Abstract
BACKGROUND
Maternal obesity is associated with adverse neurodevelopmental outcomes in children, including autism spectrum disorders, developmental delay, and attention deficit hyperactivity disorder. The underlying mechanisms remain unclear. We previously identified second trimester amniotic fluid and term cord blood gene expression patterns suggesting dysregulated brain development in fetuses of obese compared to lean women.
OBJECTIVES
We sought to investigate the biological significance of these findings in a mouse model of maternal diet-induced obesity. We evaluated sex-specific differences in fetal growth, brain gene expression signatures and associated pathways.
STUDY DESIGN
Female C57BL/6J mice were fed a 60% high-fat diet or 10% fat control diet for 12–14 weeks prior to mating. During pregnancy, obese dams continued on the high-fat diet (HFD/HFD), or transitioned to the CD (HFD/CD). Lean dams stayed on the control diet. On embryonic day 17.5, embryos were weighed and fetal brains were snap frozen. RNA was extracted from male and female forebrains (10/diet group/sex) and hybridized to whole genome expression arrays. Significantly differentially expressed genes were identified using Welch’s t-test with the Benjamini-Hochberg correction. Functional analyses were performed using Ingenuity Pathways Analysis and Gene Set Enrichment Analysis.
RESULTS
Embryos of HFD/HFD dams were significantly smaller than controls, with males more severely affected than females (p=0.01). Maternal obesity and maternal obesity with dietary change in pregnancy resulted in significantly more dysregulated genes in male versus female fetal brains (386 vs 66, p<0.001). Maternal obesity with and without dietary change in pregnancy was associated with unique brain gene expression signatures for each sex, with overlap of only one gene. Changing obese dams to a control diet in pregnancy resulted in more differentially expressed genes in the fetal brain than maternal obesity alone. Functional analyses identified common dysregulated pathways in both sexes, but maternal obesity and maternal dietary change affected different aspects of brain development in males compared to females.
CONCLUSIONS
Maternal obesity is associated with sex-specific differences in fetal size and fetal brain gene expression signatures. Male fetal growth and brain gene expression may be more sensitive to environmental influences during pregnancy. Maternal diet during pregnancy significantly impacts the embryonic brain transcriptome. It is important to consider both fetal sex and maternal diet when evaluating the effects of maternal obesity on fetal neurodevelopment.
Keywords: diet, fetal programming, growth, maternal obesity, neurodevelopment, sex differences, transcriptome
Introduction
Maternal obesity has reached epidemic proportions in the United States. More than one-third of reproductive-age women are obese at conception, with a 70% increase in pre-pregnancy obesity in recent decades.1,2 Data from large epidemiologic studies suggest an association between maternal obesity and adverse neurodevelopmental outcomes in children, including lower general cognitive capabilities,3 increased prevalence of autism spectrum disorders,4 and increased prevalence of attention deficit hyperactivity disorder (ADHD).5 Yet the mechanisms by which maternal obesity results in adverse neurodevelopmental outcomes for offspring remain unclear.
In our prior functional genomic analyses of second trimester amniotic fluid supernatant (AFS) and term umbilical cord blood in fetuses of obese versus lean women, we found gene expression patterns consistent with dysregulated brain development, and dysregulated inflammatory and metabolic signaling.6,7 We transitioned to a mouse model of maternal diet-induced obesity (DIO) to evaluate the biological significance of these findings via direct examination of the fetal brain.
In our analyses of human AFS and term cord blood, subjects were matched for fetal sex, and sex-specific effects of maternal obesity on the fetus were not evaluated. Given that some animal model studies suggest sex-specific effects of maternal obesity on offspring neurodevelopment,8–11 and different susceptibilities of male and female offspring to maternal dietary interventions,8 here we examined both male and female embryonic brains. Our objective was to determine whether maternal obesity is associated with a distinct pattern of fetal brain gene expression, and whether maternal obesity has different effects on male and female fetal brain development, by characterizing embryonic brain gene expression signatures and associated pathways in a mouse model of diet-induced obesity.
Materials and Methods
Animal Model and Genotyping
The Tufts Medical Center Institutional Animal Care and Use Committee (IACUC) approved this protocol (#B2013-82); all institutional guidelines for animal care and use were followed. Five week old female C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were fed ad libitum either a lard-based, high-fat diet (HFD) containing 60% calories from fat (N=22, Research Diets D12492, New Brunswick, NJ), or a control diet (CD) containing 10% calories from fat (N=10, Research Diets D12450J). The diets were matched for protein, fiber, sucrose, and micronutrient content (Supplementary File 1). Female mice were weighed weekly during the 12–14 week feeding period. Maternal obesity was defined as at least a 30% increase in weight compared to age-matched controls.12,13 Male C57BL/6J mice were fed the CD. Prior to breeding, male and female body composition was characterized by EchoMRI™ (EchoMRI LLC, Houston, TX).
