Abstract
Cellulose synthase, encoded by the cesA gene, is responsible for the synthesis of cellulose in nature. We show that the cell wall of the cyanobacterium Synechococcus sp. PCC 7002 naturally contains cellulose. Cellulose occurs as a possibly laminated layer between the inner and outer membrane, as well as being an important component of the extracellular glycocalyx in this cyanobacterium. Overexpression of six genes, cmc–ccp–cesAB–cesC–cesD–bgl, from Gluconacetobacter xylinus in Synechococcus sp. PCC 7002 resulted in very high-yield production of extracellular type-I cellulose. High-level cellulose production only occurred when the native cesA gene was inactivated and when cells were grown at low salinity. This system provides a method for the production of lignin-free cellulose from sunlight and CO2 for biofuel production and other biotechnological applications.
Keywords: synthetic biology, photosynthesis, cellulose, cell wall, bioethanol
Introduction
Cellulose is the most abundant biopolymer in nature. It accounts for about half of the CO2 fixed through photosynthesis and is critically important to our society because of its uses in the wood, paper, and fiber industries. Cellulose utilization as a biofuel is increasing rapidly [1–4], and it also has important uses in biomedical applications [5]. Cellulose is a long-chain polymer of D-glucose molecules joined by β-1,4-glycosidic bonds, and it is an important structural component of the cell walls of higher plants, algae, fungi, and some bacteria [6]. Cellulose forms crystalline microfibrils that group to form bundles, which in turn form ‘ribbons’. A pellicle of cellulose is formed from a random assemblage of fibrils (<130 nm wide), which are composed of a bundle of much finer microfibrils (2–4 nm in diameter). Bacterial cellulose is distinguished from plant cellulose by having a high crystallinity index (>60%). There are two common forms of crystalline cellulose, denoted type I and II, which can be distinguished by X-ray diffraction and other spectroscopic methods. Type-I cellulose is formed from laterally and unidirectionally aligned, parallel β-1,4-glucan chains; the glucan chains in type-II cellulose are arranged in an antiparallel manner [6].
Cellulose is synthesized by cellulose synthases [7–10], which form terminal complexes and are composed of at least five polypeptides (BcsABCD and Ccp) in the bacterium Gluconacetobacter xylinus [11, 12]. The genes encoding cellulose synthases, variously denoted as ces, acs, or bcs, and related proteins are often organized in operons (for example, cmc–ccp–cesAB–cesC–cesD–bgl for G. xylinus) [13–15]. Terminal complexes span the inner and outer membranes and are usually organized in rows along the longitudinal axis of the cell [16, 17]. In Rhodobacter sphaeroides a BcsA–BcsB heterodimer catalyzes and regulates cellulose synthesis, and the structure of this complex was recently determined at 3.25-Å resolution together with a translocating glycan chain [18]. The glycosyltransferase domain and active site occurs in the cytoplasm, and the membrane-intrinsic portion forms a cellulose-conducting channel. BcsA contributes eight transmembrane α-helices to this channel, whereas the periplasmic protein BcsB contributes one transmembrane α-helix that anchors this protein to the membrane. The structure suggests a model in which cellulose is synthesized at the cytoplasmically localized active site and translocated through the cytoplasmic membrane one glucose residue at a time [18]. The cellulose-synthesizing activity of BcsA is strongly stimulated by cyclic-di-GMP, which binds to a PilZ domain that occurs at the C terminus of BcsA and which lies close to the glycosyltransferase active site [18].
In G. xylinus, CesC and CesD are involved in cellulose secretion and crystallization, which may be the rate-limiting steps of cellulose synthesis [11–15, 18, 19]. Additional proteins, including periplasmic endoglucanases, also have roles in cellulose synthesis and secretion [9, 12, 14, 20, 21]. Higher plant genomes encode multiple copies of cesA, but no homologs of the other bacterial ces genes and related proteins have been identified [10, 22]. Several cyanobacteria, which are oxygenic photoautotrophs like plants, have cesA genes encoding proteins with sequence similarity to bacterial and plant cellulose synthases, but similar to plants, no other ces/bcs genes have been identified to date in cyanobacterial genomes [23]. Cellulose microfibers, which may have a role in cellular aggregation, have been reported in cyanobacteria [23, 24]. However, little biochemical and genetic evidence directly connects cyanobacterial CesA with cellulose synthesis.
To characterize CesA function in the euryhaline cyanobacterium Synechococcus sp. PCC 7002 (hereafter Synechococcus 7002) and to explore the potential of cyanobacteria for cellulose production for biotechnological applications, we constructed and characterized a cesA (SynPCC7002_A2118) null mutant. We report here that cellulose is an important component of the cell wall and glycocalyx of Synechococcus 7002. We additionally show that high-level production of type-I cellulose can be achieved in Synechococcus 7002 by heterologous overexpression of a six-gene cassette from G. xylinus under appropriate environmental conditions.
Results
Cellulose layer in the cell wall of Synechococcus 7002
The genome of Synechococcus 7002 encodes an open-reading frame, SynPCC7002_A2118, that predicts a protein with strong sequence similarity to previously characterized cesA gene products, which have been shown to be cellulose synthases. SynPCC7002_A2118 predicts a protein of 788 amino acids, which is slightly larger than the characterized cellulose synthase of Nostoc sp. PCC 7120 (693 amino acids; 54% identical, 70% similar) [23] because of an insertion of ~90 amino acids. However, this CesA is identical in size to the BcsA protein of R. sphaeroides, whose X-ray structure is known [18] and is ~30% identical and ~45% similar in sequence. Similar proteins were found in about 20 other cyanobacterial genomes, as well as in the genomes of numerous bacteria known to synthesize bacterial cellulose. Many of the cyanobacteria are soil, benthic, or colonial organisms, in which cellulose might have a structural role in mat or colony formation. Interestingly, the mat-forming organisms Chloroflexus spp. and Roseiflexus spp. (phylum Chloroflexi), as well as the purple non-sulfur bacterium, R. sphaeroides (phylum Proteobacteria), are other commonly studied chlorophototrophic bacteria that have cesA/celA/bcsA genes and thus presumably have the ability to synthesize cellulose. However, most purple bacteria do not have a gene for cellulose synthase and thus cannot synthesize cellulose.
