Summary
FLRTs are cell-adhesion molecules with emerging functions in cortical development and synapse formation. Their extracellular regions interact with LPHNs to mediate synapse development, and with UNC5/Netrin receptors to control the migration of neurons in the developing cortex. Here, we present the crystal structures of FLRT3 in isolation and in complex with LPHN3. The FLRT3/LPHN3 structure reveals that LPHN3 binds to FLRT3 at a distinct site from UNC5. Structure-based mutations specifically disrupt FLRT3/LPHN3 binding, but do not disturb their interactions with other proteins or their cell-membrane localization. Thus, they can be used as molecular tools to dissect the functions of FLRTs and LPHNs in vivo. Our results suggest that UNC5 and LPHN3 can simultaneously bind to FLRT3 forming a trimeric complex and that FLRT3 may form trans-synaptic complexes with both LPHN3 and UNC5. These findings provide molecular insights for understanding the role of cell-adhesion proteins in synapse function.
Keywords: attention deficit hyperactivity disorder, synaptic junction, neural development, neuronal communication, adhesion-type GPCR
Introduction
During neural development, immature neurons migrate from their birthplaces in the embryo to their final positions and complete synaptic circuits. Outgrowth of axons and dendrites from neurons, guidance of the motile growth cone through the embryo towards postsynaptic partners and finally the generation of synapses between these axons and their postsynaptic partners are essential landmarks of neural development that are each mediated by a complex interaction network of cell-surface proteins on the nerve cells. Distinct properties of each neuron and its synaptic connections are also a function of the cell-adhesion molecules expressed on its surface and the interactions these molecules are involved in. An imperfection in any of these steps may lead to malformations or inappropriate connectivity of the brain and is believed to be involved in many neurodevelopmental disorders such as attention deficit hyperactivity disorder, autism and brain cancers (Asherson and Gurling, 2012; Sudhof, 2008). Although the role of binary protein-protein interactions are commonly studied and understood, it is likely that the presence or absence of ternary, quaternary and even higher-order protein-protein interactions are determinants for the build-up of a network as complex as is in the brain. However, the molecules and the underlying mechanisms of such complex phenomena are unclear.
Fibronectin leucine-rich repeat transmembrane (FLRTs) proteins are cell-surface molecules that contribute to early embryonic, vascular, and neural development (Egea et al., 2008; Leyva-Diaz et al., 2014; Maretto et al., 2008; Muller et al., 2011; O’Sullivan et al., 2012; Yamagishi et al., 2011). FLRTs are type-I membrane proteins with extracellular regions consisting of a leucine-rich repeat (LRR) domain with ten LRR repeats, a fibronectin type 3 domain followed by a juxtamembrane linker (Lacy et al., 1999) (Figure 1A). FLRTs are expressed in many tissues, including brain (Bottcher et al., 2004; Lacy et al., 1999), and different FLRT isoforms (FLRT1-3) have different cell type-specific expression patterns in the hippocampus and cortex (Allen Mouse Brain Atlas, 2009). FLRTs interact with the axonal guidance receptors UNC5B and UNC5D proteins (Yamagishi et al., 2011). Their ectodomains are suggested to shed from neurons to act as repulsive cues in axon guidance and neuron migration (Sollner and Wright, 2009; Yamagishi et al., 2011). FLRTs have also been identified as high affinity endogenous ligands for latrophilins (LPHNs) and were suggested to play a role in glutamatergic synapse development (O’Sullivan et al., 2012; O’Sullivan et al., 2014). Moreover, FLRT proteins interact with each other and may promote homotypic cell adhesion, and are additionally implicated in FGF (fibroblast growth factor) signaling during development (Karaulanov et al., 2006) (Bottcher et al., 2004) (Maretto et al., 2008). The N-terminal LRR domain of FLRTs is reported to be involved in all reported interactions of FLRTs (Karaulanov et al., 2009; Karaulanov et al., 2006). The involvement of LRR domains of FLRTs in numerous heterophilic interactions suggest a central role for FLRTs in neural development and raises the necessity to identify the specific binding sites for each of LRR domain interactions (such as Latrophilin, UNC5 and homophilic FLRT binding sites on FLRT) in order to unambiguously dissect the role of each interaction for FLRT function.
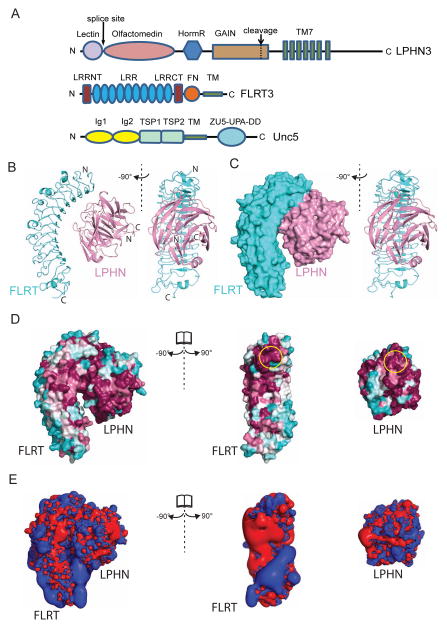
Figure 1. Crystal structure of the FLRT3/ LPHN3 complex.
(A) Schematic diagram of vertebrate LPHN3, FLRT3, and UNC5D showing their domains. (B) Ribbon diagram of the FLRT3/LPHN3 heterodimer. (C) Surface representation of the FLRT3/LPHN3 heterodimer. (D) The structure of the FLRT3/LPHN3 complex is shown in surface representation on which the conservation of residues are mapped from most conserved (magenta) to least conserved (cyan) (using the ConSurf server (Landau et al., 2005)). The LPHN-binding site and the FLRT-binding site, where they come closest to each other, are indicated by yellow circles. (E) Positive (blue) and negative (red) electrostatic isosurfaces calculated at contours ±2 kT/e using PDB2QPR and APBS for LPHN3, FLRT3, and the complex structures. The structures in C, D and E are shown in a similar orientation as in B. See also Figure S1.
Latrophilins (LPHN1-3) are cell-surface molecules that belong to the adhesion-type G-protein coupled receptor (GPCR) family (Krasnoperov et al., 1996; Lelianova et al., 1997). LPHN1 was identified as the calcium-independent receptor for α-latrotoxin, a black widow spider toxin that triggers massive neurotransmitter release from neurons and neuroendocrine cells (Deak et al., 2009; Krasnoperov et al., 1997; Lelianova et al., 1997; Sudhof, 2001; Sugita et al., 1999). Mutations of LPHNs have been linked to attention deficit hyperactivity disorder as well as numerous cancers (Arcos-Burgos et al., 2010; Kan et al., 2010; O’Hayre et al., 2013). LPHNs are highly expressed in the brain (Sugita et al., 1998), and were shown to function as heterophilic cell adhesion molecules in processes such as synapse formation or maintenance. They are the only adhesion-type GPCRs besides flamingo-like CESLR proteins that are conserved between vertebrates and invertebrates. In C. elegans, LPHN1 homolog Lat-1 is required for the alignment of cell division planes to the anterior-posterior axis during development (Langenhan et al., 2009). In Drosophila, Latrophilin/dCIRL sensitizes the chordotonal neurons to modulate the perception of mechanical signals (Scholz et al., 2015). In vertebrates, LPHN3 and FLRT3 were reported to interact in trans through their ectodomains to mediate cell adhesion, an interaction that promotes the development of glutamatergic synapses (O’Sullivan et al., 2012). LPHNs have large extracellular sequences that contain an N-terminal lectin domain, a central olfactomedin (Olf) domain, a serine/threonine-rich region, a hormone-binding domain, and a C-terminal conserved GPCR Autoproteolysis Inducing (GAIN) domain that mediates autoproteolysis (Arac et al., 2012; Sugita et al., 1998) (Krasnoperov et al., 1996; Lelianova et al., 1997) (Figure 1A). The LPHN Olfactomedin (Olf) domain is required for its synapse-promoting function and also for FLRT binding (O’Sullivan et al., 2014). Similar to FLRTs, LPHNs are also involved in numerous interactions. In addition to FLRTs, LPHNs interact with the members of the teneurin/ODZ family proteins (Boucard et al., 2014; Levine et al., 1994; Silva et al., 2011) and with neurexins (Boucard et al., 2012).
