Abstract
Cytochrome P450 (P450) protein-protein interactions have been shown to alter their catalytic activity. Furthermore, these interactions are isoform specific and can elicit activation, inhibition, or no effect on enzymatic activity. Studies show that these effects are also dependent on the protein partner cytochrome P450 reductase (CPR) and the order of protein addition to purified reconstituted enzyme systems. In this study, we use controlled immobilization of P450s to a gold surface to gain a better understanding of P450-P450 interactions between three key drug-metabolizing isoforms (CYP2C9, CYP3A4, and CYP2D6). Molecular modeling was used to assess the favorability of homomeric/heteromeric P450 complex formation. P450 complex formation in vitro was analyzed in real time utilizing surface plasmon resonance. Finally, the effects of P450 complex formation were investigated utilizing our immobilized platform and reconstituted enzyme systems. Molecular modeling shows favorable binding of CYP2C9-CPR, CYP2C9-CYP2D6, CYP2C9-CYP2C9, and CYP2C9-CYP3A4, in rank order. KD values obtained via surface plasmon resonance show strong binding, in the nanomolar range, for the above pairs, with CYP2C9-CYP2D6 yielding the lowest KD, followed by CYP2C9-CYP2C9, CYP2C9-CPR, and CYP2C9-CYP3A4. Metabolic incubations show that immobilized CYP2C9 metabolism was activated by homomeric complex formation. CYP2C9 metabolism was not affected by the presence of CYP3A4 with saturating CPR concentrations. CYP2C9 metabolism was activated by CYP2D6 at saturating CPR concentrations in solution but was inhibited when CYP2C9 was immobilized. The order of addition of proteins (CYP2C9, CYP2D6, CYP3A4, and CPR) influenced the magnitude of inhibition for CYP3A4 and CYP2D6. These results indicate isoform-specific P450 interactions and effects on P450-mediated metabolism.
Introduction
Cytochrome P450s (P450s) are a superfamily of mono-oxygenases responsible for metabolizing 75% of commercial pharmaceuticals and other xenobiotics and endobiotics (Guengerich, 2006). They are found primarily in the liver, where they are membrane bound via their hydrophobic N terminus to the smooth endoplasmic reticulum (Szczesna-Skorupa and Kemper, 1993). Their mechanism of action is dependent on the sequential transfer of two electrons, each requiring cytochrome P450 reductase (CPR) (Guengerich and Johnson, 1997). The second transfer is facilitated by cytochrome b5 but cytochrome b5 is not required for P450-mediated metabolism (Yamazaki et al., 1997; Shimada et al., 2005; Locuson et al., 2006). In microsomes, P450 levels exceed those of CPR by at least 5-fold (Estabrook et al., 1971; Reed et al., 2011), suggesting that enzyme mobility is required (Kawato et al., 1982; Gut et al., 1983) or that multiple P450s use a single CPR (Peterson et al., 1976; Taniguchi et al., 1979; Eyer and Backes, 1992).
It has been suggested that P450s are clustered and not uniformly distributed in the endoplasmic reticulum (Matsuura et al., 1978) and that zonal differences may exist (Jungermann, 1995). In vitro, it has been shown that P450s interact (Kaminsky and Guengerich, 1985; Alston et al., 1991; Davydov, 2011), modulating P450-mediated metabolism (Backes and Kelley, 2003; Reed and Backes, 2012). This effect has been seen in many reconstituted enzyme systems, including human CYP2C9-CYP2D6 (Subramanian et al., 2009), human CYP2C9-CYP3A4 (Subramanian et al., 2010), human CYP2C9-CYP2C19 (Hazai and Kupfer, 2005), human CYP3A-CYP2E1 (Davydov et al., 2015), rabbit CYP1A2-CYP2B4 (Backes et al., 1998; Davydov et al., 2001; Reed et al., 2013), rabbit CYP1A2-CYP2E1 (Kelley et al., 2006), and human CYP3A4-CYP1A1 and CYP3A4-CYP1A2 (Yamazaki et al., 1997). P450-P450 interactions have also been observed in microsomal preparations (Kaminsky and Guengerich, 1985; Cawley et al., 2001), triple expression systems (Cawley et al., 1995; Tan et al., 1997; Li et al., 1999), and homomeric oligomerization of CYP2E1 (Jamakhandi et al., 2007), CYP2B4 (Davydov et al., 1992), and CYP3A4 (Davydov et al., 2005, 2010).
Although P450-P450 interactions in vitro have been found to affect metabolism, ambiguity remains because some studies have reported that P450-P450 interactions do not affect metabolism. Dutton, et al. (1987) showed no effect on P450-mediated testosterone metabolism in the presence of other P450s in rat microsomes (Dutton et al., 1987). Alston et al. (1991) showed the formation of heterodimers by some P450s, suggesting that there is some selectivity for the formation of heteromeric P450 complexes. Elucidating the underlying factors controlling P450-P450 interactions is essential for accurately predicting in vivo P450 substrate clearance rates from in vitro data. Predictions of clearance are often based on single-isoform reconstituted systems, which lack the possibility of heteroaggregate formation-based effects. In addition, although oligomerization does occur in solution, the extent and characterization of how P450s interact in vivo has yet to be fully examined. To this end, several groups have investigated P450 incorporation into membranes using molecular modeling approaches (Cojocaru et al., 2011; Liu et al., 2013). In addition, in vitro studies establishing P450-P450 membrane interactions using crosslinking (Alston et al., 1991; Hu et al., 2010), fluorescence resonance energy transfer (Szczesna-Skorupa et al., 2003), and luminescence resonance energy transfer (Davydov et al., 2013) have provided insight on the degree of P450 oligomerization in membranes.
There are three main mechanisms by which a P450 can affect the metabolic rate of another, including competition for CPR, complex formation with altered affinity for CPR, and formation of a complex with altered substrate binding or turnover rate (Backes and Kelley, 2003; Reed and Backes, 2012). Controlled immobilization of P450s has recently been used to allow better understanding of metabolism (Davydov et al., 2005; Gannett et al., 2006; Mak et al., 2010; Fantuzzi et al., 2011; Panicco et al., 2011; Wollenberg et al., 2012; Bostick et al., 2015) and the formation of homomeric/heteromeric complexes (Backes and Kelley, 2003). We developed a scheme whereby soluble P450 is covalently attached to a self-assembled monolayer (SAM) on a gold film. The result retains full metabolic activity compared with the same amount of lipid-reconstituted P450 (Supplemental Fig. 1) and uses the normal protein partners and cofactors to metabolize substrates (Gannett et al., 2006). This platform models how P450 is bound in vivo and allows for control of P450 aggregation. Here, we assess the interaction of CYP2C9 with three major P450 isoforms (CYP2C9, CYP3A4, and CYP2D6) and with CPR, demonstrate the formation of homomeric/heteromeric P450 complexes in real time, and determine the consequences of these complexes on P450-mediated metabolism, all supported by molecular modeling studies of the P450 complexes.
Materials and Methods
Chemicals and Reagents.
All chemicals were used as purchased. Potassium phosphate, HEPES, sodium chloride, 11-mercaptoundecanoic acid (MUA), 1-octanethiol (OT), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide sodium salt (NHS), (S)-flurbiprofen (flurbiprofen), 4′-hydroxyflurbiprofen, dilauroylphosphatidylcholine (DLPC), NADPH, and cetyltrimethylammonium bromide were purchased from Sigma-Aldrich (St. Louis, MO). Truncated histidine-tagged CYP2C9 was prepared by expression in an Escherichia coli system, isolated, and purified as described previously (Cheesman et al., 2003; Hummel et al., 2005; Locuson et al., 2006). Human CPR was purchased from Invitrogen (Carlsbad, CA). All solutions were prepared in 18 MΩ-cm water unless otherwise noted. Phosphate buffer (40 mM, pH 7.4) contained 150 mM sodium chloride [phosphate-buffered saline (PBS)]. HEPES buffer (10 mM, pH 7.4) contained 150 mM sodium chloride, 3 mM EDTA, and 0.005% (v/v) Surfactant P20. Titanium and gold pellets were purchased from Kurt J. Lesker Company (Jefferson Hills, PA). Cryobuffer was made from PBS and glycerol [80:20 (v/v)].
Molecular Modeling.
Computational modeling was employed to determine possible enzyme binding orientations and the relative binding energy of the enzyme pairs CYP2C9-CYP2C9, CYP2C9-CYP3A4, CYP2C9-CYP2D6, and CYP2C9-CPR. To account for conformational changes affecting the binding strength and binding site, several CYP2C9 crystal structures were examined, including 1OG2 (without substrate), 1OG5 (with bound warfarin, closed) (Williams et al., 2003), and 1R9O (with bound flurbiprofen, partially open) (Wester et al., 2004). Structural similarity was assessed using the MultiSeq module (Eargle et al., 2006) of Visual Molecular Dynamics. P450 structures, crystallized without substrate, were used to match the experimental conditions: CYP2D6 [Rowland et al., 2006; Protein Data Bank (PDB) identifier 2F9Q] and CYP3A4 [Yano et al., 2004; PDB identifier 1TQN]. The crystal structure selected for CPR is locked into a conformation adopted when binding to a P450 enzyme due to a TGEE truncation in the hinge region (Hamdane et al., 2009; PDB identifier 3ES9 chain A). GRAMM-X (Tovchigrechko and Vakser, 2006) was used to computationally predict binding modes and the 10 lowest potential energy models per enzyme pair were selected for analysis. All homodimers and heterodimers were examined in Visual Molecular Dynamics (Humphrey et al., 1996) with respect to residues defining areas of interest as reported in the literature, including the active site and solvent tunnel entrances (Cojocaru et al., 2011), the CPR binding site (Shen and Kasper, 1995; Hazai et al., 2005; Hamdane et al., 2009), and the truncated N terminus. The relative binding energy for the lowest-energy GRAMM-X models for each binding pair was calculated using the molecular mechanics (MM)/generalized born surface area (GBSA) method (Miller et al., 2012) after minimization, equilibration, and 2 ns molecular dynamics at 300 K using the Amber software suite (Roe and Cheatham, 2013).
