Abstract
Enterococcus faecalis is an opportunistic pathogen that ranks among the leading causes of biofilm-associated infections. We previously demonstrated that the endocarditis- and biofilm-associated pili (Ebp) of E. faecalis play a major role in biofilm formation, adherence to abiotic surfaces and experimental infections. In this study, derivatives of E. faecalis strain OG1 were engineered to further characterize functions of Ebp pili. Loss of pili resulted in a 36-fold decrease in the number of closely associated cells when OG1RFΔebpABC was mixed with OG1SSpΔebpABC, compared with mixing the Ebp+ parental strains. In addition, using the Ebp+ parental strains as donor and recipient, we found a statistically significant increase (280–360 %, P < 0.05) in the frequency of plasmid transfer versus using Ebp− mutants in the conjugation experiments. These results demonstrate a previously unrecognized role of Ebp pili, namely, as important contributors to microscale cell aggregation and horizontal spread of genetic material.
Introduction
The endocarditis- and biofilm-associated pili (Ebp) are surface-associated filamentous structures considered to play a pivotal role in Enterococcus faecalis virulence (Nallapareddy et al., 2006, 2011b; Kemp et al., 2007; Singh et al., 2007; Nielsen et al., 2012; Sillanpää et al., 2013). Ebp pili are encoded by the ubiquitous ebpABC-bps cluster, which codes for the cell wall anchor pilin EbpB, the major shaft pilin EbpC and the fibre tip pilin EbpA. A pilus-specific class C sortase (Bps, for biofilm- and pilus-associated sortase), catalyses the assembly of the structural subunits into pili, before the housekeeping sortase A (SrtA) covalently binds the elongated pili to the cell wall via EbpB (Nallapareddy et al., 2006; Sillanpää et al., 2013). Studies on ebp regulation identified the upstream ebpR gene (Bourgogne et al., 2007) and the rnjB gene (Gao et al., 2010) as activators at the mRNA level, while the Fsr quorum-sensing system was described as a weak repressor (Bourgogne et al., 2006). In addition, a recent report by Montealegre et al. (2015) demonstrated that E. faecalis uses the rare initiation codon ATT, found in all E. faecalis strains sequenced, as the start codon of the tip pilin EbpA and that this start codon results in reduced expression, at the translational level, relative to an engineered ATG, and negatively affects Ebp-associated functions (Montealegre et al., 2015).
We previously reported that Ebp pili contribute to biofilm formation, adherence to abiotic surfaces (Nallapareddy et al., 2006; Sillanpää et al., 2013) and adherence to platelets (Nallapareddy et al., 2011a), fibrinogen and collagen (Nallapareddy et al., 2011b) of E. faecalis, thus supporting the establishment and persistence of this bacterium in clinically important infections. Pili were found to be immunogenic in the human host during infection (Sillanpää et al., 2004), and their contribution to vascular tissue colonization by E. faecalis was demonstrated in a rat model of infective endocarditis (Nallapareddy et al., 2006). In addition, deletion of the ebp locus resulted in a diminished capacity of E. faecalis OG1RF to colonize kidneys and bladders in a murine model of ascending urinary tract infection (UTI) and in experimental cathether-associated UTI (CAUTI) (Singh et al., 2007; Nielsen et al., 2012). Monoclonal antibodies raised specifically against the major pilin component EbpC, as well as polyclonal anti-EbpA antibodies, were shown to provide protection against E. faecalis CAUTI in mice and infective endocarditis in rats (Flores-Mireles et al., 2014; Pinkston et al., 2014).
Despite the clear role of Ebp in attachment and infection, no studies have yet assessed their contribution to micro-scale cell aggregation or dissemination of genetic material. We previously noted that attenuation of OG1RFΔebpABC in rat infective endocarditis appeared to be less when using mixed inocula vs mono-inocula, suggesting that piliated cells might entrap non-piliated ones in the vegetation so that more mutant cells were present than when used alone for infection (Nallapareddy et al., 2006). In this study, we employed ebpABC isogenic deletion mutants of E. faecalis OG1-derivatives and evaluated the effect of this deletion on the ability of E. faecalis to interact with neighbouring cells and to facilitate lateral gene transfer by conjugation.