Obese and lean females were bred with control males. The presence of a vaginal plug was defined as embryonic day 0.5 (e0.5). Females were weighed at pregnancy days 0 (P0), P10 and P15. To separate the fetal effects of pre-pregnancy obesity from the effect of exposure to a high-fat diet during pregnancy, there were three study groups:
C57BL/6J female mice fed a CD throughout (CD group).
C57BL/6J female mice fed a HFD to induce a DIO phenotype, then continued on a HFD during gestation (HFD/HFD).
C57BL/6J female mice fed a HFD to induce a DIO phenotype, then switched at e0.5 to a CD for gestation (HFD/CD).
At embryonic day e17.5, pregnant mice were euthanized with isoflurane followed by decapitation. Embryos were rapidly dissected from the uterine horns and placed in ice-cold phosphate buffered saline (PBS 1x) containing an RNA preservative (RNALater, Qiagen). Crown-rump lengths and weights were recorded. Sex genotyping was performed on tail snip DNA using real time PCR with specific probes for the Sry gene (Transnetyx, Cordova, TN). Colony statistics were performed using GraphPad Prism 6 (GraphPad Software, San Diego, CA). Male and female embryos were analyzed separately, with differences between the three diet groups determined using Kruskal Wallis testing, followed by pairwise comparisons if significance was identified (Bonferroni-corrected p < 0.05).
Forebrain RNA isolation and microarray methods
Forebrains were rapidly dissected from skulls and snap frozen in liquid nitrogen. Total RNA was isolated using the NucleoSpin™RNA/protein kit (Macherey-Nagel, Düren, Germany). RNA purity, integrity, and quantity were assessed using the NanoDrop ND-8000 (NanoDrop, Wilmington, DE) and the Bioanalyzer system (Agilent 2100; Agilent Technologies Inc, Palo Alto, CA).
RNA samples were processed using the Affymetrix GeneChip WT PLUS kit and hybridized to Mouse Gene 1.0 ST Arrays (Affymetrix, Santa Clara, CA). Ten arrays per sex per experimental group were used, with each array corresponding to one animal. Four to six litters were represented in each diet group to minimize litter effects. Quality control and normalization were performed using the pipeline at the www.arrayanalysis.org website (Maastricht University, the Netherlands).14 Normalization was performed using the Robust Multichip Average (RMA) algorithm15 and the default Affymetrix Chip Description File (CDF) for this chip. The 27619 main probe sets were used for further analysis; probe sets corresponding to Affymetrix controls or unmapped sequences were discarded after normalization.
Bioinformatics analysis of microarray data
Statistical analyses were performed using R software version 3.1.2. Male and female gene expression data were analyzed separately. Welch’s t-tests were used to identify differentially expressed genes (DEGs) between diet groups. P-values were corrected for multiple testing by calculating the Benjamini and Hochberg false discovery rate (FDR).16 Probe sets with an FDR < 20% were considered significantly differentially expressed. The number of DEGs per comparison was visualized as a Venn diagram. Gene expression changes were further visualized as a heatmap combined with hierarchical clustering analysis using Euclidean distance and Ward linkage. Principal Component Analyses (PCA) were performed using R.
Further in silico functional analysis was performed on the top 1% of up- or downregulated genes using Ingenuity Pathway Analysis (IPA, Ingenuity® Systems, Core Analysis build version 338830M, content version 23814503). Statistical significance within IPA was determined according to recommended thresholds (p < 0.05 or bias-corrected absolute Z score ≥ 2).17–19 Only pathways containing 3 or more genes were considered. The microRNA Target Filter tool on IPA was used to predict genes and pathways affected by dysregulated miRNAs. Only “high confidence” predictions were considered.
Whole-genome analysis of functional gene set regulation was determined by Gene Set Enrichment Analysis (GSEA),20 using gene sets from the Developmental FunctionaL Annotation at Tufts (DFLAT) database on fetal development (parameters as previously described).21 Gene sets were considered significantly regulated if the FDR q-value was < 0.20. Within each sex, analyses were performed for the following diet group comparisons: HFD/HFD vs. CD; HFD/CD vs. CD; HFD/CD vs. HFD/HFD.
