Abstract
Gamma-amino butyric acid (GABA) and glutamate are the major inhibitory and excitatory neurotransmitters in the mammalian central nervous system, respectively, and have been associated with suicidal behavior and major depressive disorder (MDD). We examined the relationship between genotype, brain transcriptome, and MDD/suicide for 24 genes involved in GABAergic and glutamatergic signaling. In part 1 of the study, 119 candidate SNPs in 24 genes (4 transporters, 4 enzymes, and 16 receptors) were tested for associations with MDD and suicidal behavior in 276 live participants (86 nonfatal suicide attempters with MDD and 190 non-attempters of whom 70% had MDD) and 209 postmortem cases (121 suicide deaths of whom 62% had MDD and 88 sudden death from other causes of whom 11% had MDD) using logistic regression adjusting for sex and age. In part 2, RNA-seq was used to assay isoform-level expression in dorsolateral prefrontal cortex of 59 postmortem samples (21 with MDD and suicide, 9 MDD without suicide, and 29 sudden death non-suicides and no psychiatric illness) using robust regression adjusting for sex, age, and RIN score. In part 3, SNPs with subthreshold (uncorrected) significance levels below 0.05 for an association with suicidal behavior and/or MDD in part 1 were tested for eQTL effects in prefrontal cortex using the Brain eQTL Almanac (www.braineac.org). No SNPs or transcripts were significant after adjustment for multiple comparisons. However, a protein coding transcript (ENST00000414552) of the GABA A receptor, gamma 2 (GABRG2) had lower brain expression postmortem in suicide (P = 0.01) and evidence for association with suicide death (P = 0.03) in a SNP that may be an eQTL in prefrontal cortex (rs424740, P = 0.02). These preliminary results implicate GABRG2 in suicide and warrant further investigation and replication in larger samples.
Keywords: GABA, glutamate, gene expression, suicidal behavior, major depressive disorder
INTRODUCTION
Major depressive disorder (MDD) and suicidal behavior have a high prevalence and incidence in the population [Borges et al., 2010; Gelenberg, 2010]. According to the World Health Organization (WHO), between 10% and 15% of the general population will experience a depressive episode in their lifetime, and 5% of men and 9% of women will experience a depressive disorder in a given year [Hirschfeld, 2012]. In 2013, the overall suicide rate in the United States was 13/100,000 [National Center for Injury Prevention and Control (NCIPC), 2013], and there are an estimated 800,000 suicides per year [World Health Organization, 2014] and about 10 times that number of nonfatal suicide attempts.
Gamma-amino butyric acid (GABA) and glutamate are, respectively, the major inhibitory and excitatory neurotransmitters in the mammalian central nervous system [Fonnum, 1984], and are thereby involved directly or indirectly in most aspects of normal brain function including cognition, mood regulation, memory, and learning. GABA deficit as an explanatory hypothetical model of major depression has found increasing support [Mohler, 2012; Mann et al., 2014]. GABA may affect monoamine levels in the brain [Petty, 1995] and in humans, it has been shown to lower stress, and is implicated in depression [Sanacora and Saricicek, 2007; Levinson et al., 2010]. The role of glutamate in mood regulation and disorders also has been increasingly recognized, since the observation that several existing antidepressants exert effects on various glutamatergic mechanisms [Owen, 2012]. Glutamate is implicated in MDD through studies that show that ketamine, an NMDA antagonist, produces a rapid and pronounced antidepressant effect through enhanced AMPA receptor signaling [Zarate et al., 2006]. Conversely, sustained higher levels of glutamate may mediate stress-induced neuronal atrophy in hippocampus [Sapolsky, 2000]. Thus, the GABA-glutamatergic systems are potentially involved in the pathogenesis of major depression and studying these neurotransmitter systems may also uncover possible antidepressant treatment targets.
Genes involved in these systems have been investigated to a limited extent in MDD and suicidal behavior in genomic studies. Some studies have reported an association of some variants in genes related to glutamate and GABA systems with suicidal behavior [Kertes et al., 2011; Chiesa et al., 2012; Myung et al., 2012] and MDD [Murphy et al., 2011], while other studies found no association [Sokolowski et al., 2013]. Postmortem microarray studies in suicide and/or MDD report differential expression of GABA receptor subunits, glutamate-ammonia ligase (GLUL), or glutamate receptors in prefrontal and/or limbic brain regions [Choudary et al., 2005; Kim et al., 2007; Klempan et al., 2009; Sequeira et al., 2007, 2009; Tripp et al., 2012]. Other groups have applied a Convergent Functional Genomics (CFG) approach to identify and prioritize differentially expressed gene biomarkers of relevance to suicide and suicidal behavior. CFG integrates multiple independent lines of evidence from both genetic and functional genomic data, applying a Bayesian strategy for identifying and prioritizing findings, thereby reducing the false-positives and false-negatives inherent in each individual approach [Le-Niculescu et al., 2013].
Whereas some applied a whole-genome approach to examine suicidality in patients with bipolar disorder, no study of the genes in the GABAergic/glutamatergic systems to date, to our knowledge, has combined the examination of associations with both gene variant and gene expression in MDD and suicidal behavior. We therefore examine 24 genes related to the glutamate and GABA systems in relation to major depression and suicidal behavior, including 4 transporters, 4 enzymes, and 16 receptors. We examined SNPs covering the regulatory regions and exons for each gene. We also used RNA-seq to quantify the transcriptome from these genes in prefrontal cortex postmortem from three groups that included: MDD dying from causes other than suicide to examine relationships to MDD, MDD dying by suicide to determine the additional effects due to suicide (DSUI), and non-psychiatric sudden death controls (NPC).
Twin studies show that the diathesis for nonfatal suicidal behavior is heritable to a comparable degree to death by suicide. Postmortem sample sizes are smaller especially when getting a psychiatric diagnosis with a psychological autopsy because the structured clinical interviews with significant others in the life of the deceased represents a great deal of work. Therefore we expanded the sample size by adding nonfatal suicide attempters. The live genetic sample was included as an additional line of independent evidence to increase sensitivity in detecting putative genetic markers associated with both suicide attempt and completion because we expect the genes to be largely the same ones.
Since it has been shown that most trait-associated SNPs are expression quantitative trait loci (eQTL) [Nicolae et al., 2010], thereby influencing gene expression, we used a large open-source repository of eQTL analysis results (www.braineac.org). This repository contains data from postmortem brain of over 100 individuals without psychiatric diagnosis. We examined whether SNPs showing evidence for an association with MDD and/or suicide were also eQTL.
MATERIALS AND METHODS
Participants
Data from three samples were available for this study. Two separate samples of subjects were genotyped, one live and other postmortem. RNA-seq data were available for a third sample of 59 postmortem subjects, partially overlapping with the genotyped postmortem sample. All samples were of Caucasian ancestry. Descriptive statistics for the three samples are included in Table I.
TABLE I.
Description of Live and Postmortem Subjects
| Psychiatric diagnosis (N) |
Age (years) |
Sex (N) |
||||||
|---|---|---|---|---|---|---|---|---|
| Sample group | Group | MDD | Other diagnosisa | No diagnosis | Mean | SD | M | F |
| Genotype sample | Live subjects | |||||||
| Suicide attempter attempter | 86 | 0 | 0 | 38.78 | 11.25 | 30 | 56 | |
| Non-attempter | 133 | 0 | 57 | 41.19 | 13.73 | 73 | 117 | |
| Total | 219 | 0 | 57 | 40.44 | 13.04 | 103 | 173 | |
| Postmortem | ||||||||
| Suicide death | 75 | 46 | 0 | 50.24 | 18.96 | 80 | 41 | |
| Non-suicide death | 10 | 11 | 67 | 50.85 | 17.46 | 66 | 22 | |
| Total | 85 | 57 | 67 | 50.5 | 18.31 | 146 | 63 | |
| RNA sampleb | Postmortem | |||||||
| Suicide death | 21 | 0 | 0 | 52.19 | 21.73 | 13 | 8 | |
| Non-suicide death | 9 | 0 | 29 | 46.92 | 20.78 | 29 | 9 | |
| Total | 30 | 0 | 29 | 48.80 | 21.08 | 42 | 17 | |
The other diagnosis category includes subjects with primary diagnosis other than MDD (see the methods section for a description).
This sample overlaps with the postmortem subjects from the genotype sample above, sharing n = 38 subjects.