Methods
Bacterial strains, construction of mutants and growth conditions
Bacterial strains, mutants and plasmids are listed in Table 1. Unless otherwise specified, E. faecalis strain OG1RF (Ebp+) (Bourgogne et al., 2008) and its derivative were cultivated in brain heart infusion (BHI) (Becton Dickinson) broth supplemented, when appropriate, with 25 mg fusidic acid l− 1 (Sigma-Aldrich). E. faecalis strain OG1SSp (Ebp+) (Dunny et al., 1978) and derivatives were cultivated in BHI broth supplemented, when appropriate, with 500 mg spectinomycin l− 1 and 2000 mg streptomycin l− 1. Liquid cultures were grown statically at 37 °C.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description† | Reference(s) |
| E. faecalis | ||
| OG1RF | Chromosomal FusR RifR; Ebp+ | Bourgogne et al. (2008); Murray et al. (1993) |
| OG1SSp | Chromosomal SpcR StrR; Ebp+ | Dunny et al. (1978) |
| TX5608 | OG1RFΔebpABC; Ebp− | Nallapareddy et al. (2011b) |
| SD234 | OG1RF harbouring a chromosomally inserted gfp gene under control of the malM promoter; OG1RF : : gfp | DebRoy et al. (2012) |
| TX5756 | OG1SSpΔebpABC; ebpABC isogenic deletion mutant of OG1SSp; Ebp− | This study |
| TX5761 | OG1RF : : gfpΔebpABC; ebpABC deletion mutant of SD234; Ebp− | This study |
| TX5758 | OG1RF : : pAMβ1; OG1RF harbouring the plasmid pAMβ1; EryR FusR RifR | This study |
| TX5760 | OG1RFΔebpABC : : pAMβ1; OG1RFΔebpABC harbouring the plasmid pAMβ1; EryR FusR RifR | This study |
| TX5755 | OG1SSp : : pAMβ1; OG1SSp carrying the plasmid pAMβ1; EryR SpcR StrR | This study |
| TX5757 | OG1SSpΔebpABC : : pAMβ1; OG1SSpΔebpABC carrying the plasmid pAMβ1; EryR SpcR StrR | This study |
| pTEX5606 | Vector for ebpABC deletion; carries a mutated pheS* gene that renders cells susceptible to p-chloro-phenylalanine; EryR | Nallapareddy et al. (2011b) |
| pAMβ1 | Vector employed for conjugation experiments; EryR | Clewell et al. (1974) |
† Ery, Erythromycin; Fus, fusidic acid; Rif, rifampicin, Spc, spectinomycin; Str, streptomycin. Superscript ‘R’ designates resistance.
For construction of in-frame ebpABC deletion mutants of E. faecalis OG1SSp and SD234 (OG1RF : : gfp) (DebRoy et al., 2012), we used the previously constructed pTEX5606 vector as described by Nallapareddy et al. (2011b).
E. faecalis cell-to-cell aggregation experiments
As serum is an environmental condition that elicits pili expression (Nallapareddy et al., 2006, 2011a), E. faecalis cells for the cell-aggregation experiments were cultivated at 37 °C in BHI broth supplemented with 40 % horse serum (BHI-S) (Sigma-Aldrich). Cells in mid-exponential phase were mixed 1 : 1, allowed to interact for 2 h at 37 °C and dilutions were then plated for single colonies on BHI agar. Ninety-six apparently single colonies were randomly picked into the wells of 96-well plates containing 200 μl BHI broth, grown overnight and then replica plated onto BHI agar supplemented with 25 mg fusidic acid l− 1 and onto BHI agar supplemented with 500 mg spectinomycin l− 1 and 2000 mg streptomycin l− 1 to identify colonies that were actually a mixture of OG1RF and OG1SSp cells. A mixed colony was defined as an apparent single colony able to grow on both selective media, indicating that the ‘single’ colony arose from a mixture of cells of each phenotype.
In a separate experiment, the presence of mixed colonies was evaluated by combining an equal amount (108 c.f.u. ml− 1) of BHI-S-grown GFP-tagged OG1RF (OG1RF : : gfp) and OG1RF : : gfpΔebpABC cells with either piliated or non-piliated OG1SSp. After 2 h, the mixture was serially diluted, plated on BHI agar and grown overnight. Colonies were imaged using a Gel Doc 2000 System (Bio-Rad) equipped with an UV lamp for GFP detection.
