing CFIm and CFIIm). Pre-incubation of the remixed cleavage activities with CIP prevents processing of both substrates (lanes 6–11).
3' cleavage reconstituted from the three fractionated activities was also susceptible to inhibition by CIP treatment. As shown in Figure 3, lanes 6–8, the cleavage of the SV40L containing pre-mRNA was completely inhibited by CIP pretreatment of the combined fractions, just as it was when nuclear extract was similarly treated. This demonstrated that the relevant CIP target copurified with at least one of the cleavage activities on the DEAE-sepharose column.
While the SV40L poly(A) site is routinely used as a model pre-mRNA in reconstituted 3' processing assays, it is unique in that it does not require poly(A) polymerase for the cleavage reaction,14 and may therefore not be entirely representative of the majority of poly(A) signals. To test the generality of dephosphorylation-mediated cleavage inhibition, the adenovirus 2 L3 poly(A) sequence (Ad2 L3) was used with the separated cleavage factor activities. As shown in Figure 3, lanes 9–11, the cleavage of this substrate was also inhibited by pretreatment of the mixed factors with CIP. The two different 5' fragments normally observed39 were affected equally. Thus, the loss of cleavage activity upon dephosphorylation of preparations containing the cleavage factors appears to be general with respect to the pre-mRNA substrate.
Next, to learn which cleavage factor activity is susceptible to dephosphorylation, the separated cleavage activities were treated individually with CIP. To ensure that only one factor per experiment was exposed to the active phosphatase, the pretreatment was stopped by the addition of EDTA prior to addition of the other two untreated factors. As shown in Figure 4, lanes 9 and 19, dephosphorylation of the CFm fraction led to the loss of cleavage activity, whereas pretreatment of CPSF and CstF did not significantly alter their respective activities. Processing of both the SV40L and Ad2 L3 pre-mRNA substrates was inhibited when CFm was treated with CIP. Thus, CFm, the fraction containing cleavage factors Im and IIm, lost its activity when treated with this non-specific phosphatase, implying that one or more of CFm’s subunits requires phosphorylation to function or, alternatively, that DEAE-separated CFm contains a component which, when dephosphorylated, becomes a 3' cleavage suppressor.
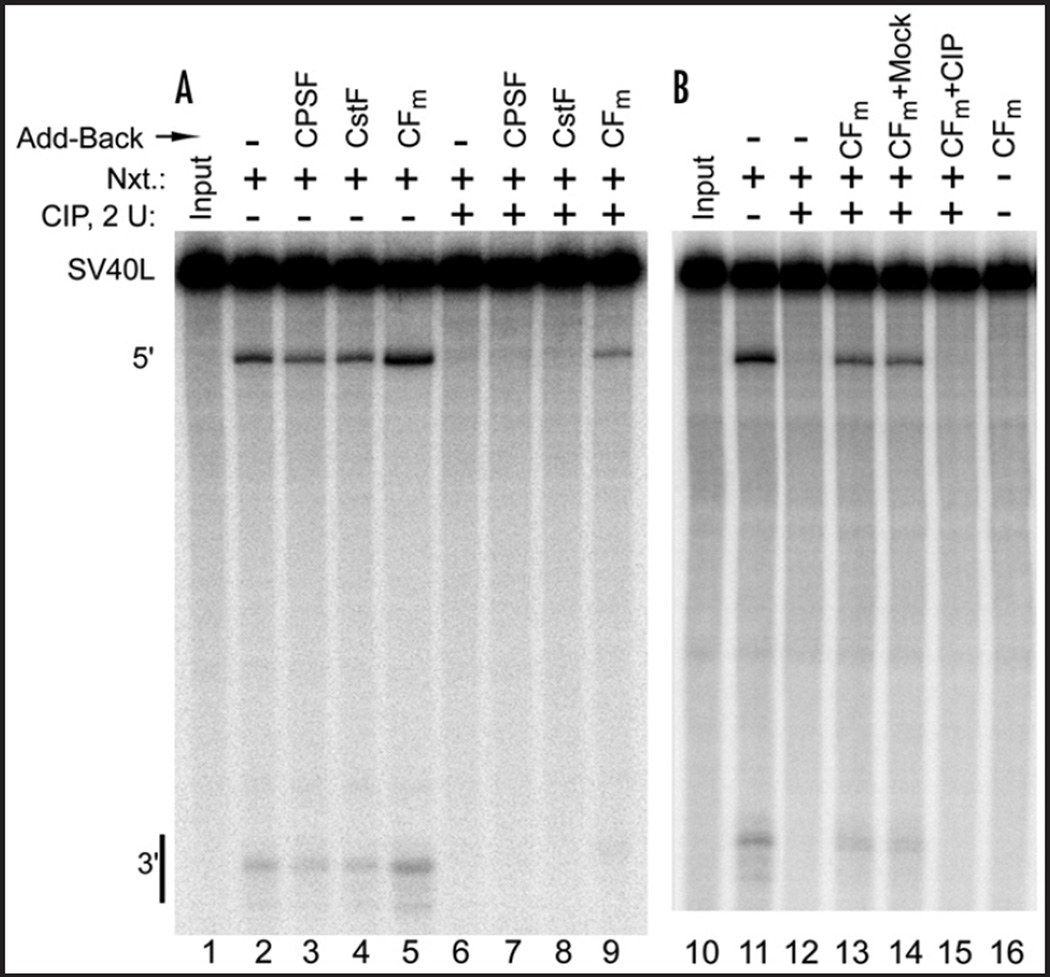
The results shown in Figure 4 suggest that the phosphatase-susceptible target in nuclear extract is contained within the DEAE CFm fraction. To further test this possibility, the DEAE-fractionated activities were added back individually to CIP-treated nuclear extract after the CIP had been inactivated with EDTA. As shown in Figure 5A, only the CFm fraction (lane 9) was able to restore 3' cleavage activity to the CIP-treated extract. However, when the CFm to be added back was itself first treated with CIP, it was unable to restore activity (Fig. 5B, lane 15). These results demonstrate that the activity lost during nuclear extract