Genotyped live sample
The live sample consisted of 276 unrelated individuals: 86 MDD suicide attempters (31%), 133 subjects (48%) with MDD but no suicide attempt, and 57 healthy volunteers (21%), recruited between 1991 and 2011 in New York and Pittsburgh. Participants gave written informed consent as required by the relevant Institutional Review Boards. Psychiatric diagnosis was established using the Structured Clinical Interview for DSM IV (SCID-I). The definition of suicide attempt was: a self-injurious act that has at least partial intent to end one's life [Posner et al., 2007]. The number, method, and medical damage of past suicide attempts were recorded on the Columbia Suicide History Form [Oquendo et al., 2003]. A group with MDD and no history of a suicide attempt was a psychiatric control group. Unrelated healthy volunteers were recruited through advertising. They were assessed by psychiatrists or clinical psychologists and evaluated using the SCID-NP to rule out axis I diagnoses, cluster B personality disorder, substance use disorder, and lifetime history of a suicide attempt. Exclusion criteria for all participants included mental retardation, dementia, and acute psychosis.
Genotyped postmortem sample
All postmortem subjects, suicides and non-suicides, died suddenly. This sample consisted of 209 postmortem subjects: 121 died of suicide and 88 were (non-suicide) sudden death victims. Cause of death for postmortem subjects, excluding suicide, was determined by the coroner or medical examiner. Suicide was determined by the research team using the Columbia Classification Algorithm for Suicide Assessment [Posner et al., 2007]. Diagnosis of major psychiatric disorder was determined using the SCID I [Spitzer et al., 1992] and our validated psychological autopsy method as previously described [Kelly and Mann, 1996]. All of the suicides and 24% of the non-suicide deaths had at least one psychiatric diagnosis. In particular, 62% of the suicides and 11% of the non-suicide deaths had a lifetime MDD diagnosis. For those subjects without MDD, other diagnoses were bipolar disorder, depression NOS, schizophrenia, schizoaffective disorder, delusional disorder, psychotic disorder, adjustment disorder senile dementia NOS, organic mood syndrome, conduct disorder, pathological gambling, and substance abuse/dependence. Biological samples were obtained from the Medical Examiner's Office in accordance with local regulations and protocols approved by the appropriate Institutional Review Board. All brains were free of gross neuropathology and had negative brain toxicology for psychoactive and neurotoxic drugs. Brain samples were dissected from Brodmann Area 9 as previously reported [Sibille et al., 2004]. Peripheral toxicology and antemortem medication history for 3 months ruled out recent exposure to psychotropic medication and illicit drugs.
Postmortem sample for RNA sequencing
An additional sample of postmortem subjects (N = 59), with RNA sequencing data available, consisted of N = 21 suicides with MDD diagnosis, nine sudden death victims with MDD, and 29 sudden death victims without psychiatric diagnosis. Some of the postmortem subjects with RNAseq data also had been included in the genotyped sample (n = 38 out of 59). Procedures for brain collection and psychological autopsy, psychiatric diagnosis, inclusion and exclusion criteria are as described above for the genotyped postmortem sample. Postmortem intervals (PMI), RIN scores, and pH for the postmortem samples were within acceptable ranges and are reported elsewhere [Galfalvy et al., 2015; Pantazatos et al., 2015].
Postmortem sample in an online dataset
Our sample of sudden death subjects with both genotype and gene expression data was too small to allow for an association analysis between the two. To identify associations between genotype and gene expression in a healthy population, we used the public online database of UK Brain Expression Consortium (UKBEC), which studied regulation and alternative splicing of gene expression in multiple tissues from human brains (http://www.braineac.org/). This dataset comprised 134 brains from individuals free of neurodegenerative disorders. Up to 10 brain regions were extracted per brain in parallel, for mRNA quantification. DNA was also collected to enable genotyping and expression quantitative trait loci (eQTL) analysis with exon-level expression. Gene-level expression was also estimated for 26,000 genes by calculating the Windsorised mean (below 10% and above 90%) signal of all probe sets corresponding to each gene (see https://ukbec.wordpress.com/ for more details).
Genotyping and RNA Sequencing
We focused our analyses on the most important and abundant transporters for removal of glutamate in the brain: glial high-affinity glutamate transporters SLC1A2 (GLT-1) and SLC1A3 (GLAST), and the major GABA transporters: SLC6A1 (GAT1) and SLC6A11 (GAT3) [Zhou and Danbolt, 2013]. We also targeted seven kinds of glutamate receptors (glutamate receptor, ionotropic, N-methyl D-aspartate 2A, GRIN2A; glutamate receptor, ionotropic, AMPA 1–4, GRIA1, GRIA2, GRIA3, GRIA4; glutamate receptor, ionotropic, kainate 1, GRIK1; and glutamate receptor, metabotropic 3, GRM3). For GABA receptors, we include nine subunits of GABAA receptors (α1, GABRA1; α4, GABRA4; α5, GABRA5; β1, GABRB1; β3, GABRB3; δ, GABRD; γ1, GABRG1; γ2, GABRG2; and ρ1, GABRR1), one GABAA receptor-associated protein like one (GABARAPL1) and one GABAB receptor subunit (GABBR2). We also include some important enzymes (glutamate decarboxylase 1 and 2, GAD1, and GAD2; glutamate-ammonia ligase, GLUL and glutaminase, GLS). Detailed information of all the candidate genes are shown in Supplementary Table SI.
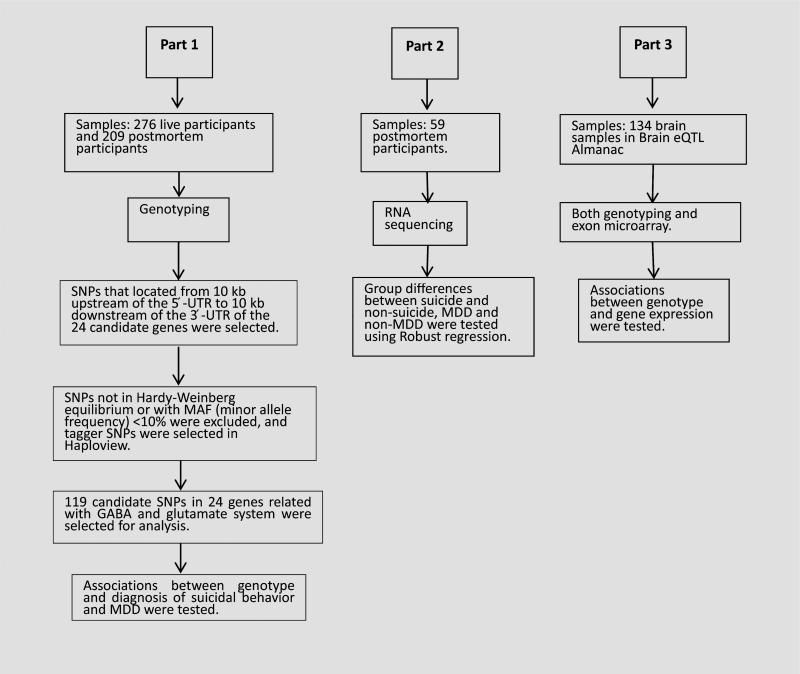
The flowchart for genotyping and sequencing of the samples is displayed in Figure 1. The first sample was genotyped as part of a larger study. Genomic DNA was isolated from peripheral blood (n = 276 live subjects) or from postmortem brain tissue (n = 209 postmortem subjects) using standard protocols [Huang et al., 1999]. Subjects were genotyped using the HumanOmni-1-Quad Beadchip (Illumina, San Diego, CA). Selection of tagger SNPs was performed in Haploview version 2.04 (Whitehead Institute for Biomedical Research, Cambridge, MA).
FIG. 1.
Schematic of workflow.
The 24 candidate genes examined in this study were selected based on evidence of their involvement in GLU/GABA systems and previous associations with major depressive disorder and suicidal behavior [Fiori and Turecki, 2010; Tripp et al., 2012; Mandelli and Serretti, 2013]. We filtered SNPs to include those with locations 10 kb upstream of the 5′-UTR to 10 kb downstream of the 3′-UTR, which are relative coordinates where many regulators are thought to be located [Pu et al., 2013]. SNPs were then selected according to the following criteria: first, the SNPs whose blocks were located in known gene exons or promoter regions were selected; second, a few additional SNPs in GAD2 previously reported to be associated with MDD [Unschuld et al., 2009] were also included; third, SNPs must be in Hardy–Weinberg equilibrium and with minor allele frequency (MAF) values of ≥10% in both live control and postmortem control. Finally, a total of 119 tagger SNPs were identified from the SNPs above in Haploview version 2.04 as candidate markers, and genotypes were generated from the original GWAS study. See Supplementary Table SII for more details regarding the SNPs used in this study.