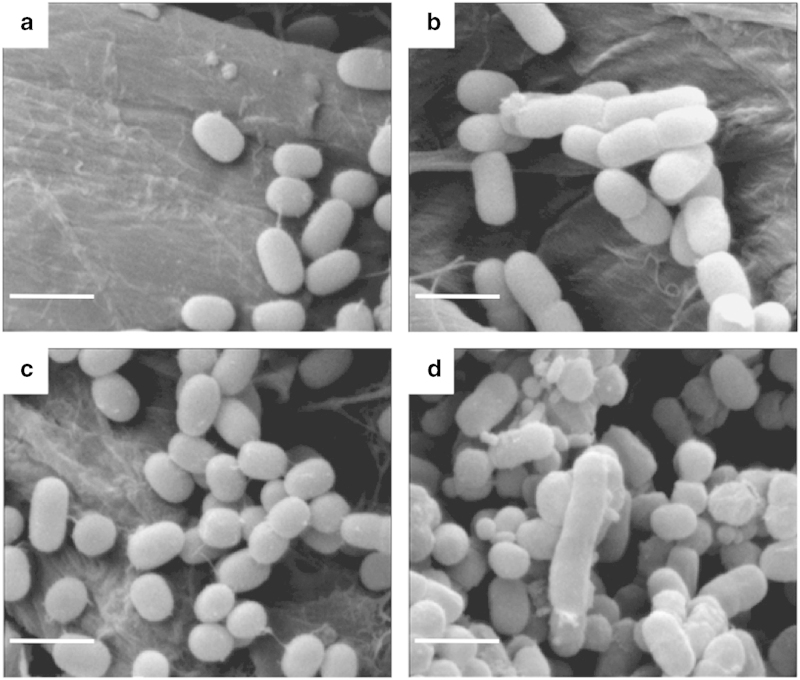
To study the function of CesA in Synechococcus 7002, we constructed a cesA null mutant and compared its properties with those of the wild type (WT) under different growth conditions. The WT grew equally well (doubling time=4.5±0.2 h) at low salinity (that is, in medium BG-11) or in a high salinity, artificial seawater medium (medium A+) [25]. The doubling time for the cesA mutant was the same as the WT in medium A+; however, the cesA mutant grew much more slowly (doubling time=9.8±0.4 h) than the WT at low salinity. Scanning electron microscopy showed that the cesA mutant formed numerous elongated cells and minicells when grown at low salinity (Figure 1d), which accounted for 5% (±0.4%) and 23% (±1.4%) of total cell numbers, respectively. These cells were not observed for WT (Figures 1a and b) or when the cesA mutant was grown at higher salinity (Figure 1c).
Figure 1.
Scanning electron microscopy (SEM) study of cell morphology of wild type (a, b) and cesA mutant (c, d) strains of Synechococcus 7002 grown in A+ medium (a, c) or BG-11 medium (b, d). Scale bar, 2 μm.
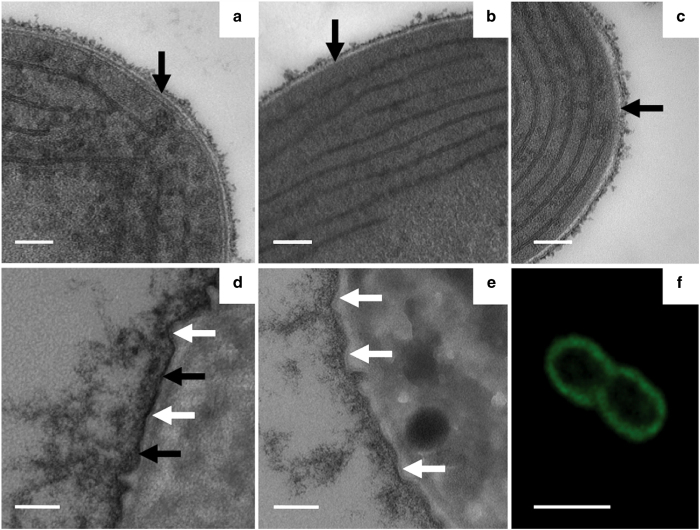
Cyanobacteria have a Gram-negative cell wall, and previous studies had suggested that their peptidoglycan (PG) layer is thicker than that of most other Gram-negative bacteria [26]. We used transmission electron microscopy to study the cell walls of Synechococcus 7002 strains. A cell wall layer between the cytoplasmic and outer membranes was clearly observed in thin sections of the WT (Figure 2a), but this layer was absent in the cesA mutant (Figure 2b). This layer also disappeared when WT cells were incubated with cellulase in the presence of 2 mM EDTA (Figure 2c), which suggested that this wall contained cellulose whose synthesis was dependent on the presence of functional CesA. This suggestion is further supported by the finding that the periplasm and cell surface could specifically be labeled with a cellulose-binding domain protein detected by immunogold labeling (Figure 3). No labeling was observed when cesA mutant cells were subjected to the same treatment (data not shown). When ultrathin sections of WT cells were stained with ruthenium red [27], this layer was visible as a distinct layer (Figure 2d), which we interpret to be laminated, but this layer was not observed in cell walls of a cesA mutant (Figure 2e). A careful analysis of these images suggested that this layer was located between the PG layer (Figure 2d, white arrows) and the outer membrane. Ruthenium red staining also revealed polysaccharides on the outer surface of the outer membrane in WT cells (Figure 2d) that were visibly much less abundant in the cesA mutant (Figure 2e). The distinctive cellulose layer in the cell wall was fully restored when the cesA mutant was complemented in trans with cesA (Figures 4a and c). When the yfp gene, encoding the yellow fluorescent protein, was fused to the cesA gene and transformed into a cesA mutant of Synechococcus 7002, the fusion gene also restored the cell wall structure to WT, although the cells obviously had much more surface-associated cellulose (note the intense ruthenium red staining in the periplasm as well as the extracellular glycocalyx in Figure 4c). The CesA–YFP fusion protein was localized on the cytoplasmic membranes by confocal microscopy (Figure 2f). Little or no fluorescence from YFP was observed in the cytoplasm in these confocal images, although fluorescence from chlorophyll a in the cytoplasm was readily apparent in the central regions of cells (data not shown). Consistent with our interpretation of the localization of YFP, the fluorescence from chlorophyll a did not overlap with that of YFP in merged images (data not shown). The uniform, non-punctate distribution of CesA–YFP on the cytoplasmic membrane, together with the fact that no other ces genes have been identified in this cyanobacterium, suggest that CesA may not be organized into terminal complexes in Synechococcus 7002.
Figure 2.
Transmission electron microscopy (TEM) study of cell walls in thin sections of Synechococcus 7002. (a) Wild type (WT). (b) cesA mutant. (c) WT cells incubated in a cellulase solution (200 μg/ml in the presence of 2 mM EDTA) for 30 min at 37 °C. The arrow in a indicates the electron-dense layer, and the arrows in b and c indicate its absence. Note that there is more cell-surface-associated glycocalyx material in a than in b or c. (d, e) TEM images of thin sections of Synechococcus 7002 cells stained with ruthenium red showing the changes in the cell wall and glycocalyx of WT (d) and a cesA deletion mutant (e), respectively. The black arrows indicate the possibly laminated, CesA-dependent cellulose layer, and the white arrows indicate the thin peptidoglycan layer. Although it was easier to see on lower-magnification images (data not shown), these images show less ruthenium red staining for the cesA mutant (e), more staining for the WT (d), and much more staining for a strain overproducing CesA (see Figure 4c). These data show that the cesA mutant had the least surface-associated polysaccharides and that the strain overproducing CesA had the most. An electron-opaque, laminated layer near the outer membrane in WT (black arrows in d) was missing in the cesA mutant (e). This layer apparently corresponds to the cellulose layer observed between the peptidoglycan (white arrows in d and e) and outer membrane layers in thin sections in a. (f) Confocal fluorescence microscopy showing spectrally deconvoluted, Z-stack image of Synechococcus 7002 cells expressing CesA–YFP. This image shows the absence of YFP fluorescence in the cytoplasm and thylakoid membranes, which could easily be visualized by fluorescence from chlorophyll a (data not shown). Scale bars, 100 nm (a-e) and 2 μm (f).