Gold-OT/MUA-CYP2C9 Chip Fabrication.
Two different materials were used as test platforms for the experiments described: gold-coated silicon wafers for generation of metabolites and gold-coated glass coverslips for surface plasmon resonance (SPR) detection of binding. Both were prepared for surface modification using a Temescal BJD-2000 system (Edwards Vacuum, Phoenix, AZ) with an Inficon XTC/2 deposition controller (Inficon, East Syracuse, NY). Evaporation occurred at a voltage of 10.0 kV. The current was set so that metal vaporization occurred at a rate of 0.03–0.10 nM/s (approximately 50 mA for titanium and 100 mA for gold). Thin films composed of titanium, 5-nm thick, and then gold, approximately 50-nm thick, were deposited on silicon chips or glass coverslips. Silicon wafer platforms were diced into 4 mm × 6 mm chips and then were sonicated in a Bransonic 220 (Bransonic, Danbury, CT) (5 minutes) and rinsed in succession with acetone, isopropyl alcohol, and water to ensure a clean surface. Glass platforms were cut into 10 mm × 12 mm rectangles, sonicated (15 minutes) with ethanol. A SAM was then formed on the gold surface by first rinsing it with ethanol, followed by soaking it in an ethanolic solution of OT (7.5 mM) and MUA (2.5 mM), overnight under argon. Excess thiol was removed by rinsing, in succession, with absolute ethanol, 95% ethanol, and water (three times each). Chips were dried under a gentle stream of nitrogen.
SPR Binding Analysis.
Glass platforms coated with gold-SAM as described above were loaded into a Biacore X100 SPR (GE Healthcare, Piscataway, NJ) for further surface modification and analysis. The surface was then activated with EDC (0.4 M) and NHS (0.1 M) in PBS running buffer as per Biacore instructions. CYP2C9 (100 nM) in PBS running buffer was flowed (10 μl/min) over the test surface (480 seconds), capped with ethanolamine (1 M) in PBS running buffer, and then rinsed with PBS running buffer alone (480 seconds). The control surface was prepared by directly capping it with ethanolamine. Immobilization levels reaching 70–200 response units above the control surface, after running buffer rinse, were considered acceptable. CYP2C9 was immobilized and the binding partner flowed over, without substrate, conditions that favor the partially open conformation. Analysis of binding between immobilized and soluble enzymes (0, 5, 10, 50, 100, and 500 nM) was conducted in PBS running buffer at 30 μl/min. After stabilization, contact times for soluble enzyme were 180 seconds followed by 900-second dissociation. Flow cells were regenerated with cetyltrimethylammonium bromide (3.9 mM). Two concentrations of the soluble enzyme (50 nM and 100 nM) were measured twice to ensure reproducibility of the data. Each P450 binding pair was run at least three times. Fitting of kinetic data was accomplished using both Biacore X100 Evaluation software and Scrubber 2.0 (BioLogic Software, Campbell, ACT, Australia). CYP2C9, CYP3A4, and CPR were fit using Biacore X100 Evaluation software. Biacore X100 Evaluation was unable to fit CYP2D6 curves, and Scrubber 2.0 was employed instead. Crossanalysis between fitting software demonstrated consistency.
Immobilization of CYP2C9 Enzyme on Gold/Silicon Platform.
To bond the CYP2C9 to the SAM, the SAM-coated slides were first activated by immersion in an aqueous solution of EDC (2 mM) and NHS (5 mM) [1:1 (v/v)], each in water, for 2 hours at room temperature. Activated chips were rinsed with PBS, then soaked (24 hours, room temperature) in a solution containing CYP2C9 (100 nM), flurbiprofen (40 µM), and dapsone (40 µM) in PBS, under argon, to covalently bond CYP2C9 to the NHS esters. We previously showed that inclusion of substrates flurbiprofen and dapsone preserves CYP2C9’s active site integrity and allows retention of enzymatic activity through immobilization (Supplemental Fig. 1) (Gannett et al., 2006). Chips bearing the bonded enzyme were rinsed with PBS to remove substrates from the active site prior to placement in cryobuffer solution and were stored at −80°C for at least 8 hours prior to use.
Reconstituted CYP2C9 Enzyme Incubation.
Reconstituted CYP2C9 incubations were run in triplicate. Incubations contained CYP2C9 (0.25 nM), saturating CPR (8 nM), DLPC (10 µg), and flurbiprofen (160 µM) in PBS (40 mM, pH 7.4) for a total volume of 250 µl. The amount of lipid per incubation is the same as other literature studies (Locuson et al., 2006; Subramanian et al., 2009, 2010). Because of the high amount of lipid to P450 in our study, controls were run at a DLPC/CYP2C9 ratio of 100:1, and the measured rates of metabolism were lower than those obtained with the conditions used in this study (data not shown). Immobilized CYP2C9 (CYP2C9i) incubations were conducted using two 4 mm × 6 mm gold chips placed back to back (gold coating facing outward) inside a 1.5-ml microcentrifuge tube. The total volume of 250 µl was sufficient to completely cover the chips and expose CYP2C9i to reaction components. No DLPC was added to incubations containing CYP2C9i, and additional nonimmobilized P450s added to these systems were not provided with lipids, creating a lipidless system in which metabolism only occurs through CYP2C9i. Coverage of CYP2C9 on gold chips was estimated based on previous work (Gannett et al., 2006; Yang et al., 2009). Ratios of CYP2C9i to CYP2C9 in solution (CYP2C9s) ranged from 1:1 to 1:4. Ratios of CYP2C9/CYP2D6 and CYP2C9/CYP3A4 ranged from 1:0.25 to 1:8 and 1:1 to 1:12, respectively, to mimic possible in vivo ratios of these enzymes (Meijerman et al., 2007). Incubations were placed on ice during the addition of precooled solutions of reactants and then preheated (3 minutes) at 37°C before initiation of the incubation by the addition of NADPH (200 µM, final concentration) and then incubated at 37°C overnight (16 hours). Reactions were terminated by the addition of 10 µl 500 ng/ml 2-fluoro-4-biphenyl acetic acid (internal standard) and 30 µl 50% phosphoric acid. Incubation mixtures were centrifuged (10 minutes, 13,400g) to pellet proteins, and supernatant (135 µl) was collected and loaded into liquid chromatography vials for analysis.
The effect of the order of mixing of P450s and CPR was investigated by 1) preincubating CYP2C9 (4 nM) or CYP2C9i (0.25 nM) with CPR (5 minutes) prior to the addition of CYP3A4 or CYP2D6 or 2) preincubating CYP2C9 (4 nM) or CYP2C9i (0.25 nM) with CYP3A4 or CYP2D6 (5 minutes) prior to addition of CPR. In studies investigating the order of addition in lipid-reconstituted systems, CPR was lowered with a P450/CPR ratio less than 1:2. CPR levels for CYP2C9i studies were the same as used for the previously explained immobilized metabolism setup, saturating CPR, to ensure sufficient metabolite generation. Incubations were placed on ice during the addition of precooled solutions of reactants and then preheated (3 minutes) at 37°C before initiation of the incubation by the addition of NADPH (200 µM, final concentration) and then incubated at 37°C for 1 hour for lipid-reconstituted CYP2C9 or overnight (16 hours) for CYP2C9i incubations. Reactions were terminated by the addition of 10 µl 500 ng/ml 2-fluoro-4-biphenyl acetic acid (internal standard) and 30 µl 50% phosphoric acid. Incubation mixtures were centrifuged (10 minutes, 13,400g) to pellet proteins, and supernatant (135 µl) was collected and loaded into liquid chromatography vials for analysis. Linearity of flurbiprofen metabolism to its 4-hydroxy metabolite as catalyzed by CYP2C9 was evaluated using study parameters for lipid-reconstituted enzyme with respect to time and was found to be nonlinear for the time courses used in our study (data not shown). Insufficient metabolite production resulted from the immobilized protein experiments to be measurable within any timeframe that might have resulted in linear formation. All experiments were performed three times on separate days.
High-Performance Liquid Chromatography Assay of 4′-Hydroxyflurbiprofen.
Formation of the flurbiprofen metabolite, 4′-hydroxyflurbiprofen, was measured by high-performance liquid chromatography with fluorescence detection. The high-performance liquid chromatography system consisted of an Alliance 2695XE pump/autosampler and a 2495 fluorescence detector (Waters, Milford, MA) set at an excitation wavelength of 280 nm and an emission wavelength of 310 nm. The mobile phase consisted of potassium phosphate (20 mM, pH 7.4) and acetonitrile [50:50 (v/v)], a 0.75 ml/min flow rate, and a 4.6 × 150 mm Zorbax C18 column (Agilent, Santa Clara, CA). The retention times of 4′-hydroxyflurbiprofen and the internal standard were 4.1 and 7.7 minutes, respectively. Metabolite concentration was determined using a calibration curve of 0–50 ng 4′-hydroxyflurbiprofen.