The second postmortem sample underwent RNA sequencing. For a detailed description of the RNA-seq methodology as previously described [Pantazatos et al., 2015]. Briefly, RNA was extracted from dorsal prefrontal cortex (BA9) using previously established protocols [Sibille et al., 2004] and conducted according to NIH Roadmap Epigenomics Mapping Consortium (REMC) guidelines. Paired-end, strand specific sequencing for total RNA was performed on Illumina HiSeq 2500 with 100 bp read lengths. Preprocessing and differential expression analyses used Tophat and Cufflinks (v.2.2.0) software [Trapnell et al., 2012] Briefly, for each clinical sample, raw RNA-seq reads were aligned and mapped to the human reference genome and assembled using Tophat v2.0.9. These assemblies were then merged together to provide a uniform basis for calculating gene and transcript expression in each sample using Cufflinks v2.2.0 (see Pantazatos et al. [2015] Supplementary Table SII for read mapping statistics). Cuffquant v2.2.1 (4237) was then used to quantify gene and isoform-level expression values, and the output CXB files were then input to Cuffnorm v2.2.1 (4237), which produced normalized expression values for each sample. These normalized expression values were subsequently analyzed using SPSS v20.0.0 and R (http://www.r-project.org/).
Statistical Analysis
The data analysis plan consisted of three parts: Part 1 tested the association between genotypes and suicidal behavior and/or a diagnosis of MDD. Analysis for live and postmortem subjects was conducted separately to identify the risk factors for suicide attempt and suicide death. We also tested the association between genetic risk and suicide attempt separately in MDD subjects only, as a sensitivity analysis. Logistic regressions were performed in R (http://www.r-project.org/) for each screened SNP (see above) with genotype as categorical predictor and an indicator variable for suicidal behavior history (yes/no) or MDD (yes/no) as response. Age and sex were included as covariates in the model. Depending on the SNP, up to 21 postmortem subjects and 20 live subjects were excluded due to missing values. For the SNPs showing sub-threshold (P < 0.05, uncorrected) association with the outcome variable, odds ratios were estimated from the logistic regression models. Haploview version 2.04 was used to determine tagger SNPs. All P-values were corrected using Benjamini–Hochberg method for false discovery rate (P < 0.1).
Part 2 of the data analysis tested for group differences in total (gene-level) and isoform-level expression of the 24 genes. Since the distribution of gene expression was very skewed with several outliers for each isoform, robust regression models were fit—in R using the function “lmrob” from the library robustbase, to compare gene expression differences between MDD and non-MDD subjects, and suicide and sudden death controls, adjusting for age, sex, and RIN score. Two subjects (one DSUI and other NPC) were missing RIN scores due to technical issues, therefore these analyses included a total of 57 subjects. The brain pH values for the three groups were very consistent (DSUI: 6.41 ± 0.30, DC: 6.41 ± 0.32, NPC: 6.49 ± 0.33), and there were no significant differences in the post-mortem interval (PMI) of the three groups (DS: 16.08 ± 7.00, DC: 15.06 ± 4.38, NPC: 13.21 ± 4.61). Transcripts were screened before being entered into the analysis, and those with expression levels that were not significantly different from 0, or those for which the robust regression model failed to converge, were excluded. Significance levels were not adjusted for multiple testing, since this was an exploratory analysis. All P-values were corrected using Benjamini–Hochberg method for false discovery rate (P < 0.1).
The third step in the analysis examined whether the genotypes associated with suicide, suicide attempt, or MDD had evidence for meaningful effects on gene expression based on findings reported in the Brain eQTL Almanac, a public online dataset providing eQTL analysis results from 10 brain regions in 134 individuals postmortem. We restricted eQTL results to the SNPs with sub-threshold suicide or MDD effects from Part 1 and in prefrontal cortex only (to keep results consistent with Part 2). We extracted P-values for the association of these candidate risk SNPs and expression of all the exon-specific probesets on the same gene (www.braineac.org). As this was an exploratory analysis, we listed the minimum P-value for each SNP in the main text and did not adjust for multiple testing. P-values for associations with all exon-level probesets for each gene are listed in the supplement (see results section).
RESULTS
Part 1: Association Between Genotypes and Suicidal Behavior, Suicide Completion, and/or MDD
In this analysis, 119 SNPs in 24 genes were tested in SNP-wise logistic regressions with genotype, age, and sex as predictors, and suicide death, attempted suicide, or MDD diagnosis as outcomes. No SNPs had a significant effect after adjustment for multiple testing. An exploratory analysis using uncorrected P-values (Tables II and III) identified 19 SNPs in 15 genes with subthreshold associations (P < 0.05, uncorrected). The 15 genes included seven GABA receptors (GABRA4, GABBR2, GABRA5, GABRB3, GABRD, GABRG1, and GABRG2), three glutamate receptors (GRIA2, GRIK1, and GRIN2A), two glutamate transporters (SLC1A3 and SLC1A2), one GABA transporter (SLC6A1) and two enzymes (GAD2 and GLS) exhibited potential associations with suicidal behavior and/or MDD. Of the 15 genes, 13 (GABRA4, GABBR2, GABRA5, GABRB3, GABRG1, GABRG2, GRIA2, GRIN2A, SLC1A3, SLC1A2, SLC6A1, GAD2, and GLS) showed an association with suicide; seven genes (GABRA4, GABBR2, GABRD, GRIA2, GRIK1, GRIN2A, and SLC6A1) were associated with MDD; and five genes (GABBR2, GABRA4, GRIA2, GRIN2A, and SLC6A1) were associated with both suicide and MDD (Table II). GABRA4, GABBR2, GABRG1, GABRG2, GLS, GRIA2, GRIN2A, SLC1A2, SLC6A1, SLC1A3, and GAD2 showed homozygous risk genotypes in suicidal behavior. Genotype GG of rs6447520 (GABRA4), genotype CC of rs2808536 (GABBR2), and genotype AA of rs424740 (GABRG2), rs6849345 (GRIA2), rs4780886 (GRIN2A), and rs1062246 (SLC6A1) were risk genotypes for suicide death, while genotype AA of rs6447520 (GABRA4), rs10836356 (SLC1A2), and rs1529461 (SLC1A3), genotype GG of rs13035504 (GLS), and rs2236418 (GAD2) were risk genotypes for attempted suicide. Genotype GG of rs10739615 (GABBR2) and rs1062246 (SLC6A1), and genotype AA of rs13303016 (GABRD), rs363527 (GRIK1), and rs3785185 (GRIN2A) were risk genotypes for MDD (Table III). Of these 19 SNPs, 6 were located in 5-UTR, 10 were located in 3-UTR, and 3 were located in exons (Table SII).
TABLE II.
Summary Table
| Association between genotype and disorders (n = 485) |
Expression alteration in disorders (n = 59) |
Association between genotype and expression (public online results; n = 134) |
|||||
|---|---|---|---|---|---|---|---|
| SNPs |
SNPs with MDD effect |
SNPs with suicide effect |
Isoforms with MDD effect |
Isoforms with suicide effect |
Significant SNPs in FCTX |
||
| Type | Gene | N(SS/TS) | Name | Name | Name | Name | Name |
| GABA RE | GABARAPL1 | 0/3 | |||||
| GABBR2 | 2/7 | rs10739615 | rs2808536 | ||||
| GABRA1 | 0/4 | ||||||
| GABRA4 | 2/8 | rs13151759 | rs12506608447520 | ||||
| GABRA5 | 1/4 | rs140685 | |||||
| GABRB1 | 0/5 | total expression↓, GABRB1-001↓, 003↓ | GABRB1-001↓ | ||||
| GABRB3 | 1/6 | rs8041610 | GABRB3-006↓ | ||||
| GABRD | 1/2 | rs13303016 | |||||
| GABRG1 | 1/2 | rs976156 | |||||
| GABRG2 | 1/2 | rs424740 | GABRG2-003↓ | rs424740 | |||
| GABA TR | SLCGA1 | 1/8 | rs1062246 | rs1062246 | |||
| SLCGA11 | 0/6 | SLC6A11-002↓ | |||||
| GLU RE | GRIA1 | 0/4 | |||||
| GRIA2 | 1/2 | rs6849345 | rs6849345 | ||||
| GRIA4 | 0/4 | ||||||
| GRIK1 | 1/8 | rs363527 | |||||
| GRIN2A | 3/8 | rs1420040, rs3785185 | rs4780886 | ||||
| GRM3 | 0/2 | ||||||
| GLU TR | SLC1A2 | 1/11 | rs10836356 | SLC1A2-004↓, 007↓ | rs10836356 | ||
| SLC1A3 | 1/10 | rs1529461 | |||||
| Enzyme | GAD2 | 1/5 | rs2236418 | rs2236418 | |||
| GAD1 | 0/2 | ||||||
| GLS | 1/3 | rs13035504 | |||||
| GLUL | 0/3 | GLUL-002↓ | |||||
Gray indicates genes with subthreshold (P < 0.05 uncorrected) effects in all three analytic parts. GABA RE, GABA receptor; GLU RE, glutamate receptor; GABA TR, GABA transporter; GLU TR, glutamate transporter; (SS/TS), (significant SNPs/tested SNPs); FCTX, frontal cortex.
