The past decade has witnessed considerable advancements in sequencing technologies, which have allowed comprehensive investigation of genetic variation of individuals at a moderate cost and within a reasonable time frame. Personalized disease risk and drug response predictions based on genomic sequences are a cornerstone of preventive precision medicine, and have also been successful at informing therapeutic decisions. However, genomics is limited in predicting the onset of most common diseases (i.e., cancer, diabetes, and cardiovascular disorders) because genetic information is mostly static and does not account for dynamic environmental (i.e., diet and lifestyle) or gut microbiota influences.
Metabolomics, the study of a large collection of metabolites, offers the advantage to measure the functional readout of activity and phenotype encoded in the genome. Hence, combining genetic and metabolic information provides a unique opportunity to gain further insights on how the genetic program is translated into biological function through metabolites, and how alterations in the program associate with the onset of diseases. This approach has already proven very useful at diagnosing and understanding the pathogenesis of rare inherited metabolic disorders. Furthermore, metabolic profiles are influenced by the environment and the gut microbes; thus metabolomics has the potential to unravel the impact of nongenetic factors on disease onset as well as reveal early biomarkers that may improve risk assessment and diagnosis of complex diseases. Thanks to rapid improvements in technology, mass spectrometry–based metabolomics can now robustly profile a broad spectrum of metabolites at a relatively low cost (1).
The concept that genetic variations can be captured at the metabolite level in a population was first demonstrated in 2008 by Gieger et al. (2). That study gave a glimpse of the usefulness of combining genetic information with metabolic traits to understand the pathogenesis of common diseases and the influence of environment. In 2015, Guo et al. propelled this concept further by showing that integration of personal genome and metabolome information could enhance precision medicine by improving the prediction of disease risks and therapeutic decisions in a clinical setting (3). Specifically, they provided several interesting examples that metabolic profiles can be successfully used to (a) measure the penetrance of mutations of known pathogenicity, (b) uncover potentially damaging genetic variants, and (c) detect early signs of disease onset and drug response.
In the study of Guo et al., the genetic information of 80 adults of normal health was captured by whole exome sequencing, and matching plasma metabolic profiles were measured by nontargeted mass spectrometry. To account for the diversity of metabolites contained in plasma samples, ranging from hydrophilic amino acids to hydrophobic lipids, multiple complementary LC-MS and GC-MS methods were used. This platform enabled the profiling of close to 600 metabolites, covering all the major biochemical pathways. To identify functional links between genes and metabolites, the authors focused on 3 aspects: (a) metabolites likely affected by damaging genetic variants (common variants were filtered out), (b) mutations predicted to be damaging on genes that could explain deregulated metabolites, and (c) metabolites derived from drugs taken at the time of the study.
Using this approach, Guo et al. were able to trace key metabolite alterations back to potential causative genetic variants. For instance, they noted a remarkable increase of fructose and sorbitol (4- and 34-fold of cohort medians, respectively) in an individual with no known history of metabolic disease. These changes—often indicative of fructose intolerance—were correlated to a damaging mutation in ALDOB (aldolase, fructose-bisphosphate B), which encodes the fructose-bisphosphate aldolase in the sorbitol degradation pathway. Interestingly, this observation resulted in dietary recommendations to avoid longterm liver and kidney damage caused by persistent fructose intolerance. Similarly, abnormal concentrations of acylcarnitines/long-chain fatty acids and bile acids were observed in conjunction with rare mutations predicted to be damaging in genes encoding a transporter and an enzyme, respectively. Although the study provides intriguing examples of deregulated metabolites that helped identify putatively damaging mutations, the links between said genes and metabolites remain correlative at this time. Further experimental evidence at the molecular level, such as demonstrated by enzymatic assays on mutant proteins, will be needed to establish the causative nature of the links.
Another goal of precision medicine is to predict individual response to medications including efficacy and toxicity. Interestingly, the nontargeted metabolomic approach used by Guo et al. not only monitored endogenous metabolites, but also detected drugs and drug degradation products. For example, antidiabetic drugs were observed in the blood of 2 individuals under well-controlled diabetes care. It was possible to gain insights into the efficacy of the treatments by looking at the concentrations of the metabolites having known association with diabetes (i.e., energy metabolism including branched-chain amino acids and glucose) in these volunteers compared with the rest of the cohort. Also, the toxicity of other commonly used drugs such as acetaminophen could be inferred by looking at plasma concentrations, as well as the amount of bile acids, which are known to correlate with drug-induced liver damage.
