Abstract
Hantaviruses are emerging zoonotic viruses that cause human diseases. In this study, sera from 642 mammals from La Réunion and Mayotte islands (Indian Ocean) were screened for the presence of hantaviruses by molecular analysis. None of the mammals from La Réunion island was positive, but hantavirus genomic RNA was discovered in 29/160 (18 %) Rattus rattus from Mayotte island. The nucleoprotein coding region was sequenced from the liver and spleen of all positive individuals allowing epidemiological and intra-strain variability analyses. Phylogenetic analysis based on complete coding genomic sequences showed that this Murinae-associated hantavirus is a new variant of Thailand virus. Further studies are needed to investigate hantaviruses in rodent hosts and in Haemorrhagic Fever with Renal Syndrome (HFRS) human cases.
Viruses belonging to the genus Hantavirus, family Bunyaviridae, are negative-sense tri-segmented RNA viruses, with L, M and S segments encoding the RNA-dependent RNA polymerase, glycoproteins Gc and Gn, and nucleoprotein, respectively. Hantaviruses are known to circulate in small mammals (rodents, shrews, moles and bats) in Europe, Asia, America and, more recently discovered, Africa. Infection of humans occurs through inhalation of rodents' aerosolized excreta and can cause two severe pathologies: Haemorrhagic Fever with Renal Syndrome (HFRS) in Asia and Europe, and Hantavirus Cardiopulmonary Syndrome (HCPS), in the Americas (Kruger et al., 2015).
An extensive capture of almost 4000 wild and domestic mammals was conducted in La Réunion, Maurice and Mayotte islands between 2006 and 2007, during Chikungunya virus (CHIKV) outbreaks. Although a low seroprevalence against CHIKV was detected in several non-human primates and rats (Vourc'h et al., 2014), the sample collection was used for virus hunting. Here, we report on an investigation of hantaviruses in 642 small wild mammals captured in La Réunion (193 Rattus rattus, 44 Rattus norvegicus, 67 Mus musculus, 133 Suncus murinus, 45 Tenrec ecaudatus) and Mayotte (160 Rattus rattus) islands.
Pools of five individual sera were prepared for the collections from both La Réunion and Mayotte islands.
RNA was extracted from the pools using the MagAttract Viral RNA M48 kit and BioRobot M48 (Qiagen). The following reverse transcription (RT)-PCRs were performed for hantavirus screening: (i) consensus RT-PCR using degenerate primers targeting the L gene (Klempa et al., 2006) for all samples; and (ii) specific RT-PCRs for insectivore hantaviruses targeting both the L and S segments (Kang et al., 2009) for insectivore samples. Sera from positive pooled RNA were subsequently screened independently by RT-PCR. Although none of the samples from La Réunion was positive for hantavirus RNA, we observed hantavirus L gene amplicons for 18 % (29/160) of Rattus rattus samples from Mayotte island. Rat species identification was confirmed by a RT-PCR targeting the cytochrome c oxidase gene and using the RodentSEA Identification Tool (http://www.ceropath.org/barcoding_tool/rodentsea) for three representative animals (Fig. S1, available in the online Supplementary Material).
The characteristics of the 29 positive individuals are described in Table 1. Thirteen positive individuals (44.8 %) were males and 16 (55.2 %) were females, reflecting the gender proportion within all captured animals (44 and 56 %, respectively). No significant association was observed between gender and infection (P = 0.923), therefore male and female black rats were equally susceptible to hantavirus infection (13/70 : 18.6 % and 16/89 : 18.0 %, respectively). Of the 28 individuals with known age, 27 (96.4 %) were adults, which was significantly higher (P < 0.05) than the proportion of adults within the whole sampled population (58.8 %). Despite the limited number of individuals, our results support the hypothesis that hantavirus infection is not acquired vertically or during the neonatal stage, as demonstrated previously experimentally (Taruishi et al., 2008).
Table 1. Description of hantavirus positive Rattus rattus in Mayotte island.