TABLE III.
SNP Associations With Suicidal Behavior and MDD in Live and Postmortem Subjects (Uncorrected P-Values)
| Y = attempted suicide/suicide death |
Y = MDD |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Postmortem |
Live |
Live MDD |
Live |
|||||||
| Suicide death (n = 121) versus sudden death (n = 88) |
Suicide attempters (n = 86) versus non-attempters (n = 190) |
Attempters with MDD (n = 86) versus Attempters without MDD (n = 132) |
MDD (n = 218) versus no diagnosis (n = 56) |
|||||||
| Gene | SNP | Alleles | P-value | Risk genotype | P-value | Risk genotype | P-value | Risk genotype | P-value | Risk genotype |
| GABBR2 | rs10739615 | A:G | 0.49 | 0.52 | 0.93 | 0.04 | GG | |||
| rs2808536 | C:A | 0.02 | CC | 0.28 | 0.22 | 0.41 | ||||
| GABRA4 | rs12506608 | A:G | 0.02 | AG | 0.51 | 0.61 | 0.75 | |||
| rs6447520 | G:A | 0.13 | 0.01 | AA | 0.01 | AA | 0.44 | |||
| GABRA5 | rs140685 | A:G | 0.04 | AG | 0.64 | 0.67 | 0.75 | |||
| GABRB3 | rs8041610 | A:C | 0.01 | AC | 0.47 | 0.79 | 0.51 | |||
| GABRD | rs13303016 | A:G | 0.87 | 0.30 | 0.57 | 0.04 | AA | |||
| GABRG1 | rs976156 | G:A | 0.02 | GG | 0.81 | 0.73 | 0.76 | |||
| GABRG2 | rs424740 | T:A | 0.03 | AA | 0.55 | 0.51 | 0.64 | |||
| GLS | rs13035504 | G:C | 0.98 | 0.03 | GG | 0.02 | GG | 0.81 | ||
| GRIA2 | rs6849345 | A:G | 0.04 | AA | 0.52 | 0.34 | 0.04 | AG | ||
| GRIK1 | rs363527 | G:A | 0.10 | 0.73 | 0.99 | 0.03 | AA | |||
| GRIN2A | rs1420040 | A:G | 0.43 | 0.79 | 0.64 | 0.02 | AG | |||
| rs3785185 | G:A | 0.79 | 0.06 | 0.08 | 0.01 | AA | ||||
| rs4780886 | G:A | 0.02 | AA | 0.51 | 0.61 | 0.64 | ||||
| SLC1A2 | rs10836356 | G:A | 0.53 | 0.03 | AA | 0.09 | 0.11 | |||
| SLC1A3 | rs1529461 | G:A | 0.72 | 0.35 | 0.04 | AA | 0.44 | |||
| SLC6A1 | rs1062246 | A:G | 0.01 | AA | 0.49 | 0.36 | 0.02 | GG | ||
| GAD2 | rs2236418 | A:G | 0.73 | 0.03 | GG | 0.04 | GG | 0.66 | ||
Gene associations with suicidal behavior and depression were all adjusted by age and sex.
Part 2: Group Differences in Gene Expression
Summary statistics for the expression levels of all the transcripts that passed the screening phase are presented in Table IV for the three groups of postmortem subjects: non-psychiatric controls who died by sudden but non-suicide cause of death (NPC), depressed non-suicides (DNS) who died by sudden but non-suicide cause of death, and depressed suicides (DSUI). Significance levels were computed for two comparisons: one for subjects who died by suicide versus those who died by other causes of sudden death; and the second comparing subjects with and without an MDD diagnosis. Following robust regression no isoforms showed group differences after multiple comparisons correction.
TABLE IV.
Gene Expression Summary Statistics for Subthreshold Results (P < 0.05, Uncorrected) for Two Comparisons: MDD Versus Non-MDD Subjects (DSUI + DNS vs. NPC) and Suicide Versus Non-suicide (DSUI vs. DNS + NPC) Subjects
| NPC (n = 29) |
DNS (n = 9) |
DSUI (n = 21) |
MDD versus non-MDD |
SUI versus non-SUI |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Transcript ID | Name | Biotype | M | Q | M | Q | M | Q | P-value | P-value |
| GABRB1 | XL0C_GABRB1 | Total expression | 50.15 | 7.12 | 42.01 | 13.45 | 46.83 | 7.67 | 0.01 | 0.05 | |
| ENST00000295454 | GABRB1-001 | Protein coding | 37.33 | 6.33 | 32.00 | 10.25 | 33.50 | 6.34 | 0.01 | 0.03 | |
| ENST00000510909 | GABRB1-003 | Nonsense mediated decay | 0.86 | 1.43 | 0.67 | 1.62 | 0.29 | 0.83 | 0.04 | 0.05 | |
| GABRB3 | ENST00000400188 | GABRB3-00G | Protein coding | 0.95 | 2.06 | 1.04 | 1.74 | 0.60 | 0.95 | 0.11 | 0.03 |
| GABRG2 | ENST00000414552 | GABRG2-003 | Protein coding | 4.23 | 1.99 | 4.03 | 1.67 | 3.12 | 1.97 | 0.06 | 0.01 |
| GLUL | ENST0000033952G | GLUL-002 | Protein coding | 102.21 | 53.33 | 80.27 | 32.18 | 84.97 | 75.46 | 0.02 | 0.07 |
| SLC1A2 | ENST00000395750 | SLC1A2-004 | Protein coding | 2.18 | 1.62 | 1.18 | 1.09 | 1.50 | 2.29 | 0.01 | 0.15 |
| ENST00000531G28 | SLC1A2-007 | Protein coding | 7.21 | 8.76 | 10.83 | 13.52 | 6.58 | 10.14 | 0.03 | 0.33 | |
| SLCGA11 | ENST00000454147 | SLCGA11-002 | Protein coding | 1.30 | 0.49 | 1.15 | 0.45 | 1.08 | 0.49 | 0.04 | 0.23 |
Bold values indicate P < 0.05 uncorrected.
Adjusted by age, sex, and RIN. NPC, non-psychiatric controls; DNS, depressed non-suicides; DSUI, depressed suicides (DSUI); M, median; Q, interquartile range.
An exploratory analysis using uncorrected thresholds identified nine RNA isoforms or variants from six genes with subthreshold group differences in MDD versus non-MDD and/or in suicide versus non-suicide (P < 0.05, uncorrected). Six genes had lower expression in MDD versus non-MDD (total gene-level expression of GABRB1: B = –4.43, SE = 1.70, t = –2.61, P = 0.01; GABRB1-001: B = –3.74, SE = 1.43, t = –2.61, P = 0.01; GABRB1-003: B = –0.44, SE = 0.21, t = –2.08, P = 0.04; GLUL-002: B = –29.98, SE = 12.01, t = –2.50, P = 0.02; SLC1A2-004: B = –0.78, SE = 0.30, t = –2.62, P = 0.01; SLC6A11-002: B = –0.20, SE = 0.09, t = –2.15, P = 0.04), while one RNA isoform had higher expression in MDD than non-MDD (SLC1A2-007: B = 3.17, SE = 1.42, t = 2.24, P = 0.03) (see Tables II and IV). Three GABRB transcript isoforms were lower in suicide versus non-suicide (GABRB1-001: B = –3.15, SE = 1.45, t = –2.18, P = 0.03; GABRB3-006: B = –0.49, SE = 0.22, t = –2.21, P = 0.03; GABRG2-003: B = –1.21, SE = 0.46, t = –2.62, P = 0.01, see Tables II and IV). All analyses were adjusted for age, sex, and RIN scores. Supplementary Table SIII lists result for all tested isoforms. Three genes (GABRB3, GABRG2, and SLC1A2) showed evidence both for altered brain gene expression as well as an association with MDD or suicide in part 1 (see Table II).