Conjugation experiments
For the conjugation assays, donor cells carrying the plasmid pAMβ1 (Clewell et al., 1974), a non-aggregation substance-producing plasmid, were harvested by centrifugation from exponential phase cultures in BHI-S supplemented with 25 mg erythromycin l− 1. Experiments were done first using OG1RF derivatives as donors and OG1SSp derivatives as recipients. Assays were then repeated using OG1SSp derivatives as donor and OG1RF derivatives as recipients.
After three washes in saline solution to remove the antibiotic, OG1(RF or SSp) : : pAMβ1 or OG1(RF or SSp)ΔebpABC : : pAMβ1 were mixed in a 1 : 10 ratio with either OG1(SSp or RF) or OG1(SSp or RF)ΔebpABC and allowed to conjugate in BHI broth for 5 h at 37 °C. The mating mixture was then serially diluted and plated onto selective agar to enumerate transconjugants.
Results and Discussion
Previous studies have shown that sortase-assembled pili support bacterial co-aggregation in Actinobacteria (Yeung 2000; Turroni et al., 2013). We therefore evaluated the possible role played by E. faecalis Ebp pili in mediating cell–cell interactions. As seen in Fig. 1, when Ebp+ OG1RF cells were mixed with Ebp+ OG1SSp cells, 18 % of the resulting apparently ‘single’ colonies were actually ‘mixed’, that is, composed of both OG1RF and OG1SSp cells, while the other colonies were composed of only one of these derivatives. On the other hand, when non-piliated TX5608 (OG1RFΔebpABC) were mixed with non-piliated TX5756 (OG1SSpΔebpABC), only 0.5 % of the resulting single (in appearance) colonies had both RF-resistant cells and SSp-resistant cells (P = 0.0009), thus indicating a 36-fold reduction for the Ebp− Ebp− mixture versus the Ebp+ Ebp+ mixture. When Ebp+ cells were combined with cells lacking ebpABC (either OG1RF+TX5756 or TX5608+OG1SSp), approximately 8 % of apparently single colonies were in fact a mixture of RF- and SSp-resistant cells (P < 0.05 vs Ebp+ OG1RF+Ebp+ OG1SSp). Since the ebpABC deletion mutants exhibited no differences in growth (data not shown), these results indicate that the lack of pili affects the ability of E. faecalis cells to co-aggregate.
Fig. 1.
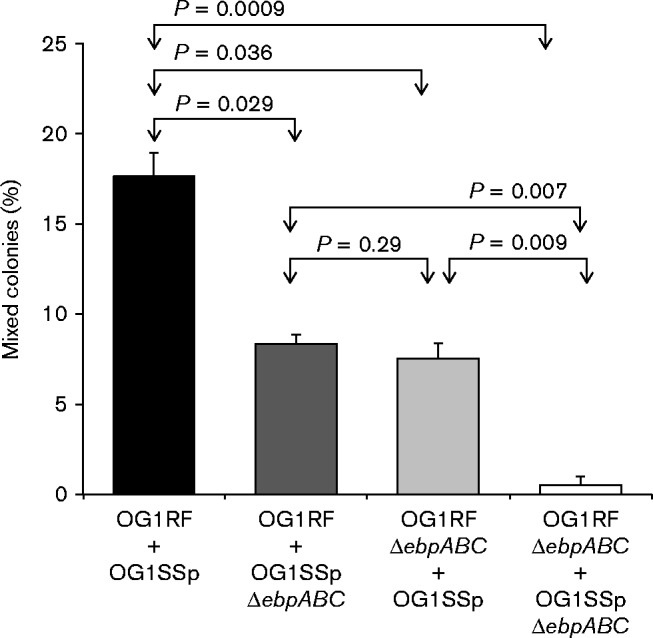
Contribution of Ebp pili to E. faecalis intercellular aggregation leading to mixed colony formation. OG1RF and its Ebp− isogenic mutant (TX5608; OG1RFΔebpABC) were tested for the ability to form mixed colonies (co-aggregates) with OG1SSp and a non-piliated OG1SSp derivative (TX5756; OG1SSpΔebpABC). Mixed colonies were identified by replica plating from BHI broth-grown cells inoculated from ‘apparent’ single colonies onto BHI agar supplemented with 25 mg fusidic acid l− 1, and onto BHI agar supplemented with 2000 mg streptomycin l− 1 and 500 mg spectinomycin l− 1. Values represent means ± sd of three independent experiments. Statistical analyses were performed by unpaired t-test.