Epidemiological information, geo-coordinates of captures and molecular analysis (RT-PCR; sequencing) are indicated for each individual.
| R. rattus ID | Sex | Age* | Capture area† | RT-PCR (L)‡ | RT-PCR (S)/sequences§ | ||||||
| Latitude | Longitude | Serum | Spleen | Liver | Spleen | Liver | GenBank accession numbers | ||||
| 463 | M | a | SE – 1 | − 12.98468 | 45.18128 | + | + | + | + | + | KT719351–KT719352 |
| 468 | M | a | SE – 1 | − 12.98468 | 45.18128 | + | − | + | + | + | KT719353–KT719354 |
| 469 | M | j | SE – 1 | − 12.98468 | 45.18128 | + | + | + | + | + | KT719355–KT719356 |
| 470 | F | a | SE – 1 | − 12.98468 | 45.18128 | + | + | + | + | + | KT719357–KT719358 |
| 90 | F | a | SE – 2 | − 12.97011 | 45.16034 | + | + | + | + | + | KT719366–KT719367 |
| 412 | F | a | SE – 4 | − 12.91093 | 45.1904 | + | + | + | + | + | KT719329–KT719330 |
| 422 | M | a | SE – 4 | − 12.91093 | 45.1904 | + | + | + | + | + | KT719331–KT719332 |
| 423 | M | a | SE – 4 | − 12.91093 | 45.1904 | + | + | + | + | + | KT719333–KT719334 |
| 96 | M | a | SE – 5 | − 12.92727 | 45.177 | + | + | + | − | + | KT719368 |
| 99 | M | a | SE – 5 | − 12.92727 | 45.177 | + | + | + | + | + | KT719369–KT719370 |
| 102 | F | a | SE – 5 | − 12.92727 | 45.177 | + | − | + | − | + | KT719320 |
| 425 | F | a | SE – 5 | − 12.92727 | 45.177 | + | + | + | + | − | KT719335 |
| 426 | F | a | SE – 5 | − 12.92727 | 45.177 | + | + | + | − | + | KT719336 |
| 428 | F | a | SE – 5 | − 12.92727 | 45.177 | + | + | + | + | + | KT719337–KT719338 |
| 429 | F | na | SE – 5 | − 12.92727 | 45.177 | + | + | + | + | − | KT719339 |
| 376 | M | a | W – 6 | − 12.83119 | 45.13716 | + | + | + | + | + | KT719321–KT719322 |
| 379 | M | a | W – 6 | − 12.83119 | 45.13716 | + | + | + | + | + | KT719323–KT719324 |
| 382 | M | a | W – 6 | − 12.83119 | 45.13716 | + | + | − | +‖‖ | +‖‖ | KT719325–KT719326 |
| 408 | F | a | W – 7 | − 12.87164 | 45.11745 | + | + | + | + | + | KT719327–KT719328 |
| 441 | M | a | N – 8 | − 12.70017 | 45.12168 | + | + | + | + | + | KT719340–KT719341 |
| 444 | F | a | N – 8 | − 12.70017 | 45.12168 | + | + | − | + | − | KT719342 |
| 445 | M | a | N – 8 | − 12.70017 | 45.12168 | + | + | + | + | + | KT719343–KT719344 |
| 449 | F | a | N – 8 | − 12.70017 | 45.12168 | + | + | + | + | + | KT719345–KT719346 |
| 456 | F | a | N – 8 | − 12.70017 | 45.12168 | + | + | + | + | + | KT719347–KT719348 |
| 457 | F | a | N – 8 | − 12.70017 | 45.12168 | + | + | + | +¶ | + | KT719349–KT719350 |
| 76 | M | a | W – 9 | − 12.76478 | 45.10652 | + | + | − | + | − | KT719363 |
| 82 | F | a | W – 9 | − 12.76478 | 45.10652 | + | + | + | + | + | KT719364–KT719365 |
| 493 | F | a | W – 9 | − 12.76478 | 45.10652 | + | + | + | +¶ | + | KT719359–KT719360 |
| 494 | F | a | W – 9 | − 12.76478 | 45.10652 | + | + | + | + | + | KT719361–KT719362 |
a, Adult; j, juvenile; na, information not available.
SE, South-east; N, north; W, west.
Different sequences from spleen and liver of the same individual. E382: C/T spleen/liver at nt 945 (corresponding to nt 957 of the coding region) of the S segment. This polymorphism is a silent mutation.
Only half the length of the sequence obtained (562 nt vs 1005 nt).
