Abstract
Glioblastoma multiforme (GBM) is the most common and malignant form of primary brain tumors. It is highly invasive and current treatment options have not improved the survival rate over the past twenty years. Novel approaches and technologies from systems biology have the potential to identify biomarkers that could serve as new therapeutic targets for GBM. This study employed lipid profiling technology to investigate lipid biomarkers in ectopic and orthotopic human GBM xenograft models. Primary patient cell lines, GBM10 and GBM43, were injected into the flank and the right cerebral hemisphere of NOD/SCID mice. Tumors were harvested from the brain and flank and proteins, metabolites, and lipids extracted from each sample. Reverse phase based high performance liquid chromatography coupled with Fourier transform ion cyclotron resonance mass spectrometry (LC-FTMS) was used to analyze the lipid profiles of tumor samples. Statistical and clustering analyses were performed to detect differences. Over 500 lipids were identified in each tumor model and lipids with the greatest fold effect in the comparison of ectopic versus orthotopic tumor models fell predominantly into four main classes of lipids: glycosphingolipids, glycerophoshpoethanolamines, triradylglycerols, and glycerophosphoserines. Lipidomic analysis revealed differences in glycosphingolipid and triglyceride profiles when the same tumor was propagated in the flank versus the brain. These results underscore the importance of the surrounding physiological environment on tumor development and are consistent with the hypothesis that specific classes of lipids are critical for GBM tumor growth in different anatomical sites.
Keywords: Biomarkers, cancer, glioblastoma multiforme, lipid profiling, mass spectrometry
INTRODUCTION
Glioblastoma Multiforme (GBM) is the most common and aggressive form of primary brain tumor [1], and is one of the most invasive and malignant cancers described. Usually, by the time of diagnosis a primitive subpopulation of GBM tumor cells has already migrated from the primary tumor site and infiltrated the distant brain parenchyma, making it impossible to cure [2]. GBM can occur at all ages but is found more frequently in older adults [3]. Although advanced diagnostic modalities, surgical techniques, and adjuvant treatment strategies have been developed over the last 30 years, the prognosis of GBM has not significantly changed over several decades [4]. GBM has a median survival period of approximately 12 to 15 months from diagnosis with a two-year survival rate of only 5 – 15% [5]. Due to the extremely aggressive nature of this cancer, current treatment options focus primarily on extending the lifespan of the patient [6].
Systems biology approaches using genomics and proteomics have been employed to investigate the characteristics of cancer progression and discover potential biomarkers and new therapeutic targets. Progression of gliomas is due to genetic and epigenetic alterations, including the loss of tumor suppressor gene function and activation of oncogenic pathways [2]. More than 500 human GBM tumors have been sequenced and characterized [7] and whole proteome analyses of gliomas have been performed using various human samples [8]. Lipidomics offers another important avenue to further understand the molecular mechanisms controlling GBM progression and may yield new therapeutic targets for GBM, complementary to the existing knowledge base generated by efforts in genomics and proteomics.
Lipidomics was introduced in 2003 by Han and Gross [9] and is a relatively new field [8]. Due to the membrane organizing properties of lipids, lipids play many important roles in a cell, tissue, and organ physiology. The main biological functions of lipids are quite diverse and critical for cell growth. Lipids function as structural components of cellular membranes and in energy storage. Changes in cell membrane composition can affect cell signaling, endocrine actions, membrane trafficking, and regulation of membrane-associated proteins [10]. Involvement of lipids may be more critical in the brain function since the human brain has the highest lipid content of all organs. The dry weight of the human brain contains 50% to 60% lipids [11]. Recent studies have shown that lipid metabolism plays an important role in cancer initiation and progression and is regulated by oncogenic signaling pathways [12]. The role of lipids in cancer, however, is still poorly understood due to biological and technical difficulties [10, 13]. Most analyses of lipids in cancer have been indirect, using the examination of enzymes in lipid biosynthesis pathways to predict the potential composition and function of lipids within the cell. In addition, information gathered from genomics and proteomics analyses has not been well integrated with lipidomics analyses.
We hypothesize that the lipid composition of GBM tumors is critical to tumor maintenance and progression. As a first step in addressing this hypothesis, we utilized two types of primary human GBM tumor cell lines (GBM10 and GBM43) in ectopic and orthotopic xenograft mouse models and examined the lipid profile in these models. Our overall objective was to determine if different anatomic sites (flank versus intracranial) resulted in distinct lipid profiles. This study also represents a multidisciplinary collaborative effort to improve the efficiency of computational pipelines. Development of appropriate infrastructure and pipelines that provide high quality and clinically relevant tissue are imperative to the analytics, visualization, identification, and interpretation of the “omics” data; such studies regularly serve as case studies for pipeline advancement efforts and identification of therapeutic biomarkers [14].
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Parental primary human GBM xenograft lines GBM10 and GBM43 were surgically removed and provided by Dr. Jann Sarkaria (Mayo Clinic, Rochester, Minnesota). Both cell lines are resistant to temozolomide and molecular properties have been described and validated in low passage lines (GBM10: wildtype EGFR, wild-type p53, CDKN2A/p16deleted, wild-type PTEN, and MGMT positive; GBM43: mutant p53, CDKN2A/p16deleted, wild-type PTEN, and MGMT positive [15]). GBM10 and GBM43 cells were expanded as flank tumors, harvested and maintained in 2.5% FBS for 4 days on matrigel-coated plates to remove murine fibroblasts. Cells were expanded in DMEM/F12 (Gibco #11965 Dulbecco’s Modified Eagle Medium; 4.5g/L D-glucose and L-Glutamine) with 10% FBS (Atlanta Biologicals Advantage FBS #S11050) for less than 2 weeks and used for set up of intracranial models. Cells were checked for mycoplasma prior to injection into mice and were negative. In each mouse, 3×105 cells were surgically implanted into right cerebral hemisphere and 3 × 106 cells subcutaneously injected into the right flank. An overview of biological sample preparation workflow is shown in (Fig. 1).