Consistent with these data, we observed a similar pattern in the percentage of mixed colonies when combining piliated or non-piliated variants of both OG1SSp and gfp-tagged OG1RF cells (Table 2), that is, a higher percentage of colonies in which part of the colony was Gfp+ and the rest was not (Fig. 2). Hence, these results corroborate the importance of Ebp pili in mediating intercellular aggregation of E. faecalis.
Table 2. Evaluation of the role of Ebp pili in E. faecalis cell–cell adherence by fluorescence imaging of plates containing GFP-tagged and non-fluorescent colonies.
GFP-tagged OG1RF (SD234) and OG1RF : : gfpΔebpABC (TX5761) cells were mixed with either piliated or non-piliated OG1SSp (TX5756). Values indicate the mean percentage ± sd.
| Cells mixed | Mixed colonies (%) |
| OG1RF : : gfp+OG1SSp | 12.85 ± 1.2 |
| OG1RF : : gfp+OG1SSpΔebpABC | 7.1 ± 1.6* |
| OG1RF : : gfpΔebpABC+OG1SSp | 6.8 ± 1.3* |
| OG1RF : : gfpΔebpABC+OG1SSpΔebpABC | 1.8 ± 0.9* |
P < 0.05 vs OG1RF : : gfp+OG1SSp.
Fig. 2.
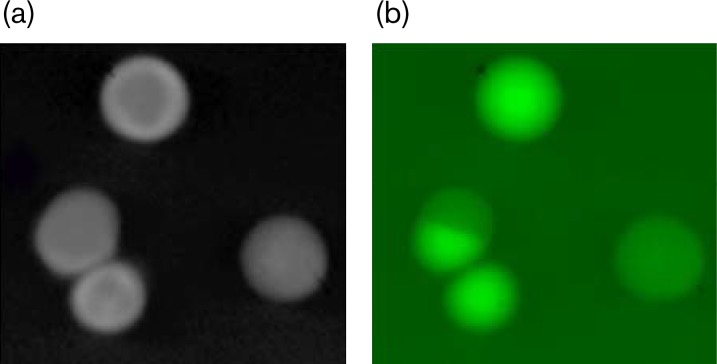
Detection of E. faecalis mixed colonies by fluorescence imaging. Colonies were obtained by subculturing from a BHI-S broth culture of the gfp-tagged strain (SD234; OG1RF : : gfp) grown with a non-gfp tagged strain (OG1SSp). Plates were imaged using a Gel Doc 2000 System (Bio-Rad) equipped with an UV lamp for GFP detection. (a) Black and white photo of four colonies and (b) fluorescent picture of the same colonies showing two Gfp+ colonies, one Gfp− colony and one mixed colony with both Gfp+ and Gfp− cells.
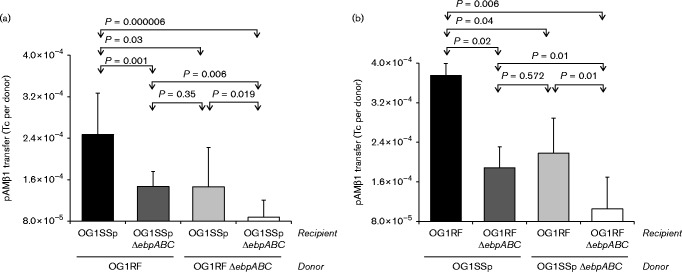
To assess whether co-aggregation mediated by Ebp pili facilitates the transfer of genetic material between cells, we introduced the vector pAMβ1, which lacks the aggregation substance that causes cell clumping, into E. faecalis OG1RF (resulting in TX5758), and evaluated its transfer to OG1SSp or OG1SSpΔebpABC in 5 h broth mating assays. Transfer of pAMβ1 from Ebp+ OG1RF cells into the Ebp− OG1SSP derivative was 170 % less than transfer into Ebp+ OG1SSp (P = 0.001) (Fig. 3a). A similar reduction was observed when pAMβ1-carrying OG1RFΔebpABC cells (TX5760) were combined with Ebp+ OG1SSp (P = 0.03). Most strikingly, absence of pili on the surface of both donor (pAMβ1-carrying OG1RFΔebpABC) and recipient (OG1SSpΔebpABC) resulted in a 280 % reduction in frequency of pAMβ1 transfer compared with the conjugation frequency of the isogenic piliated variants (P = 0.000006). In addition, we observed similar differences in pAMβ1 transfer frequencies (Fig. 3b) when the donor and recipient were reversed, i.e. when piliated and non-piliated OG1SSp cells were used as donors and OG1RF or OG1RFΔebpABC as recipients. In particular, a 360 % increase in conjugation was observed when piliated OG1SSp/RF Ebp+ Ebp+ were mixed compared with the OG1SSp/RF Ebp− Ebp− mixing. On the basis of these results, we conclude that Ebp pili promote plasmid transfer, presumably by facilitating micro-scale cell aggregation, i.e. attachment of cells in close proximity, and by the stabilization of the mating pair.