Part 3: Association Between Genotype and Gene Expression
Only the SNPs with subthreshold associations (P-values <0.05, uncorrected) with suicide and/or MDD were examined in the Brain eQTL Almanac UKBEC (http://www.braineac.org/) (N = 134) for association with gene expression in the prefrontal cortex (FCTX, Brodmann Areas 9 and 46, see Trabzuni et al. [2011]). No SNPs showed a significant effect after adjustment for multiple comparisons. Out of all exon-specific probesets from the same gene, the one with minimum (uncorrected) P-value is reported in Table V. An exploratory analysis identified genotype AA of rs424740 (GABRG2, suicide effect in Parts 1 and 2), genotype AA of rs10836356 (SLC1A2, suicide effect in Part 1, MDD effect in Part 2) and genotype GG of rs2236418 (GAD2, suicide effect in Part 1) with subthreshold eQTL effects with exon-specific probesets on the same gene in the frontal cortex (P-value = 0.02, 0.04, 0.01, respectively) (Table V and Supplementary Figs. S1–3). Supplementary Table SIV lists eQTL results for all tested SNPs.
TABLE V.
Association Between Candidate Risk SNPs (SNPs With Subthreshold Effects on Suicidal Behavior, Suicide Death, and/or MDD in Part 1 of This Project) and Expression of All the Exon-Specific Probesets on the Same Gene in the Frontal Cortex in UKBEC (a Distinct Postmortem Sample, n = 134; Only the Minimal P-Values for Each SNP Are Listed Below)
| Gene | SNP | tID | exprID | Chromosome | P-value |
|---|---|---|---|---|---|
| GABBR2 | rs10739615 | 3217242 | 3217252 | chr9 | 0.08 |
| rs2808536 | 3217242 | 3217244 | chr9 | 0.13 | |
| GABRA4 | rs12506608 | 2768056 | 2768077 | chr4 | 0.32 |
| rs6447520 | 2768056 | 2768078 | chr4 | 0.10 | |
| GABRA5 | rs140685 | 4039130 | 4039145 | chr15 | 0.34 |
| GABRB3 | rs8041610 | 3614534 | 3614564 | chr15 | 0.07 |
| GABRD | rs13303016 | 4054481 | 4054487 | chr1 | 0.25 |
| GABRG1 | rs976156 | 2767932 | 2767935 | chr4 | 0.05 |
| GABRG2 | rs424740 | 2838462 | 2838490 | chr5 | 0.02 |
| GLS | rs13035504 | 2520291 | 2520317 | chr2 | 0.15 |
| GRIK1 | rs363527 | 3928211 | 3928280 | chr21 | 0.09 |
| GAD2 | rs2236418 | 3239667 | 3239676 | chr10 | 0.01 |
| SLC1A2 | rs10836356 | 3369366 | 3369378 | chr11 | 0.04 |
| GRIA2 | rs6849345 | 2749222 | 2749255 | chr4 | 0.40 |
| GRIN2A | rs1420040 | 3679812 | 3679818 | chr16 | 0.13 |
| rs3785185 | 3679812 | 3679833 | chr16 | 0.11 | |
| rs4780886 | 3679809 | 3679811 | chr16 | 0.15 | |
| SLC1A3 | rs1529461 | 2806643 | 2806657 | chr5 | 0.30 |
| SLC6A1 | rs1062246 | 2610631 | 2610649 | chr3 | 0.07 |
Bold values indicate P < 0.05 uncorrected.
tID, The tID is the transcript cluster ID from Affymetrix; exprID, The exprID is the Probe Set ID from Affymetrix.
DISCUSSION
In this study, we chose 119 candidate SNPs from possible functional regions of 24 genes related to GABA and glutamate systems: 4 transporters, 4 enzymes, and 16 receptors. We found no relationship between genotype or gene expression with suicidal behavior and MDD after correction for multiple tests. However, our exploratory analyses suggest that GABRG2 may be associated with suicide across multiple independent lines of evidence including genetic association, gene expression association, and eQTL analyses. This finding is consistent with a previous study, which found genetic variation within GABRG2 was associated with history of suicide attempt in schizophrenia [Zai et al., 2014], as well as studies, which found altered GABRG2 brain gene expression (prefrontal cortex) in suicide and/or depression [Choudary et al., 2005; Klempan et al., 2009]. For each type of analysis, we also reported exploratory findings at lower thresholds that are provisional pending replication in independent samples.
Part 1: GABAergic and Glutamatergic Genetic Associations With Suicidal Behavior and MDD: Results From Exploratory Analyses
We found 19 variants with sub-threshold associations with suicidal behavior and/or MDD. At the gene level, we found 15 genes with sub-threshold associations. Thirteen of them (GABRA4, GABBR2, GABRA5, GABRB3, GABRG1, GABRG2, GRIA2, GRIN2A, SLC1A3, SLC1A2, SLC6A1, GAD2, and GLS) showed an association with suicide; seven genes (GABRA4, GABBR2, GABRD, GRIA2, GRIK1, GRIN2A, and SLC6A1) were associated with MDD; and five genes (GABBR2, GABRA4, GRIA2, GRIN2A, and SLC6A1) were associated with both suicidal behavior and MDD.
Some studies have reported associations of the above genes with suicidal behavior and/or MDD, in agreement with our findings. The allelic variability in SLC1A2 may be associated with the underlying diathesis for suicidal acts [Murphy et al., 2011]. The GABRG2 polymorphisms are associated with suicidal behavior in schizophrenia with alcohol dependence or abuse [Zai et al., 2014]. GABRD variants are associated with childhood-onset mood disorders [Feng et al., 2010] and antidepressant response [Pu et al., 2013], and polymorphisms within the GAD2 locus have been associated with mood disorders [Unschuld et al., 2009). A marginal association between two GRIA2 variants and age of onset of MDD is reported [Chiesa et al., 2012], but no association with MADRS (Montgomery–Åsberg Depression Rating Scale) improvement scores or other clinical/sociodemographic variables in MDD patients [Chiesa et al., 2013]. There are reported associations of variations within GABBR2, GRIN2A, and GABRG1 with alcohol dependence [Domart et al., 2012; Ittiwut et al., 2012; Terranova et al., 2013]. Many studies have demonstrated associations between alcohol use disorder and MDD and suicidal behavior [Briere et al., 2014, Shoval et al., 2014], suggesting these phenotypes may share a common etiology in the GABA and glutamate system.
Part 2: GABAergic and Glutamatergic Brain Gene Expression in Suicide and MDD: Results From Exploratory Analyses
Over 90% of multi-exon genes undergo alternative splicing or RNA editing [Wang et al., 2008, Nilsen and Graveley, 2010]. We used RNA sequencing in brain tissue from the dorsal lateral prefrontal cortex (BA 9) to quantify isoform-level expression of these genes to further understand their potential role in psychiatric disorders. Growing evidence has supported alterations in both of these neurotransmitter systems in MDD and suicide [Stewart and Reid, 2002; Brambilla et al., 2003; Merali et al., 2004; Sanacora and Saricicek, 2007; Levinson et al., 2010; Milak et al., 2015]. Most previous studies using microarrays focused on a few isoforms and/or total gene expression and there are very few studies that have examined alternative splice variants related to GABA and glutamate systems in psychiatric disorders [Tao et al., 2012]. Although our results did not survive correction for multiple comparisons, altered expression of these isoforms in suicide and MDD suggests they may play a role in the pathogenesis of these disorders, and warrant further investigation and replication in larger and/or independent samples.
The dorsolateral prefrontal cortex is involved in the neurobiology of suicidal behavior and major depression [van Heeringen et al., 2011]. Other than one isoform of SLC1A2, which showed higher expression in MDD versus non-MDD, we found generally lower gene-level expression of GABRB1 and some isoforms of GABRB1, SLC1A2, SLC6A11, and GLUL in MDD versus non-MDD, and lower isoform-level expression of GABRB1, GABRB3, and GABRG2 in suicide versus non-suicide. Such a finding of lower expression is consistent with observations of overall altered DNA methylation in the brain of depressed suicides [Labonte et al., 2013; Haghighi et al., 2014]. Lower expression of these genes may thus contribute to GABAergic deficits hypothesized to underlie mood disorders such as MDD as well as suicide [Brambilla et al., 2003].