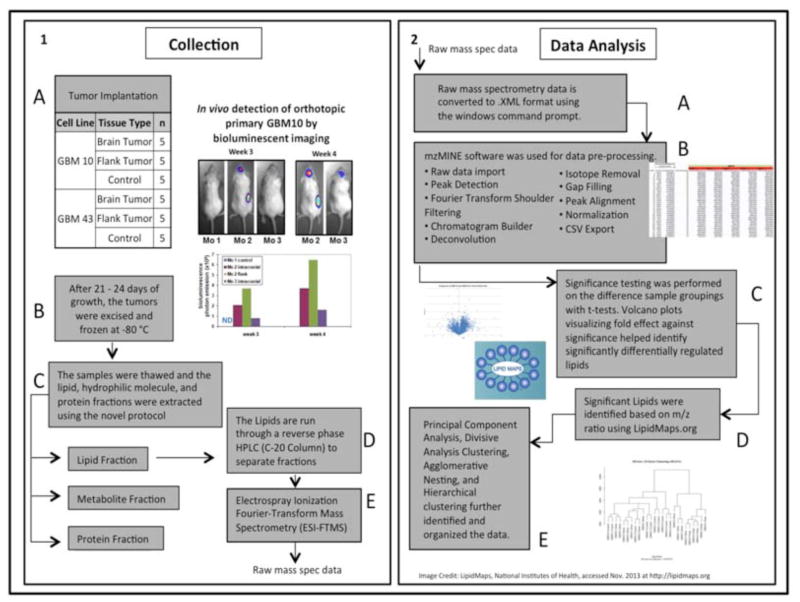
Fig. 1.
Overview of lipidomic analysis of GBM workflow. (1 A) Number of brain and flank tumors of GBM10 and GBM43. (1 B) In vivo tumor harvest information and mouse xenograft model validation via bioluminescence imaging. (1 C) Biomolecule extraction procedure. (1 D–E) Separation and ionization of lipid method for mass spectrometry. (2 A–B) Data processing of mass spectrometry data using MzMine2. (2 C) Statistical analysis (2 D) Lipid identification using the database, LIPID MAPS. (2 E) Validation of the data by performing hierarchical clustering.
Human GBM Xenograft Models
NOD.CB17-Prkdcscid/J (NOD/Scid) mice were obtained from the onsite breeding colony in the In Vivo Therapeutics Core at the Indiana University School of Medicine. Five mice were utilized per GBM cell line. GBM10 and GBM43 cells were implanted in the right flank in matrigel at 3 × 106 cells per mouse. Flank and intracranial tumors were implanted in the same mice. For intracranial implantation, a digitalized stereotaxic delivery system was utilized (David Kopf Instruments, Model 5000 microinjection unit, Tujunga, CA) as described previously [16, 17]. For stereotaxic delivery of tumor cells, mice were placed under general anesthesia (ip injection of 16 mg/kg xylazine and 120 mg/kg ketamine) and positioned in the stereotaxic device. A digitalized drill assembly was used to bore a hole 0.3 mm in depth and 0.8 mm diameter in the cranium at a position 0.5 mm anterior and 1.2 mm lateral to the bregma anatomical landmark. Tumor cells (3 × 105) in 10 μl of PBS) were introduced slowly using a 10 μl Hamilton syringe at a depth of 3.5 mm at a rate of 2 μl/min. Once injection was complete, the needle was kept in place for at least 5 minutes and then slowly removed and the hole sealed with bone wax. The incision was closed with 3M vetbond Tissue Adhesive (3M Animal Care Products). In previous validation studies, bioluminescence imaging of xenograft tumors that express a luciferase-enhanced green fluorescent protein (EGFP) fusion protein was performed in order to detect the tumors in the brain and flank sites (Fig. 1B). Survival analysis using pre-death endpoint criteria was also previously performed and the median survival is typically 35–55 days post-implantation of tumor cells for GBM10 and 30–40 days for GBM43 in NOD/Scid mice (unpublished observations, Wang and Pollok). Mice with flank tumors were euthanized once tumors reached ~250 mm3. Tumors were excised and flash frozen and stored at −80°C. For mice with intracranial tumors, the mice were observed twice daily starting at 2 weeks and prior to reaching the pre-death endpoint were euthanized. Tumor tissue was carefully excised from the right cerebrum and control tissue from the left cerebrum; tissues were immediately flash frozen, and stored at −80°C until analysis.
Lipid Extraction
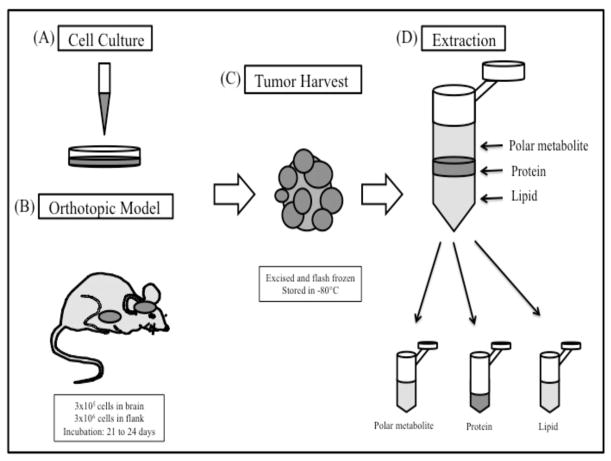
Sample preparation of the mouse tumors prior to mass spectrometry was performed using a novel biomolecule extraction method to simultaneously harvest the proteins, metabolites, and lipids in one simple and fast procedure visualized in (Fig. 2). Proteins and hydrophilic metabolites of the mice tissues were collected and stored for the future analyses. Ten to 100 mg of in vivo mouse GBM tumor tissues were placed in a low retention 2.0 ml microcentrifuge tubes and placed on ice. These tumor samples were mixed with 200 μl of 75% MetOH in 0.15M NaCl. Approximately 50 μl of grinding balls (ZrO; diameter ~0.5 mm) were added to each tube in order to homogenize the tumor tissues using Next Advance Bullet Blender for two minutes. This step was repeated until the tissues were completely homogenized. After the tissue homogenization, 20 μl of suspension from each tube was transferred into the new 1.5 ml microcentrifuge tubes and mixed with 180 μl of 0.15M NaCl and 1 ml of chloroform/methanol (2:1) with 0.01% BHT by vortex for two minutes, then incubated at room temperature for 30 minutes. After the incubation, the tubes were centrifuged for five minutes at 7,800xg and 250 μl of the lower chloroform phase of the mixtures was transferred into the new tubes and labeled as ‘lipid’ fraction. These lipid fractions were stored in −80°C until LC-MS analysis.