Fig. 3.
Transfer frequencies of pAMβ1 in 5 h matings in broth. (a) Piliated or non-piliated OG1RF cells carrying the plasmid pAMβ1 (TX5758 and TX5760, respectively) were mixed with either piliated or non-piliated OG1SSp. (b) Piliated or non-piliated OG1SSp cells carrying the plasmid pAMβ1 (TX5755 and TX5757, respectively) were mixed with either piliated or non-piliated OG1RF. Transfer frequencies are expressed as the number of transconjugants per donor cell (Tc per Donor). Histograms depict the mean and sd of at least three independent experiments. Statistical analyses were performed by unpaired t-test.
Previous studies have shown that Ebp pili are major contributors to the ability of E. faecalis to adhere to components of the extracellular matrix and to form biofilm, phenotypes linked to various enterococcal infections including endocarditis and UTIs (Sillanpää et al., 2013). When part of a biofilm community, bacteria are less susceptible to the host immune system and to antibiotic treatment; in addition, biofilms are optimal environments for genetic material exchange (Parsek & Singh, 2003). While we previously showed that pili are important for primary attachment to abiotic surfaces (Nallapareddy et al., 2006), our results here imply that a further contribution of pili is the promotion of cell-to-cell adherence at a micro-scale level, so that the piliated cells can attach both piliated and non-piliated ones. Thus, even though E. faecalis pili display a bi-phasic expression pattern, with only a portion of the population piliated at any given time (Sillanpää et al., 2013; Pinkston et al., 2014), the presence of pili on at least some cells can still promote accumulation of bacteria at a given site. In such settings, we speculate that the interactions mediated by Ebp pili may represent a driving force for intercellular attachment, colonization of new sites and biofilm formation and for the mobilization of plasmids carrying selective traits, such as antibiotic resistance or virulence factors, therefore providing E. faecalis or other clinically important species with fitness and survival advantages in natural environments. Taking these findings together, the presence of Ebp pili on the surface of E. faecalis cells likely has an important impact not only on colonization and adherence but also on bacterial pathogenicity and the spread of antibiotic resistance.
Acknowledgements
We thank Karen Jacques-Palaz and Chungyu Chang for their technical assistance. This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (NIAID) (R01 AI047923) to B.E.M.
Abbreviations:
- Ebp
endocarditis- and biofilm-associated pili
- UTI
urinary tract infection.