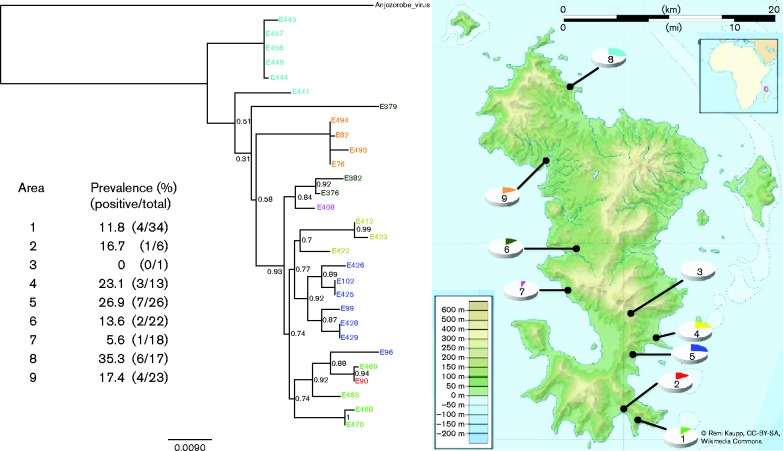
A precise location of each capture according to geo-coordinate (latitude and longitude) values (Table 1) allowed the differentiation of nine geographical areas across Mayotte island according to the virus circulation (Fig. 1). Five areas were in the south-east: areas 1 (prevalence 11.8 %), 2 (16.7 %), 3 (0 %), 4 (23.1 %) and 5 (26.9 %); three were in the west of the island: areas 6 (13.6 %), 7 (5.6 %) and 9 (17.4 %); and one was in the north: area 8 (35.3 %). We have noticed two main prevalence foci in the north (area 8) and south-east (area 5) of the island. However, the relatively low number of samples per area did not allow statistical comparison tests to be applied to the regional prevalence data. Additional samples and further ecological analysis are required to fully elucidate hantavirus transmission and circulation in Mayotte island.
Fig. 1.
Hantavirus prevalence and phylogeography in Rattus rattus from Mayotte island. Prevalence and intra-strain variability of MAYOV hantavirus according to localization of the captured R. rattus. Phylogenetic analysis was done using partial S segment sequences (nt 13–1017). Anjozorobe virus, previously detected in Madagascar (GenBank accession no. KC490916) was used as an outgroup. Bar, mean number of nucleotide substitutions per site. Colour codes correspond between the map and the tree.
To explore further the hantavirus genetic diversity in Rattus rattus from Mayotte island, RNA was extracted (QIAamp Viral RNA Extraction kit and QIAcube; Qiagen) from homogenized (TissueLyser; Qiagen) liver and spleen from the 29 positive animals. The majority of the spleen (27/29 = 93.1 %) and liver (26/29 = 90.0 %) samples tested positive using the nested RT-PCR targeting the L segment (Table 1). Both degenerate and specific primers were designed along the S segment by multiple alignments of the Murinae-associated hantavirus sequences (primers available on request) to obtain the nucleoprotein coding sequence from at least one organ per animal. Sequences from both organs (22/26) were identical to each other, except for one individual (E382) where a C/T polymorphism was observed (spleen/liver) at position 957 of the coding region on the S segment (Table 1), this mutation being silent at the amino acid level. The determined partial N gene coding sequences (1005 nt) within the S segment (nt 13–1017), were used. The method used was the maximum-likelihood method with PhyML v.3.0 (Guindon et al., 2010) under the GTR (general time reversible) substitution model with a γ-distribution model among site rate heterogeneity and a proportion of invariant sites (GTR+G+I), as determined by mega v.6.0 (Tamura et al., 2013) and with a statistical approximate likelihood ratio test of branch support. Fig. 1 shows the phylogenetic pattern merged with the virus distribution across the island. The genetic cluster of most of the hantavirus sequences appeared to follow a geographical trajectory with a probable gradient of the virus distribution from north to south. However, individuals E96 and E379 did not follow this pattern, branching out of their group.