Results
Maternal diet-induced obesity mouse model
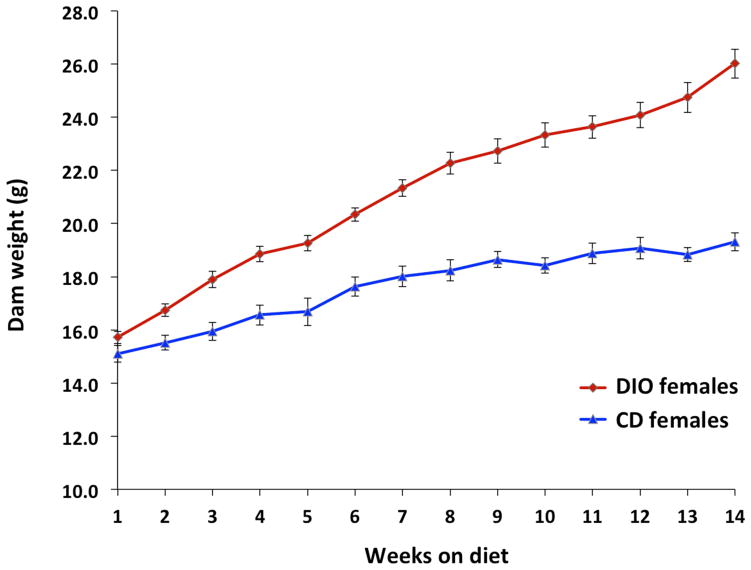
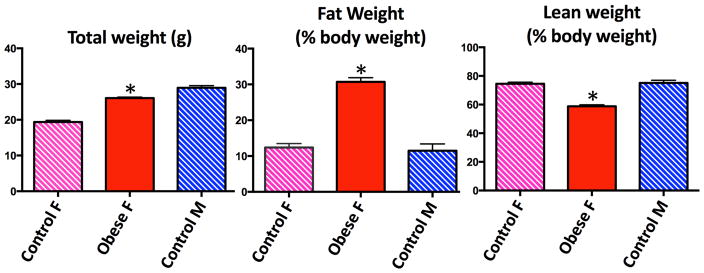
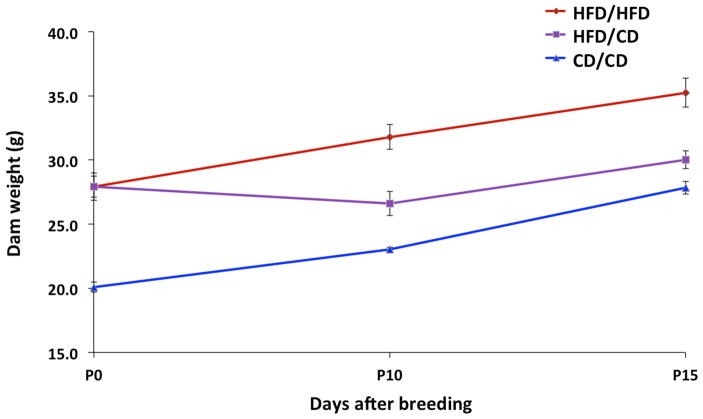
Dam weight trajectories prior to pregnancy by diet group are depicted in Figure 1. Of 22 dams fed the HFD, 18 became obese after 14 weeks of feeding. Four dams did not meet criteria for obesity and were excluded. HFD-fed dams had significantly higher percent body fat and significantly lower percent lean body weight than control females or males by EchoMRI™ analysis (Figure 2). Descriptive data for the dams and embryos by sex and diet group are reported in Table 1. There were no significant differences between study groups with respect to litter size. There were significant differences between groups with respect to dam weight gain in pregnancy. Control dams gained the most weight in pregnancy, while HFD/CD dams gained the least (Figure 3).
Figure 1. Dam weight trajectories prior to pregnancy.
Diet-induced obese females were significantly heavier than their control diet counterparts at the end of the pre-breeding feeding period.
DIO, diet-induced obesity; CD, control diet
Figure 2. Adult body composition prior to breeding.
Diet-induced obese females had significantly higher percent body fat and lower percent lean body weight compared to control females and males.
F, female; M, male
Table 1.
Dam and embryo characteristics by sex and diet group
| Characteristic | Sex | Diet Group | P-valuea | ||
|---|---|---|---|---|---|
| HFD/HFDb | CD/CDc | HFD/CDd | |||
| Embryo weight (g, mean +/− SEM) | Male | 0.85 (0.04) | 1.02 (0.04) | 0.95 (0.04) | 0.01 |
| Female | 0.86 (0.04) | 0.95 (0.04) | 0.88 (0.03) | 0.26 | |
| Combined | 0.85 (0.03) | 0.98 (0.03) | 0.91 (0.03) | 0.007 | |
| Embryo length (mm, mean +/− SEM) | Male | 19.58 (0.36) | 21.00 (0.31) | 21.02 (0.42) | 0.02 |
| Female | 19.49 (0.36) | 20.53 (0.35) | 19.87 (0.35) | 0.28 | |
| Combined | 19.54 (0.25) | 20.78 (0.23) | 20.37 (0.28) | 0.01 | |
| Dam weight at breeding (g, mean +/− SEM) | N/A | 28.63 (0.64) | 20.32 (0.35) | 28.03 (0.99) | 0.002 |
| Dam weight gain in pregnancy (g, mean +/−SEM) | N/A | 13.18 (2.2) | 15.68 (0.96) | 8.57 (1.35) | 0.01 |
| Litter size (N, mean +/− SEM) | N/A | 7.25 (1.03) | 7.83 (0.40) | 6.33 (1.12) | 0.44 |
P-value determined by Kruskal-Wallis testing;
HFD/HFD, high-fat diet/high-fat diet,
CD, control diet;
HFD/CD, high-fat diet/control diet.
Figure 3. Pregnancy weight trajectories by diet group.