Fig. 2.
GBM biological sample preparation for omics study. (A) GBM10 and GBM43 cells were expanded as flank tumors and harvested. Human GBM10 and GBM43 tumor cells were cultured and expanded in vitro prior to implantation. (B) GBM cells were then implanted into the flank (3×105 cells/flank) and the right cerebrum (3×106 cells/brain) of each mouse and tumors were allowed to develop for 21 to 24 days. (C) Tumor samples from intracranial and flank sites were harvested by excision. The tumors were flash frozen immediately and stored in −80°C. (D) Proteins, metabolites, and lipids of GBM were extracted. The 3 fractions of the biomolecules (polar metabolite, protein, and lipid) are indicated.
LC-MS Analysis
All analyses were carried out using an Agilent 1200 Series HPLC, ACE 5 C8-300 column (2.1 × 100 mm) and linear gradient elution at a flow rate of 0.1 mL/min. Injection volume was 4 μL. The mobile phases were composed of (A) 0.1% formic acid and 10 mM ammonium acetate in H2O, and (B) 0.1% formic acid, 10 mM ammonium acetate in ACN/Isopropanol (50/50; v/v). Separation of lipids was achieved at the following gradient: T=0 min: 30% B; T=1 min: 30% B; T=25 min: 100% B; T=45 min: 100% B; T=47 min: 30% B; and T=60 min: 30% B (column re-equilibration).
HPLC system was directly coupled to a Bruker solariX 70 Hybrid FTMS instrument equipped with electrospray ionization source (ESI). The system was controlled by HyStar v.3.4.8.0 software (Bruker Daltonics). MS data was collected with resolving power of 78,000 (at m/z 400) in positive or negative mode under following conditions: a capillary voltage of (+/−) 4,500 V and an end plate offset of −500 V. The dry temperature was set at 180 °C. Dry gas flow was maintained 4 L/min. Acquisition range was 244 – 1,800 m/z with 0.2 s ion accumulation time.
Data Processing & Statistical Analysis
LC-MS data was converted into mzXML format using CompassXport v. 3.0.6. (Bruker Daltonics) and processed by the mzMine v.2.10 data analysis software. Data processing involved mass detection, chromatographic peak detection and deconvolution, gap filling, isotopic peaks grouping, normalization and peak alignment. To determine the most significant changes, a series of homoscedastic t-tests comparing GBM10 and GBM43 brain, flank, and control tumors in all possible permutations was performed. The corresponding lipids were tentatively identified using LIPID MAPS, a web-based lipid database, that uses a textual and ontological search to identify sub class, class, and common structure of a lipid within a variable m/z tolerance of 0.02 m/z, representing the closest catalogued lipid. Additional structure was not elucidated. (LIPID MAPS Lipidomics Gateways, 2014).
Ethics Approval
All studies were reviewed and approved by the Indiana University Animal Care and Use Committee (IACUC #10103) or the Purdue University Institutional Biosafety Committee (IBC 10-003).
RESULTS
Mass Spectrometry Data Overview
Flank and brain tumor samples derived from two primary human GBM tumor cell lines, GBM10 and GBM43, were collected and analyzed. Lipid profiles of intracranial tumors versus control brain tissue and intracranial tumors versus flank tumors were determined. Among all 26 samples, which were actually extracted and analyzed, 11,218 unique positive-ion mode peaks were quantified by mzMine2, representing an m/z ratio range of 244 to 1,800. To filter out insignificant peaks, a truncated data set was created by removing any peak detected in fewer than five samples, as there were five replicate samples in each model grouping. This yielded 4,422 positive-ion mode peaks. A similar process for data ran in the negative-ion mode yielded 725 unique peaks.
Positive and negative ion mode of electrospray ionization (ESI) mass spectra was used to distinguish different charge state distribution of the ions in macromolecules. Generally, the positive ion mode detects the ion peaks in different protonation states [18]. As described in the material and methods, GBM10 and GBM43 cells were implanted in the flank and right cerebral hemisphere in each mouse on study. Sufficient sample size of 10 mg or greater was required for analysis and was obtained in all GBM10 and GBM43 intracranial tumors and for GBM10 flank tumors (n=5 per group). For GBM43 implanted mice, intracranial tumors progressed faster than the flank GBM43 tumors. Therefore it was necessary to euthanize GBM43-implanted mice at a time point when only four out of five mice had measurable flank tumors; this was necessary in order to capture high quality intracranial GBM43 tumor tissue for extraction and subsequent lipid analyses. At this time point, three out of four GBM43 flank tumors were too small (1–2 mg) for extraction. Lipid profiling was completed on the GBM43 flank tumor of sufficient size. In order to compare the tumor lipid profiles against the normal brain lipid profiles, t-test was performed between the tumor and normal brain profiles of GBM10 (both flank and intracranial tumors) and GBM43 (intracranial tumors only). Of the positive-ion mode data, 368 lipids were identified to be significantly different in lipid levels between GBM10 brain tumor tissue and the control brain tissue, 305 lipids between GBM43 brain tumor tissue and the control brain tissue, and 1926 lipids between GBM10 flank tumor tissue and control brain tissue. Interestingly, GBM10 flank tumor profiles showed a higher number of lipid differences compared to the brain tumor profiles. Of the negative-ion mode data, 149 lipids were expressed at significantly different levels between GBM10 brain tumor tissue and the control brain tissue, 233 lipids between GBM43 brain tumor tissue and the control brain tissue, and 157 lipids between GBM10 flank tumor tissue and the control brain tissue. All these lipids had a fold effect change of one or greater and p-value less than 0.05 from the t-test. A summary of the number of significant lipids from the tumors propagated at different anatomical sites (flank versus intracranial sites) and the ratio of significantly decreased and increased lipids compared to control brain tissue is shown in Table 1. More than 500 lipid species were detected at significantly different levels in tissues derived from the flank versus the brain, and more than 90% of these lipids were decreased in both GBM10 and GBM43 brain and flank tumors when compared to control brain tissue. The data supports that GBM tumor tissue contains dramatically lower levels of lipids and a different lipid composition than normal brain tissue.