dephosphorylation can be replaced by the DEAE CFm fraction, unless it too has been dephosphorylated. This result suggests that CFIm or CFIIm contains the CIP-targeted component required for the 3’ cleavage reaction, and that when added back, this putative phosphoprotein can regain its place in CFIm or CFIIm by exchanging back into these or any other complexes requiring it for 3' cleavage. This experiment also supports the idea that the unknown phosphoprotein is an active CFm subunit, and not an inhibitory protein unrelated to 3' processing that is activated by dephosphorylation, since adding back more of a masked inhibitor should not restore activity. Furthermore, this experiment provides us with a straightforward means to track future purification of CFm, not by following its role in the basal cleavage reaction, as has been done elsewhere,16,40 but by following its ability to restore cleavage activity to CIP-inhibited nuclear extract.
Figure 5.
The CFm fraction restores cleavage activity to nuclear extract that has been dephosphorylated by CIP. (A) HeLa cell nuclear extract was pretreated with CIP (lanes 6–9), or CIP dialysis buffer (lanes 2–5), before EDTA treatment. The partially purified cleavage factors were then added individually, as indicated, followed by a brief incubation to allow for protein complex exchange before processing. (B) Similar to A., except the added back CFm was itself first treated with CIP (lane 15) (or CIP dialysis buffer, lane 14), and the CIP was then inactivated by EDTA before the CFm was added to the CIP-inhibited nuclear extract. Processing was then carried out as in (A).
Discussion
The experiments described here were undertaken to investigate the hypothesis that creatine phosphate may fortuitously stimulate in vitro 3' pre-mRNA cleavage by inhibiting an otherwise active inhibitor of the reaction, in particular a negatively regulating protein phosphatase. Such a mode of action for creatine phosphate is reasonable since at high concentrations many low molecular weight phosphate compounds competitively inhibit phosphatases,35 and creatine phosphate is reported to be a substrate, and therefore a competitive inhibitor at high concentrations, of at least two protein phosphatases, prostatic acid phosphatase41 and alkaline phosphatase derived from cow’s milk and intestine.42 Used here as unregulated surrogates for a hypothetical cleavage-regulating phosphatase, calf intestinal alkaline phosphatase (CIP), lambda phosphatase and PP1 were each found to inhibit the 3' cleavage activity of HeLa cell nuclear extract, despite the presence of creatine phosphate during subsequent processing. In contrast, a representative tyrosine phosphatase did not inhibit 3' cleavage. The effect of the phosphatase treatment suggests that at least one of the cleavage factors requires phosphorylation on serine or threonine to function in 3' pre-mRNA cleavage. This conclusion raises the possibility that basal cleavage factor phosphorylation may be a previously undetected means of regulating mammalian gene expression at the level of pre-mRNA 3' cleavage.