The study represents the most recent effort to integrate genomic sequence with metabolic traits in the context of precision medicine (Fig. 1). Undoubtedly, metabolomics will improve the overall disease diagnosis and risk assessment for patient care by giving access to biochemical information in the context of observed genetic variations. Nonetheless, a number of improvements are still needed to reveal the full potential of integrated multiomic profiles in the context of precision medicine.
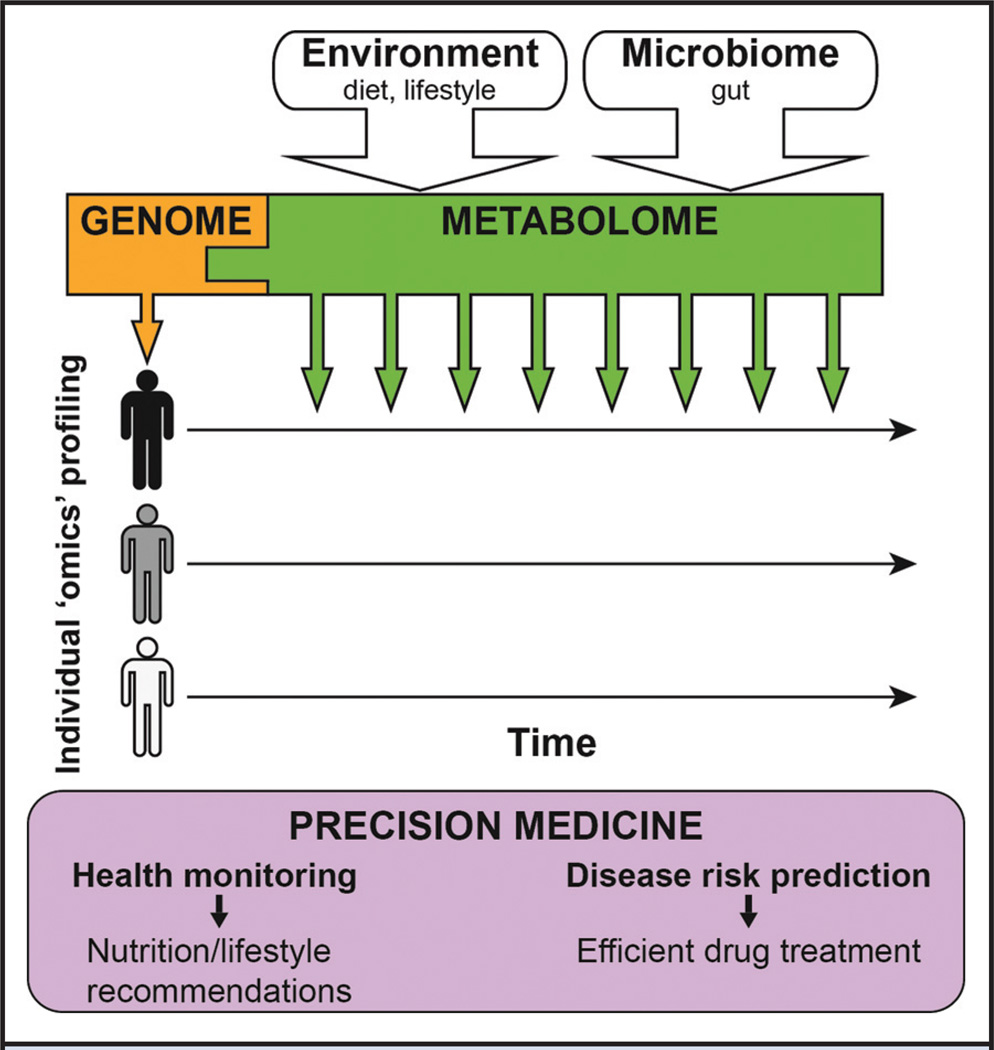
Fig. 1. Combination of personal genetic information and metabolic profiles for precision medicine.
Static DNA sequences inform on inherited genetic variations, whereas dynamic metabolites—influenced by the environment and the microbiome—give access to the phenotypic readout of the genome. After defining an individual health baseline, regular metabolome measurements (indicated by the multiple green down arrows) allow health monitoring as well as disease risk prediction and can result in nutrition/lifestyle recommendations to prevent the onset of a disease or increase the efficiency of drug treatments.