HFD/HFD and CD/CD dams gained weight steadily throughout pregnancy. HFD/CD dams initially lost weight at P10, but by P15 had gained an average of 2.1 grams over their pre-pregnancy weight.
HFD/HFD, high-fat diet pre-pregnancy and during pregnancy; HFD/CD, high-fat diet pre-pregnancy, control diet during pregnancy; CD/CD, control diet pre-pregnancy and during pregnancy.
Sex-specific differences in embryo size
Male embryos exposed to maternal obesity were significantly (approximately 17%) smaller in weight than their corresponding controls (Table 1, p= 0.01). Female embryos demonstrated milder embryo size reduction in the setting of maternal obesity that did not achieve statistical significance (9.5%, p= 0.26).
Sex-specific differences in embryonic brain gene expression profiles
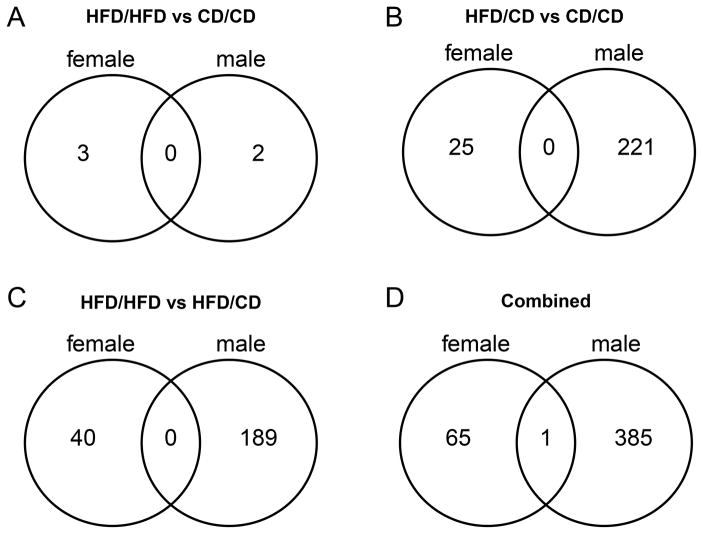
Maternal obesity and maternal obesity with dietary change in pregnancy (herein after referred to as “dietary change”) resulted in more dysregulated genes in male compared to female fetal brains (386 in males vs 66 in females, p<0.001). These numbers reflect the total number of unique DEGs in males and females for each of the diet group comparisons, including HFD/HFD vs. CD, HFD/CD vs. CD, and HFD/HFD versus HFD/CD. To consider only those genes differentially regulated by maternal obesity with or without dietary change in pregnancy we summed the HFD/HFD vs. CD and HFD/CD vs. CD comparison, again finding a significant difference between the sexes in the number of dysregulated genes (223 in males vs. 28 in females, p< 0.001). Figures 4A–D depict Venn diagrams for the number of unique and common differentially expressed genes in males versus females for each comparison of maternal obesity and dietary change in pregnancy. Maternal obesity and dietary change are associated with distinct brain gene expression signatures for each sex, with overlap of only one gene (Nuclear encoded rRNA 5S 170, function unknown) across all comparisons (4D). Maternal obesity with dietary change during pregnancy resulted in more DEGs in the fetal brain than continuation on the high-fat diet. Supplementary Files 2 and 3 show the list of significant DEGs for males and females for each diet group comparison.
Figure 4. A–D. Number of significantly differentially regulated brain genes in females versus males in maternal obesity with and without dietary change in pregnancy.
Maternal obesity alone and with dietary change in pregnancy was associated with distinct brain gene expression signatures for females and males, with overlap of only one gene across all comparisons.
HFD/HFD, high-fat diet pre-pregnancy and during pregnancy; HFD/CD, high-fat diet pre-pregnancy, control diet during pregnancy; CD/CD, control diet pre-pregnancy and during pregnancy.
Gene expression changes in males and females compared to their corresponding controls were further visualized as a heatmap (Figure 5A). The hierarchical clustering and expression intensity confirm stronger gene expression changes in males versus females in the setting of maternal obesity and dietary change. Within these DEGs, a block of 37 differentially-expressed stem-loop microRNAs (miRNAs) were identified (Figure 5B). For these miRNAs, the stronger effects of maternal obesity and dietary change on male fetuses are even more pronounced.
Figure 5. A/B: Heatmap of differentially regulated brain genes (A) and microRNAs (B) by sex and diet group.
Maternal obesity and dietary change in pregnancy had a stronger effect on male brain gene expression. Red indicates higher expression, green indicates lower expression, yellow is neutral. Heat map values correspond to expression ratios relative to the matched control diet for that sex (1 = unchanged, 2 = 2-fold increased expression compared to controls, 0.5 = 2-fold decreased expression compared to controls).
FCC, female control group; FHC, female high-fat diet pre-pregnancy, control diet during pregnancy; FHH, female high-fat diet pre-pregnancy and during pregnancy; MCC, male control group; MHC, male high-fat diet pre-pregnancy, control diet during pregnancy; MHH, male high-fat diet pre-pregnancy and during pregnancy.