Figure 3.
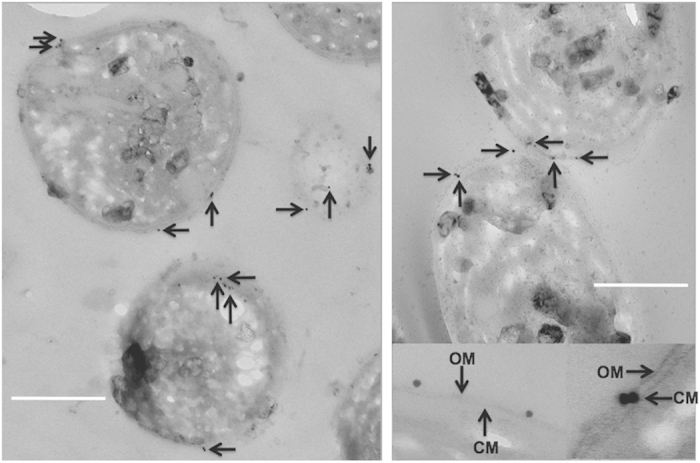
Immunolocalization of cellulose in cesA mutant complemented with cesA–yfp expressed from the P cpcAB promoter of Synechocystis sp. PCC 6803 (see Materials and Methods for details). The recombinant fusion protein rCBDs-Protein L (Sigma) was used to bind to cellulose selectively, and the cellulose-binding protein was detected using goat anti-mouse IgG conjugated to 10-nm colloidal gold particles (Sigma, St Louis, MO, USA) as the second antibody. Gold particles (arrows) were predominantly observed directly over the periplasmic space and at the peripheral surface of the outer membrane (OM) of the cell wall (associated with the glycocalyx). The insets at the lower right portion of the right panel show enlargements of gold particles associated with the periplasmic space and the glycocalyx just beyond the OM. The arrows in the inset show the OM and the cytoplasmic membrane (CM). Scale bar, 500 nm.
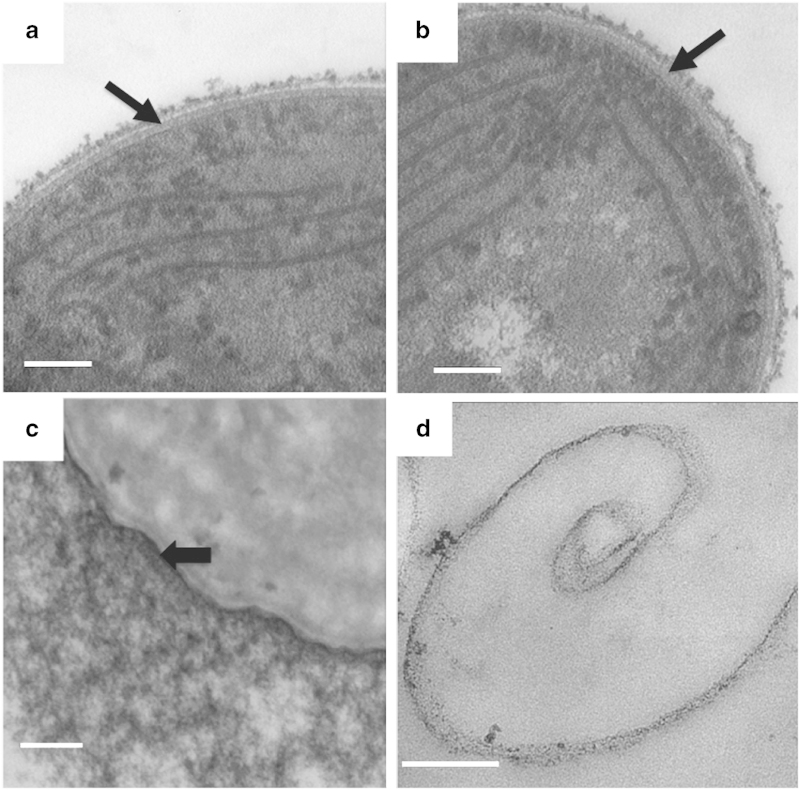
Figure 4.
Transmission electron microscopy (TEM) analysis of thin sections of a cesA insertion mutant that was complemented with a wild-type cesA gene (a) and bacterial artificial chromosome clone 38 (b) (see main text for details), respectively. The black arrows indicate the cellulose layer that has been restored in both cases. (c) Thin-section electron micrograph after ruthernium red staining of the cesA deletion mutant of Synechococcus 7002 complemented with cesA::yfp under the control of the P cpcBA promoter of Synechocystis sp. PCC 6803. The black arrow indicates the possibly laminated cellulose layer has been restored by complementation with the cesA::yfp fusion gene. Note the increased amount of ruthenium-red-stainable polysaccharide on the external surface of the cell compared with wild type or the cesA mutant (compare with Figures 2d and e). (d) TEM image of isolated peptidoglycan from the cesA mutant. Scale bar, 100 nm.
Although it was difficult to observe the PG layer directly in cesA mutant cells by transmission electron microscopy (however, see white arrows in Figures 2d and e), the following evidence suggests that a PG layer was still present. The cesA mutant cells were viable, could still grow and divide, and were still sensitive to lysozyme treatment; moreover, a lysozyme-sensitive PG fraction could be isolated from the mutant cells (Figure 4d). Mass spectrometry showed that N-acetylglucosamine was present in the hydrolyzate from this fraction (Supplementary Figure S1). These results suggested that the cell wall layer between the outer membrane and plasma membrane in Synechococcus 7002 is composed of a thin inner PG layer and a thicker, possibly laminated, outer cellulose layer. A major component of this outer layer is cellulose produced by CesA (Figure 5). Cellulose, which was more abundant in a strain overproducing CesA (Figure 4c), was also apparently associated with the glycocalyx outside the outer membrane. Because at least 20 cyanobacterial genomes encode homologs of cesA, cellulose is probably a component of many but not all cyanobacterial cell walls.
Figure 5.