Table 1.
Differentially expressed significant lipids in different types of GBM tissues and control brain tissues.
| GBM10 Brain | GBM43 Brain | GBM10 Flank | ||
|---|---|---|---|---|
| Positive ion mode | Over-expressed | 26 | 21 | 360 |
| Under-expressed | 342 (92.9%) | 284 (93.1%) | 1566 (81.3%) | |
| Negative ion mode | Over-expressed | 2 | 0 | 4 |
| Under-expressed | 147 (98.7%) | 233 (100%) | 153 (97.5%) | |
Comparison of Lipid Profiles in Orthotopic Versus Ectopic Xenograft Tumors
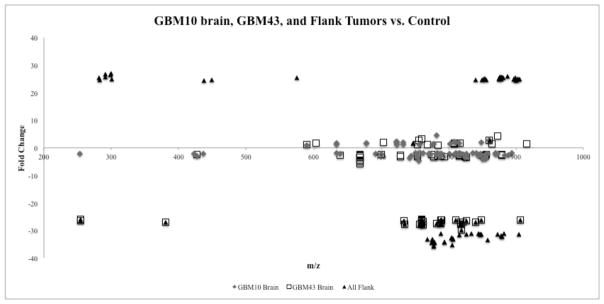
We next investigated differences in overall lipid composition between tumor tissues derived from the same primary human GBM cell lines but located in distinct anatomical sites. A t-test was used to compare the lipid profiles of the all brain tumors and all flank tumors in the same manner as the comparison between the tumor and the control profiles. 1960 lipids were differentially expressed on the positive-ion mode profile of the flank tumor compared to the brain tumor profiles and 206 lipids were differentially expressed on the negative-ion mode profile (Table 2). All these lipids also had the fold effect change of one or greater and p-value less than 0.05 based on the t-test. As shown in Table 2, the majority of these lipids were decreased in flank tumor tissues compared to brain tumor tissues. Differentially expressed lipid types between brain and flank tumors support the pattern in (Fig. 3), which expresses m/z range and different fold effect change among the brain tumors and the flank tumors. In general, lipidomic profiles of the flank tumors of GBM expressed a greater fold change when compared to the brain tumor profiles. The range of the lipid fold change of the flank tumors was between approximately 20 to 30 and −35 to −20. In contrast, brain tumors of both GBM10 and GBM43 showed a fold effect change range between −5 and 5.
Table 2.
Differentially expressed significant lipids of flank tumors compared to brain tumors.
| Number of lipids | ||
|---|---|---|
| Positive ion mode | Negative ion mode | |
| Over-expressed | 267 | 7 |
| Under-expressed | 1693 | 199 |
Fig. 3.
m/z and fold effect comparison of each tissue type. The m/z and fold effect range of differentially expressed lipids between brain and flank tumors are indicated. In general, lipidomic profiles of the flank tumors of GBM expressed a greater fold change (between −35and −20) when compared to the brain tumor profiles. m/z value of the identified lipids were mostly concentrated in the range of 750 and 900.
Significantly Different Lipid Levels and Composition in GBM
Most of the 500 significant lipids identified were from the lipid classes of glycosphingolipids, glycerophosphoethanolamines, triradylglycerols, glycerophosphocholines, and glycerophosphoserines. The thirty lipids with the highest differences in fold effect from the comparison of GBM43 brain tumor versus the control, GBM10 brain tumor versus the control, and flank tumor versus the control were selected from the larger group for closer examination. These lipids fell mostly into four main lipid classes: glycosphingolipids, glycerophosphoethanolamines, triradylglycerols, and glycerophosphoserines. There were three compounds among top 30 significant lipids identified from each tumor type that were identified in both positive and negative mode. These identified lipids were Glycerophosphoglycerols (mass= 788.59, PG(38:2)/PG(38:1)), Glycerophosphocholines/Glycerophosphoethanolamines (mass=753.55, PC(35:5)/PC(34:4), PE(38:4)/PE(37:4)), and Glycerophosphocholines (mass=873.71, PC(42:0)). Fig. (4) is a visual comparison of the differences in lipid concentration among the tissue types.
Fig. 4.
Decreased trend of thirty significantly expressed lipids and lipid classes. Thirty most significantly identified lipids from each tissue type (all flank, GBM10 brain, and GBM43 brain tumors) were plotted and grouped in four lipid species. These lipids include glycerophosphoserines, glycerophosphocholines, glycerophosphoethanolamines, and triradylglycerols. * symbol on the lipid structure represents the lipids that are identified as more than one lipid classes.
The positive and negative ion profiles revealed differences in the level and composition of lipid classes among brain and flank tumors. The positive ion profiles of glycerophosphoserines showed decreased levels of lipids in GBM43 brain tumor tissue and flank tumor tissue. There were only a few increased levels of lipids from GBM10 and GBM 43 tumor tissue (Fig. 4A). The negative ion profile of glycerophosphoserine did not show any increased levels of lipids and the majority of the decreased lipids were expressed in flank tumor tissue (Fig. 4B). Glycerophosphocholines were identified as the most frequently identified lipids in the negative ion profiles among different tissue sources and also showed a decreased pattern of lipid levels (Fig. 4D). Some of the flank tumor lipids from the positive ion profiles of glycerophosphocholine were increased in comparison to the control, while the negative ion profiles showed a similar distribution of lipids for all three tumor tissue types: GBM10 brain tumors, GBM43 brain tumors, and all flank tumors. Glycerophosphocholines in flank tumor tissue, however, had higher fold change than brain tumor tissue (Fig. 4C). The profiles of glycerophosphoethanolamines had a similar trend of decreased level of lipids. The level of glycerophosphoethanolamines in GBM10 brain tumor tissue often increased in the positive ion profiles, while GBM43 and flank tumor tissue showed decreased fold change (Fig. 4E). The negative ion profiles of glycerophosphoethanolamines were similar to glycerophosphocholines (−) with decreased levels of lipids (Fig. 4F). The positive ion profiles of triradylglycerol plot showed lipids primarily from flank tumor tissue. Some lipids of GBM10, GBM43, and flank tumor were decreased, yet overall, the majority of the levels of lipids that were detected from flank tumor tissue were increased (Fig. 4G). Several lipid classes and structures were significant in both the positive and negative mode, namely glycerophosphoserines, glycerophosphocholines, and glycerophosphoethanolamines (Fig. 4).