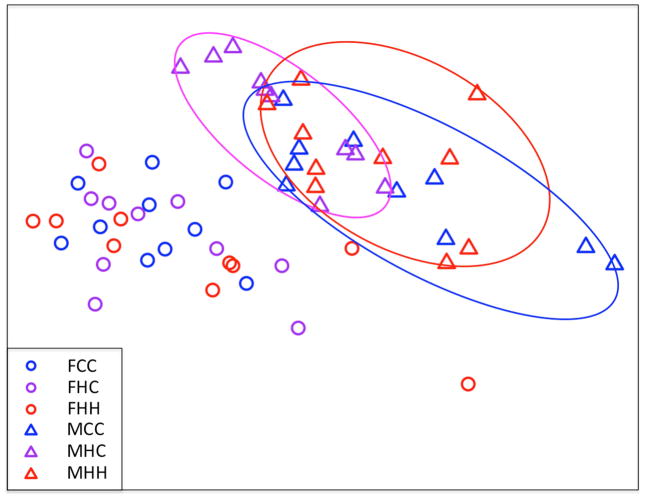
A principal component analysis (PCA) was performed to identify the dominant sources of variation in the gene expression data. The most pronounced clustering was by sex (Figure 6). Subtle clustering by diet group was observed in the male but not the female brain gene expression data. There were no significant differences in within-group variability between males and females, or between any of the diet groups. The average relative variation in gene expression for each diet group was as follows: Female CD/CD: 14.9%; Female HFD/CD: 15.3%; Female HFD/HFD: 17.4%; Male CD/CD: 15.9%; Male HFD/CD: 14.9%; Male HFD/HFD: 14.9%. Principal component analysis also demonstrated that gene expression in the samples cannot be distinguished by litter within male or female diet groups (i.e. no evidence of litter effects).
Figure 6. Principal component analysis of significantly differentially regulated brain genes.
Males are represented by triangles and females by circles. Gene expression segregates most strongly on the basis of fetal sex, with subtle clustering of gene expression by diet group noted in males but not females. On the x-axis is principal component 1, which accounts for 28.9% of the variance in gene expression data. On the y-axis is PC 2, which accounts for 7.8% of the variance.
FCC, female control group; FHC, female high-fat diet pre-pregnancy, control diet during pregnancy; FHH, female high-fat diet pre-pregnancy and during pregnancy; MCC, male control group; MHC, male high-fat diet pre-pregnancy, control diet during pregnancy; MHH, male high-fat diet pre-pregnancy and during pregnancy.
Pathway analyses
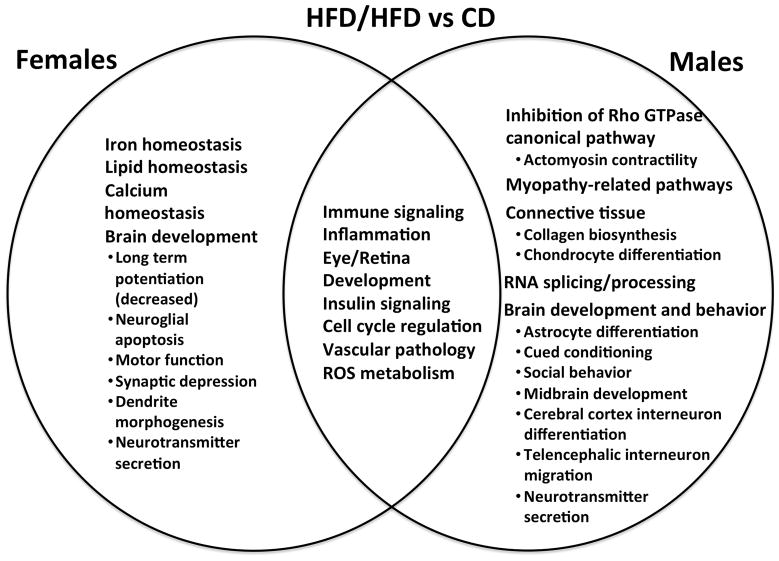
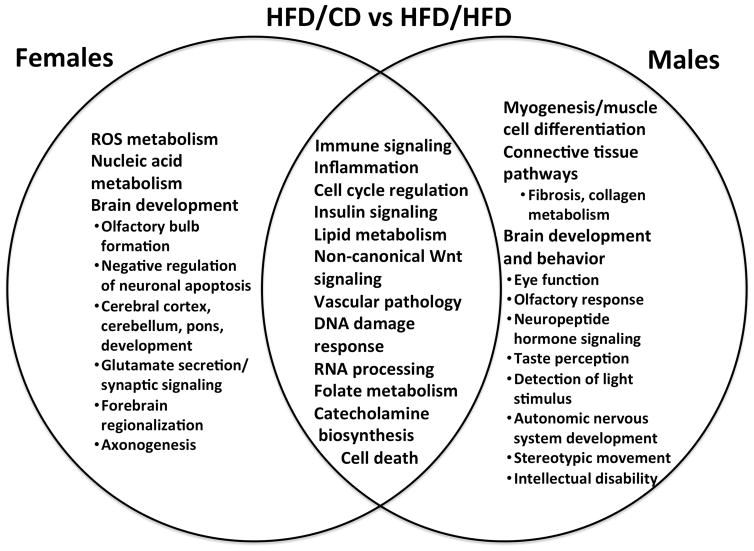
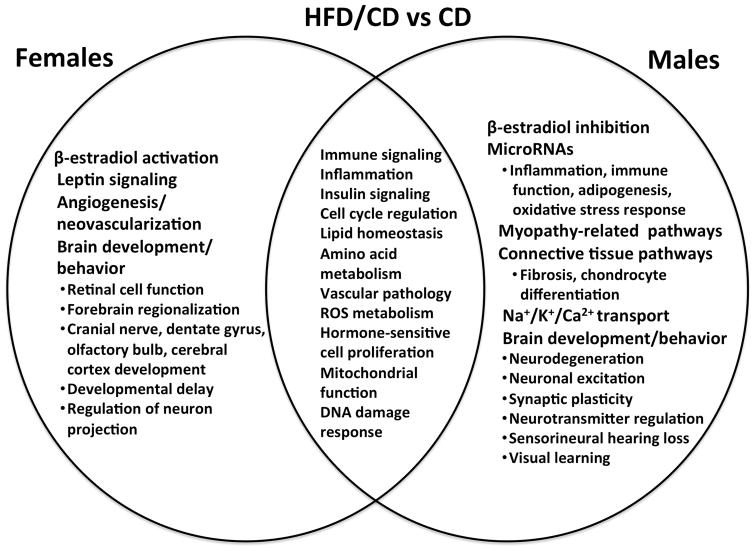
Pathway analyses were performed using IPA and GSEA/DFLAT. The working files for the IPA analyses may be found in Supplementary File 4. Dysregulated biological processes and pathways in males, females, and both are depicted for each diet group comparison in Figures 7–9. Pathway analyses demonstrated that male and female fetal brains had several dysregulated processes in common across all three comparisons (immune/inflammatory signaling, cell cycle regulation, insulin signaling, and vascular pathology). Biological processes that were specifically dysregulated sin females included iron homeostasis, leptin signaling, and angiogenesis. Pathways that were dysregulated specifically in males included those