In support of the hypothesis that creatine phosphate may work as a phosphatase inhibitor, we found that at the concentrations typically used in the cleavage reaction, creatine phosphate inhibited protein phosphatase 1 (PP1). Interestingly, Glc7, the sole yeast ortholog of PP1, was recently found to play a role in 3' processing.19 PP1 typically works in tandem with a regulatory subunit, many of which have an RVxF motif that helps to bind PP1.43 Among the cleavage factors, CstF-77 and the RNA Pol II largest subunit each have two such sequences. However, many proteins that have the RVxF sequence do not interact with PP1,44 and we have not yet verified an association between PP1 and the 3' cleavage factors. Because two specific PP1 inhibitors, okadaic acid and Inhibitor-2, did not stimulate in vitro cleavage in place of creatine phosphate, this compound does not appear to work by inhibiting endogenous PP1. This result does not rule out the involvement of related phosphatases, such as PP2A and PP2B, whose catalytic domains are approximately 50% homologous with PP1,45 or other phosphatases, any of which might be inhibited by high concentrations of low molecular weight phospho compounds such as creatine phosphate.
Individual CIP treatment of the separated cleavage factor activities identified the CFm fraction as the sole cleavage activity susceptible to CIP dephosphorylation, and the complementation experiments shown in Figure 5 confirmed this result. Though crudely purified, CFm contributes only the composite CFI/IIm activity to the reconstituted reaction, ruling out the possibility that CPSF or CstF is the target of CIP. In accordance with this result, the CPSF and CstF preparations were unable to restore cleavage activity to CIP-treated nuclear extract. These results do not rule out the possibility that factors other than those contained in CFm are phosphorylated. Indeed, CPSF-100 and CPSF-160 have been found in phosphorylated form in HeLa cell nuclear extract.46 However, by the results presented here such phosphates do not appear to be of functional significance to the 3’ in vitro cleavage reaction.
Why would it be necessary or advantageous to regulate the basal 3' cleavage reaction at all? CPSF has been found to associate with model transcription units from beginning47 to end,7 and also to associate with the transcription machinery.48 Furthermore, the 73 kD subunit of this factor was recently identified as the 3' cleavage endonuclease, during which study the isolated protein was found to have intrinsic RNA degradation activity.49 The tethering of the nuclease to the RNA synthesis machinery may have guided the evolution of reversible phosphorylation of one of the cleavage factors as a safety mechanism to keep the nuclease switched off until needed, in order to prevent cleavage of the pre-mRNA as it is made. Such a switch could also be envisioned to be of use during cotranscriptional selection among alternative poly(A) sites.
In summary, the results presented here demonstrate that a component of the mammalian CFm activity fraction contains a functional phosphoprotein whose dephosphorylation leads to loss of in vitro cleavage activity. Current work in our lab is directed towards identifying the CIP-susceptible component of CFm, and the enzymes that modify it, in order to learn how this post-translational modification influences mammalian gene expression through the modulation of pre-mRNA 3' cleavage activity.
Supplementary Material
Acknowledgments
This work was funded by the City College of New York Science Division, with additional support from NIH Research Centers in Minority Institutions Grant number G12RR- 03060. I thank Frida Kleiman for helpful comments and suggestions. I also thank Patricia J. Kim for western blotting.