Uncoordinated-5 (UNC5 A–D) receptors are type-I membrane proteins with extracellular regions containing two immunoglobulin domains and two thrombospondin domains (Figure 1A). UNC5 receptors bind to secreted Netrin ligands that act as repulsive cues during the development of the neural system to regulate neuronal migration (Sun et al., 2011). However, the migration of UNC5D-expressing neurons in the developing cortex, where netrin is not expressed, is regulated by FLRTs (Yamagishi et al., 2011). The crystal structure of UNC5D in complex with the LRR domain of FLRT2 has revealed the binding interface, and it was suggested that during cortical development, the repulsive FLRT-UNC5 interaction mediates radial (upwards) migration of neurons, whereas the adhesive homophilic FLRT-FLRT interaction is important for the tangential (sideways) migration of neurons (Seiradake et al., 2014). However, the effect of other interactions on the function of these proteins, for example a possible simultaneous interaction of FLRT with LPHN3 was not clarified. In particular, the crystal structure suggests that FLRT dimers may exist in cis, casting doubt on the notion that hemophilic cell-adhesion can be mediated by FLRTs.
FLRTs and LPHNs work together with other proteins such as UNC5s to support the formation of proper neuronal connections. Although the FLRT/UNC5 crystal structure is available, the FLRT/ LPHN structure and well-characterized mutations that abolish a single interaction of each protein while maintaining the other interactions are not known. The presence of multiple binding partners and their compatibilities with each other, the formation of cis, trans or both interactions between proteins, and the pre-synaptic or post-synaptic localization of each protein are additional complications that need to be addressed in order to decipher the mechanism of action of these proteins in neural development. We determined the three-dimensional structures of FLRT3 in isolation and in a complex with LPHN3 by X-ray crystallography. The structure of the complex reveals a large LPHN3-binding interface on the concave surface of FLRT3, which was previously reported as the dimerization surface. We designed multiple mutations on the LPHN3-binding surface of FLRT3 and on the FLRT3-binding surface of LPHN3 to specifically disrupt the interaction between them. We confirmed the wild type-like folding and trafficking of the mutant FLRT3s and mutant LPHN3s. The mutations that disrupt the FLRT3/ LPHN3 interaction without interfering with folding or trafficking confirm the binding interface revealed by the FLRT3/ LPHN3 complex structure. These mutants can be used as molecular tools to dissect the FLRT3/ LPHN3 interaction in vivo. We also studied the previously reported FLRT3 mutants called “FLRT3 dimerization mutant” and “UNC5 binding mutant” (Seiradake et al., 2014), and found that the FLRT3 dimerization mutant that was reported to abolish tangential (sideways) migration of neurons during cortex development is indeed defective in LPHN3 binding more severely than in dimerization. Our flow cytometry experiments showed that FLRT3, LPHN3 and UNC5 form a trimeric complex. FLRT3 binds the other two proteins simultaneously and bridges them, although LPHN3 and UNC5 do not directly interact with each other. Finally, we demonstrate that FLRT/LPHN pair as well as FLRT/UNC5 pair can interact in trans and cause cell-aggregation, suggesting that these protein-protein complexes are trans-synaptic complexes.
Results
Structure of the FLRT3/ LPHN3 complex
To determine the structure of the FLRT3/ LPHN3 complex, the extracellular LRR domain of human FLRT3 (residues K29-D357, corresponding to the N-terminal cap, LRR repeats and C-terminal cap) and the olfactomedin domain of LPHN3 (residues V132-G392) were produced in insect cells using the baculovirus expression system (residue numbering includes signal peptide even when it is absent). After purification of individual proteins, the complex was formed, and purified by size exclusion chromatography. The formation of the complex is independent of calcium as observed by native gel analysis (data not shown). Crystals of the complex were obtained in space group P43 with eight FLRT3/ LPHN3 complexes in the asymmetric unit, and diffracted to dmin = 3.6 Å (Table 1). The complex structure was obtained by molecular replacement using the available mouse FLRT3 structure (PDB ID 4V2E) and the mycolin olfactomedin structure (PDB ID 4WXQ, kindly provided by Raquel Lieberman before its release) and was further refined by the recently released human LPHN3 olfactomedin/lectin structure (PDB ID 5AFB) (Donegan et al., 2015; Jackson et al., 2015; Seiradake et al., 2014).
Table 1.
Data collection and refinement statistics
| FLRT3 | FLRT3/LPHN3 Complex | |
|---|---|---|
| Data collection | ||
| Wavelength | 0.96638 | 1.00883 |
| Space group | P21 | P43 |
| Cell dimensions | ||
| a, b, c (Å) | 76.140 106.581 84.152 | 102.268 102.268 419.23 |
| α β γ (°) | 90 90.31 90 | 90 90 90 |
| Resolution (Å) | 50 - 2.601 (2.64 - 2.60)* | 73.2 - 3.60 (3.66-3.60)* |
| Rsym | 0.144 (0.640) | 0.262 (0.634) |
| CC1/2 | 0.763 (high res shell) | 0.671(high res shell) |
| Completeness (%) | 95.7 (73.1) | 98.3 (79.3) |
| Redundancy | 8.8 (1.6) | 8.9 (1.5) |
| Refinement | ||
| Resolution (Å) | 45 - 2.60 (2.67 - 2.60) | 73.20 - 3.60 (3.71 – 3.60) |
| No. reflections | 39414 (1492) | 45263 (1507) |
| Rwork | 0.2344 (0.2973) | 0.2004 (0.2508) |
| Rfree | 0.2956 (0.4364) | 0.2621 (0.3460) |
| No. atoms | ||
| Protein | 10,300 | 18360 |
| Ligand/ion | 56/0 | 56/4 |
| B-factors | ||
| Protein | 48.138 | 117.938 |
| Ligand/ion | 40.311 | 118.261 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.0123 | 0.006 |
| Bond angles (°) | 1.330 | 0.983 |
| Ramachandran statistics | ||
| Favored (%) | 90.28 | 93.5% |
| Outliers (%) | 0 | 0 |
Values in parentheses are for highest-resolution shell.
We also determined the crystal structure of the isolated human FLRT3 at a space group different than the previously published mouse FLRT3 (Figure S1). Crystals of FLRT3 were obtained in space group P2 with four molecules in the asymmetric unit and diffracted to dmin=2.6 Å. The structure was solved by molecular replacement using the available mouse FLRT3 structure as a model (PDB ID 4V2E) (Table 1). As described previously, FLRT3 has a horseshoe shape with dimensions ~83Å × 32Å × 40Å and contains ten LRR repeats (Seiradake et al., 2014).
The crystal structure of the FLRT3/LPHN3 complex comprises a heterodimer in which the five-bladed beta-propeller LPHN3 olfactomedin domain (dimensions ~52 Å × 43 Å × 43 Å) docks into the groove on the concave surface of the horseshoe-shaped FLRT3 LRR domain, creating a large buried surface area of 1550 Å2 (Figure 1B,C). There were no other significant hetero-typic interactions in the crystal lattice (Figure S1D,E, see Figure S1A–C for a discussion of homotypic interactions). Apart from sidechain rotamer changes, no major conformational changes are observed when the complex structure is compared with the individual structures of FLRT3 or LPHN3. N-linked glycosylation is observed at one (residue N226) glycosylation site on FLRT3, and three disulfide bonds (C31–C37, C35–C44, C309–C334) are observed in the FLRT3 LRR domain. A single disulfide bond (C135–C317) and no glycosylation is observed in LPHN3 Olfactomedin domain. The relative orientation of LPHN3 to FLRT3 is consistent with the previous prediction based on mutagenesis (Jackson et al., 2015). However, numerous differences exist in the fine details of the molecular interactions.
The LPHN3-binding region on FLRT3 is located at the top of the inside concave surface of the LRR domain close to the N-terminal cap (Figure 1B,C). This region is away from the UNC5-binding region on FLRT3 (Seiradake et al., 2014). However, it is at the same side as the previously reported dimerization surface (see further below for the compatibility of these interactions with each other). The FLRT3-binding region on LPHN3 is located at the top/side surface of the olfactomedin domain and is mediated by the conserved loops primarily in the second and third blades. The blades of the olfactomedin domain are asymmetrical, and the fifth blade from where the N- and C-termini emerge is larger than the other blades, spanning as much space as almost two blades. This type of beta-propeller structure was recently observed in the structure of olfactomedin domain of gliomedin as well (Han and Kursula, 2015).
There is electron density at the same position as of the Ca2+ ion in the isolated Olf-Lectin structure (PDB ID 5AFB). The FLRT3 binding site is opposite to the N- and C-termini of the olfactomedin domain which connect to the lectin and STP/HormR/GAIN/TM domains, respectively. The alternatively spliced sequence (residues K127–K131) between the lectin and olfactomedin domains of LPHN3 is located immediately N-terminal of the olfactomedin domain, distant from the FLRT3 binding region, explaining why its presence does not affect FLRT3 binding to LPHN3 (Boucard et al., 2014). The FLRT3/ LPHN3 complex structure also explains why FLRT binding is mediated by only the olfactomedin domain (O’Sullivan et al., 2014), as opposed to teneurin binding that is mediated by both the lectin and olfactomedin domains and the splice insert between them (Boucard et al., 2014). As both the N- and C-termini of the olfactomedin domain reside next to each other, it is not possible to conclude from the FLRT/LPHN structure whether the FLRT3/ LPHN3 interaction is in trans or cis.