Statistical Analysis.
The results for metabolism studies are expressed as the means ± S.E. and were analyzed by a one-way analysis of variance followed by Dunnett’s test for multiple comparisons with the positive control group. One-way analysis of variance with Dunnett's post-test was performed using GraphPad Prism software (version 6 for Windows, www.graphpad.com; GraphPad Software, La Jolla, CA).
Results
Computationally Predicted Dimers.
P450s are known to form dimers and multimers and their formation is believed to affect metabolic activity. Here, modeling approaches were used to predict the likely binding poses of homodimers and heterodimers, to infer the consequences of dimer formation on metabolic activity, and to estimate the stability of the dimeric complexes relative to the monomer forms. The enzyme pairs examined were CYP2C9-CYP2C9, CYP2C9-CYP3A4, CYP2C9-CYP2D6, or CYP2C9-CPR. For each pair, only the CYP2C9 residues involved in binding were considered, including those involved in CPR binding (Shen and Kasper, 1995; Hamdane et al., 2009), the solvent access channel, and the substrate channel entrance (Cojocaru et al., 2007, 2011) (Supplemental Table 1).
It has been demonstrated that P450s exist in both open and closed conformations (Scott et al., 2004). The closed conformation is favored upon substrate binding (Williams et al., 2000, 2003; Yano et al., 2004) and alters the affinity toward enzyme partners (Hazai et al., 2005; Roberts et al., 2010). To account for the potential conformational changes on the modeling results, dimer formation was examined with an open and a closed form of CYP2C9. To select these structures, three CYP2C9 crystal structures were considered: 1OG2 (without substrate), 1OG5 (with bound warfarin) (Williams et al., 2003), and 1R9O (with bound flurbiprofen) (Wester et al., 2004). The 1OG2 and 1OG5 models, that represent open and closed states, respectively, were more conformationally similar to each other than to 1R9O, a partially open conformation (Supplemental Fig. 2). Therefore, further analysis was conducted only on the 1OG5 (closed) and 1R9O (partially open) models.
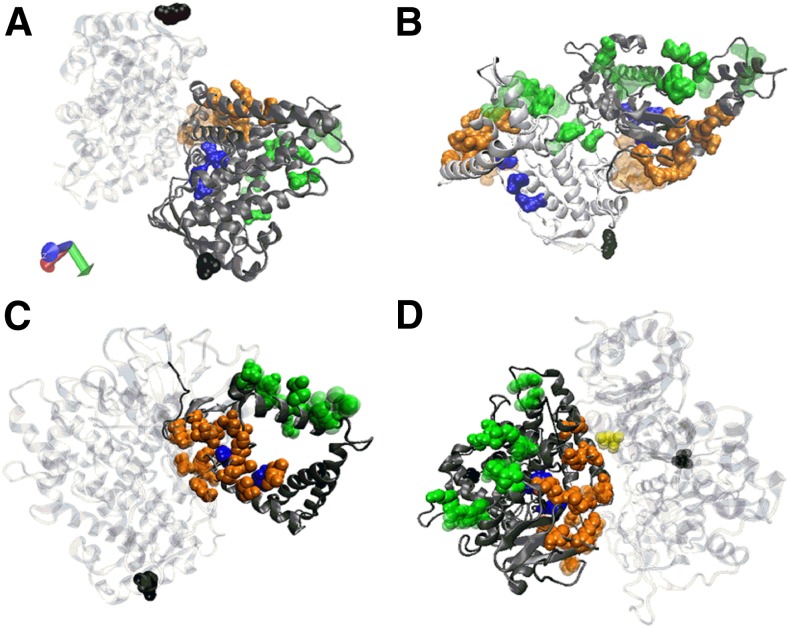
For the purpose of generating dimer structures, blind docking, using GRAMM-X was performed for each dimer combination. Each of the 10 models returned by GRAMM-X for the homodimers and heterodimers were examined for binding in defined regions of interest (Supplemental Table 1), and the occurrence of each interaction type and frequency is shown in Table 1. Dimerization involved the solvent or substrate channels 25% of the time and the CPR binding site approximately 7.5% of the time in predicted models (Table 1). Dimerization most frequently involved the solvent access channel region of CYP2C9. In the case of the closed CYP2C9 (1OG5), this location was heavily favored. In either case, the CPR binding site was the least preferred site. The distinctions between bindings for the three sites were greatest for the closed model, the top pairs for which are shown for in Fig. 1.
TABLE 1.
Dimer binding sites predicted by GRAMM-X
Each dimer was composed of either 1R90 or 1OG5 and the indicated binding partner.
| CYP2C9 Model | Binding Partnera | Active Site Access Channelb | Solvent Access Channelsb | CPR Binding Siteb |
|---|---|---|---|---|
| n | ||||
| 1R9O | CYP2C9 | 1 | 2 | 2 |
| CYP3A4 | 2 | 2 | 1 | |
| CYP2D6 | 2 | 2 | 1 | |
| CPR | 3 | 4 | 0 | |
| 1OG5 | CYP2C9 | 2 | 6 | 0 |
| CYP3A4 | 0 | 5 | 1 | |
| CYP2D6 | 0 | 3 | 1 | |
| CPR | 1 | 6 | 0 | |
The binding partner structures were 19RO.pdb for CYP2C9, 1TQN.pdb for CYP3A4, 3TBG for CYP2D6, or 3ES9.pdb for CPR.
Numbers shown indicate the number of models out of the top 10 that displayed binding in specified region of the CYP2C9 model (1R9O or 1OG5). Interactions for each of the top 10 binding models was considered to occur if residues in the specified regions, as shown in Supplemental Table 1, were involved. Dimerization involved the solvent or substrate channels 25% of the time and the CPR binding site approximately 7.5% of the time in predicted models. The percentage of residues in specified regions varied by dimer complex (Supplemental Table 2). The CPR binding site consisted of residues as described in Supplemental Table 3.
Fig. 1.
Representation of the top-scored interaction predicted by GRAMM-X for 1R9O bound to CYP3A4, CYP2C9, CYP2D6, and CPR. Residues defining the CPR binding site (green), the substrate access channel entrance (orange), the solvent channel entrance (blue), and the N terminus (black) are highlighted, and other residues have been removed from view. Partner molecules are light gray. (A) 1R9O-CYP3A4 solvent channel interface area. Binding as depicted by this model could affect the substrate turnover rate. (B) 1R9O-CYP2C9 CPR binding site interface area. In addition, the partially blocked surface could affect the strength of the CYP2C9-CPR binding interaction for the CYP2C9 (white) molecule. (C) 1R9O-CYP2D6 active site area. Binding as depicted by this model could affect substrate entry and exit into the active site. (D) 1R9O-CPR substrate access channel area. Dimerization as depicted in this model could affect substrate recognition, binding, and turnover rate.
Binding Free Energy In Silico.
Computational modeling was employed to determine the relative binding energy between four enzyme pairs: CYP2C9-CYP2C9, CYP2C9-CYP3A4, CYP2C9-CYP2D6, and CYP2C9-CPR. Molecular dynamics were run on these structures (neutralized and fully solvated with explicit waters). Binding free energies were calculated using the MM/GBSA method, which has been shown to be more successful in ranking binding affinities than the MM/Poisson–Boltzmann surface area method, although it is less successful in calculating absolute energy values (Hou et al., 2011). Resultant energies are reported in kilocalories per mole in Table 2. Negative values for total binding free energy suggest favorable protein-protein complexes. The rank order of the dimers is, from strongest to weakest, CYP2C9-CPR, CYP2C9-CYP2D6, CYP2C9-CYP2C9, and CYP2C9-CYP3A4.
TABLE 2.
MM/GBSA binding free energies for CYP2C9 dimers
Calculated binding free energies for top-scored interactions predicted by GRAMM-X for 1R9O bound to CPR, CYP2D6, CYP2C9, and CYP3A4. Values in parentheses are standard deviations. All values are based on at least 50 snapshots from a single trajectory spaced by 10 ps to avoid correlation between models.
| Energy Component | CPR | CYP2D6 | CYP2C9 | CYP3A4 |
|---|---|---|---|---|
| kcal/mol | ||||
| VDW | −124.2 (6.9) | 223.9 (9.9) | −157.5 (6.4) | −115.7 (6.15) |
| EEL | −500.0 (43.2) | −88.9 (28.3) | −224.3 (34.6) | −238.0 (35.3) |
| EGB | 548.2 (38.9) | 249.8 (28.7) | 332.9 (33.5) | 324.7 (32.6) |
| ESURF | −17.23 (0.8) | −28.2 (1.2) | −20.6 (0.7) | −14.5 (0.6) |
| ΔG (gas) | −624.3 (42.0) | −312.7 (32.8) | −381.8 (34.8) | −353.7 (35.3) |
| ΔG (sol) | 531.0 (38.7) | 221.6 (28.0) | 312.4 (33.3) | 310.2 (32.7) |
| ΔG (total) | −93.26 (6.9) | −91.1 (8.2) | −69.4 (5.9) | −43.5 (6.28) |
ΔG (gas), VDW + EEL; ΔG (sol), EGB + ESURF; ΔG (total), ΔG (gas) + ΔG (sol); ΔG (total), binding free energy [(ΔG complex) – (ΔG receptor + ΔG ligand)]; EEL, electrostatic energy; EGB, electrostatic contribution to the solvation free energy calculated by the generalized Born approach; ESURF, nonpolar contribution to the solvation free energy calculated by an empirical model; VDW, van der Waals contribution from the molecular mechanical energy.