References
- Bourgogne A., Hilsenbeck S. G., Dunny G. M., Murray B. E. (2006). Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease J Bacteriol 188 2875–2884 10.1128/JB.188.8.2875-2884.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A., Singh K. V., Fox K. A., Pflughoeft K. J., Murray B. E., Garsin D. A. (2007). EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF J Bacteriol 189 6490–6493 10.1128/JB.00594-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A., Garsin D. A., Qin X., Singh K. V., Sillanpaa J., Yerrapragada S., Ding Y., Dugan-Rocha S., Buhay C., other authors (2008). Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF Genome Biol 9 R110 10.1186/gb-2008-9-7-r110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. (1974). Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance J Bacteriol 117 283–289 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S., van der Hoeven R., Singh K. V., Gao P., Harvey B. R., Murray B. E., Garsin D. A. (2012). Development of a genomic site for gene integration and expression in Enterococcus faecalis J Microbiol Methods 90 1–8 10.1016/j.mimet.2012.04.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G. M., Brown B. L., Clewell D. B. (1978). Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone Proc Natl Acad Sci U S A 75 3479–3483 10.1073/pnas.75.7.3479 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Mireles A. L., Pinkner J. S., Caparon M. G., Hultgren S. J. (2014). EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice Sci Transl Med 6 254ra127 10.1126/scitranslmed.3009384 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Pinkston K. L., Nallapareddy S. R., van Hoof A., Murray B. E., Harvey B. R. (2010). Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation J Bacteriol 192 5489–5498 10.1128/JB.00725-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp K. D., Singh K. V., Nallapareddy S. R., Murray B. E. (2007). Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection Infect Immun 75 5399–5404 10.1128/IAI.00663-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montealegre M. C., La Rosa S. L., Roh J. H., Harvey B. R., Murray B. E. (2015). The Enterococcus faecalis ebpA pilus protein: attenuation of expression, biofilm formation, and adherence to fibrinogen start with the rare initiation codon att MBio 6 e00467–e00415 10.1128/mBio.00467-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Singh K. V., Ross R. P., Heath J. D., Dunny G. M., Weinstock G. M. (1993). Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function J Bacteriol 175 5216–5223 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy S. R., Singh K. V., Sillanpää J., Garsin D. A., Höök M., Erlandsen S. L., Murray B. E. (2006). Endocarditis and biofilm-associated pili of Enterococcus faecalis J Clin Invest 116 2799–2807 10.1172/JCI29021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy S. R., Sillanpää J., Mitchell J., Singh K. V., Chowdhury S. A., Weinstock G. M., Sullam P. M., Murray B. E. (2011a). Conservation of Ebp-type pilus genes among Enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets Infect Immun 79 2911–2920 10.1128/IAI.00039-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy S. R., Singh K. V., Sillanpää J., Zhao M., Murray B. E. (2011b). Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF Infect Immun 79 2901–2910 10.1128/IAI.00038-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H. V., Guiton P. S., Kline K. A., Port G. C., Pinkner J. S., Neiers F., Normark S., Henriques-Normark B., Caparon M. G., Hultgren S. J. (2012). The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection MBio 3 e00177–e00112 10.1128/mBio.00177-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M. R., Singh P. K. (2003). Bacterial biofilms: an emerging link to disease pathogenesis Annu Rev Microbiol 57 677–701 10.1146/annurev.micro.57.030502.090720 . [DOI] [PubMed] [Google Scholar]
- Pinkston K. L., Singh K. V., Gao P., Wilganowski N., Robinson H., Ghosh S., Azhdarinia A., Sevick-Muraca E. M., Murray B. E., Harvey B. R. (2014). Targeting pili in enterococcal pathogenesis Infect Immun 82 1540–1547 10.1128/IAI.01403-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää J., Xu Y., Nallapareddy S. R., Murray B. E., Höök M. (2004). A family of putative MSCRAMMs from Enterococcus faecalis Microbiology 150 2069–2078 10.1099/mic.0.27074-0 . [DOI] [PubMed] [Google Scholar]
- Sillanpää J., Chang C., Singh K. V., Montealegre M. C., Nallapareddy S. R., Harvey B. R., Ton-That H., Murray B. E. (2013). Contribution of individual Ebp Pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection PLoS One 8 e68813 10.1371/journal.pone.0068813 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K. V., Nallapareddy S. R., Murray B. E. (2007). Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection J Infect Dis 195 1671–1677 10.1086/517524 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F., Serafini F., Foroni E., Duranti S., O'Connell Motherway M., Taverniti V., Mangifesta M., Milani C., Viappiani A., other authors (2013). Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions Proc Natl Acad Sci U S A 110 11151–11156 10.1073/pnas.1303897110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M. K. (2000). Actinomyces: surface macromolecules and bacteria-host interactions. In Gram-Positive Pathogens, pp. 583–593. Edited by Fischetti V. A., Novick R. P., Ferretti J. J., Portnoy D. A., Rood J. I. Washington, DC: American Society for Microbiology. [Google Scholar]