The sequence identities between FLRT LRR domains and the sequence identity between the LPHN Olfactomedin domains are high (Table S1,S2). Although invertebrate LPHN homologs lack an olfactomedin domain, it is the most conserved extracellular domain of LPHNs in vertebrates, suggesting an essential function. Similarly, no direct FLRT homolog is readily identifiable in invertebrates. To visualize conserved and variable regions of FLRT LRR domains and LPHN Olfactomedin domains, we mapped the conservation of residues on the FLRT3/LPHN3 complex structure, and colored residues from most conserved to least conserved. The binding surfaces of FLRTs and LPHNs to each other correspond to one of the most conserved regions (yellow ovals in Figure 1D). Visualization of the electrostatic surface potential on the complex structure shows that the inside concave surface of the LRR domain is positively and negatively charged on separate sides, suggesting that charge-mediated interactions are likely to be mediated by this concave surface (Figure 1E). The binding interface of FLRT and LPHN is not highly charged.
Mutations at the binding surface disrupt the interaction of FLRT3 and LPHN3
The high affinity of the FLRT3/ LPHN3 complex (Kd = 220 nM-260 nM as determined by Bio-layer interferometry (BLITZ), Figure S2, (O’Sullivan et al., 2012)) is achieved by a combination of interactions, comprised of hydrogen bonds, salt bridges, hydrophobic interactions and long-range electrostatic interactions (Figure 2A). The docking of the complementary surfaces of the round-shaped Olfactomedin domain into the concave surface of the LRR domain creates a large interface. Our observed binding affinity and the observed interface area of the LPHN3/FLRT3 complex are in line with a study that demonstrates correlation of the binding affinity with interface area (Chen et al., 2013). Numerous polar residues are at the binding interface (Figure 2A).
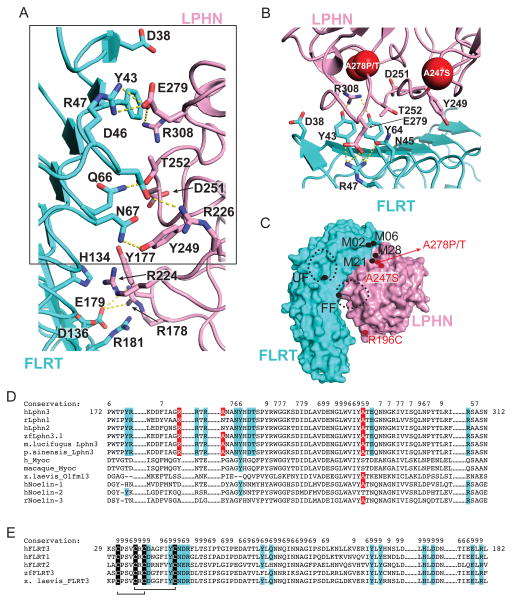
Figure 2. Binding interface between FLRT3 and LPHN3.
A) Ribbon diagram showing the entire interface between FLRT3 and LPHN3. Polar interactions are shown by dashed lines. Residues at the binding interface are shown as sticks. B) Close-up view of the binding interface between FLRT3 and LPHN3 (box in A). Labeled residues correspond to the residues that were mutated in the selected M02, M06, M21 and M28 mutants. Red spheres show the locations of the attention deficit hyperactivity disorder and cancer mutations on LPHNs. (C) Schematic drawing of locations of mutations studied in this manuscript (labeled black) and three disease mutations (labeled red). Dashed black spheres indicate the N-linked glycosylation moieties introduced as a result of mutagenesis. Carbohydrates on FLRT3-FF mutant clashes with LPHN3. (D) Sequence alignment of the FLRT binding site on different olfactomedin domain sequences. The key residues involved in binding to FLRT are highlighted in cyan. Disease mutations are highlighted in red. The residues essential for binding to FLRTs are conserved only in LPHNs. Conservation of each residue is labeled. Value 9 corresponds to highest conservation. (E) Sequence alignment of the LPHN binding site on FLRT sequences. The conserved cysteines are highlighted in black. The key residues involved in binding to LPHN are highlighted in cyan. Disulfide bonds are indicated by black lines. See also Figure S2.
Mapping the residues that are mutated in some attention deficit hyperactivity disorder (A247S in human LPHN3 (Domene et al., 2011)) and cancer cases (R196C, A278P and A278T in human LPHN2 (Kan et al., 2010; O’Hayre et al., 2013)) on the FLRT3/ LPHN3 structure reveals that A247S and A278P/T mutations map close to the binding interface, and thus may cause a direct defect in binding to FLRT3 (Figure 2B,C). The R196C mutation is at the distant edge of the binding interface. Interestingly, the A278P/T mutation that is very close to the binding interface is mutated to two different residues in cancers and is reported by two different studies (Kan et al., 2010; O’Hayre et al., 2013).
In order to specifically abolish a single interaction without interfering with other interactions or cell-surface localization of FLRT3 and LPHN3, and to confirm the validity of the binding interface that is observed in the FLRT/LPHN complex structure, we designed surface mutations on full-length FLRT3 (FL-FLRT3) and full length LPHN3 (FL-LPHN3) that change only a few atoms on the protein surface rather than introduce large posttranslational modifications (Figure 2B). We designed mutations on LPHN3 to abolish binding to FLRT3 and mutations on FLRT3 to abolish binding to LPHN3. In designing all of these mutants, we tried to avoid any interference with the folding of the proteins. We also studied the previously published FLRT3 dimerization mutant (“FF”: R181N, D183T) and the FLRT3 mutant that abolishes UNC5 binding (“UF”: H165N). (See Table 2 for a full list of all mutations on the complex structure; see Figure 2C for a schematic representation of select important mutations; see Figure 2D,E for conservation of the residues at the binding sites).
Table 2.
Summary of mutations and their characteristics
| hLPHN3 | |||
|---|---|---|---|
| Mutant name | Mutations | Cell Surface Localization | FLRT3 LRR Binding |
| wild type | +++ | +++ | |
| M19 | Y249A | +++ | +++ |
| M20 | Y249A, D251A, T252A | +++ | + |
| M21* | Y249A, D251A, T252A, R308A | +++ | − |
| M22 | Y177A, Y249A, R205A | ++ | + |
| M23 | N248A, Y249A | + | − |
| M24 | N248A, Y249A, E279A | + | − |
| M27 | D251A, T252A | ++ | + |
| M28* | Y249A, D251A, T252A, E279A, R308A | +++ | − |
| hFLRT3 | ||||
|---|---|---|---|---|
| Mutant name | Mutations | Cell Surface Localization | LPHN3 Olf Binding | Dimerization |
| wild type | +++ | +++ | +++ | |
| M01 | Y89A, Y91A | ++ | − | |
| M02 | Y43A, Y64A | +++ | − | +++ |
| M03 | Y43A, Y64A, Y89A, Y91A | + | − | |
| M04 | Y64A, Q66A, N67A | − | − | |
| M05 | Y43A, N45A, R47A | +++ | − | |
| M06 | Y43A, N45A, R47A, D38A | +++ | − | |
| M08 | R117A, E113A | +++ | ++ | +++ |
| M11 | R181A, D183A | +++ | ||
| M26 | Q66A, N67A | + | − | |
| M35 | Y43A, Y89A, Y91A | + | − | |
| FLRT UF | H165N | +++ | +++ | +++ |
| FLRT FF | R181N, D183T | +++ | − | + |
We first examined the expression levels and surface transport of all FL-FLRT3 and FL-LPHN3 mutants in order to eliminate misfolded mutants that are likely to be poorly expressed, and unlikely to reach the cell surface. The FL-FLRT3 and FL-LPHN3 had extracellular N-terminal Myc and Flag tags, respectively, to allow for detection of expression levels and cell-surface localization of wild type and mutant proteins. We expressed wild-type and mutant full-length FLRT3 and LPHN3 in transfected HEK293 cells. Cells were stained without detergent permeabilization (to label only the cell-surface localized protein), with an antibody suitable to react with the extracellular tag on the proteins, and the amount of surface-exposed FL-FLRT3 or FL-LPHN3 was detected by indirect immunofluorescence using flow cytometry (Figures 3,4,S3,S4,S5 and Table S3).