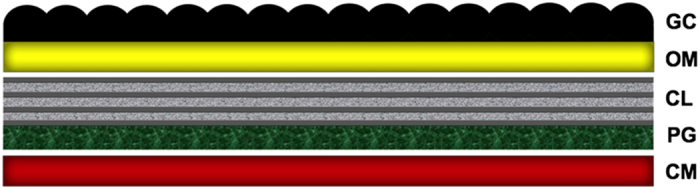
Schematic of cell wall structure of the cyanobacterium Synechococcus 7002. CL, possibly laminated layer with cellulose as a major component; CM, cytoplasmic membrane; GC, glycocalyx layer; OM, outer membrane; PG, peptidoglycan.
High-yield production of cellulose in Synechococcus 7002
In an attempt to overproduce extracellular cellulose in photoautotrophic Synechococcus 7002, we first tried to express the native cesA gene under a strong PcpcBA promoter from Synechocystis sp. PCC 6803. Although cells appeared to produce more surface-associated polysaccharides than WT (compare Figure 4c with Figures 2d and e), cellulose production did not increase markedly. Likewise, no significant extracellular cellulose production was observed when the cesAB–cesC–cesD operon from G. xylinus ATCC 53582 was inserted into the chromosome or the high-copy plasmid pAQ1 [28] in either the WT or cesA mutant strains of Synechococcus 7002. We next constructed a bacterial artificial chromosome (BAC) library of genomic DNA from G. xylinus using a BAC vector that can stably integrate foreign DNA into the Synechococcus 7002 chromosome. We transformed both the WT and cesA mutant with the BAC clones and screened for high-level production of cellulose among the transformants by staining with Calcofluor white, a dye that detects polymers with β-1,4-glycosidic bonds [29]. A positive strain (strain 56) was identified among transformants derived from the cesA mutant, and transmission electron microscopy revealed that the cellulose layer had been restored in the cell walls (Figure 4b). The corresponding BAC clone (clone 38) was isolated and sequenced, and the cloned fragment included the cesAB–cesC–cesD operon together with its flanking regions.
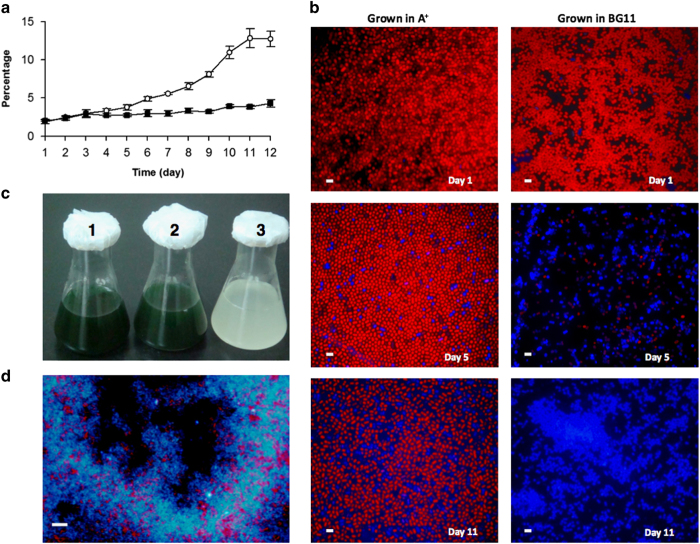
To improve the cellulose yield, various combinations of the ces operon and its flanking genes were inserted onto plasmid pAQ1 [28]. Transformants were tested under different growth conditions, and only one strain (CM12) in the cesA mutant background exhibited greatly enhanced cellulose production. Strain CM12 contained two upstream genes (cmc and ccp (cellulose complementing protein)) and one downstream gene (bgl) that flank the cesAB–cesC–cesD operon; this DNA fragment was named the cellulose synthesis module (CSM) (Supplementary Figure S2). Cellulose production by strain CM12 was strongly stimulated by transferring the cells from medium A+ to medium BG-11. Under the latter conditions, the total cellulose content reached ~14% of the cell dry weight within 12 days (Figure 6a). Fluorescence microscopy revealed that cells accumulating cellulose exhibited strong blue fluorescence when excited with ultraviolet light in the presence of Calcofluor white. The typical red fluorescence from the photosynthetic pigments was nearly completely obscured (Figure 6b), and the color of the culture gradually changed from blue-green to white (Figure 6c). The white cultures contained cell aggregates (Figure 6d), and the cells of such cultures eventually bleached and died (Supplementary Figures S3 and S4).
Figure 6.
Cellulose production by strain CM12 after introduction of the CSM genes into the cesA mutant strain of Synechococcus 7002. (a) Time course of cellulose production at high salinity (medium A+, filled circles) or low salinity (medium BG-11, open circles). Each point is the average of three measurements, and the bars show the s.e. Cellulose content was measured as glucose content and expressed as percentage of the dry weight of isolated cells (indicated as ‘percentage’ on the y axis of the figure). (b) Fluorescence microscopy of the Calcofluor-stained cells of strain CM12 grown at high salinity (medium A+, left panels) or low salinity (medium BG-11, right panels) for the times indicated. Fluorescence was excited with ultraviolet (UV) light. The red fluorescence emission emanates from photosynthetic pigments and the bright blue fluorescence emission comes from Calcofluor cellulose. Scale bar, 5 μm. (c) Batch cultures of the wild type (1) and strain CM12 at high salinity (medium A+) (2) or after transfer from high to BG-11 medium (low salinity) (3) for 2 weeks. The cultures (1) and (2) were both in stationary phase and their optical densities at 750 mm were ~2. (d) Fluorescence of the cells in flask (3) of c. Fluorescence in the absence of Calcofluor white was also excited with UV light. Scale bar, 20 μm. CSM, cellulose synthesis module.
The reason(s) for cell death is not clear, but it might be related to severe depletion of intracellular carbon reserves because of excessive cellulose synthesis. Alternatively, it might be due to light limitation imposed when cells were encapsulated in cellulose fibers. Because the cells bleach, it is also possible that hyperproduction of cellulose may trigger the formation of reactive oxygen species or damage the cell membranes. Surprisingly, WT cells transformed with the CSM produced very little cellulose even when grown in hyposaline medium. Thus, maximal cellulose production from the heterologous expression of the CSM genes by Synechococcus 7002 required that two conditions were satisfied: (i) the endogenous, native cesA gene had to be inactivated, and (ii) cells had to be grown under hyposaline conditions. Hyposaline growth conditions might allow cells to divert metabolic resources that would otherwise be used for the production of compatible solutes toward cellulose synthesis [30, 31]. An intriguing possibility is that hyposaline conditions might cause intracellular levels of cyclic-di-GMP levels to increase, which could stimulate CesAB activity by binding to its PilZ domain [32].