implicated in actomyosin contractility, neuromuscular disorders, and connective tissue development. Males had significantly more dysregulated miRNAs (36 vs 1, p < 0.001). These miRNAs have been implicated in pro-inflammatory and immune signaling, cell death, adipogenesis, and oxidative stress response, among other functions. A description of the stem-loop miRNAs, corresponding mature miRNAs, and putative dysregulated target genes and pathways appears in Supplementary File 5.
Figure 7. Common and unique dysregulated pathways in the female and male embryonic brain in maternal obesity.
This Venn diagram compares common and unique pathways dysregulated in male and female brains exposed to maternal obesity and a high-fat diet in utero (HFD/HFD group) against a reference group of male or female controls (CD). Males and females were each compared to their corresponding controls. Pathways reflect results from both Ingenuity Pathways Analysis (IPA) and Gene Set Enrichment Analysis with Developmental FunctionaL Annotation at Tufts (GSEA/DFLAT).
Figure 9. Common and unique dysregulated pathways in the female and male embryonic brain in maternal obesity with dietary change in pregnancy.
This Venn diagram compares common and unique pathways dysregulated in male and female brains exposed to maternal obesity and a control diet in utero (HFD/CD group) against a reference group of males or females exposed to maternal obesity and a high-fat diet in utero (HFD/HFD group). Pathways reflect results from both Ingenuity Pathways Analysis (IPA) and Gene Set Enrichment Analysis with Developmental FunctionaL Annotation at Tufts (GSEA/DFLAT).
Notably, different aspects of brain development were altered in males versus females for each of the three comparisons. Brain development terms implicated in both sexes included pathways related to developmental delay, cerebral cortex development, and neurotransmitter signaling. Brain development terms dysregulated in only females included negative regulation of neuronal and glial apoptosis, cranial nerve and dentate gyrus formation, forebrain regionalization, and axonogenesis. Terms dysregulated in males only included neurodegeneration, astrocyte differentiation, midbrain development, synaptic plasticity, neuropeptide hormone signaling, visual learning and memory, cued conditioning, and social behavior. Significantly dysregulated pathways and gene sets identified by IPA and GSEA/DFLAT for each comparison are presented in Supplementary Files 6 and 7.