Hierarchical Clustering of GBM Tumors
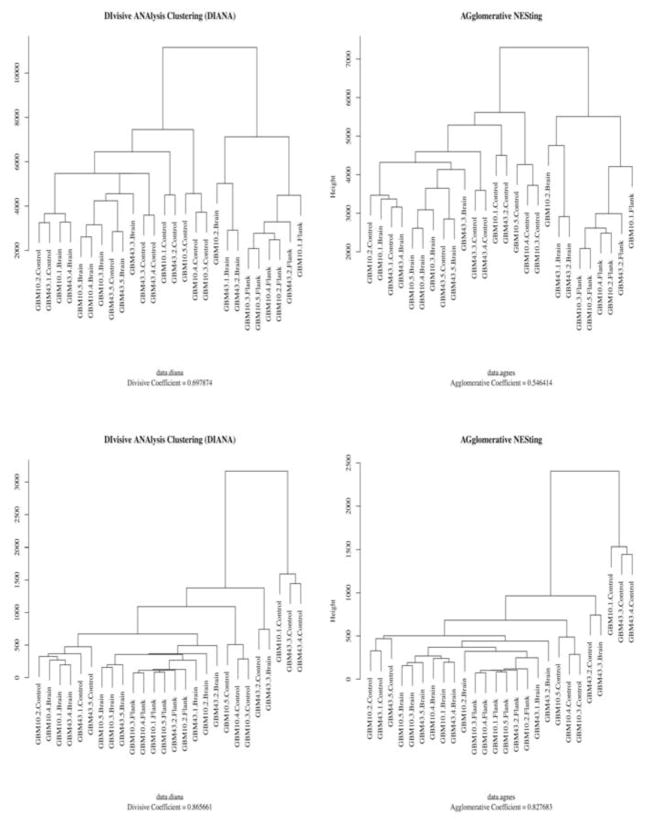
For quality assurance purposes, a series of analytical approaches were employed in order to identify any definite peculiarities associated with the results. In particular, special attention was paid to determine whether the data for the samples demonstrated affinities consistent with the grouping of the samples. For this purpose, an analysis using a set of clustering algorithms, Divisive ANAlysis (DIANA) and AGglomerative NESting Hierarchical Clustering (AGNES), was performed. Clustering is a widely adopted technique that is unsupervised and data-dependent in that it explores the relationships between all of the variables and not simply the targeted ones [19], and we chose clustering to avoid introducing bias into our quality assurance efforts. To generate the clusters of the samples, m/z values and peak intensities from MzMine2 data were compared. As a result, the output from DIANA and AGNES showed similar patterns of clusters while these two algorithms have distinctly different paths to generate the output [20] and have a history of use with similar data [14].
As is evident from (Fig. 5), these disparate approaches produced closely related results. All the figures generated from DIANA and AGNES showed a propensity for the flank subjects to clearly cluster away from the other tissue types while the brain and control groups demonstrate a more closely clustered set of results. However, even in the brain and control groups, each group tended to segregate in the expected cohorts. Based on the evidence rendered from using DIANA and AGNES, it is likely that the results achieve the aforementioned affinities, and accordingly the pattern mining efforts help to establish the validity of the experimental protocol.
Fig. 5.
Hierarchical clustering output of xenograft tissues. Two different hierarchical clustering methods (DIANA and AGNES) were utilized to validate the data analysis method. Clusters were created by m/z values and peak intensity from the data generated by MzMine2. Top two graphs were generated from the positive ion mode data and bottom two graphs were generated from the negative ion mode data.
DISCUSSION
GBM Cell Lines and Tumor Size
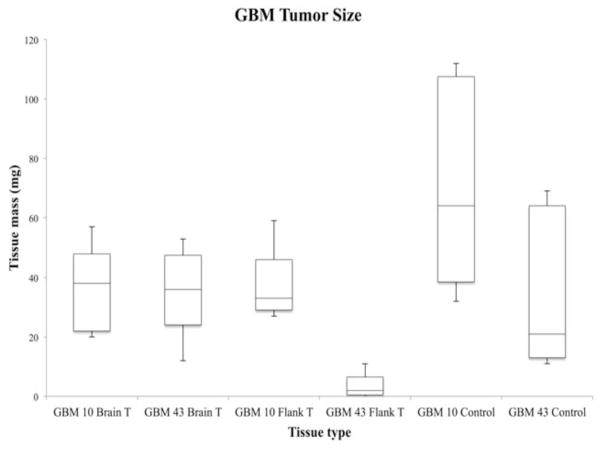
Two human primary GBM cell lines, GBM10 or GBM43, were used in ectopic and orthotopic mouse xenograft models and harvested after adequate tumor growth and expansion. In addition to the lipid profile differences described in this study, qualitative differences were also observed among the tumors derived from either GBM10 or GBM43. Fig. (6) was generated to compare the tumor and normal brain tissue sizes obtained from the different tissue types. All tissue types contained sample size of five (n=5) each. The box plot of the tumor size shows that the size of GBM43 flank tumors was significantly smaller compared to other tumor tissues. The analysis of lipid profiling was done in one out of the four GBM43 flank tumors and the results were consistent with observations in the GBM10 model in that different lipid profiles exist overall in tumors propagated in the flank versus the brain. Flank tumor tissues were also more difficult to homogenize during the extraction procedure when compared to brain tumors and control samples. Control brain and brain tumor tissues were mostly homogenized on the first attempt. However, most of the flank tumors from GBM10 and GBM43 were homogenized up to five times in order to adequately break down the tissue for subsequent analysis. Observed morphological differences in the tumors may be due to molecular differences between GBM10 and GBM43. Previous studies have reported that GBM10 is wildtype for p53 but GBM43 is mutant for p53 [15]. p53 is a tumor suppressor protein that has many functions in cancer cells, including lipid metabolism [21].