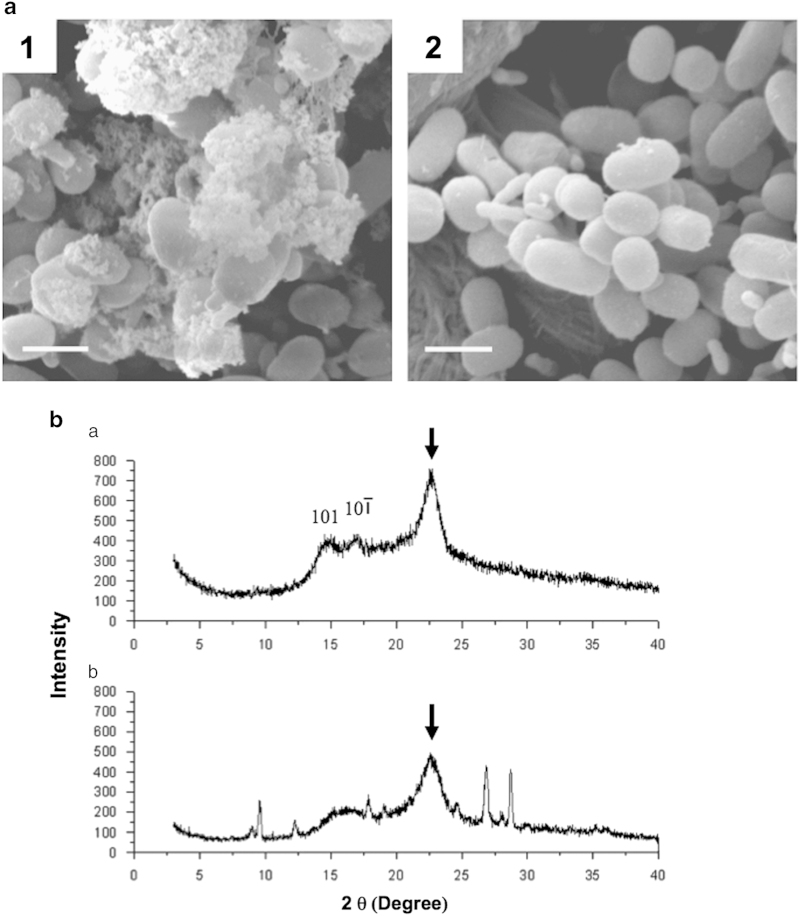
When cells of strain CM12 were examined by scanning electron microscopy, a very large amount of microfibrillar (μm size range) material was observed, which was absent after cells had been treated with cellulase (Figure 7a). This observation strongly implied that this fibrous material was cellulose. Normal cellular morphology was not fully restored in strain CM12, which still formed minicells. The fibrous extracellular material was isolated and analyzed by X-ray diffraction. Its X-ray diffraction pattern (Figure 7b) was very similar to the pattern obtained for type-I cellulose produced by G. xylinus [33, 34]. Although peaks 101, 10ī, and 040 were broadened, the peak at 23° corresponded well with diffraction peak 002 of type-I cellulose. This peak, and the overall similarities of the diffraction patterns, led us to conclude that, even though some impurities were present in the sample as indicated by the peaks at 27° and 28.5°, the isolated material contained type-I cellulose. The crystallinity index of the celluloses produced by G. xylinus and strain CM12 were calculated [35] and they were found to be 79% and 65%, respectively. Thus, more than half of the cellulose isolated from Synechococcus 7002 strain CM12 was in crystalline and/or microcrystalline form, but it contained less type-I cellulose than the cellulose fibers isolated from G. xylinus.
Figure 7.
Analysis of cellulose fibers produced by strain CM12 of Synechococcus 7002. (a) Scanning electron microscopy (SEM) analysis of extracellular cellulose fibers. Strain CM12 cells were grown in medium BG-11 for 12 days and were prepared for SEM examination without (1) or with cellulase treatment (2) for 30 min. Scale bar, 2 μm. (b) X-ray diffraction analysis of the cellulose isolated from G. xylinus (a) and from Synechococcus 7002 strain CM12 (b). Arrows indicate the position of peak 002.
Discussion
In this study we show that the cesA (SYNPCC7002_A2118) gene product is required for the production of a distinctive cell wall layer in Synechococcus 7002. This layer is most likely localized between the PG layer and the outer membrane and is possibly laminated. CesA also contributed to the production of polysaccharides associated with the glycocalyx. Three lines of evidence indicate that this polysaccharide is cellulose. First, Synechococcus 7002 CesA is required for the production of this polysaccharide, and this protein has strong sequence similarity to bacterial cellulose synthases as well as to CesA from Nostoc sp. PCC 7120, a cyanobacterium that has previously been shown to synthesize cellulose [23]. Second, in the presence of EDTA, some (but not all) of the extracellular glycocalyx as well as the periplasmic layer could be digested with cellulase, which indicates that the polysaccharides found in the digestible fraction contain β-1,4-glucosidic bonds. Third, the polysaccharide in the periplasmic space and the glycocalyx could be specifically labeled with a recombinant cellulose-binding domain (Figure 3) and was missing in a cesA mutant. Collectively, these data establish that the cell walls and glycocalyx of Synechococcus 7002 contain cellulose that is produced by the action of CesA.
To obtain very high levels of cellulose production in Synechococcus 7002, it was necessary to introduce six genes, cmc–ccp–cesAB–cesC–cesD–bgl, from G. xylinus into this cyanobacterium. Polymerization and crystallization are coupled in cellulose synthesis in G. xylinus and are thought to be the rate-liming steps in these processes [9, 36]. Because the strains with only the cesAB–cesC–cesD operon did not produce detectable extracellular cellulose, the high-yield production of extracellular crystalline cellulose in strain CM12 strongly suggested that the genes flanking the cesAB–cesC–cesD operon are essential for cellulose synthesis in Synechococcus 7002. Previous studies have shown that the products of the cmc (endo-β-1,4-glucanase) and ccp genes upstream of cesA and the downstream cesD and bgl genes are involved in cellulose production and/or crystallization [11, 12, 14, 20, 21, 37, 38]. When only the cesAB genes were introduced into a cyanobacterium, only amorphous cellulose was produced [39]. These observations further support the idea that genes other than those encoding the cellulose synthase (that is, other than cesAB/bcsAB) [18] are important for high-yield production of cellulose in bacteria. Therefore, the cluster of six genes (cmc–ccp–cesAB–cesC–cesD–bgl) from G. xylinus can be regarded as a complete CSM module for bacterial cellulose synthesis, even though we cannot completely rule out the possibility that some factors could be provided by Synechococcus 7002 for this process. For example, it is known that cyclic-di-GMP is required for bacterial cellulose synthesis [8]. The Synechococcus 7002 genome predicts three diguanylate cyclases and one enzyme for degradation of cyclic-di-GMP, and it is therefore highly likely that this cyanobacterium can produce the cyclic-di-GMP required to activate the cellulose synthase of G. xylinus. As noted above, it is possible that low-salinity growth conditions might activate the production of cylic-di-GMP in Synechococcus 7002, which could in turn activate the cellulose synthase via its PilZ domain.