Binding Affinity In Vitro.
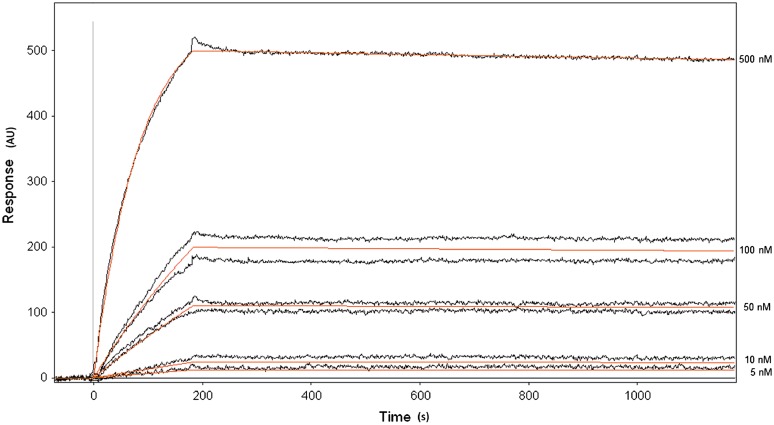
CYP2C9 was immobilized to a planar gold surface in a SPR flow cell via a SAM (Supplemental Fig. 3), reproducibly achieving immobilization levels of 70–200 response units relative to the ethanolamine-capped control surface. This low immobilization level was targeted to allow kinetic measurements of the system (Karlsson et al., 1991; Myszka, 1999). CYP2C9i was then exposed to soluble CYP2C9, CYP2D6, or CYP3A4 to analytically determine equilibrium dissociation constants for each dimer pair. A control flow cell was used to subtract out any nonspecific interactions between the solution enzyme and the SAM. A representative sensorgram obtained for CYP2C9i and CYP2D6 is shown in Fig. 2, along with the best fits, which assumed a dimer formation (1:1). Attempts to fit with higher-order oligomers (e.g., 1:2) resulted in poorer fits, implying that only dimers were formed in the SPR experiment. Therefore, the binding equilibrium dissociation constants were calculated based on a 1:1 binding model (Table 3). The rank order of binding affinities, from strongest to weakest, is the enzyme pairs CYP2C9-CYP2D6, CYP2C9-CYP2C9, CYP2C9-CPR, and CYP2C9-CYP3A4.
Fig. 2.
SPR sensorgram obtained from a system composed of soluble CYP2D6 binding to CYP2C9i. Both the observed response (black) and the Scrubber 2.0 fitted lines (orange) are shown for CYP2D6 concentrations of 5, 10, 50, 100, and 500 nM. Concentrations of 50 and 100 nM were repeated to ensure reproducibility. CYP2D6 in PBS was flowed over the surface from 0 to 180 seconds and then PBS only was flowed for 180 to 900 seconds at a flow rate of 30 μl/min. CYP2C9i was bonded to the surface of the sample cell and ethanolamine-capped SAM served as the control, in the control cell, at 30 μl/min.
TABLE 3.
SPR-derived CYP2C9 equilibrium dissociation constants
Average equilibrium dissociation constants (KD) ± S.E.M. are given for the binding of CYP2C9 with the indicated partner. Values were calculated by fitting the SPR sensorgrams with both Biacore Evaluation or Scrubber 2.0 software and assuming 1:1 stoichiometry. SPR runs were performed in triplicate on separate days.
| Partner | CYP2D6 | CYP2C9 | CYP3A4 | CPR |
|---|---|---|---|---|
| nM | ||||
| KD | 1.1 ± 0.5 | 2.6 ± 1.0 | 18.1 ± 3.0 | 7.3 ± 2.2 |
CYP2C9 Effect on CYP2C9i-Mediated Flurbiprofen Metabolism.
We next investigated the effects of P450-P450 interactions on metabolism utilizing our immobilized platform. The amount of immobilized enzyme was estimated as previously described (Gannett et al., 2006; Yang et al., 2009) coupled with the SPR immobilization response. This gave a coverage of 41 pg/mm2. The effect of various amounts of solution CYP2C9 coincubated with CYP2C9i on flurbiprofen metabolism was then determined.
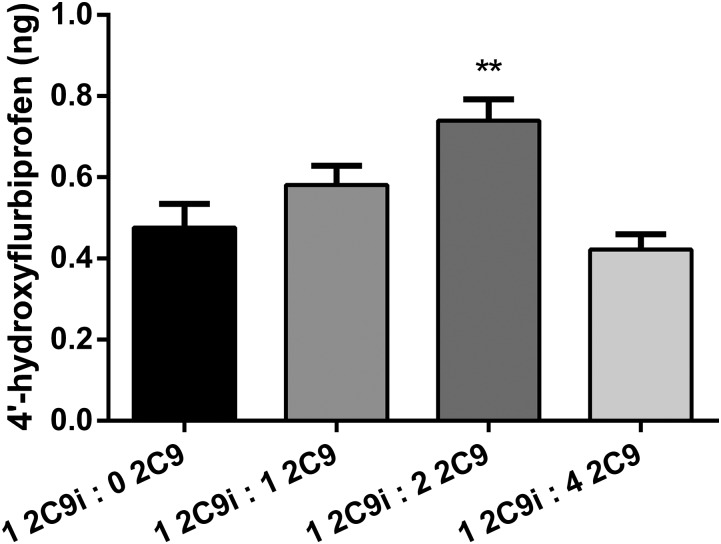
Figure 3 shows the amounts of metabolite formed from CYP2C9i-mediated metabolism of flurbiprofen in the presence of three concentrations of lipidless CYP2C9 in solution at a total P450/CPR ratio of 1:4. Metabolism by lipidless CYP2C9 alone did not significantly differ from the negative control (data not shown); thus, metabolite formation is due entirely to CYP2C9i. CPR was added prior to the addition of lipidless CYP2C9, followed by the addition of NADPH. With a ratio of CYP2C9i/CYP2C9 of 1:2, significant activation of flurbiprofen metabolism was observed. CYP2C9i-mediated metabolism of flurbiprofen was not significantly different from the positive control at ratios above 1:2 (Fig. 3).
Fig. 3.
Effect of solution CYP2C9 on CYP2C9i-mediated metabolism of flurbiprofen at three ratios of 2C9i to lipidless CYP2C9 and saturating CPR [P450(total)/CPR, 1:4], incubated for 16 hours. A significant activation (P < 0.01) is seen at a 2C9i/2C9 ratio of 1:2. Single-factor analysis of variance was used for statistical comparisons. 2C9, CYP2C9; 2C9i, immobilized CYP2C9. **P < 0.01.
Effect of CYP3A4 and CYP2D6 on CYP2C9-Mediated Metabolism of Flurbiprofen.
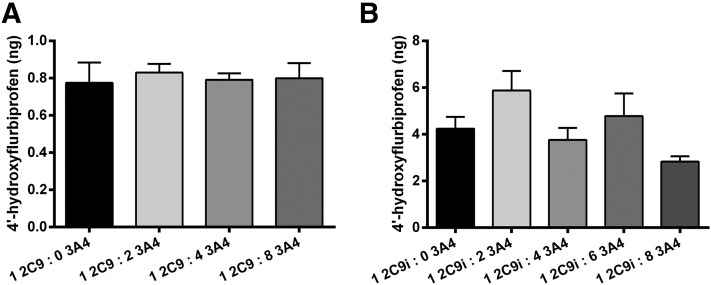
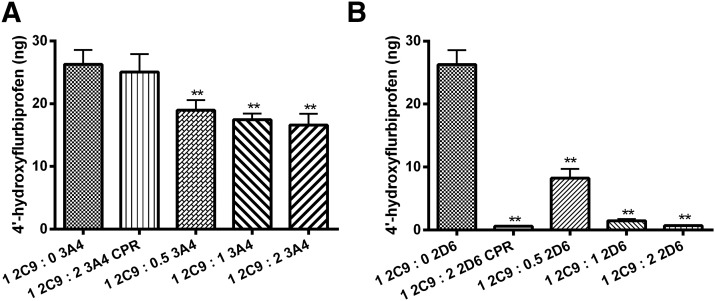
The effects of various amounts of CYP3A4 and CYP2D6 on CYP2C9i-mediated flurbiprofen metabolism are presented in Figs. 4 and 5. The presence of CYP3A4 had no effect on CYP2C9-mediated metabolism of flurbiprofen when CYP2C9 was in solution with lipid (Fig. 4A) or when CYP2C9 was immobilized (Fig. 4B) with P450(total)/CPR ratios of 1:8. In both cases, CPR was added prior to the addition of CYP3A4.
Fig. 4.