First, despite the capability to generate robust omics data, the biologic interpretation of such profiles is still lagging. More comprehensive functional annotation of genetic variants in databases and more sophisticated computational algorithms are necessary to improve the prediction of pathogenic variants. This is particularly true for genomics data generated by whole genome sequencing, which includes regulatory elements, such as promoters and enhancers. Furthermore, metabolomics is hampered by the lack of comprehensive databases that contain human endogenous and exogenous metabolites as well as their biological functions. This need is illustrated by the fact that up to 50% of the signal measured in plasma samples by nontargeted metabolomics approaches results from unknown metabolites (i.e., not yet described in databases). These metabolites are likely derived from food and gut microbes and may have great impact on human health (4). Hence, identifying unknown molecules and understanding their origin and biological functions will be critical for a comprehensive understanding of human biology and disease. In addition to an increased knowledge of biologic implications of genetic mutations and metabolite concentrations, more systematic bioinformatic procedures need to be developed to efficiently integrate genomic and metabolomic data.
Second, multiomics studies should be designed very carefully to reach sufficient statistical power. Although Guo et al. uncovered interesting examples of significantly deregulated metabolites, the identification of abnormal chemicals was impaired by the relatively small number of subjects in a heterogeneous cohort not matched for any traits (e.g., sex, body mass index, and age) other than a healthy status. The huge interindividual variability of metabolic profiles illustrates the need to statistically define health by systematically profiling the metabolites of thousands of people and tracking their long-term phenotypic outcomes to readily detect abnormalities. Well-documented health records will be essential for this initiative to link metabolic profiles with clinically defined health and disease states.
Third, contrary to the genetic information (which is mostly static), the metabolome is dynamic and varies with time owing to environmental fluctuations. For example, we are beginning to appreciate that even in blood drawn on fasting individuals in the morning, the concentration of some metabolites (such as acylcarnitines) will be influenced by the composition of the last evening meal as well as physical activity. For this reason, longitudinal metabolic profiles are critical to obtain a robust quantitative measurement of personal metabolomes (Fig. 1). In this context, we recently showcased that individual longitudinal omics profiles were beneficial to uncover dynamic molecular and pathway changes across healthy and prediseased states, which resulted in the actionable diagnosis of medical risks (5).
Finally, as is the case for whole genome sequencing, ethical and legal issues arise from nontargeted metabolic profiling. For instance, data protection is crucial, because state-of-the art metabolomics is able to reveal xenobiotics in the blood of research participants (e.g., antidepressants and recreational drugs). Also, excessive care should be taken in returning individual research results and particularly incidental findings, because they could lead to unnecessary medical interventions. Hence, establishing actionability thresholds—supported by integrated genomics and metabolomics information—is a prerequisite to distinguish abnormalities that will likely lead to medical outcomes and make these technologies efficient and cost-effective in a clinical setting.
Although numerous challenges remain, the combined use of metabolomic and genomic data has the potential to revolutionize personal health management by improving personal disease risk assessment, diagnosis, and drug treatment. In the future, we can envision that personal genomes will be sequenced and metabolomes (as well as potentially other omes such as transcriptome and proteome) will be measured to define a personal health baseline (Fig. 1). The integration of routine metabolic profiles (e.g., monthly) with genetic information and metadata (nutrition, exercise, and medical records) will allow the detection of early signs of a condition and will result in nutrition/lifestyle recommendations and efficient drug treatments.
Footnotes
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: M. Snyder, Personalis, Genapsys, and SensOmics.
Consultant or Advisory Role: M. Snyder, Personalis, Genapsys, and SensOmics.
Stock Ownership: M. Snyder, Abcam.
Honoraria: None declared.
Research Funding: None declared.
Expert Testimony: None declared.
Patents: None declared.
This is an un-copyedited authored manuscript copyrighted by the American Association for Clinical Chemistry (AACC). This may not be duplicated or reproduced, other than for personal use or within the rule of 'Fair Use of Copyrighted Materials' (section 107, Title 17, U.S. Code) without permission of the copyright owner, AACC. The AACC disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The final publisher-authenticated version of the article will be made available at http://www.clinchem.org 12 months after its publication in Clinical Chemistry.
References
- 1.Contrepois K, Jiang L, Snyder M. Optimized analytical procedures for the untargeted metabolomic profiling of human urine and plasma by combining hydrophilic interaction (HILIC) and reverse-phase liquid chromatography (RPLC)-mass spectrometry. Mol Cell Prot. 2015;14:1684–1695. doi: 10.1074/mcp.M114.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo L, Milburn MV, Ryals JA, Lonergan SC, Mitchell MW, Wulff JE, et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc Nat Acad Sci USA. 2015;112:E4901–E4910. doi: 10.1073/pnas.1508425112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 5.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]