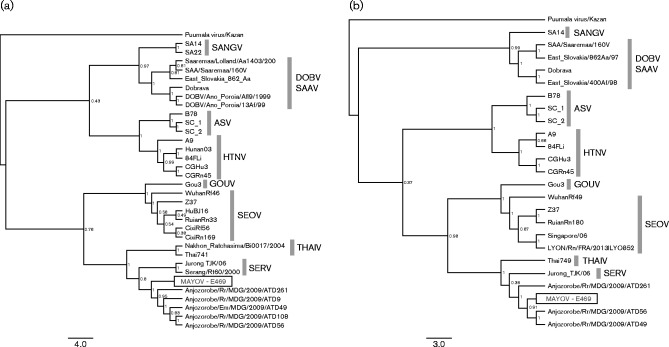
More exhaustive genetic information for this newly observed hantavirus was obtained from a representative individual (E469) by Next-Generation Sequencing (NGS) (Illumina MiSeq; Illumina). RNA was extracted and depleted of genomic DNA and ribosomal RNA using a protocol optimized for RNA viruses (Marston et al., 2013). Double-stranded cDNA and subsequent library preparation for Illumina sequencing were undertaken as described previously (Marston et al., 2013). Host sequences were removed by mapping to the Rattus rattus reference genome (BWA-mem v.0.7.5). blast analysis of the assembled contigs of non-host reads (Velvet v.1.2.10) was used to select the most similar reference sequence [Anjozorobe virus (ANJV)]. Iterative mapping and intermediate consensus alignment was used to generate the final consensus sequence. RT-PCR targeting with Sanger sequencing was undertaken to fill missing gaps in the sequence. Complete protein coding sequences (S, M and L) of the newly identified virus isolate, named Mayotte virus (MAYOV), were obtained. Sequences from the S and M segments were used for phylogenetic analysis (Fig. 2a, b), following the methodology described above. This analysis showed that MAYOV clusters within the Thailand hantavirus clade closer to the isolates circulating in the Indian Ocean, South-East Asia (Hugot et al., 2006; Plyusnina et al., 2009; Johansson et al., 2010) and Madagascar (Reynes et al., 2014), rather than the Murinae-associated hantaviruses circulating in continental Africa (Klempa et al., 2006, 2012; Witkowski et al., 2014). The data suggested that both MAYOV and Anjozorobe virus, the closest variant within the Thailand hantavirus clade, may have resulted from a westward expansion of an ancestral South-East Asian hantavirus introduced with R. rattus (Cheke & Hume, 2008; Tollenaere et al., 2010). Pairwise sequence identities between their coding regions, calculated with the blastn tool available online at http://blast.ncbi.nlm.nih.gov/Blast.cgi, were 91/99 % (nucleotide/amino acid identity) for the S segment, 90–91/97–98 % (nucleotide/amino acid identity) for the M segment (for ATD49 and AT261 Anjozorobe virus strains, respectively) and 90/99 % (nucleotide/amino acid identity) for the L segment. It is likely that exchanges between the Indian Ocean islands may have occurred as indicated by the phylogenetic trees in Fig. 2: Fig. 2(a) indicates that MAYOV and Anjozorobe virus have a common ancestor from which MAYOV diverged first, whereas Fig. 2(b) is less definitive and shows only the common ancestor of these two viral strains.
Fig. 2.
Phylogenetic analysis of Murinae-associated hantaviruses including MAYOV. Phylogenetic analysis was performed for the S (a) and M (b) segments among the Murinae-associated hantaviruses. MAYOV sequences were obtained by next-generation sequencing (S, 142 reads; M, 306 reads) and Sanger sequencing. The sequences used for phylogenetic analysis were obtained from GenBank: Sangassou virus (SANGV) (N: JQ082303, JQ082300; Gc-Gn: JQ082301); Dobrava virus (DOBV)/Saarema virus (SAAV) (N: AJ269550, AJ009773, AJ616854, L41916, AJ410619, NC005233; Gc-Gn: AY168578, AJ009774, L33685, AY168577); Amur/Soochong virus (ASV) (N: AB127997, AY675349, AY675350; Gc-Gn: AB127994, AY675353, DQ056293); Hantaan virus (HTNV) (N: AF329390, AF366568, JN712306, EU363809, EU092221; Gc-Gn: AF035831, AF366569, EU363818, EU092225); Gou virus (GOUV) (N: AF184988; Gc-Gn: AF145977); Seoul virus (SEOV) (N: JQ665919, AF187082, FJ803202, FJ803207, GQ279380, FJ803215; Gc-Gn: JQ665895, AF187081, GU592931, GQ274942, KF387724); Thailand virus (THAIV) (N: AM397664, AB186420; Gc-Gn: L08756); Serang virus (SERV) (N: AM998808, GQ274941; Gc-Gn: GQ274939); Anjozorobe (N: KC490914-KC490918; Gc-Gn: KC490919-KC490921). The Kazan strain of Puumala virus (Z84204, Z84205) was used as an outgroup for both trees. Bars, mean number of nucleotide substitutions per site.