Effect of CYP3A4 on CYP2C9-mediated metabolism of flurbiprofen. (A) Metabolite formation in solution incubations containing CYP3A4 and CYP2C9, reconstituted in lipid, and with saturating CPR [P450(total)/CPR, 1:8], incubated for 16 hours. (B) Metabolite formation in incubations containing CYP3A4 (solution), CYP2C9i-mediated flurbiprofen metabolism at saturating CPR [P450(total)/CPR, 1:8], incubated for 16 hours. Single-factor analysis of variance was used for statistical comparisons. 2C9, CYP2C9; 2C9i, immobilized CYP2C9; 3A4, CYP3A4.
Fig. 5.
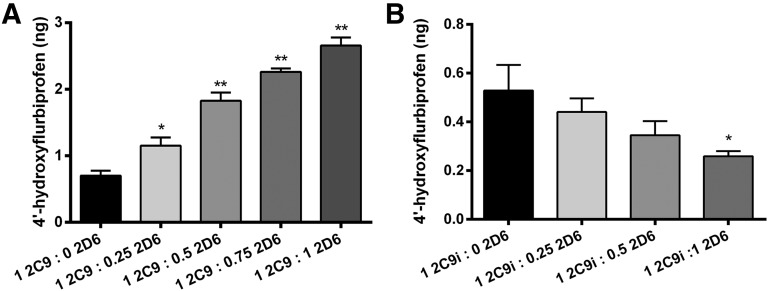
Effect of CYP2D6 on CYP2C9-mediated flurbiprofen metabolism. (A) Effect of CYP2D6 on solution metabolism of CYP2C9 reconstituted in lipid with saturating CPR [P450(total)/CPR, 1:8], incubated for 16 hours. Significant activation is observed at a 2C9/2D6 ratio of 1:0.25 (P < 0.05) and for 2C9/2D6 ratios higher than 1:0.25 (P < 0.01). (B) Effect of CYP2D6 on CYP2C9i-mediated flurbiprofen metabolism at saturating CPR [P450(total)/CPR, 1:8], incubated for 16 hours. Significant inhibition (P < 0.05) is seen for a 2C9i/2D6 ratio of 1:1. Single-factor analysis of variance was used for statistical comparisons. 2C9, CYP2C9; 2C9i, immobilized CYP2C9; 2D6, CYP2D6. *P < 0.05; **P < 0.01.
In contrast, the presence of CYP2D6 significantly affected CYP2C9-mediated metabolism of flurbiprofen, as suggested by the data shown in Fig. 5. In particular, in solution, CYP2C9-mediated metabolism of flurbiprofen was enhanced by the presence of CYP2D6 and in a concentration-dependent manner when CPR was present at saturating concentrations [P450(total)/CPR, 1:8] and added prior to the addition of CYP2D6 (Fig. 5A). This activation saturated at a CYP2C9/CYP2D6 ratio of 1:4 and increased metabolite production by approximately 400% (Supplemental Fig. 4). In contrast, CYP2C9i-mediated metabolism of flurbiprofen in the presence of CYP2D6 resulted in a concentration-dependent inhibition of CYP2C9-mediated metabolism of flurbiprofen (Fig. 5B). CYP2D6 inhibition of CYP2C9i metabolism saturated at 75% inhibition and did not change up to a ratio of CYP2C9i/CYP2D6 of 1:8 (Supplemental Fig. 5).
Influence of the Order of Addition of Enzymes on CYP2C9-Mediated Metabolism.
Figure 6 displays the amount of metabolite formed as a function of the order of addition of either CYP3A4 or CYP2D6 and CPR from CYP2C9-mediated metabolite production. Figure 6A shows that there is no effect of CYP3A4 on CYP2C9-mediated metabolism when CPR is added first and allowed to preincubate, which is in agreement with the results presented in Fig. 4A. In contrast, when CYP3A4 is allowed to preincubate with CYP2C9, a significant inhibition (up to 34%) of CYP2C9-mediated metabolism of flurbiprofen at a CYP2C9/CYP3A4 ratio of 1:2 is observed. The effect on metabolite production by CYP2D6 is depicted in Fig. 6B, which shows that the presence of CYP2D6 strongly inhibits CYP2C9 metabolism (up to 98%) regardless of the order in which the components are added prior to initiating the incubation. There is no significant difference between metabolite production at the 1:2 CYP2C9/CYP2D6 ratio when CPR is added before or after CYP2D6.
Fig. 6.
Order of addition of CPR and CYP3A4 or CYP2D6 effect on CYP2C9-mediated flurbiprofen metabolism. (A) Effect of CYP3A4 on solution metabolism of CYP2C9 reconstituted in lipid-lowered CPR concentration [P450(total)/CPR, 1:1.5], incubated for 1 hour. The column labeled “1 2C9 : 2 3A4 CPR” represents preincubation of CYP2C9 and CPR for 5 minutes prior to CYP3A4 addition. All other columns represent CYP3A4 preincubation for 5 minutes prior to addition of CPR. A significant inhibition (P < 0.01) is seen at all 2C9/3A4 ratios when CYP3A4 is preincubated but not when CPR is preincubated. (B) Effect of CYP2D6 on solution metabolism of CYP2C9 reconstituted in lipid with lowered CPR concentration [P450(total)/CPR, 1:1.5], incubated for 1 hour. The column labeled “1 2C9 : 2 2D6 CPR” represents preincubation of CYP2C9 and CPR for 5 minutes prior to CYP2D6 addition. All other columns represent CYP2D6 preincubation for 5 minutes prior to addition of CPR. Significant inhibition (P < 0.01) is seen in the presence of CYP2D6. There was no significant difference between preincubating the CPR or CYP2D6 with CYP2C9 at a 2C9/2D6 ratio of 1:2. Single-factor analysis of variance was used for statistical comparisons. 2C9, CYP2C9; 2D6, CYP2D6; 3A4, CYP3A4. **P < 0.01.
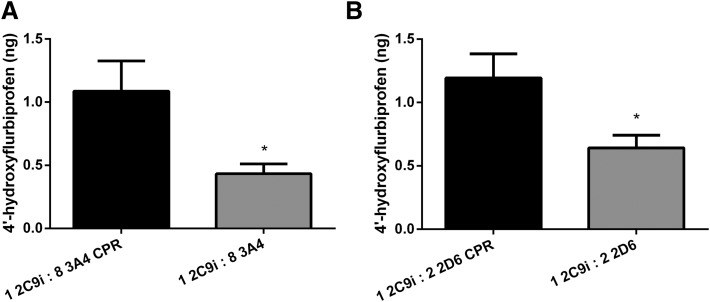
In comparison, Fig. 7 represents the same order-of-addition studies as mentioned earlier using CYP2C9i. Figure 7A shows that there is a significant decrease (up to 60%) of CYP2C9i-mediated metabolism when CYP3A4 is allowed to preincubate prior to the addition of CPR compared with when CYP2C9i is preincubated with CPR prior to the addition of CYP3A4. This result is in agreement with what is observed in order-of-addition studies of CYP2C9 and CYP3A4 in solution (Fig. 6A). When CYP2D6 was allowed to preincubate, we also saw a significant decrease (up to 46%) of CYP2C9i-mediated metabolism. This result is in contrast with solution studies that showed that order of addition had no effect on CYP2D6 inhibition of CYP2C9-mediated metabolism (Fig. 6B), and these findings may be attributable to a different binding conformation of CYP2C9-CYP2D6 between solution and immobilized studies.
Fig. 7.
Order of addition of CPR and CYP3A4 or CYP2D6 effect on CYP2C9i-mediated flurbiprofen metabolism. (A) Effect of CYP3A4 on CYP2C9i metabolism in incubations containing CYP3A4 (solution) at saturating CPR [P450(total)/CPR, 1:8], incubated for 16 hours. The column labeled “1 2C9i : 8 3A4 CPR” represents preincubation of CYP2C9 and CPR for 5 minutes prior to CYP3A4 addition. The column labeled “1 2C9i : 8 3A4” represents CYP3A4 preincubation for 5 minutes prior to addition of CPR. Preincubation of CYP3A4 results in a significant inhibition (P < 0.05) compared with CPR preincubation. (B) Effect of CYP2D6 on CYP2C9i metabolism in incubations containing CYP2D6 (solution) at saturating CPR [P450(total)/CPR, 1:8], incubated for 16 hours. The column labeled “1 2C9i : 2 2D6 CPR” represents preincubation of CYP2C9 and CPR for 5 minutes prior to CYP2D6 addition. The column labeled “1 2C9i : 2 2D6” represents CYP2D6 preincubation for 5 minutes prior to addition of CPR. Preincubation of CYP2D6 results in a significant inhibition (P < 0.05) compared with CPR preincubation. Single-factor analysis of variance was used for statistical comparisons. 2C9i, immobilized CYP2C9; 2D6, CYP2D6; 3A4, CYP3A4. *P < 0.05.
Discussion
Protein aggregation between P450s occurs in solution and can alter P450-mediated metabolism. Here, we show the formation of homomeric (CYP2C9-CYP2C9) and heteromeric (CYP2C9-CYP3A4, CYP2C9-CYP2D6, and CYP2C9-CPR) complexes occurs with different binding affinities. We reveal for the first time activation of CYP2C9 metabolism by homomeric dimerization. Furthermore, we show that controlling the aggregation state of CYP2C9 changes the effect that P450-P450 interactions have on metabolism. The immobilization used here offers an artificial mimic of in vivo P450 tethering (Gannett et al., 2006), which is of particular importance for more accurate insight on bound P450 function. Finally, we show that the order of addition and the concentration of CPR both modulate inhibition of CYP2C9-mediated metabolism by CYP2D6 and CYP3A4 in lipid-reconstituted and immobilized metabolism studies.