Previous studies have observed brain expression differences in many of the same genes examined in this study. One study found up-regulation of GABRB1 in the hippocampus in MDD [Sequeira et al., 2009], while another study observed lower GABRB1 protein in cerebellum in MDD [Fatemi et al., 2013], consistent with our findings. Several studies found lower GLUL expression in the prefrontal cortex [Sequeira et al., 2009], locus coeruleus [Bernard et al., 2011], anterior cingulate cortex, and left dorsolateral prefrontal cortex [Choudary et al., 2005] in MDD, which are consistent with our findings. One study found GLUL expression was not different in locus coeruleus in men with MDD [Chandley et al., 2013]. Another study showed GLUL levels in comorbid alcoholism plus MDD were lower than in alcoholics and controls, and lower in MDD subjects than in alcoholics in the left orbitofrontal cortex [Miguel-Hidalgo et al., 2010]. One study found GABRG2 was higher in dorsolateral prefrontal cortex (DLPFC) in MDD [Choudary et al., 2005], while another study found lower GABRG2 brain expression in suicide [Klempan et al., 2009]. Many studies found lower SLC1A2 mRNA expression in the dorsolateral prefrontal cortex [Oh et al., 2014], locus coeruleus [Bernard et al., 2011; Chandley et al., 2013], anterior cingulate cortex, and left dorsolateral prefrontal cortex [Choudary et al., 2005], hippocampus [Medina et al., 2013] and the left orbitofrontal cortex in MDD versus controls [Miguel-Hidalgo et al., 2010], which are consistent with our findings. Whereas we found lower GABRB3 brain expression in suicide, Choudary et al. [2005] found GABRB3 was higher in DLPFC in MDD. Differences in direction of the expression of these genes (i.e., higher in MDD vs. lower in suicides) could be due to differences in the pathophysiology of MDD and the diathesis for suicide.
Part 3: Evidence for eQTL: Results From Exploratory Analyses
In the third part, we examined the SNPs with subthreshold effects on suicide and/or MDD for evidence that they may act as eQTL using the Brain eQTL Almanac in a distinct samples of postmortem brain free of psychiatric diagnoses. Evidence suggests that single nucleotide polymorphisms (SNPs) associated with complex traits (from http://www.genome.gov/gwastudies/) are significantly more likely to be eQTLs than minor-allele-frequency–matched SNPs chosen from high-throughput GWAS platforms [Nicolae et al., 2010]. Therefore SNPs in part 1 may be more likely to be true effects if those SNPs have been found to be eQTL.
In the Brain eQTL Almanac, we found that genotype AA of rs424740 (GABRG2), genotype AA of rs10836356 (SLC1A2), and genotype GG of rs2236418 (GAD2) showed lower expression of some exon-specific probesets for the same gene in the frontal cortex. These three polymorphisms were all associated with suicidal behavior in part 1. Additionally, the expression of many isoforms of these genes appeared to be lower in suicides, suggesting these eQTL may contribute to the etiology of suicide via less GABAergic gene expression in dorsolateral prefrontal cortex. However, our postmortem sample which included both genotyping and gene expression was small and so the results require replication.
Multiple Independent Lines of Evidence Implicate GABRG2 in Suicide
GABRG2 was the only gene that showed effects (P < 0.05, uncorrected) in all three parts of our analyses. Genotype AA of rs424740 (GABRG2) (5:162154029) showed an association with suicide in part 1 (in the postmortem sample, the frequency rs424740 AA genotype was 17% (21/121) in suicide and 7% (6/88) in non-suicide), GABRG2-003 (ENST00000414552) expression was lower in dorsolateral prefrontal cortex in suicides in part 2, and genotype AA of rs424740 (GABRG2) was associated with lower expression of at least 3 exon-specific probesets targeting GABRG2 in part 3 (Supplementary Table SIV). One study found genetic variation within GABRG2 was associated with history of suicide attempt in schizophrenia [Zai et al., 2014], and Klempan et al. [2009] also found lower GABRG2 brain gene expression (ventral prefrontal cortex) in suicides. The gamma 2 subunit coded by GABRG2 was reported to affect the kinetics of GABAA channels and is critical for clustering of major postsynaptic GABAA receptor subtypes and trafficking of the intracellular GABAA receptor [Schweizer et al., 2003]. One possible pathway for the GABRG2's effect is that the genotype AA of rs424740 (GABRG2) may increase risk of suicide through down-regulating expression of the RNA isoform GABRG2-003, which is the largest GABRG2 protein coding RNA isoform (3937 bp length and 515 aa protein), thus affecting the kinetics and cellular turnover of the GABAA receptor, which could lead to GABAergic dysfunction. Future studies involving both genotyping, epigenetics that affect gene expression such as methylation, and gene expression in larger samples of suicides and controls are required to confirm this hypothesis.
We also examined GABRG2 isoform-level group expression differences in each gender separately (Supplementary Table SVI) to test whether group differences were gender-dependent. Results suggest GABRG2 ENST00000414552 isoform SUI versus non-SUI differences are present in males but not females, and conversely for MDD versus non-MDD differences in females versus males (Supplementary Table SVI). However, these results should be interpreted with caution as the total female sample size was small (6 NPC; 3 DSN; 8 DSUI).
CONCLUSION AND LIMITATIONS
In summary, we applied an integrated functional genomics approach, which included genotype and gene expression profiling of GABA and glutamate genes in order to examine potential risk markers for MDD and suicide. Our results highlight the complexity of gene variation and gene expression of GABA and glutamate systems in the human dorsolateral prefrontal cortex. Although exploratory findings in this study are largely negative when correcting for multiple comparisons, we present preliminary evidence that implicates GABRG2 in suicide. The main limitation of this study is modest sample sizes, but it is difficult to obtain larger sample sizes of postmortem cases and controls assessed in such detail clinically, neuropathologically and known to be medication and drug-free. In addition, other types of genetic variation (i.e., copy number variation) as well as epigenetic markers including DNA methylation should be examined in future studies. Variability of subjects’ psychiatric diagnoses in the non-suicide group in part 1 should optimize the design to find biological contributors to suicide independent of specific diagnoses. However, this is also a potential limitation if effects of suicide risk factors interact with diagnosis, an effect that may decrease sensitivity to observe significant associations with suicide.
In Part 2, we examined a group of MDD subjects who did not die by suicide and were not treated with antidepressants, a group also exceedingly difficult to obtain and that has not been previously examined in the literature. A second limitation is that we only examined one brain region and did not distinguish between neurons and glia. It is currently technically difficult to obtain sufficient transcript for quantification from each cell type. Given the importance of some of GABAergic and glutamatergic genes in glial metabolism, and the growing evidence pointing to astro-glial alterations in major depression [Rajkowska and Miguel-Hidalgo, 2007], future studies should investigate cell specific changes in gene expression by means of methods such as laser capture microdissection.
In our internal dataset, of the 38 subjects who had both GWAS and RNA-seq data, only three subjects had a AA genotype for rs424740. Qualitatively these three subjects had lower median GABRG2 gene and isoform-level expression values relative to AT and TT genotypes (Supplementary Table SVII), which is partially consistent in directionality with the eQTL results using BRAINEAC; however, it was not possible to conduct meaningful eQTL analyses in the internal dataset due to small sample size.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by NIMH 5R01MH082041 (PI: JJM) and a Paul Janssen Translational Neuroscience Postdoctoral Fellowship (SPP). Collection and psychiatric characterization of brain samples was supported by MH40210 (PI: VA), MH062185 (PI: JJM), and MH064168 (PI: Andrew Dwork). We would like to thank Peter L. Nagy, Stuart J. Andrews, and Jane Dunning-Broadbent for contributions to RNA sequencing, data processing, interpretation, and analysis.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher's web-site.