Comment
Principal Findings
In this study, we demonstrated that maternal obesity is associated with statistically significant sex-specific differences in fetal size and fetal brain gene expression signatures. Male embryos exposed to maternal obesity and a high-fat diet in utero were significantly smaller than controls, while weight differences were less pronounced for females. In the setting of maternal obesity and dietary change, males and females had unique brain gene expression signatures with overlap of only one gene. More dysregulated genes and stronger differential regulation were noted in male compared to female brains. Pathway analyses demonstrated significant dysregulation of the cell cycle, and of immune, inflammatory and insulin signaling in the brains of both sexes, and highlighted unique dysregulated aspects of brain development for each sex. Switching obese dams to a control diet in pregnancy had a stronger impact on embryonic brain gene expression than did continuing the high-fat diet.
These data help fill a knowledge gap. Little is known regarding the mechanisms that underlie the association between maternal obesity and adverse neurodevelopmental outcomes for offspring, and the relative contributions of the in utero versus postnatal environment. The use of a mouse model permitted direct examination of the fetal brain in obese pregnancy, which allowed us to definitively address possible in utero origins of the postnatal neurodevelopmental morbidities noted in epidemiologic studies. Few studies have examined sex effects of maternal obesity on fetal brain development and offspring behavior.8,11 To our knowledge, this is the first study to examine the effects of maternal obesity on the fetal brain in both sexes. The limited number of studies that have investigated the effects of maternal obesity on brain development focused on the postnatal period,8,11,22 on male embryos and pups only,22–24 or the fetal sex was unknown/not considered.25 A mechanistic understanding of how sex modifies the effect of maternal obesity on fetal brain development is critical to designing effective and targeted interventions.
Findings in Context
The significant sex differences noted in our study, with very little overlap between DEGs in males and females, are plausible based on prior literature. The murine placental transcriptome in the setting of maternal high-fat diet is sexually dimorphic, with very few dysregulated genes in common, and marked differences between the sexes in the biological functions of dysregulated genes.26 One other study has examined the fetal brain transcriptome in a rat model of maternal high carbohydrate diet, but had fewer subjects per group and did not consider fetal sex.25 The few studies examining neurodevelopmental sex differences in the setting of maternal obesity have focused on the postnatal period.8,10,11 In a similar murine DIO model with a shorter period of pre-breeding feeding, exposure to maternal HFD during gestation and lactation resulted in increased hyperactivity in male offspring, and increased anxiety and decreased sociability in female offspring.8 This model suggested higher postnatal sensitivity of female pups to maternal dietary change in lactation, reporting amelioration of female, but not male, neurobehavioral phenotype and neuroinflammation after switching dams to a control diet during lactation. In a rat model of maternal HFD, male offspring displayed increased anxiety, but improved spatial learning compared to female offspring and controls.11 There appear to be sex-specific effects of maternal obesity and maternal diet on fetal and offspring neurodevelopment, but it is not clear whether one sex is more adversely affected.
The finding that male fetal growth and brain gene expression may be more sensitive to environmental influences during pregnancy is also consistent with prior studies. Males have traditionally been viewed as more vulnerable to gestational and perinatal exposures compared to females.27,28 There is a higher frequency of male fetuses miscarried and stillborn,29 a lower rate of neonatal survival in males compared to females at a given gestational age,30 and an increased incidence of neurologic and non-neurologic perinatal complications in males.31 In addition, males are at increased risk for neurodevelopmental morbidity compared to females born at the same gestational age.31 A variety of hypotheses have been proposed to explain increased male neurodevelopmental vulnerability, including delayed male brain maturation in utero,27 androgen exacerbation of excitotoxic cell death,32 and sex differences in cell death pathways that may favor females.28 With respect to specific pathways and processes implicated in our data, maternal obesity is known to be an inflammatory state, and we identified gene expression patterns consistent with increased inflammation in both male and female fetal brains. The male cerebral cortex may be more vulnerable to inflammation in utero; male cortical astrocytes were noted to produce more pro-inflammatory mediators after exposure to lipopolysaccharide than did female astrocytes, an effect which appeared to be mediated in part by testosterone exposure.33 Sex differences in microglial function and subsequent neuroinflammation during development have also been proposed as a mechanism by which males are predisposed to later neurological and neurodegenerative disorders.34 In a mouse model of intrauterine inflammation, increased male brain vulnerability was described, with male adult offspring more likely to have hippocampal volume loss on MRI and cortical infiltrating monocytes on histological examination.35 Further investigations are needed, however, to determine whether the increased number of DEGs in the male brain is associated with more neurodevelopmental morbidity (see “Clinical and Research Implications”).
Clinical and Research Implications
Additional research is required before the information presented here can be translated to a human clinical setting. First, it is not clear whether the differences noted in fetal brain gene expression will translate to functional differences in offspring neurobehavior, so we plan neurobehavioral testing on offspring of both sexes from the three diet groups. Second, the human correlate of the profound effect of obese females changing to a control diet in pregnancy is unknown. It may be better if obese women undertake significant dietary and lifestyle changes prior to, rather than during, pregnancy.