Computational modeling was used to predict dimer formation and probe how the complex may affect P450-P450 modulation of metabolism (Fig. 1). Docking indicated that dimer formation rarely involved residues necessary for CPR binding (Table 1), implying that the CPR binding site is not involved in modulating metabolism as a result of P450-P450 interactions. Instead, CYP2C9 metabolism is more likely to be affected due to dimerization occurring at the solvent channel for dimers with CYP2C9, CYP3A4, and CYP2D6, thus possibly changing the binding or turnover rate. This finding is in agreement with previous literature findings of dimerization occurring with residues near the F-G loop (Ozalp et al., 2006; Hu et al., 2010).
MM/GBSA calculations suggest that, in all cases, dimer formation is energetically favorable and is most favorable for CYP2C9-CPR (Table 2). However, none of the models resulting from blind docking between CYP2C9 and CPR involved the residues required for a functional CYP2C9-CPR complex. Others have also found that blind docking fails to generate a functional P450-CPR complex and have resorted to directed docking or homology modeling to achieve a functional complex (Hazai et al., 2005; Hamdane et al., 2009). Thus, the inability to achieve a correct model by docking may be related to the differences seen in the rank order between the modeling and in vitro studies (Table 3). This demonstrates that although molecular modeling can be a useful tool, results must be confirmed in vitro.
Recent work has demonstrated that P450-P450 interactions occur in membranes utilizing crosslinking (Hu et al., 2010; Reed et al., 2010; Davydov et al., 2015), coimmunoprecipitation (Alston et al., 1991; Subramanian et al., 2010), and fluorescent resonance energy transfer studies (Davydov et al., 2001; Szczesna-Skorupa et al., 2003; Ozalp et al., 2005). It is important to quantify the rates associated with these interactions and determine whether they may be involved in modulating P450 activity. Here, we used SPR (Ivanov et al., 2001) to measure binding of an analyte P450 to CYP2C9i in real time. The KD values determined for the binding of CPR to CYP2C9 are in line with reported values of 2.3 ± 1.0 nM (Wei et al., 2007) and 32.8 ± 0.2 nM (Locuson et al., 2007), demonstrating the validity and accuracy of the method. The high affinity binding that occurs between CYP2C9-CYP2C9 and CYP2C9-CYP2D6 is likely relevant to the results of metabolism assays, because they bind to one another more strongly than either does to CPR (Table 3). We also show that CYP3A4 forms a heteromeric complex with CYP2C9, but its KD (2.5 times that of CPR; Table 3) suggests that CYP3A4 competition for the CPR binding site is unlikely to factor into metabolism effects, although it is possible that CYP3A4 may alter CYP2C9’s affinity for CPR. Furthermore, since the SPR immobilization scheme is the same as used for the metabolism studies, correlation of complex formation with metabolic effects is valid.
P450 aggregation can affect metabolism by altering CPR affinity, substrate binding, or turnover rate (Reed and Backes, 2012). Altered CPR affinity can occur by blocking association of CPR, or through conformational changes that result from dimerization. Here, we use our controlled immobilization platform to show, for the first time, the activation of CYP2C9i metabolism through homomeric complex formation (Fig. 3). Although it is possible that dimerization of CYP2C9 restores activity to CYP2C9 that is in solution (lipid free), we would expect metabolite generation to saturate at the highest level if this is the case; yet a decrease, at ratios greater than 1:2, is seen (Fig. 3). We do note, however, that the observed decrease could be a result of higher-order assemblages of CYP2C9 in solution at higher concentrations preventing interaction with CYP2C9i. Further studies are necessary to definitively prove that the increased metabolism results from activation due to CYP2C9 dimerization. Given work by Hu et al. (2010) demonstrating that CYP2C8 exists as a dimer in membranes, we believe that it is possible that dimerization of CYP2C9 is necessary for the most catalytically active species. Our modeling data (Table 1) indicate that interaction with solvent access channels may implicate dimerization promoting substrate binding and exit, which could lead to higher substrate turnover rates (Stepankova et al., 2013).
In solution, it has been shown that interactions between CYP2C9 and CYP3A4 give a protein concentration-dependent inhibition of CYP2C9 metabolism if incubated with CYP3A4 prior to CPR addition (Subramanian et al., 2010). Here, we show that CYP2C9 preincubation with CPR prevents inhibition by CYP3A4 (Fig. 4) regardless of CPR concentration (Fig. 6A). Furthermore, the work by Subramanian et al. (2010) showed that the CYP2C9-CYP3A4 interaction could be eliminated by truncation of the N terminus of either binding partner. We show that if both P450s are truncated, interaction can still occur (Table 3) and this binding can result in decreased CYP2C9 metabolism if preincubated with CYP3A4 (Fig. 6A; Fig. 7A). The order of addition of protein partners has been shown to be important when evaluating the effects of P450-P450 interactions (Gorsky and Coon, 1986; Subramanian et al., 2009). The order-of-addition effect on CYP3A4 inhibition points to the formation of a quasi-irreversible heteromeric complex (Fig. 6A; Fig. 7A).
Studies have also shown there is a protein concentration-dependent inhibition of CYP2C9 metabolism when coincubated with CYP2D6 at subsaturating CPR concentrations (Subramanian et al., 2009). Remarkably, we note drastically different effects of CYP2D6 on CYP2C9 metabolism depending on study parameters. Our result of 94% inhibition of CYP2C9 in solution by CYP2D6 at a 1:1 ratio (Fig. 6B) is in reasonable agreement with the 50%–80% reported previously (Subramanian et al., 2009). The difference in maximum inhibition could possibly be due to slight differences in lipid/protein ratios between the two studies. However, we show that at saturating CPR concentrations, metabolism by CYP2C9 in solution is activated by CYP2D6 (Fig. 5A). Even more compelling is that when CYP2C9 is immobilized, CYP2D6 is once again inhibited despite an excess of CPR allowed to preincubate with the CYP2C9i (Fig. 5B).
One possible explanation for the solution metabolism results is a dual effect of CYP2D6 on CYP2C9 metabolism. If the CYP2D6-CYP2C9 complex has a faster substrate turnover rate, but lower CPR affinity, we would expect to see activation of metabolism when CPR is in excess and a faster turnover rate dominating the kinetics, but we would expect to see inhibition when the CPR concentration is lower and electron transfer becomes a limiting step. Another factor to consider is the difference between solution and immobilized platform results (Fig. 5). Immobilization of CYP2C9 controls its aggregation, with the majority being monomeric (Gannett et al., 2006; Bostick et al., 2015). Therefore, the inhibition of CYP2C9 metabolism could be a result of the lack of competition for CYP2C9 homomeric complex (CYP2C9-CYP2C9) formation with heteromeric complex (CYP2C9-CYP2D6) formation, evidenced by the similar KD values for both interactions (Table 3). On the immobilized platform, the binding modes and conformations may also be limited or otherwise different compared with solution and CYP2D6 may prevent CPR from associating with CYP2C9, thus limiting metabolism. In this case, the lack of oligomerization when immobilized leads to more catalytically inactive CYP2C9-CYP2D6 complexes. This agrees with the hypothesis of Davydov et al. (2015) that complexed P450 species bind and metabolize differently depending on the degree of oligomeriziation. Furthermore, in solution, the order of addition had no effect on CYP2D6 inhibition of CYP2C9 metabolism (Fig. 6B), but it did result in increased inhibition of CYP2C9i metabolism (Fig. 7B). This suggests, but does not conclusively prove, that CYP2D6 binds in a different conformation that results in a quasi-irreversible complex when CYP2C9 is immobilized. Although changes in CYP2C9 metabolism through complexation are clearly observed in our study, it is important to note that the mechanisms of activation (Fig. 3; Fig. 5A) and inhibition (Fig. 5B; Figs. 6 and 7) remain ambiguous since metabolite formation was nonlinear over the extended time course of the studies.
This work demonstrates the feasibility of using an immobilized platform to better understand P450-P450 interactions. We propose that the immobilization of CYP2C9 may offer a more complete functional assessment of CYP2C9 dimers, due to its bound nature, as seen in vivo, and control of aggregation state without detergents. Being able to immobilize an enzymatically active P450, allows study of both P450-P450 complex formation and the resultant metabolic modulation. We show that CYP2C9 forms both homomeric and heteromeric complexes and that these complexes may activate or inhibit CYP2C9 metabolism. Further study is necessary to fully analyze the conformation and activity of immobilized P450s to offer complete comparison of their functions to that of in vivo P450s. Future studies using this platform may further elucidate the intricacies of P450-P450 interactions and can be expanded to probe whether the effects are due to changes in CPR affinity or substrate turnover.
Supplementary Material
Acknowledgments
The authors thank A.R. Biundo and E.L. Dolan for their assistance in running experiments. They also acknowledge the West Virginia University Shared Research Facilities for support in completing this work.
Abbreviations
- CPR
cytochrome P450 reductase
- DLPC
dilauroylphosphatidylcholine
- EDC
N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride
- GBSA
generalized born surface area, MM, molecular mechanics
- MUA
11-mercaptoundecanoic acid
- NHS
N-hydroxysulfosuccinimide sodium salt
- OT
1-octanethiol
- P450
cytochrome P450
- PBS
phosphate-buffered saline
- PDB
Protein Data Bank
- SAM
self-assembled monolayer
- SPR
surface plasmon resonance
Authorship Contributions
Participated in research design: Bostick, Hickey, Wollenberg, Gannett.