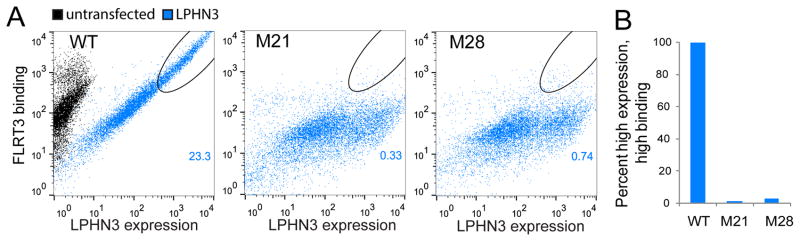
Figure 3. Mutations on LPHN3 abolish FLRT3 binding but do not interfere with proper cell-surface localization.
(A) Wild-type and mutant full-length LPHN3 proteins were tested for surface expression in HEK293 cells as well as their ability to bind soluble FLRT3 LRR domain using flow cytometry. Non-permeabilized HEK293 cells expressing N-terminally FLAG-tagged FL-LPHN3 were stained with mouse anti-FLAG primary antibody and anti-mouse FITC conjugated secondary antibody. LPHN3 surface expression was measured as green fluorescence from FITC (x-axis). FLRT3 LRR binding to FL-LPHN3-expressing cells was measured by monitoring red fluorescence of DyLight attached to neutravidin (y-axis). (refer to Figure S3B and Table S3 for detailed experimental setup and experimental conditions). Dot plot shows correlation between LPHN3 expression and FLRT3 binding in LPHN3-transfected cells (blue), or in untransfected cells (black). Black ovals on the plot show the “high LPHN3 expression and high FLRT3 binding” gate. The number on the plot represents the percent of all events that is in the high expression and high binding gate. 100nM purified wild-type His-tagged FLRT3 LRR domain was incubated with Biotin-Tris-NTA and was tetramerized with neutravidin to increase avidity before binding to cells (See setup in Figure S3B). Mutant FL-LPHN3 constructs (M21, M28) show proper surface expression (x-axis), but do not bind FLRT3 LRR (y-axis). (B) Quantification of cells that fall within the gate of “high LPHN3 expression and high FLRT3 binding” (black ovals) as indicated in A normalized to wild type. Bar height represents the percent of cells that fall within the gates shown. See also Figure S3 and S4.
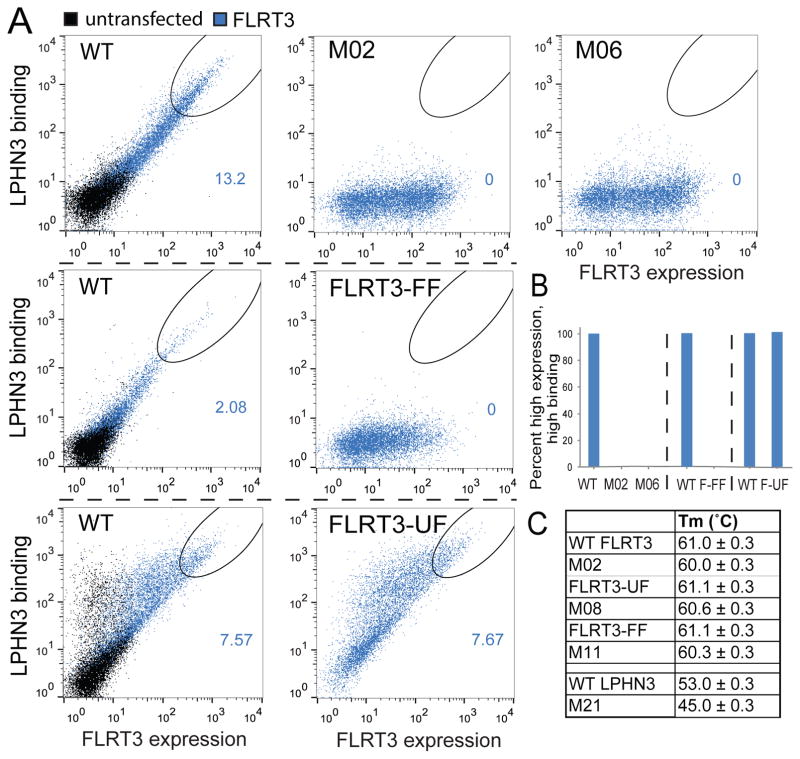
Figure 4. Mutations on FLRT3 abolish LPHN3 binding but do not interfere with proper cell-surface localization.
(A) Wild-type and mutant full-length FLRT3 proteins were tested for surface expression in HEK293 cells as well as their ability to bind soluble LPHN3 Olf domain using flow cytometry. Non-permeabilized HEK293 cells expressing N-terminally Myc-tagged FL-FLRT3 were stained with mouse anti-c-Myc primary antibody and anti-mouse FITC conjugated secondary antibody. FLRT3 surface expression was measured as green fluorescence from FITC (x-axis). Biotinylated LPHN3 Olf binding to FL-FLRT3-expressing cells was measured by monitoring red fluorescence of DyLight attached to neutravidin (y-axis). (refer to Figure S3C and Table S3 for detailed experimental setup and experimental conditions). Dot plot shows correlation between FLRT3 expression and LPHN3 binding in FLRT3-transfected cells (blue), or in untransfected cells (black). See legend for Figure 3 for details. See setup in Figure S3C. Mutant FLRT3 constructs (M2, M6, FLRT-FF and FLRT-UF) show proper surface expression (x-axis), but do not bind LPHN3 (y-axis) except FLRT3-UF. Different sets of experiments are separated with dashed black lines. Wild type data is provided for each set of experiments as positive control. (B) Quantification of cells that fall within the gate of “high FLRT3 expression and high LPHN3 binding” (black ovals) as indicated in A normalized to wild type. Bar height represents the percent of cells that fall within the gates shown. See also Figure S3. (C) Differential scanning fluorimetry (DSF) of wild-type and mutant FLRT3 LRR and LPHN3 olfactomedin domains showing the melting temperature (Tm). See also Figure S5.
We next measured soluble FLRT3 LRR binding to all FL-LPHN3 mutants, using surface binding of recombinant FLRT3 LRR domain to HEK293 cells transfected with full-length wild type or mutant LPHN3s using flow cytometry (Figure 3 and S4, see Figure S3 and Table S3 for details). To ensure the mutations completely abolish binding, low and high concentrations of wild type recombinant FLRT3 LRR were used in binding experiments. Specifically, by staining with purified FLRT3 LRR at concentrations as low as 10 nM, only high-affinity interactions can be detected. By staining at concentrations as high as 10 μM, lower affinity interactions can be detected. Additionally, by staining with tetramerized FLRT3 LRR, the avidity effect increases the effective concentration by over 100-fold (Wooldridge et al., 2009)(His-FLRT3 was tetramerized by mixing with biotin-tris-NTA (BTtrisNTA) and neutravidin -a protein that binds to biotin and tetramerizes it). Thus, by observing no binding of 100 nM tetramerized FLRT3 LRR to mutant FL-LPHN constructs, we concluded that the affinity for wild-type FLRT3 LRR for these LPHN3 mutants was very low (i.e. Kd > 10 μM).
These experiments showed that FL-LPHN3-M21 (Y249A, D251A, T252A, R308A) and FL-LPHN3-M28 (Y249A, D251A, T252A, E279A, R308A) mutants were defective in FLRT LRR binding and had no surface localization problems (Figure 3A,B). Similar experiments were performed for testing wild type recombinant LPHN3 Olf domain binding to HEK293 cells transfected with full-length wild type or mutant FLRT3s (Biotinylated LPHN3 Olf was tetramerized by mixing with neutravidin). FL-FLRT3-M02 (Y43A, Y64A) and FL-FLRT3-M06 (D38A, Y43A, N45A, R47A) mutants were defective in LPHN3 Olf binding and had no surface localization problems (Figure 4A,B, Figure S5, see Figure S3 and Table S3 for details). To further confirm the proper folding and stability of the mutants, we also performed differential scanning fluorimetry experiments with select mutants that measure the stability of proteins by monitoring the melting temperature (Figure 4C). Mutant FLRT3 LRR and mutant LPHN3 Olf proteins expressed and purified in baculovirus expression system were used for these experiments. Together, these mutants that are transported to the cell surface and exhibit either a defect in FLRT3 binding or in LPHN3 binding, or no apparent defect provide us with a toolkit to explore the function of LPHN3 and FLRT3 in the neuronal activities of these cell-adhesion proteins.
We also studied the previously reported FLRT3 mutants called “FLRT3-FF dimerization mutant” and “FLRT3-UF UNC5 binding mutant” (see Figure 2C for a schematic illustration of their location on the complex structure) (Seiradake et al., 2014). These mutations, however, were generated by introducing large N-linked carbohydrate modifications into the protein surface to disrupt interactions. In vivo studies performed with these mutants had reported that FLRT dimerization is involved in tangential (sideways) migration of neurons during cortex development, whereas repulsive FLRT-UNC5 interaction is involved in radial (upward) migration of neurons. Our further analysis of these mutants showed that FLRT3-FF decreases dimerization although it does not abolish it (see below, Figure 5A,B). Importantly, it completely abolishes the binding of LPHN3 LRR to FL-FLRT3 (Figure 4A,B) suggesting that the in vivo effect of the FLRT3-FF mutant on tangential migration of neurons is likely due to the lack of LPHN3 binding to FLRT3. The UNC5-binding mutant FLRT3-UF, on the other hand, had no detectable effect on LPHN3 binding (Figure 4A,B).