Fig. 6.
Tumor size comparison between different GBM tumors and control brain tissue. The box plot represents the tumor size of different tissue types. The mass of GBM10 brain tumors (n=5), GBM43 brain tumors (n=5), and GBM10 (n=5) tumors were similar. However, the size of GBM43 flank tumors (n=5) from the xenograft model was significantly smaller compared to other tumor tissues. One of GBM43 flank tumor did not grow, which counted as 0mg. Four tumors were harvested for the lipid extraction. However, three among these four tumors were too small for the extraction.
Biological Sample Preparation for Mass Spectrometry
The novel protocol for concurrent protein, metabolite, and lipid extraction from a single sample was designed for a sample tissue size between 10mg and 100mg and helps address current limitations in sample preparation of proteins, metabolites and lipids, thus resulting in multiple benefits. First, simple and minimal procedures can save sample preparation time, minimize degradation of metabolites, and lower sample loss during preparation period. Secondly, the bio-molecular samples isolated from this protocol include a wide range of lipids, metabolites and proteins, which is beneficial for untargeted omics studies. Thirdly, due to the simple and fast sample preparation steps, the method is easily reproducible. Systems biology approaches use a large number of samples and reproducibility is critical [22].
Data Analysis
Quantitative analysis of the detected lipids revealed patterns that indicate noteworthy differences between tumor location in the mouse xenograft model of GBM. As previously mentioned, cursory analysis using t-tests and manual identification indicated that while the general lipid profiles of flank tumors and brain tumors appeared to be in a similar m/z range, a comparison of lipid classes showed differences in the content of lipids. Hierarchical clustering indicates distinct profiles of lipids in flank tumor tissues distinguished from both brain tumor and brain control tissue, while brain tumor and brain control tissue cluster together. This evidence may suggest that control brain and brain tumor tissues were more closely related to each other than flank tumor tissues. The clustering output may also imply that certain ‘background noise’ from surrounding tissue of the tumor (i.e. the variance in lipid composition of brain tissue and flank tissue) can account for a small portion of this clustering pattern.
Beyond the hierarchical clustering of the data, lipid identification introduces an element of uncertainty into the analysis. Because the mass spectrometer returned hundreds of significantly differentially regulated lipids, many of which shared nearly identical m/z ratios, power of manual analysis is influenced by both (1) the resolution of the LipidMaps database and (2) the size of the database (containing just over 37,500 unique lipids). The potential of fragmentation and creation of adducts within the ion generator, though roughly accounted for by mzMine2, add further complication to the manual identification of lipids. The instrument used, a Bruker 7-Tesla FT-MS, utilizing the solariX platform, provides mass accuracy on the magnitude of greater than under one part per million, which greatly increases the certainty of database returns on lipid identification. Likewise, the soft-ionization of the electrospray limits fragmentation and is compatible with front-coupled RHPLC used to separate lipids prior to analysis in the spectrometer.
Lipid Studies and Decreased Level of Lipids in GBM
Our data showed that more than 90% of significantly identified lipids from brain and flank GBM tumors were decreased compared to the control brain tissue. The majority of studies exploring lipid metabolism in cancer have reported increased levels of fatty acid synthesis [23]. Lipidomic analysis is difficult due to the specificity and complexity of lipid composition and the lack of techniques for analysis [10]. In addition, the regulation and complex mechanisms of lipid composition associated with cellular homeostasis are still poorly understood [13]. There are also limitations in the use of mass spectrometry for the structural identification of lipids. It is extremely difficult to accommodate all lipid classes using existing current detection methods due to the number and variety of classes and molecular species of lipids [10]. Thus most studies focus on investigating lipids indirectly by examining the proteins involved in lipid metabolism instead of directly measuring the composition of lipids in cancer cells. Some previous studies have examined the correlation between membrane lipid composition and malignancy of brain tumors. Campanella and colleagues (1992) measured membrane lipid changes among different grades of human gliomas using HPTLC. They concluded that higher levels of malignant glioma have significantly lower levels of total plasma membrane lipids in tumor tissues [24]. More recently, Eberlin and colleagues (2012) used mass spectrometry to classify types of gliomas and showed that the total lipid abundance profile of grade IV astrocytoma was lower than the low-grade astrocytomas [25].
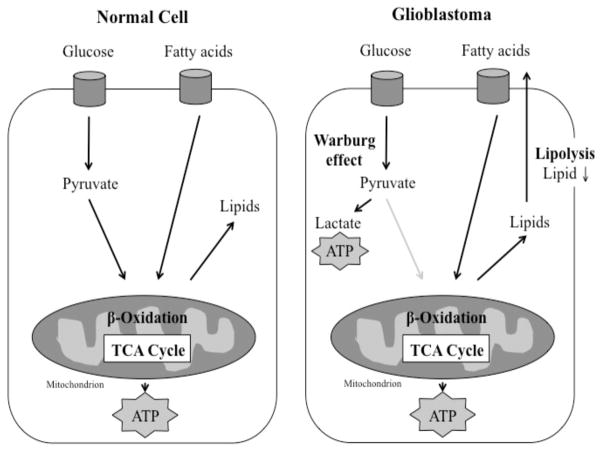
Normal cells use mainly glucose and fatty acids to generate energy and fulfill the requirements for cell growth. However, cancer cells use an altered metabolism in order to sustain rapid growth. This altered metabolism, where cancer cells use higher levels of glucose to generate energy by anaerobic glycolysis rather than aerobic glycolysis through the tricarboxylic acid (TCA) cycle, is called Warburg effect [26]. As shown in (Fig. 7), the decreased levels of lipids observed in the glioblastoma tumor tissue may suggest that GBM depends on fatty acids as a fuel source in addition to glucose from anaerobic glycolysis. Potentially, an increased level of lipolysis in the GBM cancer cell generates energy for cancer cell proliferation and results in an overall decreased level of lipids detected. Other studies have shown that fatty acids and internal structures are used as an energy source under starvation conditions, such as in a tumor environment [27] and that the lipolytic enzyme, monoacylglycerol lipase (MAGL), is highly upregulated in aggressive forms of cancer [23]. Inhibition of MAGL subsequently inhibited migration, invasion, and survival characteristics of cancer cells in both in vitro and in vivo models [23].