The high-yield production of cellulose by cyanobacteria using only light and inorganic nutrients has potential uses for bioenergy as well as traditional industries that use cellulose. Assuming that cellulose produced by Synechococcus 7002 could be converted to ethanol at 70% efficiency, this system could yield ~4 500 gallons of ethanol per acre-ft per year if the biomass were harvested every 3 weeks at a final dry weight concentration of 20 mg/l. Even if cellulose production were only feasible for 6 months per year, Synechococcus 7002 would still yield sixfold more ethanol than corn, which typically yields 300–400 gallons of ethanol per acre per year [40]. A potentially interesting application of the findings reported here would be to couple cellulose production by Synechococcus 7002 to triglyceride synthesis in a cellulose-degrading, biodiesel-producing, eukaryotic alga such as Chlamydomonas reinhardtii [41]. Alternatively, large-scale cellulose synthesis could facilitate CO2 sequestration. Marine cyanobacteria and algae are widely believed to have important roles in the carbon cycle by removing of CO2 from the atmosphere [42, 43]. Because cellulose degradation occurs more slowly in the open ocean than in coastal and terrestrial areas [44], cellulose production and/or deposition in the open ocean could lead to carbon sequestration. Extracellular cellulose might promote fixed-carbon deposition by enhancing the formation of larger, more rapidly sedimenting cell aggregates. Finally, bacterial cellulose has shown significant potential for biomedical applications, especially for wound care and the regeneration of damaged organs [5].
Materials and Methods
Strains and culture conditions
Synechococcus sp. strain PCC 7002 WT, cesA mutant strains, and engineered strains were grown in a high-salinity (marine) A+ medium or low-salinity BG-11 medium (BG-11 used in this work was always added with vitamin B12 at a final concentration of 4 μg/l) [45, 46] with appropriate antibiotics when needed at 35 °C under cool-white fluorescent light at ~150 μmol/m2/s. The cultures were sparged with air plus 1% (v/v) CO2. Gluconacetobacter xylinus ATCC 53582 was cultured according to instructions provided by American Type Culture Collections (www.atcc.org). Escherichia coli strain DH5α was used for all routine recombinant DNA procedures.
Construction of cesA insertion and deletion mutant strains
Insertional inactivation (Supplementary Figure S5) of the cesA gene (SYNPCC7002_A2118) of Synechococccus sp. PCC 7002 (Synechococcus 7002) was performed as follows: a 4.37-kb DNA fragment containing cesA plus flanking 1-kb upstream and 1-kb downstream regions was amplified by PCR using total chromosomal DNA as template with TransStart FastPfu DNA polymerase (Transgen, Beijing, China).
The primers used for the PCR were 5′-CGATATTCACACGGCTATTGGCCCG-3′ and 5′-CCAGGGAAATTGCAACAGACGGAGC-3′. The amplified fragment was cloned into the pEASY-Blunt Simple Cloning Vector (Transgen). The resultant plasmid was then used as the PCR template with the primers 5′-CAATTGCTGTGTATACTTATCAACTTCGGCTTCT-3′ and 5′-ATCGATCTCCCCAGATGTCGCTACATTGCT-3′ to generate a blunt-ended DNA fragment, which was ligated to a 1.3-kb blunt-ended DNA fragment encoding the aphA-II gene (conferring resistance to kanamycin) and transformed into E. coli. The plasmid was then isolated and transformed into Synechococcus 7002 for construction of a cesA mutant.
Complete deletion of cesA in PCC 7002 was performed as previously described [47]. Flanking regions of cesA were amplified from Synechococcus sp PCC 7002 genomic DNA using the following primers: upstream region: 5′-TAGTCTGTAAACCTTCCGATGCATCTTCAGAGGATGTAGATGT-3′ and 5′-CGCATTAATAATGTATGCTAGCGGAGTGTGACTCGTTTGT-3′ and downstream region: 5′-TCAAATATTTTTCTTTTTGGTACCAATAATGGCCTTGATCTGG-3′ and 5′-GCAATCACCAATAACAAAGCATGCTCAAGAAGACAAACATAC-3′. Restriction sites are underlined. The two PCR products were digested with NsiI, NheI and KpnI, and SphI, respectively. A chloramphenicol resistance cassette was excised from pRL409 [48] with NheI and KpnI. The plasmid pGEM-7zf was digested with NsiI and SphI. The PCR products, antibiotic resistance cassette, and the pGEM-7zf were ligated to form a new plasmid in which the upstream and downstream flanking regions of cesA flanked the cat gene. This plasmid was digested with NsiI and used to transform Synechococcus 7002. Transformants carrying the cesA deletion mutation were selected on A+ medium supplemented with chloramphenicol (10 μg/ml). Complete segregation of the WT and cesA deletion alleles was verified by PCR analysis using primers cesA-For 5′-CTATGAAGGCCATACAAGAGAAATGCG-3′ and cesA-Rev 5′-TCAAGAATCACAAATTCCTTAGACATGAGC-3′.
Construction of cesA::yfp fusion gene
Overexpression of a CesA–YFP fusion protein in a cesA deletion mutant of Synechococcus 7002 was performed according to Xu et al. [28]. Full-length cesA was amplified from genomic DNA of Synechococcus 7002 with Platinum Pfx DNA polymerase (Invitrogen, Grand Island, NY, USA) using the following primers: forward: 5′-TATTCATTTTATTTATCACATATGTTGGAACTAGTCCCTTCATTTAAAAACAAACGAGTC-3′ and reverse: 5′-TTACTTGTCTCCTAAAACCATATGTTGTTCTTCTGGCTGCCAGTAAG-3′. The TAA stop codon of the cesA gene was changed to CAT in the reverse primer. The full-length cesA gene in this PCR product started and ended with 5′-CATATG-3′ (NdeI sites). After digestion with NdeI, the PCR product was inserted into the NdeI site of the plasmid pAQ1-EX-PcpcBA::yfp, to produce pAQ1-EX-PcpcBA::cesA-yfp (Supplementary Figure S6). Plasmids with the cesA insert were sequenced to determine the orientation of the cesA insertion and to verify the cesA sequence. One plasmid with correct orientation and sequence was used to transform the cesA deletion mutant of Synechococcus 7002. Selection for the complemented transformants was performed on medium A+ supplemented with 10 μg/ml chloramphenicol and 100 μg/ml spectinomycin. Transcription of cesA::yfp in complemented mutant was confirmed by reverse transcriptase-PCR.