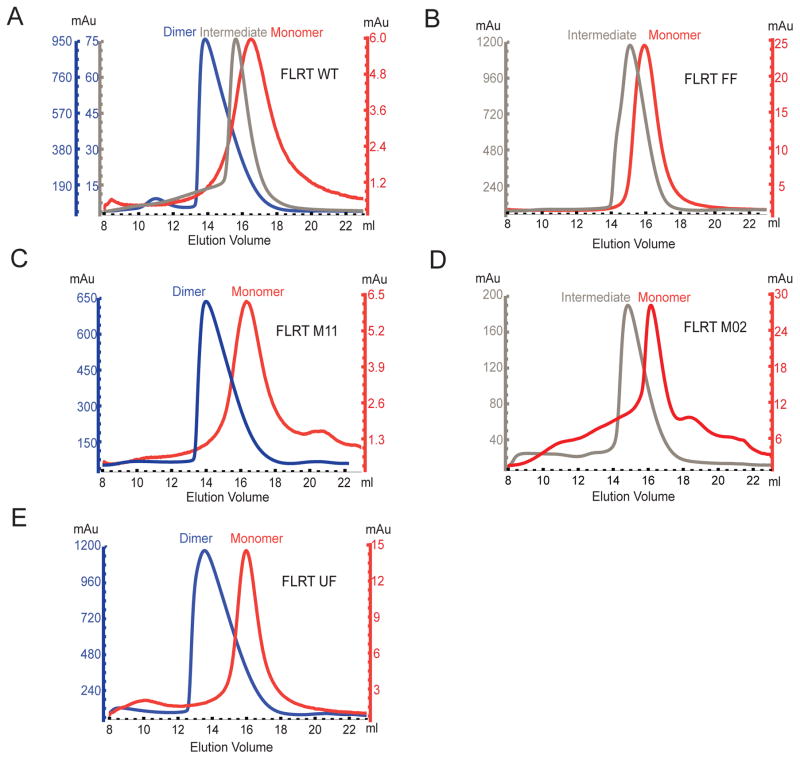
Figure 5. Oligomerization of Wild type and Mutant FLRT LRR domains.
(A)–(F) Size-exclusion gel filtration profile showing elution volumes of wild type FLRT3 LRR domain or mutants at various protein concentrations. Protein concentration is monitored by the UV absorbance at 280 nm (mAu) indicated by the y-axis. Different colors of the y-axis match the different elution peaks. Blue, gray, and red curves indicate dimer, intermediate, or monomer FLRT3 LRR, respectively. Note that M2 mutant displays wild type behavior. Due to low protein yield, the protein could not be concentrated to high enough concentrations to form dimers. However, the observed intermediate peak elutes at similar concentrations as wild type (note similar mAU in y-axis). See also Figure S6.
Effect of mutations on FLRT3 dimerization
Next, we tested the effect of the FLRT3 mutations on FLRT3 dimerization. It was previously reported that gel filtration chromatography can detect the concentration dependent dimerization of FLRT3 LRR (Seiradake et al., 2014). As expected, the size exclusion chromatogram of purified FLRT3 LRR showed that the protein concentration affects the elution volume, indicating that at high protein concentrations FLRT3 LRR forms a dimer, whereas at low protein concentrations FLRT3 LRR is a monomer (Figure 5A, notice the correlation of the elution volume with the absorbance units (mAU) reflective of protein concentration, see Figure S6 for elution profiles of gel filtration standards, dimeric FLRT3 LRR, LPHN3 Olf and the FLRT3/LPHN3 complex). At intermediate concentrations, the eluted peak was in between the dimer and monomer elution volumes. We speculate that the monomer/dimer has a fast exchange rate giving rise to a single intermediate peak rather than a mixture of monomer/dimer peaks. When dimer was diluted and reran on the gel filtration column, a monomer peak was observed. Similarly, when the monomer peak was concentrated and reran, a dimer peak was observed, showing that the dimer-monomer formation is reversible. Based on the measured protein concentrations, a low affinity in the submicromolar range is predicted for the FLRT3 LRR dimer in vitro. However, the affective affinity of two FL-FLRT3 monomers involved in a possible cis-interaction on the cell surface is likely higher.
We used gel filtration chromatography to monitor the effect of the studied FLRT mutations on the ability of FLRT LRR to dimerize. The reported FLRT dimerization mutant (FLRT3-FF) decreased dimerization but did not completely abolish it (Figure 5B). When we mutated the same residues to alanine without introducing a glycosylation site (M11, R181A, D183A instead of R181N, D183T), FLRT3 LRR dimerization was not affected (Figures 5C), suggesting that the large glycosylation moiety introduced to the surface of the concave side of FLRT3 LRR in the FLRT-FF mutant is blocking FLRT3 dimerization due to steric hindrance rather than breaking specific molecular interactions. This result demonstrates that the introduction of carbohydrates to a site can block protein interactions dramatically. In addition, we tested our FLRT3 mutant that does not bind to LPHN3 (M02) for dimerization, and showed that its dimerization ability is not affected (Figure 5D). Thus it can be used as a specific LPHN3-binding mutant in further experiments. The FLRT-UF mutant, on the other hand, showed wild-type like dimerization ability (Figure 5E).
FLRT3, LPHN3 and UNC5 form a trimeric complex
FLRT proteins are involved in heterodimeric interactions with LPHNs and with UNC5s, and in homodimeric interactions with themselves. However, whether all of these interactions are compatible is unclear. Next, we investigated whether FLRT3/UNC5 interaction is compatible with FLRT3/ LPHN3 interaction, or in other words, whether FLRT3, LPHN3 and UNC5 can form a trimeric complex. The availability of the FLRT3/LPHN3 complex structure and the previously reported FLRT3/UNC5 complex structure (Seiradake et al., 2014) enabled us to compare structures and predict, then test, the compatibility of the possible interactions of FLRT3 with each other. Intriguingly, superimposition of the FLRT3/LPHN3 structure with the FLRT3/UNC5 structure suggests that UNC5 and LPHN3 bind to distinct surfaces on FLRT3 and that there are no clashes between UNC5 and LPHN3, suggesting that LPHN3 and UNC5 can bind to FLRT3 simultaneously (Figure 6A).
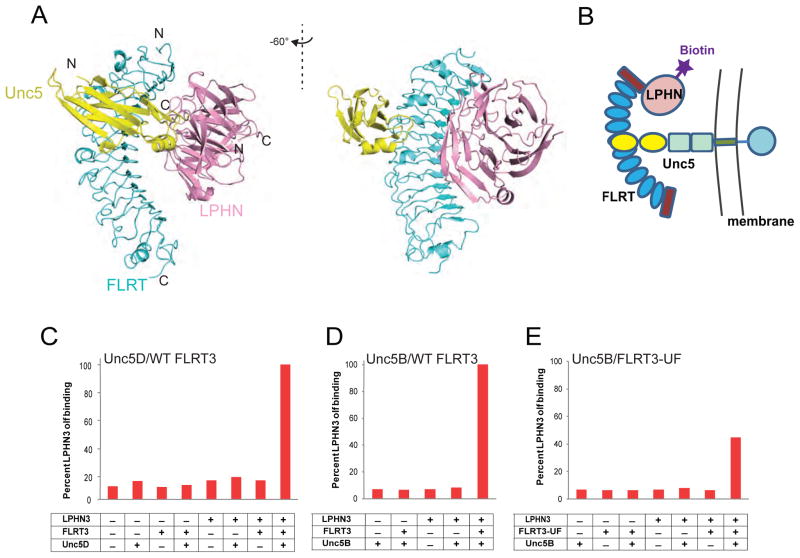
Figure 6. Formation of a trimeric complex between FLRT3, LPHN3 and UNC5 proteins.
(A) Superimposition of FLRT3/ LPHN3 and FLRT2/UNC5D (PDB ID 4V2C) structures. (B) Simple schematic diagram of the experimental design for the detection of the trimeric complex on HEK293 cells. (Refer to Figure S3D and Table S3 for detailed experimental setup and experimental conditions.)
(C) Quantification of purified biotinylated LPHN3 Olf binding to HEK293 cells transfected with FL-UNC5D. FLRT3 LRR domain (non-biotinylated), biotinylated LPHN3 Olf domain and neutravidin (allowing tetramerization) were mixed at different combinations at 5 nM or 100 nM concentrations (Figure S7D) and red fluorescence of DyLight attached to neutravidin was detected by flow cytometry. Pretetramerization of FLRT3 and LPHN3 with neutravidin increases avidity. See raw data in Figure S7A.