Conducted experiments: Bostick, Hickey, Wollenberg.
Contributed new reagents and analytic tools: Flora, Tracy.
Performed data analysis: Bostick, Hickey, Wollenberg.
Wrote or contributed to the writing of the manuscript: Bostick, Hickey, Wollenberg, Flora, Tracy, Gannett.
Footnotes
All authors are co-first authors.
This research was supported by West Virginia University via the National Science Foundation through the Experimental Program to Stimulate Competitive Research [Grant EPS-1003907]; the National Institutes of Health National Institute of General Medical Sciences [Grant R01-GM086891]; and the National Science Foundation Integrative Graduate Education and Research Traineeship Program [Grant DGE-1144676].
dx.doi.org/10.1124/dmd.115.067637.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Alston K, Robinson RC, Park SS, Gelboin HV, Friedman FK. (1991) Interactions among cytochromes P-450 in the endoplasmic reticulum. Detection of chemically cross-linked complexes with monoclonal antibodies. J Biol Chem 266:735–739. [PubMed] [Google Scholar]
- Backes WL, Batie CJ, Cawley GF. (1998) Interactions among P450 enzymes when combined in reconstituted systems: formation of a 2B4-1A2 complex with a high affinity for NADPH-cytochrome P450 reductase. Biochemistry 37:12852–12859. [DOI] [PubMed] [Google Scholar]
- Backes WL, Kelley RW. (2003) Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol Ther 98:221–233. [DOI] [PubMed] [Google Scholar]
- Bostick CD, Flora DR, Gannett PM, Tracy TS, Lederman D. (2015) Nanoscale electron transport measurements of immobilized cytochrome P450 proteins. Nanotechnology 26:155102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley GF, Batie CJ, Backes WL. (1995) Substrate-dependent competition of different P450 isozymes for limiting NADPH-cytochrome P450 reductase. Biochemistry 34:1244–1247. [DOI] [PubMed] [Google Scholar]
- Cawley GF, Zhang S, Kelley RW, Backes WL. (2001) Evidence supporting the interaction of CYP2B4 and CYP1A2 in microsomal preparations. Drug Metab Dispos 29:1529–1534. [PubMed] [Google Scholar]
- Cheesman MJ, Baer BR, Zheng Y-M, Gillam EMJ, Rettie AE. (2003) Rabbit CYP4B1 engineered for high-level expression in Escherichia coli: ligand stabilization and processing of the N-terminus and heme prosthetic group. Arch Biochem Biophys 416:17–24. [DOI] [PubMed] [Google Scholar]
- Cojocaru V, Balali-Mood K, Sansom MSP, Wade RC. (2011) Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLoS Comput Biol 7:e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V, Winn PJ, Wade RC. (2007) The ins and outs of cytochrome P450s. Biochim Biophys Acta BBA 1770:390–401. [DOI] [PubMed] [Google Scholar]
- Davydov DR. (2011) Microsomal monooxygenase as a multienzyme system: the role of P450-P450 interactions. Expert Opin Drug Metab Toxicol 7:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov DR, Davydova NY, Sineva EV, Halpert JR. (2015) Interactions among cytochromes P450 in microsomal membranes: oligomerization of cytochromes P450 3A4, 3A5, and 2E1 and its functional consequences. J Biol Chem 290:3850–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov DR, Davydova NY, Sineva EV, Kufareva I, Halpert JR. (2013) Pivotal role of P450-P450 interactions in CYP3A4 allostery: the case of α-naphthoflavone. Biochem J 453:219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. (2005) Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: heterogeneity of the enzyme caused by its oligomerization. Biochemistry 44:13902–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov DR, Knyushko TV, Hoa GH. (1992) High pressure induced inactivation of ferrous cytochrome P-450 LM2 (IIB4) CO complex: evidence for the presence of two conformers in the oligomer. Biochem Biophys Res Commun 188:216–221. [DOI] [PubMed] [Google Scholar]
- Davydov DR, Petushkova NA, Bobrovnikova EV, Knyushko TV, Dansette P. (2001) Association of cytochromes P450 1A2 and 2B4: are the interactions between different P450 species involved in the control of the monooxygenase activity and coupling? Adv Exp Med Biol 500:335–338. [DOI] [PubMed] [Google Scholar]
- Davydov DR, Sineva EV, Sistla S, Davydova NY, Frank DJ, Sligar SG, Halpert JR. (2010) Electron transfer in the complex of membrane-bound human cytochrome P450 3A4 with the flavin domain of P450BM-3: the effect of oligomerization of the heme protein and intermittent modulation of the spin equilibrium. Biochim Biophys Acta 1797:378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton DR, McMillen SK, Sonderfan AJ, Thomas PE, Parkinson A. (1987) Studies on the rate-determining factor in testosterone hydroxylation by rat liver microsomal cytochrome P-450: evidence against cytochrome P-450 isozyme:isozyme interactions. Arch Biochem Biophys 255:316–328. [DOI] [PubMed] [Google Scholar]
- Eargle J, Wright D, Luthey-Schulten Z. (2006) Multiple alignment of protein structures and sequences for VMD. Bioinformatics 22:504–506. [DOI] [PubMed] [Google Scholar]
- Estabrook RW, Franklin MR, Cohen B, Shigamatzu A, Hildebrandt AG. (1971) Biochemical and genetic factors influencing drug metabolism. Influence of hepatic microsomal mixed function oxidation reactions on cellular metabolic control. Metabolism 20:187–199. [DOI] [PubMed] [Google Scholar]
- Eyer CS, Backes WL. (1992) Relationship between the rate of reductase-cytochrome P450 complex formation and the rate of first electron transfer. Arch Biochem Biophys 293:231–240. [DOI] [PubMed] [Google Scholar]
- Fantuzzi A, Mak LH, Capria E, Dodhia V, Panicco P, Collins S, Gilardi G. (2011) A new standardized electrochemical array for drug metabolic profiling with human cytochromes P450. Anal Chem 83:3831–3839. [DOI] [PubMed] [Google Scholar]
- Gannett PM, Kabulski J, Perez FA, Liu Z, Lederman D, Locuson CW, Ayscue RR, Thomsen NM, Tracy TS. (2006) Preparation, characterization, and substrate metabolism of gold-immobilized cytochrome P450 2C9. J Am Chem Soc 128:8374–8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsky LD, Coon MJ. (1986) Effects of conditions for reconstitution with cytochrome b5 on the formation of products in cytochrome P-450-catalyzed reactions. Drug Metab Dispos 14:89–96. [PubMed] [Google Scholar]
- Guengerich FP. (2006) Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J 8:E101–E111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP, Johnson WW. (1997) Kinetics of ferric cytochrome P450 reduction by NADPH-cytochrome P450 reductase: rapid reduction in the absence of substrate and variations among cytochrome P450 systems. Biochemistry 36:14741–14750. [DOI] [PubMed] [Google Scholar]
- Gut J, Richter C, Cherry RJ, Winterhalter KH, Kawato S. (1983) Rotation of cytochrome P-450. Complex formation of cytochrome P-450 with NADPH-cytochrome P-450 reductase in liposomes demonstrated by combining protein rotation with antibody-induced cross-linking. J Biol Chem 258:8588–8594. [PubMed] [Google Scholar]
- Hamdane D, Xia C, Im SC, Zhang H, Kim JJP, Waskell L. (2009) Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J Biol Chem 284:11374–11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazai E, Bikádi Z, Simonyi M, Kupfer D. (2005) Association of cytochrome P450 enzymes is a determining factor in their catalytic activity. J Comput Aided Mol Des 19:271–285. [DOI] [PubMed] [Google Scholar]
- Hazai E, Kupfer D. (2005) Interactions between CYP2C9 and CYP2C19 in reconstituted binary systems influence their catalytic activity: possible rationale for the inability of CYP2C19 to catalyze methoxychlor demethylation in human liver microsomes. Drug Metab Dispos 33:157–164. [DOI] [PubMed] [Google Scholar]
- Hou T, Wang J, Li Y, Wang W. (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Johnson EF, Kemper B. (2010) CYP2C8 exists as a dimer in natural membranes. Drug Metab Dispos 38:1976–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel MA, Locuson CW, Gannett PM, Rock DA, Mosher CM, Rettie AE, Tracy TS. (2005) CYP2C9 genotype-dependent effects on in vitro drug-drug interactions: switching of benzbromarone effect from inhibition to activation in the CYP2C9.3 variant. Mol Pharmacol 68:644–651. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- Ivanov YD, Kanaeva IP, Karuzina II, Archakov AI, Hoa GHB, Sligar SG. (2001) Molecular recognition in the p450cam monooxygenase system: direct monitoring of protein-protein interactions by using optical biosensor. Arch Biochem Biophys 391:255–264. [DOI] [PubMed] [Google Scholar]
- Jamakhandi AP, Kuzmic P, Sanders DE, Miller GP. (2007) Global analysis of protein-protein interactions reveals multiple cytochrome P450 2E1–reductase complexes. Biochemistry (Mosc) 46:10192–10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungermann K. (1995) Zonation of metabolism and gene expression in liver. Histochem Cell Biol 103:81–91. [DOI] [PubMed] [Google Scholar]
- Kaminsky LS, Guengerich FP. (1985) Cytochrome P-450 isozyme/isozyme functional interactions and NADPH-cytochrome P-450 reductase concentrations as factors in microsomal metabolism of warfarin. Eur J Biochem 149:479–489. [DOI] [PubMed] [Google Scholar]