Fig. 7.
Normal cells primarily use glucose to generate energy and fulfill the requirements for cell growth. Cancer cells alter glucose metabolism and bypass the TCA cycle through the “Warburg effect,” in order to sustain rapid cell growth. Decreased amounts of lipids, which may occur through lipolysis in cellular organelles such as ER and Golgi, may suggest that glioblastoma cells use fatty acids as an alternative fuel source.
The central nervous system has specialized pathways for lipid synthesis and degradation related to its specialized physiology and function [28]. Two scenarios may thus result in a decrease in the lipid composition: lipid rich brain tissue replacing less lipid rich tumor cells would decrease the amount of lipids overall while tumor cells would also likely exploit normal physiological pathways related to lipid metabolism in central nervous tissue. There are numerous reports in the literature that fatty acid synthetase (FASN) is highly upregulated in a variety of cancers including GBM and may be a good therapeutic target [29, 30]. Future studies will employ more robust methods of lipid identification to confirm the findings of this work, as well as explore the mechanism related to the decrease in lipid composition by examining the metabolic profile and cellular composition of the tumors.
Lipid Function and Fatty Acid Oxidation
Cellular growth requires energy. High levels of lipids are critical for building membranes and are required in rapidly proliferating cancer cells. Cancer cells can use anaerobic glycolysis to maintain energy levels in a cancer environment through the Warburg effect [31]. Some types of cancer also use alternative energy sources, as shown in aggressive cancer cell lines that express higher levels of free fatty acids [32]. Research suggests that prostate cancer does not depend on glucose for survival and instead may depend heavily on fatty acid oxidation for cancer cell growth [33]. Fatty acid oxidation also has a critical role in the proliferation and survival of leukemia [34]. Fatty acid oxidation has also been shown to contribute to energy production and resistance to oxidative and nutrient stress in GBM [35].
Recent articles [36, 37] have cautioned about the impact of experimental conditions on the reproducibility of systems biology experiments. An important consideration for team-based science is establishing a pipeline that ensures reproducible handling and processing of samples and data along multiple steps. Through this study, we have established a pipeline for the identification of molecular changes in a model system for GBM, including a multidisciplinary team of research scientists, clinicians and statisticians with expertise in current technologies.
CONCLUSION
Overall, we found that GBM xenograft tumors had significantly lower levels of lipids when compared to the normal brain tissue. Our results suggest that GBM may use fatty acid oxidation as an additional energy source in a nutrient deprived cancer environment (Fig. 7). It is also possible that the lower level of lipids could reflect changes in the cellular composition of the tumor over time. Normal brain tissue has extensive cellular insulation that includes lipid-rich myelin. As normal neuronal cells become replaced with tumor cells, the lipid-rich myelin may decrease and subsequently the overall lipid profile in the tumor would decrease. The profile for the flank tumors also decreased, however, and thus it is likely that multiple factors contribute to the changes we observed in the lipids.
Models for cancer systems are complex and have limitations that must be balanced with experimental needs and objectives. We explored differences in tumors promoted by two human primary tumor cell lines when placed in two distinct environments, the flank or the brain, within a mouse model system for glioblastoma multiforme. Lipid expression profiles and statistical clustering analysis revealed that tumors from the flank were different from tumors in the brain, suggesting that the molecular profile of the tumors was dependent upon the environment. The ease of homogenization during the biomolecule extraction revealed differences in physical characteristics between the brain tumors and flanks tumors. The physical differences were supported by molecular profile differences in classes of significant lipids between the two xenograft tissues as flank tumors showed higher fold effects overall when compared to brain tumors. This has implications for future studies using mouse xenograft models to examine the molecular mechanisms that control the progression of GBM and the impact of potential therapeutic interventions. Studies using tumors propagated in foreign micro-environments such as the flank while informative in initial stages of exploration must be used with caution when testing the efficacy of anti-cancer therapies.
Acknowledgments
This work was supported in part by the Riley Children’s Foundation, the Indiana University Simon Cancer Center, the Jeff Gordon Research Foundation (HW, and KP), and RO1 CA138798 (HW, SC, ML, and KP), The College of Technology Purdue University (KC), Mayo Brain tumor SPORE CA108961 (JS), and Indiana Clinical and Translational Sciences Institute (KC, KP, JA and JR). This work was also supported in part by the IUPUI Signature Center Initiative for the Cure of Glioblastoma.
Footnotes
AUTHORS’ CONTRIBUTIONS
Kari Clase was responsible for leading the collaboration, analyzing data and interpreting findings within the context of the biological system and establishing the data collection and sample analysis pipeline.
Soo Jung Ha and Gordon M. Showalter conducted the sample preparation, mass spectrometry data collection and statistical data analysis.
Jiri Adamec supervised mass spectrometry data collection.
John Springer conducted statistical clustering analysis of mass spectrometry data.
Shanbao Cai, Haiyan Wang, Wei M. Liu were directly responsible for intellectual input on model set up. They also set up all flank and orthotopic model, mouse monitoring, and isolation of tumor samples.
Karen Pollok was responsible regulatory requirements, intellectual input, and all aspects of in vivo model set up, monitoring, and sample collection.
Jenna Rickus provided intellectual input.
Jann Sarkaria provided tumor cell lines and intellectual input.
Aaron Cohen-Gadol provided intellectual input.
CONFLICT OF INTEREST
The authors confirm that there is no conflict of interest with the data presented in this article.