(D) Same experiment as in C performed using UNC5B instead of UNC5D, using 100 nM pretetramerized complex. (raw data in Figure S7B)
(E) Same experiment as in D using FLRT3-UF mutant. (raw data in Figure S7C) See also Figure S7.
In order to test whether this model is correct, we expressed full-length UNC5D or full-length UNC5B in HEK293 cells and added premixed purified recombinant FLRT3 LRR, purified recombinant biotinylated LPHN3 Olf, and fluorescently labeled neutravidin (a protein that binds to biotin and tetramerizes it) onto the cells (see Figure 6B for a schematic representation of the experiment). Bound LPHN3 Olf was detected using flow cytometry. Our results showed that LPHN3 is detected on cells only when FLRT3, LPHN3 and neutravidin are all added, indicating the formation of a trimeric complex (Figure 6C,D and see Figure S7 for raw data). No LPHN3 binding was detected when FLRT3 was not added onto UNC5-expressing cells, suggesting that there is no direct interaction between UNC5 and LPHN3, instead, FLRT3 bridges the two proteins to form a trimeric complex. As mentioned above, due to avidity, by tetramerizing biotinylated LPHN3 Olf with neutravidin, the effective concentration of the FLRT3- LPHN3 affinity is more than 100-fold higher than the concentration used. Similar results were observed both with low or high protein concentrations with UNC5D. Flow cytometry experiments performed with the purified FLRT3-UF LRR mutant showed that this mutant does not completely abolish UNC5 binding consistent with the previous experiments (Figure 6E and 4A). These results suggest the formation of a trimeric complex between UNC5, FLRT and LPHN that forms in vitro.
FLRT3/ LPHN3 and FLRT3/UNC5 mediate formation of intercellular contacts
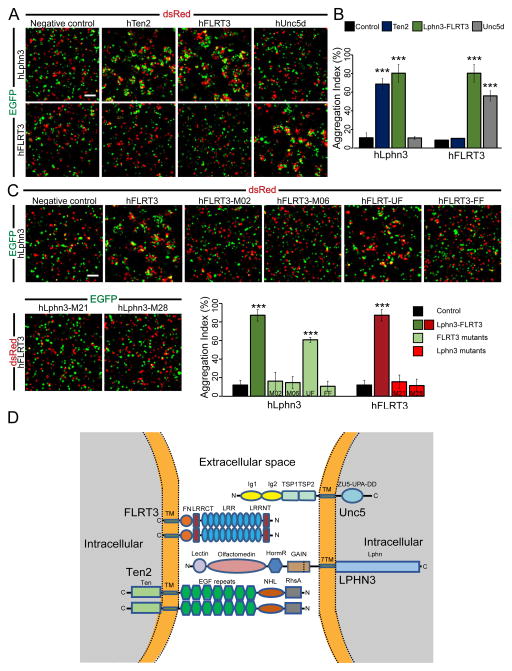
Cell-adhesion molecules may function via two different ways: Two cell-adhesion molecules expressed on the same cell might be involved in cis-interactions or two cell-adhesion molecules each expressed on one of the two neighboring cells might be involved in trans-interactions. To examine if trans-interactions between LPHN3, FLRT3, or UNC5d can support cell-cell adhesion, we performed cell aggregation assays with non-adherent HEK293 cells in which each full-length protein is expressed on different cell populations and the cells are then mixed to monitor cell aggregation (Figure 7). Previous studies have shown that LPHN1 binding to Teneurin2 in trans can promote cell aggregation (Boucard et al., 2014). Therefore, this condition was used as a positive control in cell aggregation experiments. Interestingly, we found that the binding of FL-LPHN3 to FL-FLRT3 and of FL-UNC5d to FL-FLRT3 both induce cell-cell adhesion in trans. Point mutations in both FL-LPHN3 and FL-FLRT3 in residues we found to be essential for complex formation in vitro abolish cell aggregation (Figure 7). LPHN3, FLRT3, and UNC5d form a heterotrimeric complex, where LPHN3 and UNC5d bind to distinct binding sites on FLRT3. We observed no cell aggregates mediated by homophilic interactions of any of these molecules, in particular FL-FLRT3 or FL-Ten2, suggesting that the hemophilic binding of these molecules operates in an exclusively cis configuration (Fig. 7D). Altogether, these results support the notion that LPHN3, FLRT3, and UNC5d function in trans-cellular adhesion.
Figure 7. Analysis of LPHN3/FLRT3/UNC5d trans interactions.
A, Representative images from cell aggregation assays with wild-type full-length proteins. LPHN3 induces cell aggregation with FLRT3, as well as Ten2, a previously identified trans interaction partner (Boucard et al. 2013. JBC). FLRT3 promotes trans-cellular adhesion with LPHN3 and UNC5d, but not with Ten2. B, quantification of cell aggregation assays with wild-type proteins (means ± standard deviation). C, point mutations that disrupt LPHN3-FLRT3 binding abolish cell aggregation. Cell adhesion is preserved between LPHN3 and FLRT3 when the UNC5d-binding site on FLRT3 is mutated. All experiments were performed in three independent culture preparations (*** denotes p<0.001). D, diagram of LPHN3/FLRT3/UNC5d/Ten2 at a cellular junction. LPHN3 and UNC5d are localized to the opposing membrane which contains FLRT3 and Ten2. (Note: proteins are not drawn to scale). Adhesion will cause aggregates of two or more cells and therefore an increase in aggregation index. Aggregation index was calculated using the program ImageJ.
Discussion
Neural development is a complex phenomenon that is mediated by the coordinated interactions of numerous cell-surface proteins on neurons and glial cells. Studying binary protein interactions is a required but insufficient step to understand neural development as other interaction partners of the binary interaction in question likely affect the functional outcome of the specific protein-protein interaction. This outcome also depends on whether the cell-surface proteins are presented from two different cells making a trans-interaction or they are presented on the same cell making a cis interaction – in many cases, cis- and trans-interactions are mutually exclusive in that only one is possible when the respective proteins are embedded in the plasma membrane. Studying the basics of the specific protein-protein interactions and designing binding-mutants that specifically abolish one interaction without interfering with the other interactions of a protein are essential first steps before moving forward with functional understanding of each protein.
In this study, using a combination of biophysical, biochemical and cell-based approaches, we studied the structure and specificity of the interaction between FLRT3 and LPHN3, and also considered the interaction of FLRT3 with UNC5 and with itself. The structure of the FLRT3/LPHN3 complex revealed that the olfactomedin domain was bound to the concave surface of the FLRT3 LRR horseshoe (Figure 1). The interaction surface spreads over the extensive complementary surfaces of both proteins. However, the major interaction hotspot is at the N-terminal top side of the LRR horseshoe and at the long loops emerging from the second and third blades of the LPHN3 olfactomedin beta-propeller (Figure 2). The concave surface of the FLRT3 LRR domain was previously reported to mediate FLRT3 dimerization, thus it is essential to study the effect of any mutations in this region on both LPHN3 binding and FLRT3 dimerization before commenting on the effect of the mutation on the function of the protein. This surface is spatially separated from the UNC5 binding site on FLRT3, and thus UNC5 binding and LPHN3 binding are likely not exclusive. On the other hand, the FLRT3 binding site on LPHN3 is likely distant from the teneurin binding site on LPHN3 as teneurin binding is largely mediated by the lectin domain and the splice insert, both of which are at the N-terminal side of the olfactomedin domain. Mapping of the mutations of LPHN3 genes that are linked to attention deficit/hyperactivity disorder and cancers on the FLRT3/LPHN3 complex structure reveals that most mutations are at the FLRT3/LPHN3 binding interface where the two proteins come closest to each other, and one mutation is at the edge of the binding interface, suggesting protein-protein interaction defects (Figure 2B,C).
Mapping the conserved and variable residues on the surface of the LPHN3 and FLRT3 structures and relating their location to interface between FLRT3 and LPHN3 in the FLRT3/ LPHN3 structure shows that the FLRT3 binding site on LPHN3 and the LPHN3 binding site on FLRT3 are highly conserved. This observation again suggests a critical role for the FLRT/LPHN interaction in neural development (Figure 1D). Mapping the electrostatics surface potential on the FLRT/LPHN3 structure shows a large positive and a large negative surface at the concave inside surface of the LRR horseshoe, suggesting that non-specific charge-charge interactions might be mediated via this surface, especially at high protein concentrations (Figure 1E).