Immunogold labeling
Immunogold labeling of thin sections was performed by a procedure similar to that described by Nobles et al. [23]. Cells were fixed 2% (w/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde in 0.1 M phosphate buffer (PB), pH 7.2, for 30 min and transferred to fresh fixative for another 3 h at room temperature. After washing three times with 0.1 M PB, cells were embedded in 0.5 ml of 3% (w/v) agarose dissolved in 0.1 M PB buffer at 40 °C. After cooling and solidification, the agarose was cut into ~1-mm3 blocks, which were washed and dehydrated in 50, 70, 90, and 100% (v/v) ethanol. The blocks were embedded in London Resin White and polymerized at 60 °C for 24 h. After ultrathin sectioning, the grids were washed with phosphate-buffered saline (PBS), pH 7.4, for 1 min and then incubated in 1:20 000 dilution of rCBD-Protein-L (Fluka Sigma-Aldrich, Buchs, Switzerland) in PBS at room temperature for 1 h. The grids were washed five times with PBS for 5 min and were blocked in 0.5% bovine serum albumin in PBS for 30 min. The grids were incubated in 1:50 goat anti-mouse IgG 10-nm gold conjugate (British Biocell International, Cardiff, UK) for 1 h at room temperature and were stained with 1% (w/v) glutaraldehyde for 5 min followed by washing with PBS three times and with water five times. The grids were stained with 1% (w/v) uranyl acetate for 5 min and washed with water five times. Grids were dried in air, and samples were observed with a JEM-1200EX electron microscope (JEM, Tokyo, Japan).
Construction of the BAC Library of G. xylinus
Construction of the G. xylinus BAC library was performed using the Epicentre CopyControl BAC Cloning Kit protocol (Epicentre, Madison, WI, USA, Cat. No. CCBAC1B), except that a 1.2-kb DNA fragment from Synechococcus 7002 and an erythromycin resistance cartridge was inserted to the ScaI site of the pCC1BAC Vector (Epicentre) so the resulting BAC plasmids could be integrated into the Synechococcus 7002 genome through a single recombinant event. The 1.2-kb DNA fragment from Synechococcus 7002 genome was amplified by PCR with primers 5′-CACGGTAGAATCCTCCGCCTGGGCC-3′ and 5′-TTGACTGCACCAATGGAGAAAC-3′. For cloning of a fragment encoding the six genes responsible for cellulose synthesis and extrusion from G. xylinus (cmc–ccp–cesAB–cesC–cesD–bgl, CSM), PCR was performed with the following primers: 5′-GAGATCTAGACGTTCCTTTATGTCGGTCATGGC-3′ and 5′-TATAGGATCCCCATACTCAGGGGGCCATGTT-3′. The fragment was then integrated into the pQ1 vector with the erythromycin resistance cartridge according to Xu et al. [28]. The genes were under the control of the promoter PcpcBA from Synechocystis sp. PCC 6803 [28].
Fluorescence and electron microscopy
Fluorescence microscopy was performed with an Olympus BX51 microscope (Olympus, Tokyo, Japan) and a Zeiss LSM 510 Live Confocal Microscope (Zeiss, Peabody, MA, USA). Low-temperature (77 K) fluorescence emission spectra were measured for cells grown at different light conditions using an SLM8000-based spectrofluorometer modified for computerized, solid-state operation by On-Line Instrument Systems Inc. (Bogart, GA, USA) as described previously [49]. Transmission electron microscopy of thin sections was performed with a JEM-1010 electron microscope (Jeol, Tokyo, Japan). Sample preparation and thin sectioning were carried out as described [50]. Scanning electron microscopy images were obtained using a field emission scanning electron microscope (FEI Quanta 200 F, Eindhoven, The Netherlands), and sample preparation was based on the protocol described [51].
Cell wall preparation and quantitative determination of cellulose
Cyanobacterial cell wall specimens were prepared as described [52] and quantitation of total glucose was performed using the anthrone–sulfuric acid method [53].
X-ray diffraction
The X-ray diffraction profiles of cellulose specimens were obtained by DMAX-2400 (Rigaku, Japan) with Cu Kα radiation. The X-ray diffraction pattern was collected at a continuous scan mode with step of 0.02° at the rate of 2°/min in the 2θ-range of 0–50°.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2009AA021405 and 2015CD150101) and the CAS International Partnership Program for Research Teams to JZ. Work in the laboratory of DAB was supported by Air Force Office of Scientific Research MURI grant FA9550-05-1-0365. We thank Nicole Zembower and Susan Magargee for assistance with confocal imaging, and Dr. Gang Ning and Missy Hazen at the Microscopy and Cytometry Facility of the Huck Institutes of the Life Sciences (Penn State University, University Park), Yingchun Hu at College of Life Sciences, Peking University and Jingnan Liang at Institute of Microbiology, Chinese Academy of Sciences for the help with transmission electron microscopy, Chunli Li at Institute of Microbiology, Chinese Academy of Sciences for the help with scanning electron microscopy, and Fuhui Liao at College of Chemistry & Molecular Engineering, Peking University for help with X-ray diffraction.
References
- Tilman D, Hill J, Lehman C. Carbon-negative biofuels from low-input high-diversity grassland biomass. Science 2006; 314: 1598–1600. [DOI] [PubMed] [Google Scholar]
- Goldemberg J. Ethanol for a sustainable energy future. Science 2007; 315: 808–810. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Tiller KH, Al-Ahmad H, Stewart NR, Stewart CN Jr. Plants to power: bioenergy to fuel the future. Trends Plant Sci 2008; 13: 421–429. [DOI] [PubMed] [Google Scholar]
- Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol 2009; 60: 165–182. [DOI] [PubMed] [Google Scholar]
- Czaja WK, Young DJ, Kawecki M, Brown RM Jr. The future prospects of microbial cellulose in biomedical applications. Biomacromolecules 2007; 8: 1–12. [DOI] [PubMed] [Google Scholar]
- Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev 1991; 55: 35–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 2006; 22: 53–78. [DOI] [PubMed] [Google Scholar]
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 1999; 50: 245–276. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown RM Jr. Cellulose biosynthesis: current views and evolving concepts. Ann Bot 2005; 96: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Debolt S, Persson S. Cellulose synthesis: a complex complex. Curr Opin Plant Biol 2008; 11: 252–257. [DOI] [PubMed] [Google Scholar]
- Hu SQ, Gao YG, Tajima K, et al. Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci USA 2010; 107: 17957–17961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa N, Fujiwara T, Yoda T, et al. Cellulose complementing factor (Ccp) is a new member of the cellulose synthase complex (terminal complex) in Acetobacter xylinum. J Biosci Bioeng 2013; 115: 607–612. [DOI] [PubMed] [Google Scholar]
- Wong HC, Fear AL, Calhoon RD, et al. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA 1990; 87: 8130–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Kudlicka K, Okuda K, Brown RM Jr. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol 1994; 176: 5735–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S, Tajima K, Uemori Y, et al. Cloning of cellulose synthesis related genes from Acetobacter xylinum ATCC23769 and ATCC53582: comparison of cellulose synthetic ability between strains. DNA Res 2002; 9: 149–156. [DOI] [PubMed] [Google Scholar]