Conclusions
This study was performed in the framework of a campaign aimed at elucidating potential reservoirs of CHIKV, which induced several outbreaks in the Indian Ocean between 2005 and 2007. While screening for hantaviruses, we demonstrated the prevalence (18 %) of a newly identified isolate in R. rattus on Mayotte island.
Interestingly, antibodies against hantaviruses were reported previously in rats from the Eastern Horn of Africa (Rodier et al., 1993). One hypothesis is that hantaviruses were introduced by the shipping route to East Africa through infected R. rattus, originating from South-East Asia, via the Middle East during Arabian trade in the 10th century, and then shipped in the same reservoir host to the Indian Ocean islands (Rollin et al., 1986; Brouat et al., 2014).
MAYOV could have been acquired earlier by R. rattus through a spillover infection event from other hantavirus rodent reservoirs such as Bandicota indica for Thailand virus and Rattus tanezumi for Jurong/Serang virus in South-East Asia (Thailand, Indonesia) (Hugot et al., 2006; Plyusnina et al., 2009; Johansson et al., 2010). The poor intrinsic genetic variability of MAYOV in R. rattus reflects a limited evolution and suggests a relatively recent colonization of Mayotte island. Further studies are required to explore hantavirus and rodent genetic diversity to understand better the origin of MAYOV and its adaptation to local hosts. Identifying the presence of hantavirus in Mayotte island is of great importance to evaluate its distribution in the tropical Indian Ocean region. A more extensive surveillance of rodents and /or other reservoirs is required to fully understand the circulation of these zoonotic viruses and to assess their potential risk of transmission to humans and subsequent occurrence of HFRS.
Acknowledgements
This study was supported by the EU FP7 programmes: EDENext (no. 261504), EMPERIE (no. 223498), ANTIGONE (no. 278976) and the Research Infrastructure Grant European Virus Archive(no. 19 228292). We thank Sandra Lacote (Virology Unit, Anses-Laboratoire, Lyon, France) for helping during RNA extraction. A. R. F. was supported by the Research and Policy for Infectious Disease Dynamics Program (RAPIDD), Science and Technology Directorate, US Department of Homeland Security and Fogarty International Center, US National Institutes of Health. Sampling was supported by the French Research National Agency ChikAni(ANR no. 06SEST06). We thank all the people who have participated in the collection of data in the field. We thank Dr Alexander Plyusnin for critical reading of the manuscript.
Supplementary Data
Supplementary Data
References
- Brouat C., Tollenaere C., Estoup A., Loiseau A., Sommer S., Soanandrasana R., Rahalison L., Rajerison M., Piry S., other authors (2014). Invasion genetics of a human commensal rodent: the black rat Rattus rattus in Madagascar Mol Ecol 23 4153–4167 10.1111/mec.12848 . [DOI] [PubMed] [Google Scholar]
- Cheke A., Hume J. (2008). Lost Land of the Dodo: The Ecological History of Mauritius, Réunion & Rodrigues London: T. & A.D. Poyser. [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0 Syst Biol 59 307–321 10.1093/sysbio/syq010 . [DOI] [PubMed] [Google Scholar]
- Hugot J. P., Plyusnina A., Herbreteau V., Nemirov K., Laakkonen J., Lundkvist A., Supputamongkol Y., Henttonen H., Plyusnin A. (2006). Genetic analysis of Thailand hantavirus in Bandicota indica trapped in Thailand Virol J 3 72 10.1186/1743-422X-3-72 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P., Yap G., Low H. T., Siew C. C., Kek R., Ng L. C., Bucht G. (2010). Molecular characterization of two hantavirus strains from different Rattus species in Singapore Virol J 7 15 10.1186/1743-422X-7-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J., Bennett S. N., Sumibcay L., Arai S., Hope A. G., Mocz G., Song J. W., Cook J. A., Yanagihara R. (2009). Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea) PLoS One 4 e6149 10.1371/journal.pone.0006149 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B., Fichet-Calvet E., Lecompte E., Auste B., Aniskin V., Meisel H., Denys C., Koivogui L., ter Meulen J., Krüger D. H. (2006). Hantavirus in African wood mouse, Guinea Emerg Infect Dis 12 838–840 10.3201/eid1205.051487 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B., Witkowski P. T., Popugaeva E., Auste B., Koivogui L., Fichet-Calvet E., Strecker T., Ter Meulen J., Krüger D. H. (2012). Sangassou virus, the first hantavirus isolate from Africa, displays genetic and functional properties distinct from those of other Murinae-associated hantaviruses J Virol 86 3819–3827 10.1128/JVI.05879-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger D. H., Figueiredo L. T., Song J. W., Klempa B. (2015). Hantaviruses – globally emerging pathogens J Clin Virol 64 128–136 10.1016/j.jcv.2014.08.033 . [DOI] [PubMed] [Google Scholar]
- Marston D. A., McElhinney L. M., Ellis R. J., Horton D. L., Wise E. L., Leech S. L., David D., de Lamballerie X., Fooks A. R. (2013). Next generation sequencing of viral RNA genomes BMC Genomics 14 444 10.1186/1471-2164-14-444 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnina A., Ibrahim I. N., Plyusnin A. (2009). A newly recognized hantavirus in the Asian house rat (Rattus tanezumi) in Indonesia J Gen Virol 90 205–209 10.1099/vir.0.006155-0 . [DOI] [PubMed] [Google Scholar]
- Reynes J. M., Razafindralambo N. K., Lacoste V., Olive M. M., Barivelo T. A., Soarimalala V., Heraud J. M., Lavergne A. (2014). Anjozorobe hantavirus, a new genetic variant of Thailand virus detected in rodents from Madagascar Vector Borne Zoonotic Dis 14 212–219 10.1089/vbz.2013.1359 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier G., Soliman A. K., Bouloumié J., Kremer D. (1993). Presence of antibodies to Hantavirus in rat and human populations of Djibouti Trans R Soc Trop Med Hyg 87 160–161 10.1016/0035-9203(93)90469-7 . [DOI] [PubMed] [Google Scholar]
- Rollin P. E., Mathiot C., Nawrocka E., Ravaoalimalala V. E., Coulanges P., Sureau P., McCormick J. B. (1986). [Hemorrhagic fever with renal syndrome in Madagascar. First seroepidemiologic survey of rat populations] Arch Inst Pasteur Madagascar 52 181–186 (in French) . [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). mega6: Molecular Evolutionary Genetics Analysis version 6.0 Mol Biol Evol 30 2725–2729 10.1093/molbev/mst197 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taruishi M., Yoshimatsu K., Hatsuse R., Okumura M., Nakamura I., Arikawa J. (2008). Lack of vertical transmission of Hantaan virus from persistently infected dam to progeny in laboratory mice Arch Virol 153 1605–1609 10.1007/s00705-008-0156-0 . [DOI] [PubMed] [Google Scholar]
- Tollenaere C., Brouat C., Duplantier J. M., Rahalison L., Rahelinirina S., Pascal M., Moné H., Mouahid G., Leirs H., Cosson J.-F. (2010). Phylogeography of the introduced species Rattus rattus in the western Indian Ocean, with special emphasis on the colonization history of Madagascar J Biogeogr 37 398–410 10.1111/j.1365-2699.2009.02228.x. [DOI] [Google Scholar]
- Vourc'h G., Halos L., Desvars A., Boué F., Pascal M., Lecollinet S., Zientara S., Duval T., Nzonza A., Brémont M. (2014). Chikungunya antibodies detected in non-human primates and rats in three Indian Ocean islands after the 2006 ChikV outbreak Vet Res 45 52 10.1186/1297-9716-45-52 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski P. T., Klempa B., Ithete N. L., Auste B., Mfune J. K., Hoveka J., Matthee S., Preiser W., Kruger D. H. (2014). Hantaviruses in Africa Virus Res 187 34–42 10.1016/j.virusres.2013.12.039 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data