Footnotes
Supplemental information can be found at: www.landesbioscience.com/supplement/ryanRNA4-1-sup.pdf
References
- 1.Wahle E, Ruegsegger U. 3'-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J, Hyman L, Moore C. Formation of mRNA 3’ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore CL, Sharp PA. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985;41:845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- 4.Gilmartin GM, Nevins JR. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3:2180–2190. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 5.Christofori G, Keller W. 3’ cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988;54:875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- 6.Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 7.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murthy KG, Manley JL. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 10.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: Means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 12.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 13.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3’-end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 15.Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 16.de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3’ end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 18.Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3’-end processing. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 19.He X, Moore C. Regulation of yeast mRNA 3’ end processing by phosphorylation. Mol Cell. 2005;19:619–629. doi: 10.1016/j.molcel.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 20.Johnson LN, O’Reilly M. Control by phosphorylation. Curr Opin Struct Biol. 1996;6:762–769. doi: 10.1016/s0959-440x(96)80005-4. [DOI] [PubMed] [Google Scholar]
- 21.Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 22.Dahmus ME. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995;1261:171–182. doi: 10.1016/0167-4781(94)00233-s. [DOI] [PubMed] [Google Scholar]
- 23.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zorio DA, Bentley DL. The link between mRNA processing and transcription: Communication works both ways. Exp Cell Res. 2004;296:91–97. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 25.Ryan K, Murthy KG, Kaneko S, Manley JL. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3’ cleavage. Mol Cell Biol. 2002;22:1684–1692. doi: 10.1128/MCB.22.6.1684-1692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 27.Hirose Y, Manley JL. Creatine phosphate, not ATP, is required for 3’ end cleavage of mammalian pre-mRNA in vitro. J Biol Chem. 1997;272:29636–29642. doi: 10.1074/jbc.272.47.29636. [DOI] [PubMed] [Google Scholar]
- 28.Mossner E, Boll M, Pfleiderer G. Purification of human and bovine alkaline phosphatases by affinity chromatography. Hoppe Seylers Z Physiol Chem. 1980;361:543–549. doi: 10.1515/bchm2.1980.361.1.543. [DOI] [PubMed] [Google Scholar]
- 29.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore CL. Preparation of mammalian extracts active in polyadenylation. Methods Enzymol. 1990;181:49–74. doi: 10.1016/0076-6879(90)81112-8. [DOI] [PubMed] [Google Scholar]
- 31.Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 32.Fernley HN. Mammalian alkaline phosphatases. In: Boyer PD, editor. The Enzymes. New York and London: Academic Press; 1971. pp. 417–447. [Google Scholar]
- 33.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 35.Fernley HN, Walker PG. Studies on alkaline phosphatase: Inhibition by phosphate derifvatives and the substrate specificity. Biochem J. 1967;104:1011–1018. doi: 10.1042/bj1041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chavira R, Jr, Burnett TJ, Hageman JH. Assaying proteinases with azocoll. Anal Biochem. 1984;136:446–450. doi: 10.1016/0003-2697(84)90242-2. [DOI] [PubMed] [Google Scholar]
- 37.Eun HM. Enzymology primer for recombinant DNA technology. Academic Press; 1996. [Google Scholar]
- 38.NEB and Promega product information [Google Scholar]
- 39.Gilmartin GM, McDevitt MA, Nevins JR. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev. 1988;2:578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- 40.Ruegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor Im involved in the 3’ end processing of messenger RNA precursors. J Biol Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 41.Porvari KS, Herrala AM, Kurkela RM, Taavitsainen PA, Lindqvist Y, Schneider G, Vihko PT. Site-directed mutagenesis of prostatic acid phosphatase: Catalytically important aspartic acid 258, substrate specificity, and oligomerization. J Biol Chem. 1994;269:22642–22646. [PubMed] [Google Scholar]
- 42.Morton RK. The substrate specificity and inhibition of alkaline phosphatases of cow’s milk and calf intestinal mucosa. Biochem J. 1955;61:232–240. doi: 10.1042/bj0610232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen PT. Protein phosphatase 1-targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 44.Zhao S, Lee EY. A protein phosphatase-1-binding motif identified by the panning of a random peptide display library. J Biol Chem. 1997;272:28368–28372. doi: 10.1074/jbc.272.45.28368. [DOI] [PubMed] [Google Scholar]
- 45.Cohen PT, Brewis ND, Hughes V, Mann DJ. Protein serine/threonine phosphatases; an expanding family. FEBS Lett. 1990;268:355–359. doi: 10.1016/0014-5793(90)81285-v. [DOI] [PubMed] [Google Scholar]
- 46.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3’ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 48.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 49.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3’-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.