REFERENCES
- Bernard R, Kerman IA, Thompson RC, Jones EG, Bunney WE, Barchas JD, Schatzberg AF, Myers RM, Akil H, Watson SJ. Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol Psychiatry. 2011;16:634–646. doi: 10.1038/mp.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges G, Nock MK, Haro Abad JM, Hwang I, Sampson NA, Alonso J, Andrade LH, Angermeyer MC, Beautrais A, Bromet E, Bruffaerts R, de Girolamo G, Florescu S, Gureje O, Hu C, Karam EG, Kovess-Masfety V, Lee S, Levinson D, Medina-Mora ME, Ormel J, Posada-Villa J, Sagar R, Tomov T, Uda H, Williams DR, Kessler RC. Twelve-month prevalence of and risk factors for suicide attempts in the World Health Organization World Mental Health Surveys. J Clin Psychiatry. 2010;71:1617–1628. doi: 10.4088/JCP.08m04967blu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla P, Perez J, Barale F, Schettini G, Soares JC. GABAergic dysfunction in mood disorders. Mol Psychiatry. 2003;8:721–737. 715. doi: 10.1038/sj.mp.4001362. [DOI] [PubMed] [Google Scholar]
- Briere FN, Rohde P, Seeley JR, Klein D, Lewinsohn PM. Comorbidity between major depression and alcohol use disorder from adolescence to adulthood. Compr Psychiatry. 2014;55:526–533. doi: 10.1016/j.comppsych.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandley MJ, Szebeni K, Szebeni A, Crawford J, Stockmeier CA, Turecki G, Miguel-Hidalgo JJ, Ordway GA. Gene expression deficits in pontine locus coeruleus astrocytes in men with major depressive disorder. J Psychiatry Neurosci. 2013;38:276–284. doi: 10.1503/jpn.120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa A, Crisafulli C, Porcelli S, Han C, Patkar AA, Lee SJ, Park MH, Jun TY, Serretti A, Pae CU. Influence of GRIA1, GRIA2 and GRIA4 polymorphisms on diagnosis and response to treatment in patients with major depressive disorder. Eur Arch Psychiatry Clin Neurosci. 2012;262:305–311. doi: 10.1007/s00406-011-0270-y. [DOI] [PubMed] [Google Scholar]
- Chiesa A, Lia L, Lia C, Lee SJ, Han C, Patkar AA, Pae CU, Serretti A. Investigation of possible epistatic interactions between GRIA2 and GRIA4 variants on clinical outcomes in patients with major depressive disorder. J Int Med Res. 2013;41:809–815. doi: 10.1177/0300060513477295. [DOI] [PubMed] [Google Scholar]
- Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, Myers RM, Bunney WE, Jr, Akil H, Watson SJ, Jones EG. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci USA. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domart MC, Benyamina A, Lemoine A, Bourgain C, Blecha L, Debuire B, Reynaud M, Saffroy FR. Association between a polymorphism in the promoter of a glutamate receptor subunit gene (GRIN2A) and alcoholism. Addict Biol. 2012;17:783–785. doi: 10.1111/j.1369-1600.2011.00321.x. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Rooney RJ, Thuras PD. Expression of GABAA alpha2-, beta1- and epsilon-receptors are altered significantly in the lateral cerebellum of subjects with schizophrenia, major depression, and bipolar disorder. Transl Psychiatry. 2013;3:e303. doi: 10.1038/tp.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Kapornai K, Kiss E, Tamas Z, Mayer L, Baji I, Daroczi G, Benak I, Kothencne VO, Dombovari E, Kaczvinszk E, Besnyo M, Gadoros J, Szekely J, Kovacs M, Vetro A, Kennedy JL, Barr CL. Association of the GABRD gene and childhood-onset mood disorders. Genes Brain Behav. 2010;9:668–672. doi: 10.1111/j.1601-183X.2010.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori LM, Turecki G. Gene expression profiling of suicide completers. Eur Psychiatry. 2010;25:287–290. doi: 10.1016/j.eurpsy.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Fonnum F. Glutamate: A neurotransmitter in mammalian brain. J Neurochem. 1984;42:1–11. doi: 10.1111/j.1471-4159.1984.tb09689.x. [DOI] [PubMed] [Google Scholar]
- Galfalvy H, Haghighi F, Hodgkinson C, Goldman D, Oquendo MA, Burke A, Huang YY, Giegling I, Rujescu D, Bureau A, Turecki G, Mann JJ. A genome-wide association study of suicidal behavior. Am J Med Genet B Neuropsychiatr Genet. 2015;168:557–563. doi: 10.1002/ajmg.b.32330. [DOI] [PubMed] [Google Scholar]
- Gelenberg AJ. The prevalence and impact of depression. J Clin Psychiatry. 2010;71:e06. doi: 10.4088/JCP.8001tx17c. [DOI] [PubMed] [Google Scholar]
- Haghighi F, Xin Y, Chanrion B, O’Donnell AH, GE Y, Dwork AJ, Arango V, Mann JJ. Increased DNA methylation in the suicide brain. Dialogues Clin Neurosci. 2014;16:430–438. doi: 10.31887/DCNS.2014.16.3/jmann. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld RM. The epidemiology of depression and the evolution of treatment. J Clin Psychiatry. 2012;73(Suppl 1):5–9. doi: 10.4088/JCP.11096su1c.01. [DOI] [PubMed] [Google Scholar]
- Huang YY, Grailhe R, Arango V, Hen R, Mann JJ. Relationship of psychopathology to the human serotonin1B genotype and receptor binding kinetics in postmortem brain tissue. Neuropsychopharmacology. 1999;21:238–246. doi: 10.1016/S0893-133X(99)00030-5. [DOI] [PubMed] [Google Scholar]
- Ittiwut C, Yang BZ, Kranzler HR, Anton RF, Hirunsatit R, Weiss RD, Covault J, Farrer LA, Gelernter J. GABRG1 and GABRA2 variation associated with alcohol dependence in African Americans. Alcohol Clin Exp Res. 2012;36:588–593. doi: 10.1111/j.1530-0277.2011.01637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TM, Mann JJ. Validity of DSM-III-R diagnosis by psychological autopsy: A comparison with clinician ante-mortem diagnosis. Acta Psychiatr Scand. 1996;94:337–343. doi: 10.1111/j.1600-0447.1996.tb09869.x. [DOI] [PubMed] [Google Scholar]
- Kertes DA, Kalsi G, Prescott CA, Kuo PH, Patterson DG, Walsh D, Kendler KS, Riley BP. Neurotransmitter and neuromodulator genes associated with a history of depressive symptoms in individuals with alcohol dependence. Alcohol Clin Exp Res. 2011;35:496–505. doi: 10.1111/j.1530-0277.2010.01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Choi KH, Baykiz AF, Gershenfeld HK. Suicide candidate genes associated with bipolar disorder and schizophrenia: An exploratory gene expression profiling analysis of post-mortem prefrontal cortex. BMC Genomics. 2007;8:413. doi: 10.1186/1471-2164-8-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempan TA, Sequeira A, Canetti L, Lalovic A, Ernst C, Ffrench-Mullen J, Turecki G. Alteredexpression ofgenesinvolved inATPbiosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Mol Psychiatry. 2009;14:175–189. doi: 10.1038/sj.mp.4002110. [DOI] [PubMed] [Google Scholar]
- Labonte B, Suderman M, Maussion G, Lopez JP, Navarro-Sanchez L, Yerko V, Mechawar N, Szyf M, Meaney MJ, Turecki G. Genome-wide methylation changes in the brains of suicide completers. Am J Psychiatry. 2013;170:511–520. doi: 10.1176/appi.ajp.2012.12050627. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Levey DF, Ayalew M, Palmer L, Gavrin LM, Jain N, Winiger E, Bhosrekar S, Shankar G, Radel M, Bellanger E, Duckworth H, Olesek K, Vergo J, Schweitzer R, Yard M, Ballew A, Shekhar A, Sandusky GE, Schork NJ, Kurian SM, Salomon DR, Niculescu AB., III. Discovery and validation of blood biomarkers for suicidality. Mol Psychiatry. 2013;18:1249–1264. doi: 10.1038/mp.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson AJ, Fitzgerald PB, Favalli G, Blumberger DM, Daigle M, Daskalakis ZJ. Evidence of cortical inhibitory deficits in major depressive disorder. Biol Psychiatry. 2010;67:458–464. doi: 10.1016/j.biopsych.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Mandelli L, Serretti A. Gene environment interaction studies in depression and suicidal behavior: An update. Neurosci Biobehav Rev. 2013;37:2375–2397. doi: 10.1016/j.neubiorev.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Mann JJ, Oquendo MA, Watson KT, Boldrini M, Malone KM, Ellis SP, Sullivan G, Cooper TB, Xie S, Currier D. Anxiety in major depression and cerebrospinal fluid free gamma-aminobutyric acid. Depress Anxiety. 2014;31:814–821. doi: 10.1002/da.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina A, Burke S, Thompson RC, Bunney W, Jr, Myers RM, Schatzberg A, Akil H, Watson SJ. Glutamate transporters: A key piece in the glutamate puzzle of major depressive disorder. J Psychiatr Res. 2013;47:1150–1156. doi: 10.1016/j.jpsychires.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Merali Z, Du L, Hrdina P, Palkovits M, Faludi G, Poulter MO, Anisman H. Dysregulation in the suicide brain: mRNA expression of corticotropin-releasing hormone receptors and GABA(A) receptor subunits in frontal cortical brain region. J Neurosci. 2004;24:1478–1485. doi: 10.1523/JNEUROSCI.4734-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel-Hidalgo JJ, Waltzer R, Whittom AA, Austin MC, Rajkowska G, Stockmeier CA. Glial and glutamatergic markers in depression, alcoholism, and their comorbidity. J Affect Disord. 2010;127:230–240. doi: 10.1016/j.jad.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milak MS, Proper CJ, Mulhern ST, Parter AL, Kegeles LS, et al. A pilot in vivo proton magnetic resonance spectroscopy study of amino acid neurotransmitter response to ketamine treatment of major depressive disorder. Mol. Psychiatry. 2015 doi: 10.1038/mp.2015.83. doi: 10.1038/mp.2015.83 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler H. The GABA system in anxiety and depression and its therapeutic potential. Neuropharmacology. 2012;62:42–53. doi: 10.1016/j.neuropharm.2011.08.040. [DOI] [PubMed] [Google Scholar]
- Murphy TM, Ryan M, Foster T, Kelly C, McClelland R, O'Grady J, Corcoran E, Brady J, Reilly M, Jeffers A, Brown K, Maher A, Bannan N, Casement A, Lynch D, Bolger S, Tewari P, Buckley A, Quinlivan L, Daly L, Kelleher C, Malone KM. Risk and protective genetic variants in suicidal behaviour: Association with SLC1A2, SLC1A3, 5-HTR1B &NTRK2 polymorphisms. Behav Brain Funct. 2011;7:22. doi: 10.1186/1744-9081-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung W, Song J, Lim SW, Won HH, Kim S, Lee Y, Kang HS, Lee H, Kim JW, Carroll BJ, Kim DK. Genetic association study of individual symptoms in depression. Psychiatry Res. 2012;198:400–406. doi: 10.1016/j.psychres.2011.12.037. [DOI] [PubMed] [Google Scholar]
- NCIPC. 2013 http://www.cdc.gov/violenceprevention/pdf/suicide-datasheet-a.pdf.