To confirm that the binding interface that is revealed by the FLRT/LPHN complex structure is biologically relevant, we designed FLRT3 mutants and LPHN3 mutants that specifically disrupt the FLRT/LPHN interaction without interfering with membrane localization (Table 2, Figure 3,4). Flow cytometry binding experiments showed that mutations located on the interaction surface break the interaction. Flow cytometry experiments monitoring cell-surface expression of the mutants on non-permeabilized cells showed that some mutations affect the proper folding/trafficking of the proteins (Fidue S3,4). Thus, only well-expressed and trafficked mutants were used in further experiments. All selected FLRT3 mutants were tested for their ability to dimerize to make sure only one specific interaction is broken in any given mutant (Figure 5). Thus, we generated well-characterized FLRT3 mutants and LPHN3 mutants that can be used as molecular tools in further experiments to specifically dissect the mechanism of these multi-interaction proteins.
Importantly, we studied the previously reported “FLRT3 dimerization mutant-FF” (Seiradake et al., 2014) and showed that the LPHN3-binding ability of this mutant is more severely affected than its dimerization ability (Figure 4A, 5B). Considering that previously performed in vivo studies reported the FLRT-FF mutant impairs tangential (sideways) migration of neurons during cortex development, we suggest that the lack of LPHN3 binding to the FLRT3-FF mutant might be responsible for this defect. The high-conservation of the binding interfaces and the localization of ADHD and cancer mutations at the FLRT/LPHN binding surface also suggest a critical role for this interaction. Similar considerations might apply to the mutants studied in another recent paper (Jackson et al., 2015) as all these mutations include introduction of a new N-linked glycosylation site into the protein sequence.
The availability of the FLRT3/LPHN3 complex structure allowed us to make comparisons with the FLRT2/UNC5D complex structure and predict that LPHN3 and UNC5D can simultaneously interact with FLRT (Figure 6A). Our binding experiments showed that FLRT, LPHN and UNC5 form a trimeric complex and FLRT binds the other two proteins simultaneously and bridges them where LPHN and UNC5 do not directly interact with each other (Figure 6). Our cell-adhesion assays showed that LPHN3/ FLRT3 binding induces trans-cellular adhesion. Mutations in the binding interface of LPHN3 and FLRT3 abrogated cell adhesion. Interestingly, while FLRT3 / UNC5d binding also induces cell aggregation, LPHN3 /UNC5d binding does not. Altogether, these results suggest that LPHN3 and UNC5d are localized to one side of the cellular junction in which they participate, whereas FLRT3 is localized to the other side (Figure 7D). At least LPHN3 and Flrt3 have been localized to synapses in mature brain, suggesting that the FLRT3/LPHN3 and the FLRT3/UNC5d interaction may, among others, contribute to synapse formation and/or synaptic transmission. However, the precise pre- vs. post-synaptic localization of these proteins, as well as if they are necessary and/or sufficient for synaptic adhesion, remains to be elucidated – for none of these proteins has it actually been shown directly whether they are pre- or postsynaptic. While FLRT3 appears to form a homodimer, we were unable to observe FLRT3-FLRT3 cell aggregation. Thus, this homodimer likely occurs in cis and is incapable of supporting trans-cellular adhesion. Interestingly, the concurrent trans-cellular interaction of FLRT3 with both UNC5d and LPHN3, and the likely simultaneous trans-cellular interaction of LPHN3 in turn with both FLRT3 and teneurins, creates an interaction network in which the cells are linked at an intercellular junction such as the synapse not by a simple one-to-one complex, but by a large complex composed of very different simultaneous interactions that may be independently regulated, and may transduce distinct trans-cellular signals.
In conclusion, the available high resolution structure of the FLRT3/LPHN3 complex provides the basis for further advances in understanding their mechanism of action in brain function which may lead to the treatment of diseases that are caused by mutations in these proteins.
Methods
Vectors and Cloning
For crystallization and protein purification, the LRR repeats of human FLRT3 (residues K29-D357; Uniprot ID: Q9NZU0) and the olfactomedin domain of human LPHN3 (residues V132-G392; Uniprot ID: Q9HAR2) were cloned into pAcGP67a. Similarly, the Ig-like domain of mouse Unc5D (residues G49-Q161; Uniprot ID: Q6UXZ4) was cloned into pAcGP67a. A 8XHis tag or biotin tag was added at the C terminus for affinity purification. For mammalian expression and functional analysis, full length human FLRT3 (residues S30–S649), human LPHN3 (residues F20-L1447), and human Unc5B (G27-E934; Uniprot ID: Q8IZJ1), and mouse Unc5D (S46-L884) constructs with preprotrypsin leader sequence containing N-terminal myc, FLAG, His and His-tags, respectively, were cloned into pCMV5.
Protein Expression, Purification, Crystallization and Structure Determination
Baculovirus expression system was used for expression of proteins used for crystallization and biophysical experiments. The secreted, glycosylated proteins were purified using nickel-nitrilotriacetic agarose resin (QIAGEN) and size exclusion chromatography (Superdex 200 10/300 GL; GE). Prior to crystallization, purified proteins were incubated with carboxypeptidase A and carboxypeptidase B to cleave off the C-terminal residues such as the His8 tag. Crystals of FLRT3 grew in 0.1M Tris pH 7, 50% (v/v) PEG200. Crystals of FLRT3/LPHN3 complex grew in 10% (w/v) PEG 3000, 100 mM MES pH6.0, 200 mM Lithium sulfate. Diffraction data was collected at Advanced Photon Source of the Argonne National Laboratories beamline 23-IBD and 19-BM. Data sets were processed using HKL2000. FLRT3 structure was solved by molecular replacement with Phaser-MR (Phenix) using a model of FLRT structure (PDB ID 4V2E). Similarly, FLRT3/LPHN3 structure was determined by molecular replacement using a homology model of the LPHN3 Olfactomedin domain based on the myocilin olfactomedin domain crystal structure (PDB ID 4WXQ). For both structures, refinement was performed in phenix.refine (Phenix) with nonscrystallographic symmetry (NCS) restraints. Olf structure in the complex was further refined using LPHN3 olf structure (PDB 5AFB). Since FLRT3/LPHN3 complex crystal displayed twinning, twin law of h, -k, -l was employed throughout the entire refinement process.
Flow cytometry
Full-length proteins were expressed in HEK293 mammalian expression system. To test LPHN3 WT and mutants expression, transfected cells were stained with mouse anti-FLAG M2 antibodies, 1:1000 (F3165, “Sigma”). Fluorescence was determined by incubating with anti-mouse FITC. To test FLRT3 WT and mutant expression, cells were stained with mouse anti c-Myc antibodies (9E10, “DSHB”) 1:20 and anti-mouse FITC, 1:100. For binding assays purified monomeric or tetrameric soluble proteins were added to primary antibodies. Precomplex of BTtrisNTA - NeutrAvidinDyLight 650 (NAV650) (84607, “Thermo Scientific”) or only NeutrAvidinDyLight 650 (NAV650) was used for fluorescent labeling.
Cell aggregation assays
FreeStyle HEK293 cells (Life Technologies) grown to a density of 1×106 cells/mL in a 30mL volume were co-transfected with 30μg of either pCMV-Emerald or pCMV-dsRed and 30μg of the indicated construct using FreeStyle Max reagent (Life Technologies). Live cells were imaged by dropping 100μL of cell suspension onto a glass slide (FisherBrand). Aggregation index was calculated as shown previously (Boucard et al. 2013. J. Cell Biol.).
Supplementary Material
Acknowledgments
We thank Engin Özkan for crystallographic advice and discussion, Raquel Lieberman for providing the mycolin domain structure coordinates before its release, Antony Boucard for sharing unpublished information about the FLRT binding domain of LPHN, Shohei Koide and Akiko Koide for synthesized BTTris-NTA and generous access to a flow cytometer, Katherine Leon and Katie Homa for various help for the experiments. We also extend our gratitude to Craig Ogata, and Ruslan Sanishvili, Gerold Rosenbaum and Changsoo Chang at Advanced Photon Source of the Argonne National Laboratory for their tremendous assistance in data collection (GM/CA, SBC).
Footnotes
Author Contributions
Y.C.L. designed and performed crystallography, structure determination, mutagenesis, and Bio-layer interferometry binding experiments. O.N. designed and performed HEK cell expression and flow cytometry binding assays. R.S. designed and performed cell aggregation assays. G.S designed and performed differential scanning fluorimetry assay and assisted flow cytometry binding assays and structure determination. T.C.S. designed cell aggregation assays and contributed to manuscript preparation. D.A designed all experiments and wrote the paper.
Accession Numbers:
The coordinates and diffraction data have been deposited in the RCSB Protein Data Bank (PDB) (PDB IDs 5CMN [FLRT3/LPHN3] and 5CMP [FLRT3]).