References
- 1.Pelloski CE, Gilbert MR. Current treatment options in adult glioblastoma. US Oncolog Dis. 2007:105–109. [Google Scholar]
- 2.Agnihotri S, Burrell KE, Wolf A, Jalali S, Hawkins C, Rutka JT, Zadeh G. Glioblastoma, a brief review of history, molecular genetics, animal models and novel therapeutic strategies. Archivum immunologiae et therapiae experimentalis. 2013;61(1):25–41. doi: 10.1007/s00005-012-0203-0. [DOI] [PubMed] [Google Scholar]
- 3.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lutolf UM, Kleihues P. Genetic Pathways to Glioblastoma A Population-Based Study. Cancer Res. 2004;64(19):6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 4.Oertel J, von Buttlar E, Schroeder HW, Gaab MR. Prognosis of gliomas in the 1970s and today. Neurosurgical focus. 2005;18(4):e12–e12. doi: 10.3171/foc.2005.18.4.13. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, Van Den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross G, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Eng J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Lipsitz D, Higgins RJ, Kortz GD, Dickinson PJ, Bollen AW, Naydan DK, LeCouteur RA. Glioblastoma multiforme: Clinical findings, magnetic resonance imaging, and pathology in five dogs. Veterinary Pathology. 2003;40:659–669. doi: 10.1354/vp.40-6-659. [DOI] [PubMed] [Google Scholar]
- 7.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu C, Genovese G, Shmulevich I, Barnholtz-Sloan J, Lihua Z, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niclou SP, Fack F, Rajcevic U. Glioma proteomics: status and perspectives. J Proteomics. 2010;73(10):1823–1838. doi: 10.1016/j.jprot.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry a bridge to lipidomics. J Lipid Res. 2003;44(6):1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Tripathy K. Lipidomics: A Promising area in. J Comp Sci Systems Biol. 2011;4(5):93–98. [Google Scholar]
- 11.Haag M. Essential fatty acids and the brain. Canadian journal of psychiatry Revue Canadienne De Psychiatry. 2003;48(3):195–203. doi: 10.1177/070674370304800308. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XD, Qin ZH, Wang J. The role of p53 in cell metabolism. Acta Pharmacologica Sinica. 2010;31(9):1208–1212. doi: 10.1038/aps.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11(8):593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 14.Gough E, Oh C, He J, Riley C, Buck C, Zhang X. Proteome discovery pipeline for mass spectrometry-based proteomics. BMC Bioinformatics. 2008;9(Suppl 7):21. [Google Scholar]
- 15.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance, and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Current Protocols in Pharmacology. 2011:14–16. doi: 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, Schroeder MA, James CD. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neurooncology. 2005;7(2):164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, Ballman KV, Kitange GJ, Guha A, Pandita A, James CD. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12(7):2264–2271. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 18.Konermann L, Douglas DJ. Unfolding of proteins monitored by electrospray ionization mass spectrometry: a comparison of positive and negative ion modes. J American Soci For Mass Spectrometry. 1998;9(12):1248–1254. doi: 10.1016/S1044-0305(98)00103-2. [DOI] [PubMed] [Google Scholar]
- 19.Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput Surv. 1999;31(3):264–323. [Google Scholar]
- 20.Kaufman L, Rousseeuw PJ. Finding groups in data: an introduction to cluster analysis. Vol. 344. John Wiley & Sons; New York: 2009. [Google Scholar]
- 21.Goldstein I, Rotter V. Regulation of lipid metabolism by p53–fighting two villains with one sword. Trends in Endocrinology & Metabolism. 2012;23(11):567–575. doi: 10.1016/j.tem.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Vuckovic D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography–mass spectrometry. Analytical Bioanalytical Chem. 2012;403:1523–1548. doi: 10.1007/s00216-012-6039-y. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, Du G. Dysregulated lipid metabolism in cancer. J Biol Chem. 2012;3(8):167. doi: 10.4331/wjbc.v3.i8.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campanella R. Membrane lipids modifications in human gliomas of different degree of malignancy. J Neurosurgi Sci. 1992;36(1):11. [PubMed] [Google Scholar]
- 25.Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, Cooks RG, Agar NY. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012;72(3):645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 27.Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, Singh R. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metabol. 2011;14(2):173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieberman M, Marks AD. Marks’ Basic Medical Biochemistry: A Clinical Approach. 4. Lippincott Williams & Wilkins; Philadelphia: 2012. [Google Scholar]
- 29.Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6(4):551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao BB, He H, Shi XH, Wang CL, Li WQ, Li B, Dong Y, Hu G, Hou L, Luo C, Chen J, Chen H, Yu Y, Sun Q, Lu YC. Up-regulation of USP2a and FASN in gliomas correlates strongly with glioma grade. J Clinical Neurosci. 2013;20(5):717–720. doi: 10.1016/j.jocn.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 31.Tennant DA, Durán RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10(4):267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 32.Buzzai M, Bauer DE, Jones RG, DeBerardinis RJ, Hatzivas-siliou G, Elstrom RL, Thompson CB. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid β-oxidation. Oncogene. 2005;24(26):4165–4173. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Diseases. 2006;9(3):230–234. doi: 10.1038/sj.pcan.4500879. [DOI] [PubMed] [Google Scholar]
- 34.Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, Andreeff M. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clinical Investigat. 2010;120(1):142. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 36.Kolker E, Özdemir V, Martens L, Hancock W, Anderson G, Anderson N, Aynacioglu S, Baranova A, Campagna SR, Chen R, Choiniere J, Dearth SP, Feng W, Ferguson L, Fox G, Frishman D, Grossman R, Heath A, Higdon R, Hutz MH, Janko I, Jiang L, Joshi S, Kel A, Kemnitz JW, Kohane IS, Kolker N, Lancet D, Lee E, Li W, Lisitsa A, Llerena A, MacNealy-Koch C, Marshall JC, Masuzzo P, May A, Mias G, Monroe M, Montague E, Mooney S, Nesvizhskii A, Noronha S, Omenn G, Rajasimha H, Ramamoorthy P, Sheehan J, Smarr L, Smith CV, Smith T, Snyder M, Rapole S, Srivastava S, Stanberry L, Stewart E, Toppo S, Uetz P, Verheggen K, Voy BH, Warnich L, Wilhelm SW, Yandl G. Toward more transparent and reproducible omics studies through a common metadata checklist and data publications. Omics: J Integrative Biol. 2014;18(1):10–14. doi: 10.1089/omi.2013.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genetics. 2013;14(2):89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]