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Oh D, Son H, Webster MJ, Weickert CS, Kim SH. An association between the reduced levels of SLC1A2 and GAD1 in the dorsolateral prefrontal cortex in major depressive disorder: Possible involvement of an attenuated RAF/MEK/ERK signaling pathway. J Neural Transm. 2014;121:783–792. doi: 10.1007/s00702-014-1189-z. [DOI] [PubMed] [Google Scholar]
- Oquendo MA, Friend JM, Halberstam B, Brodsky BS, Burke AK, Grunebaum MF, Malone KM, Mann JJ. Association of comorbid posttraumatic stress disorder and major depression with greater risk for suicidal behavior. Am J Psychiatry. 2003;160:580–582. doi: 10.1176/appi.ajp.160.3.580. [DOI] [PubMed] [Google Scholar]
- Owen RT. Glutamatergic approaches in major depressive disorder: Focus on ketamine, memantine, and riluzole. Drugs Today (Barc) 2012;48:469–478. doi: 10.1358/dot.2012.48.7.1832873. [DOI] [PubMed] [Google Scholar]
- Pantazatos SP, Andrews SJ, Dunning-Broadbent J, Pang J, Huang YY, Arango V, Nagy PL, John Mann J. Isoform-level brain expression profiling of the spermidine/spermine N1-Acetyltransferase1 (SAT1) gene in major depression and suicide. Neurobiol Dis. 2015;79:123–134. doi: 10.1016/j.nbd.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty F. GABA and mood disorders: A brief review and hypothesis. J Affect Disord. 1995;34:275–281. doi: 10.1016/0165-0327(95)00025-i. [DOI] [PubMed] [Google Scholar]
- Posner K, Oquendo MA, Gould M, Stanley B, Davies M. Columbia Classification Algorithm of Suicide Assessment (C-CASA): Classification of suicidal events in the FDA's pediatric suicidal risk analysis of antidepressants. Am J Psychiatry. 2007;164:1035–1043. doi: 10.1176/appi.ajp.164.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu M, Zhang Z, Xu Z, Shi Y, Geng L, Yuan Y, Zhang X, Reynolds GP. Influence of genetic polymorphisms in the glutamatergic and GABAergic systems and their interactions with environmental stressors on antidepressant response. Pharmacogenomics. 2013;14:277–288. doi: 10.2217/pgs.13.1. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Miguel-Hidalgo JJ. Gliogenesis and glial pathology in depression. CNS Neurol Disord Drug Targets. 2007;6:219–233. doi: 10.2174/187152707780619326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanacora G, Saricicek A. GABAergic contributions to the patho-physiology of depression and the mechanism of antidepressant action. CNS Neurol Disord Drug Targets. 2007;6:127–140. doi: 10.2174/187152707780363294. [DOI] [PubMed] [Google Scholar]
- Sapolsky RM. Glucocorticoids and hippocampal atrophy in neuro-psychiatric disorders. Arch Gen Psychiatry. 2000;57:925–935. doi: 10.1001/archpsyc.57.10.925. [DOI] [PubMed] [Google Scholar]
- Schweizer C, Balsiger S, Bluethmann H, Mansuy IM, Fritschy JM, Mohler H, Luscher B. The gamma 2 subunit of GABA(A) receptors is required for maintenance of receptors at mature synapses. Mol Cell Neurosci. 2003;24:442–450. doi: 10.1016/s1044-7431(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Klempan T, Canetti L, Ffrench-Mullen J, Benkelfat C, Rouleau GA, Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, Rehal S, Klempan T, Gratton A, Benkelfat C, Rouleau GA, Mechawar N, Turecki G. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS ONE. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval G, Shmulewitz D, Wall MM, Aharonovich E, Spivak B, Weizman A, Hasin D. Alcoholdependenceandsuicide-relatedideation/behaviors in an Israeli household sample, with and without major depression. Alcohol Clin Exp Res. 2014;38:820–825. doi: 10.1111/acer.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibille E, Arango V, Galfalvy HC, Pavlidis P, Erraji-Benchekroun L, Ellis SP, John Mann J. Gene expression profiling of depression and suicide in human prefrontal cortex. Neuropsychopharmacology. 2004;29:351–361. doi: 10.1038/sj.npp.1300335. [DOI] [PubMed] [Google Scholar]
- Sokolowski M, Ben-efraim YJ, Wasserman J, Wasserman D. Glutamatergic GRIN2B and polyaminergic ODC1 genes in suicide attempts: Associations and gene-environment interactions with childhood/adolescent physical assault. Mol Psychiatry. 2013;18:985–992. doi: 10.1038/mp.2012.112. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Williams JB, Gibbon M, First MB. The structured clinical interview for DSM-III-R (SCID). I: History, rationale, and description. Arch Gen Psychiatry. 1992;49:624–629. doi: 10.1001/archpsyc.1992.01820080032005. [DOI] [PubMed] [Google Scholar]
- Stewart CA, Reid IC. Antidepressant mechanisms: Functional and molecular correlates of excitatory amino acid neurotransmission. Mol Psychiatry. 2002;7(Suppl 1):S15–S22. doi: 10.1038/sj.mp.4001014. [DOI] [PubMed] [Google Scholar]
- Tao R, Li C, Newburn EN, Ye T, Lipska BK, Herman MM, Weinberger DR, Kleinman JE, Hyde TM. Transcript-specific associations of SLC12A5 (KCC2) in human prefrontal cortex with development, schizophrenia, and affective disorders. J Neurosci. 2012;32:5216–5222. doi: 10.1523/JNEUROSCI.4626-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova C, Tucci M, Sartore D, Cavarzeran F, Di Pietra L, Barzon L, Palu G, Ferrara SD. GABA receptors, alcohol dependence, and criminal behavior. J Forensic Sci. 2013;58:1227–1232. doi: 10.1111/1556-4029.12201. [DOI] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp A, Oh H, Guilloux JP, Martinowich K, Lewis DA, Sibille E. Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. Am J Psychiatry. 2012;169:1194–1202. doi: 10.1176/appi.ajp.2012.12020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unschuld PG, Ising M, Specht M, Erhardt A, Ripke S, Heck A, Kloiber S, Straub V, Brueckl T, Muller-Myhsok B, Holsboer F, Binder EB. Polymorphisms in the GAD2 gene-region are associated with susceptibility for unipolar depression and with a risk factor for anxiety disorders. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:1100–1109. doi: 10.1002/ajmg.b.30938. [DOI] [PubMed] [Google Scholar]
- van Heeringen C, Bijttebier S, Godfrin K. Suicidal brains: A review of functional and structural brain studies in association with suicidal behaviour. Neurosci Biobehav Rev. 2011;35:688–698. doi: 10.1016/j.neubiorev.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization 2014 http://www.who.int/mediacentre/factsheets/fs398/en.
- Zai CC, Zai GC, Tiwari AK, Manchia M, de Luca V, Shaikh SA, Strauss J, Kennedy JL. Association study of GABRG2 polymorphisms with suicidal behaviour in schizophrenia patients with alcohol use disorder. Neuropsychobiology. 2014;69:154–158. doi: 10.1159/000358839. [DOI] [PubMed] [Google Scholar]
- Zarate CA, Jr, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA, Charney DS, Manji HK. ArandomizedtrialofanN-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry. 2006;63:856–864. doi: 10.1001/archpsyc.63.8.856. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Danbolt NC. GABA and glutamate transporters in brain. Front Endocrinol (Lausanne) 2013;4:165. doi: 10.3389/fendo.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.