References
- Arac D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Sudhof TC, Brunger AT. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. The EMBO journal. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, Domene S, Velez JI, Karkera JD, Balog J, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Molecular psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- Asherson P, Gurling H. Quantitative and molecular genetics of ADHD. Current topics in behavioral neurosciences. 2012;9:239–272. doi: 10.1007/7854_2011_155. [DOI] [PubMed] [Google Scholar]
- Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nature cell biology. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- Boucard AA, Ko J, Sudhof TC. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. The Journal of biological chemistry. 2012;287:9399–9413. doi: 10.1074/jbc.M111.318659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Maxeiner S, Sudhof TC. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. The Journal of biological chemistry. 2014;289:387–402. doi: 10.1074/jbc.M113.504779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sawyer N, Regan L. Protein-protein interactions: general trends in the relationship between binding affinity and interfacial buried surface area. Protein science : a publication of the Protein Society. 2013;22:510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak F, Liu X, Khvotchev M, Li G, Kavalali ET, Sugita S, Sudhof TC. Alpha-latrotoxin stimulates a novel pathway of Ca2+-dependent synaptic exocytosis independent of the classical synaptic fusion machinery. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:8639–8648. doi: 10.1523/JNEUROSCI.0898-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domene S, Stanescu H, Wallis D, Tinloy B, Pineda DE, Kleta R, Arcos-Burgos M, Roessler E, Muenke M. Screening of human LPHN3 for variants with a potential impact on ADHD susceptibility. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156B:11–18. doi: 10.1002/ajmg.b.31141. [DOI] [PubMed] [Google Scholar]
- Donegan RK, Hill SE, Freeman DM, Nguyen E, Orwig SD, Turnage KC, Lieberman RL. Structural basis for misfolding in myocilin-associated glaucoma. Human molecular genetics. 2015;24:2111–2124. doi: 10.1093/hmg/ddu730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea J, Erlacher C, Montanez E, Burtscher I, Yamagishi S, Hess M, Hampel F, Sanchez R, Rodriguez-Manzaneque MT, Bosl MR, et al. Genetic ablation of FLRT3 reveals a novel morphogenetic function for the anterior visceral endoderm in suppressing mesoderm differentiation. Genes & development. 2008;22:3349–3362. doi: 10.1101/gad.486708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Kursula P. The olfactomedin domain from gliomedin is a beta-propeller with unique structural properties. The Journal of biological chemistry. 2015;290:3612–3621. doi: 10.1074/jbc.M114.627547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson VA, Del Toro D, Carrasquero M, Roversi P, Harlos K, Klein R, Seiradake E. Structural Basis of Latrophilin-FLRT Interaction. Structure. 2015;23:774–781. doi: 10.1016/j.str.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- Karaulanov E, Bottcher RT, Stannek P, Wu W, Rau M, Ogata S, Cho KW, Niehrs C. Unc5B interacts with FLRT3 and Rnd1 to modulate cell adhesion in Xenopus embryos. PloS one. 2009;4:e5742. doi: 10.1371/journal.pone.0005742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaulanov EE, Bottcher RT, Niehrs C. A role for fibronectin-leucine-rich transmembrane cell-surface proteins in homotypic cell adhesion. EMBO reports. 2006;7:283–290. doi: 10.1038/sj.embor.7400614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnoperov VG, Beavis R, Chepurny OG, Little AR, Plotnikov AN, Petrenko AG. The calcium-independent receptor of alpha-latrotoxin is not a neurexin. Biochem Biophys Res Commun. 1996;227:868–875. doi: 10.1006/bbrc.1996.1598. [DOI] [PubMed] [Google Scholar]
- Krasnoperov VG, Bittner MA, Beavis R, Kuang Y, Salnikow KV, Chepurny OG, Little AR, Plotnikov AN, Wu D, Holz RW, et al. alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron. 1997;18:925–937. doi: 10.1016/s0896-6273(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Lacy SE, Bonnemann CG, Buzney EA, Kunkel LM. Identification of FLRT1, FLRT2, and FLRT3: a novel family of transmembrane leucine-rich repeat proteins. Genomics. 1999;62:417–426. doi: 10.1006/geno.1999.6033. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhan T, Promel S, Mestek L, Esmaeili B, Waller-Evans H, Hennig C, Kohara Y, Avery L, Vakonakis I, Schnabel R, et al. Latrophilin signaling links anterior-posterior tissue polarity and oriented cell divisions in the C. elegans embryo. Developmental cell. 2009;17:494–504. doi: 10.1016/j.devcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelianova VG, Davletov BA, Sterling A, Rahman MA, Grishin EV, Totty NF, Ushkaryov YA. Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. The Journal of biological chemistry. 1997;272:21504–21508. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- Levine A, Bashan-Ahrend A, Budai-Hadrian O, Gartenberg D, Menasherow S, Wides R. Odd Oz: a novel Drosophila pair rule gene. Cell. 1994;77:587–598. doi: 10.1016/0092-8674(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Leyva-Diaz E, del Toro D, Menal MJ, Cambray S, Susin R, Tessier-Lavigne M, Klein R, Egea J, Lopez-Bendito G. FLRT3 is a Robo1-interacting protein that determines Netrin-1 attraction in developing axons. Current biology : CB. 2014;24:494–508. doi: 10.1016/j.cub.2014.01.042. [DOI] [PubMed] [Google Scholar]
- Maretto S, Muller PS, Aricescu AR, Cho KW, Bikoff EK, Robertson EJ. Ventral closure, headfold fusion and definitive endoderm migration defects in mouse embryos lacking the fibronectin leucine-rich transmembrane protein FLRT3. Developmental biology. 2008;318:184–193. doi: 10.1016/j.ydbio.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Muller PS, Schulz R, Maretto S, Costello I, Srinivas S, Bikoff E, Robertson E. The fibronectin leucine-rich repeat transmembrane protein Flrt2 is required in the epicardium to promote heart morphogenesis. Development. 2011;138:1297–1308. doi: 10.1242/dev.059386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Gutkind JS. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nature reviews Cancer. 2013;13:412–424. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR, 3rd, Ghosh A. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan ML, Martini F, von Daake S, Comoletti D, Ghosh A. LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural development. 2014;9:7. doi: 10.1186/1749-8104-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz N, Gehring J, Guan C, Ljaschenko D, Fischer R, Lakshmanan V, Kittel RJ, Langenhan T. The Adhesion GPCR Latrophilin/CIRL Shapes Mechanosensation. Cell reports. 2015;11:866–874. doi: 10.1016/j.celrep.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Seiradake E, del Toro D, Nagel D, Cop F, Hartl R, Ruff T, Seyit-Bremer G, Harlos K, Border EC, Acker-Palmer A, et al. FLRT structure: balancing repulsion and cell adhesion in cortical and vascular development. Neuron. 2014;84:370–385. doi: 10.1016/j.neuron.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JP, Lelianova VG, Ermolyuk YS, Vysokov N, Hitchen PG, Berninghausen O, Rahman MA, Zangrandi A, Fidalgo S, Tonevitsky AG, et al. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12113–12118. doi: 10.1073/pnas.1019434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner C, Wright GJ. A cell surface interaction network of neural leucine-rich repeat receptors. Genome biology. 2009;10:R99. doi: 10.1186/gb-2009-10-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. alpha-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annu Rev Neurosci. 2001;24:933–962. doi: 10.1146/annurev.neuro.24.1.933. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Ichtchenko K, Khvotchev M, Sudhof TC. alpha-Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G-protein-linked receptors. G-protein coupling not required for triggering exocytosis. The Journal of biological chemistry. 1998;273:32715–32724. doi: 10.1074/jbc.273.49.32715. [DOI] [PubMed] [Google Scholar]
- Sugita S, Khvochtev M, Sudhof TC. Neurexins are functional alpha-latrotoxin receptors. Neuron. 1999;22:489–496. doi: 10.1016/s0896-6273(00)80704-7. [DOI] [PubMed] [Google Scholar]
- Sun H, Le T, Chang TT, Habib A, Wu S, Shen F, Young WL, Su H, Liu J. AAV-mediated netrin-1 overexpression increases peri-infarct blood vessel density and improves motor function recovery after experimental stroke. Neurobiology of disease. 2011;44:73–83. doi: 10.1016/j.nbd.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L, Lissina A, Cole DK, van den Berg HA, Price DA, Sewell AK. Tricks with tetramers: how to get the most from multimeric peptide-MHC. Immunology. 2009;126:147–164. doi: 10.1111/j.1365-2567.2008.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi S, Hampel F, Hata K, Del Toro D, Schwark M, Kvachnina E, Bastmeyer M, Yamashita T, Tarabykin V, Klein R, et al. FLRT2 and FLRT3 act as repulsive guidance cues for Unc5-positive neurons. The EMBO journal. 2011;30:2920–2933. doi: 10.1038/emboj.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.