- Karlsson R, Michaelsson A, Mattsson L. (1991) Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods 145:229–240. [DOI] [PubMed] [Google Scholar]
- Kawato S, Gut J, Cherry RJ, Winterhalter KH, Richter C. (1982) Rotation of cytochrome P-450. I. Investigations of protein-protein interactions of cytochrome P-450 in phospholipid vesicles and liver microsomes. J Biol Chem 257:7023–7029. [PubMed] [Google Scholar]
- Kelley RW, Cheng D, Backes WL. (2006) Heteromeric complex formation between CYP2E1 and CYP1A2: evidence for the involvement of electrostatic interactions. Biochemistry 45:15807–15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DN, Pritchard MP, Hanlon SP, Burchell B, Wolf CR, Friedberg T. (1999) Competition between cytochrome P-450 isozymes for NADPH-cytochrome P-450 oxidoreductase affects drug metabolism. J Pharmacol Exp Ther 289:661–667. [PubMed] [Google Scholar]
- Liu J, Tawa GJ, Wallqvist A. (2013) Identifying cytochrome P450 functional networks and their allosteric regulatory elements. PLoS One 8:e81980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locuson CW, Gannett PM, Tracy TS. (2006) Heteroactivator effects on the coupling and spin state equilibrium of CYP2C9. Arch Biochem Biophys 449:115–129. [DOI] [PubMed] [Google Scholar]
- Locuson CW, Wienkers LC, Jones JP, Tracy TS. (2007) CYP2C9 protein interactions with cytochrome b(5): effects on the coupling of catalysis. Drug Metab Dispos 35:1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak LH, Sadeghi SJ, Fantuzzi A, Gilardi G. (2010) Control of human cytochrome P450 2E1 electrocatalytic response as a result of unique orientation on gold electrodes. Anal Chem 82:5357–5362. [DOI] [PubMed] [Google Scholar]
- Matsuura S, Fujii-Kuriyama Y, Tashiro Y. (1978) Immunoelectron microscope localization of cytochrome P-450 on microsomes and other membrane structures of rat hepatocytes. J Cell Biol 78:503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijerman I, Sanderson LM, Smits PHM, Beijnen JH, Schellens JHM. (2007) Pharmacogenetic screening of the gene deletion and duplications of CYP2D6. Drug Metab Rev 39:45–60. [DOI] [PubMed] [Google Scholar]
- Miller BR, 3rd, McGee TD, Jr, Swails JM, Homeyer N, Gohlke H, Roitberg AE. (2012) MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput 8:3314–3321. [DOI] [PubMed] [Google Scholar]
- Myszka DG. (1999) Survey of the 1998 optical biosensor literature. J Mol Recognit 12:390–408. [DOI] [PubMed] [Google Scholar]
- Ozalp C, Szczesna-Skorupa E, Kemper B. (2005) Bimolecular fluorescence complementation analysis of cytochrome P450 2c2, 2e1, and NADPH-cytochrome P450 reductase molecular interactions in living cells. Drug Metab Dispos 33:1382–1390. [DOI] [PubMed] [Google Scholar]
- Ozalp C, Szczesna-Skorupa E, Kemper B. (2006) Identification of membrane-contacting loops of the catalytic domain of cytochrome P450 2C2 by tryptophan fluorescence scanning. Biochemistry 45:4629–4637. [DOI] [PubMed] [Google Scholar]
- Panicco P, Dodhia VR, Fantuzzi A, Gilardi G. (2011) Enzyme-based amperometric platform to determine the polymorphic response in drug metabolism by cytochromes P450. Anal Chem 83:2179–2186. [DOI] [PubMed] [Google Scholar]
- Peterson JA, Ebel RE, O’Keeffe DH, Matsubara T, Estabrook RW. (1976) Temperature dependence of cytochrome P-450 reduction. A model for NADPH-cytochrome P-450 reductase:cytochrome P-450 interaction. J Biol Chem 251:4010–4016. [PubMed] [Google Scholar]
- Reed JR, Backes WL. (2012) Formation of P450 · P450 complexes and their effect on P450 function. Pharmacol Ther 133:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Cawley GF, Backes WL. (2011) Inhibition of cytochrome P450 1A2-mediated metabolism and production of reactive oxygen species by heme oxygenase-1 in rat liver microsomes. Drug Metab Lett 5:6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Cawley GF, Backes WL. (2013) Interactions between cytochromes P450 2B4 (CYP2B4) and 1A2 (CYP1A2) lead to alterations in toluene disposition and P450 uncoupling. Biochemistry 52:4003–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Eyer M, Backes WL. (2010) Functional interactions between cytochromes P450 1A2 and 2B4 require both enzymes to reside in the same phospholipid vesicle: evidence for physical complex formation. J Biol Chem 285:8942–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AG, Cheesman MJ, Primak A, Bowman MK, Atkins WM, Rettie AE. (2010) Intramolecular heme ligation of the cytochrome P450 2C9 R108H mutant demonstrates pronounced conformational flexibility of the B-C loop region: implications for substrate binding. Biochemistry 49:8700–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe DR, Cheatham TE., 3rd (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9:3084–3095. [DOI] [PubMed] [Google Scholar]
- Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, et al. (2006) Crystal structure of human cytochrome P450 2D6. J Biol Chem 281:7614–7622. [DOI] [PubMed] [Google Scholar]
- Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. (2004) Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem 279:27294–27301. [DOI] [PubMed] [Google Scholar]
- Shen AL, Kasper CB. (1995) Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J Biol Chem 270:27475–27480. [DOI] [PubMed] [Google Scholar]
- Shimada T, Mernaugh RL, Guengerich FP. (2005) Interactions of mammalian cytochrome P450, NADPH-cytochrome P450 reductase, and cytochrome b(5) enzymes. Arch Biochem Biophys 435:207–216. [DOI] [PubMed] [Google Scholar]
- Stepankova V, Khabiri M, Brezovsky J, Pavelka A, Sykora J, Amaro M, Minofar B, Prokop Z, Hof M, Ettrich R, et al. (2013) Expansion of access tunnels and active-site cavities influence activity of haloalkane dehalogenases in organic cosolvents. ChemBioChem 14:890–897. [DOI] [PubMed] [Google Scholar]
- Subramanian M, Low M, Locuson CW, Tracy TS. (2009) CYP2D6-CYP2C9 protein-protein interactions and isoform-selective effects on substrate binding and catalysis. Drug Metab Dispos 37:1682–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Tam H, Zheng H, Tracy TS. (2010) CYP2C9-CYP3A4 protein-protein interactions: role of the hydrophobic N terminus. Drug Metab Dispos 38:1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Kemper B. (1993) An N-terminal glycosylation signal on cytochrome P450 is restricted to the endoplasmic reticulum in a luminal orientation. J Biol Chem 268:1757–1762. [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Mallah B, Kemper B. (2003) Fluorescence resonance energy transfer analysis of cytochromes P450 2C2 and 2E1 molecular interactions in living cells. J Biol Chem 278:31269–31276. [DOI] [PubMed] [Google Scholar]
- Tan Y, Patten CJ, Smith T, Yang CS. (1997) Competitive interactions between cytochromes P450 2A6 and 2E1 for NADPH-cytochrome P450 oxidoreductase in the microsomal membranes produced by a baculovirus expression system. Arch Biochem Biophys 342:82–91. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Imai Y, Iyanagi T, Sato R. (1979) Interaction between NADPH-cytochrome P-450 reductase and cytochrome P-450 in the membrane of phosphatidylcholine vesicles. Biochim Biophys Acta 550:341–356. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA. (2006) GRAMM-X public web server for protein-protein docking. Nucleic Acids Res 34:W310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Locuson CW, Tracy TS. (2007) Polymorphic variants of CYP2C9: mechanisms involved in reduced catalytic activity. Mol Pharmacol 72:1280–1288. [DOI] [PubMed] [Google Scholar]
- Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. (2004) The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-A resolution. J Biol Chem 279:35630–35637. [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. (2000) Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell 5:121–131. [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Ward A, Angove HC, Matak Vinković D, Jhoti H. (2003) Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature 424:464–468. [DOI] [PubMed] [Google Scholar]
- Wollenberg LA, Jett JE, Wu Y, Flora DR, Wu N, Tracy TS, Gannett PM. (2012) Selective filling of nanowells in nanowell arrays fabricated using polystyrene nanosphere lithography with cytochrome P450 enzymes. Nanotechnology 23:385101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Gillam EMJ, Dong M-S, Johnson WW, Guengerich FP, Shimada T. (1997) Reconstitution of recombinant cytochrome P450 2C10(2C9) and comparison with cytochrome P450 3A4 and other forms: effects of cytochrome P450-P450 and cytochrome P450-b5 interactions. Arch Biochem Biophys 342:329–337. [DOI] [PubMed] [Google Scholar]
- Yang M, Kabulski JL, Wollenberg L, Chen X, Subramanian M, Tracy TS, Lederman D, Gannett PM, Wu N. (2009) Electrocatalytic drug metabolism by CYP2C9 bonded to a self-assembled monolayer-modified electrode. Drug Metab Dispos 37:892–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem 279:38091–38094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.