- Brown RM Jr, Willison JH, Richardson CL. Cellulose biosynthesis in Acetobacter xylinum: visualization of the site of synthesis and direct measurement of the in vivo process. Proc Natl Acad Sci USA 1976; 73: 4565–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Chen HP, Saxena IM, Brown RM Jr, Itoh T. Localization of c-di-GMP-binding protein with the linear terminal complexes of Acetobacter xylinum. J Bacteriol 2001; 183: 5668–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 2013; 493: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Hay ID, Li C, et al. Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci USA 2011; 108: 13083–13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonouchi N, Tahara N, Kojima Y, et al. A beta-glucosidase gene downstream of the cellulose synthase operon in cellulose-producing Acetobacter. Biosci Biotechnol Biochem 1997; 61: 1789–1790. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Lin FC, Brown RM Jr. Identification of a new gene in an operon for cellulose biosynthesis in Acetobacter xylinum. Plant Mol Biol 1991; 16: 947–954. [DOI] [PubMed] [Google Scholar]
- Richmond T. Higher plant cellulose synthases. Genome Biol 2000; 1: REVIEWS3001.1–3001.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles DR, Romanovicz DK, Brown RM Jr. Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase?. Plant Physiol 2001; 127: 529–542. [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Saotome T, Ochiai Y, Katayama M, Narikawa R, Ikeuchi M. Cellulose accumulation and a cellulose synthase gene are responsible for cell aggregation in the cyanobacterium Thermosynechococcus vulcanus RKN. Plant Cell Physiol 2011; 52: 957–966. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Bryant DA. Synechococcus sp. strain PCC 7002 transcriptome: acclimation to temperature, salinity, oxidative stress, and mixotrophic growth conditions. Front Microbio 2012; 3: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiczyk E, Hansel A. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriol 2000; 182: 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M, Floodgate GD. An electron-microscopic demonstration of an acidic polysaccharide involved in the adhesion of a marine bacterium to solid surfaces. J Gen Microbiol 1973; 74: 325–334. [Google Scholar]
- Xu Y, Alvey RM, Byrne PO, Graham JE, Shen G, Bryant DA. Expression of genes in cyanobacteria: adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. Methods Mol Biol 2011; 684: 273–293. [DOI] [PubMed] [Google Scholar]
- Herth W, Schnepf E. The fluorochrome, calcofluor white, binds oriented to structural polysaccharide fibrils. Protoplasma 1980; 105: 129–133. [Google Scholar]
- Xu Y, Guerra LT, Li Z, Ludwig M, Dismukes GC, Bryant DA. Altered carbohydrate metabolism in glycogen synthase mutants of Synechococcus sp. strain PCC 7002: cell factories for soluble sugars. Metab Engin 2013; 16: 56–67. [DOI] [PubMed] [Google Scholar]
- Guerra LT, Xu Y, Bennette N, McNeely K, Bryant DA, Dismukes GC. Natural osmolytes are much less effective substrates than glycogen for catabolic energy production in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J Biotechnol 2013; 166: 65–75. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Komoda K, Sakurai N, Tajima K, Tanaka I, Yao M. The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A. Biochem Biophys Res Commun 2013; 431: 802–807. [DOI] [PubMed] [Google Scholar]
- Atalla RH, Vanderhart DL. Native cellulose: a composite of two distinct crystalline forms. Science 1984; 223: 283–285. [DOI] [PubMed] [Google Scholar]
- Haigler CH, Brown RM Jr, Benziman M. Calcofluor white ST alters the in vivo assembly of cellulose microfibrils. Science 1980; 210: 903–906. [DOI] [PubMed] [Google Scholar]
- Segal L, Creely JJ, Martin AE Jr, Conrad CM. An empirical method for estimating the degree of crystallinity of native cellulose using the X-ray diffractometer. Tex Res J 1962; 29: 786–794. [Google Scholar]
- Benziman M, Haigler CH, Brown RM Jr, White AR, Cooper KM. Cellulose biogenesis: polymerization and crystallization are coupled processes in Acetobacter xylinum. Proc Natl Acad Sci USA 1980; 77: 6678–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo HM, Song SH, Pyun YR, Kim YS. Evidence that a beta-1,4-endoglucanase secreted by Acetobacter xylinum plays an essential role for the formation of cellulose fiber. Biosci Biotechnol Biochem 1998; 62: 2257–2259. [DOI] [PubMed] [Google Scholar]
- Nakai T, Nishiyama Y, Kuga S, Sugano Y, Shoda M. ORF2 gene involves in the construction of high-order structure of bacterial cellulose. Biochem Biophys Res Commun 2002; 295: 458–462. [DOI] [PubMed] [Google Scholar]
- Nobles DR, Brown RM Jr. Transgenic expression of Gluconacetobacter xylinus strain ATCC 53582 cellulose synthase genes in the cyanobacterium Synechococcus leopoliensis strain UTCC 100. Cellulose 2008; 15: 691–701. [Google Scholar]
- Wallington TJ, Anderson JE, Mueller SA, et al. Corn ethanol production, food exports, and indirect land use change. Environ Sci Technol 2012; 46: 6379–6384. [DOI] [PubMed] [Google Scholar]
- Blifernez-Klassen O, Klassen V, Doebbe A, et al. Cellulose degradation and assimilation by the unicellular phototrophic eukaryote Chlamydomonas reinhardtii. Nat Commun 2012; 3: 1214. [DOI] [PubMed] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 1998; 281: 237–240. [DOI] [PubMed] [Google Scholar]
- Falkowski PG. The ocean's invisible forest. Sci Am 2002; 287: 54–61. [DOI] [PubMed] [Google Scholar]
- Strand SE, Benford G. Ocean sequestration of crop residue carbon: recycling fossil fuel carbon back to deep sediments. Environ Sci Technol 2009; 43: 1000–1007. [DOI] [PubMed] [Google Scholar]
- Stevens SE Jr, Patterson COP, Myers J. The production of hydrogen peroxide by blue-green algae: A survey. J Phycol 1973; 9: 427–430. [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 1979; 111: 1–61. [Google Scholar]
- Frigaard NU, Sakuragi Y, Bryant DA. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol Biol 2004; 274: 325–340. [DOI] [PubMed] [Google Scholar]
- Elhai J, Wolk CP. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 1988; 68: 119–138. [DOI] [PubMed] [Google Scholar]
- Shen G, Bryant DA. Characterization of a Synechococcus sp. Strain PCC 7002 mutant lacking photosystem I. Protein assembly and energy distribution in the absence of the photosystem I reaction center core complex. Photosynth Res 1995; 44: 41–53. [DOI] [PubMed] [Google Scholar]
- Ohki K, Fujita Y. Photoregulation of phycobilisome structure during complementary chromatic adaptation in the marine cyanophyte Phormidium sp. C86. J Phycol 1992; 28: 803–808. [Google Scholar]
- Albertano P, Urzi C. Structural interactions among epilithic cyanobacteria and heterotrophic microorganisms in Roman hypogea. Microbial Ecol 1999; 38: 244–252. [DOI] [PubMed] [Google Scholar]
- York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol 1986; 118: 3–40. [Google Scholar]
- Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem 1969; 32: 420–424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.