Strengths and Limitations
Strengths of our study include the whole transcriptome approach, which does not preselect for specific gene transcripts or pathways based on prior knowledge. Another strength is our comprehensive functional analysis with multiple bioinformatics resources, including a resource annotated specifically for the fetus (DFLAT).21 Prior work has demonstrated the importance of using more than one systems biology resource for the most complete understanding of dysregulated biological processes.36 The weight differences between the obese and lean dams prior to breeding are more significant in our study than in most other studies that have used a DIO model to examine the effects on offspring neurodevelopment.8,11,22,24 Ensuring substantial weight differences between the HFD-fed and CD-fed dams allowed us to examine the effects of true maternal pre-pregnancy obesity and not just maternal HFD on fetal brain development. Finally, our examination of the fetal effects of maternal obesity separately from maternal high-fat diet in pregnancy has not been performed in other studies, and permitted the detection of the significant effect of dietary change in pregnancy on fetal brain gene expression.
As a limitation, the study was designed with three diet groups to isolate pre-pregnancy maternal obesity as the variable influencing fetal brain gene expression in the HFD/CD group. We did not envision the HFD/CD group as a “dietary intervention” group. The unexpected finding that obese females eating a control diet in pregnancy was associated with more differentially-expressed fetal brain genes than maternal obesity and continuation on a high-fat diet suggests that the change to a control diet in pregnancy might represent more of an “intervention” than a return to the baseline situation. This is a limitation, as we are not able to tease apart the relative contributions of maternal obesity and the dietary switch on fetal brain gene expression. The two variables may act synergistically to impact fetal brain gene expression, or there may be a third unmeasured effect (such as maternal stress) that is driving the changes. The fact that both males and females exposed to dietary change had annotations related to dysregulated catecholamine compound biosynthesis (including the neurotransmitters adrenaline, noradrenaline and dopamine) may suggest that maternal/fetal stress could be playing a role in dysregulated brain gene expression. The HFD/CD dams did not appear unhealthy or malnourished during their pregnancy. Although they lost an average of 1.3 grams by P10 of their pregnancy, they gained an average of 2.1 grams by P15, suggesting the dietary change was not consistent with “starvation”-type conditions. Formal behavioral or biochemical evaluations of maternal stress after switching to the control diet in pregnancy were not performed as these findings were not anticipated. Whether the significant fetal effects of dietary change in pregnancy for the obese dams (or the relatively few dysregulated genes in the setting of continuation of maternal HFD) have a direct human parallel is unclear. Even so, these data are important as they highlight the key consideration of maternal pregnancy diet as a separate variable in studies such as these.
The relatively small fold-change differences between groups might be perceived as a limitation. While the effect size of the fetal brain gene expression differences due to maternal obesity and maternal dietary change in pregnancy are smaller than one would expect to find in gene expression studies of conditions such as cancer or infection, these modest effect sizes are not only biologically plausible, but could result in a distinct neurobehavioral phenotype. A recent study of fetal brain gene expression in a mouse model of Down syndrome found similarly modest fold changes, and an abnormal neurobehavioral phenotype in pups.37
Conclusions
Maternal obesity and dietary change in pregnancy are associated with distinct fetal brain gene expression signatures for males and females, and more dysregulated genes in the male brain. Pathway analysis suggests different aspects of brain development are dysregulated in males versus females. Male growth and male brain gene expression may be more sensitive to environmental influences during pregnancy, such as maternal obesity and maternal diet. It is important to consider both fetal sex and maternal diet when evaluating the effects of maternal obesity on neurodevelopment.
Supplementary Material
Composition of maternal diets
Significantly differentially regulated genes in female embryonic brains for each diet group comparison
Significantly differentially regulated genes in male embryonic brains for each diet group comparison
Working files uploaded to Ingenuity Pathways Analysis for males and females, all diet group comparisons
Significantly differentially regulated microRNAs and putative dysregulated target genes and pathways
Significantly dysregulated functional annotations identified by Ingenuity Pathways Analysis for males and females by diet group comparison
Significantly dysregulated terms identified by GSEA/DFLAT for males and females by diet group comparison
Figure 8. Common and unique dysregulated pathways in the female and male embryonic brain in maternal obesity with dietary change in pregnancy.
This Venn diagram compares common and unique pathways dysregulated in male and female brains exposed to maternal obesity and a control diet in utero (HFD/CD group) against a reference group of male or female controls (CD). Males and females were each compared to their corresponding controls. Pathways reflect results from both Ingenuity Pathways Analysis (IPA) and Gene Set Enrichment Analysis with Developmental FunctionaL Annotation at Tufts (GSEA/DFLAT).
Acknowledgments
Source of financial support: This work was supported by the Reproductive Scientist Development Program (RSDP) and Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) (2K12HD000849), and the March of Dimes
Role of the funding sources: The funding sources had no involvement in study design; collection, analysis and interpretation of data; the writing of the report; or the decision to submit the article for publication.
Footnotes
The authors report no conflicts of interest
Presentation at a scientific meeting: This work has been accepted for presentation at the 36th Annual Meeting of the Society for Maternal-Fetal Medicine, Atlanta, GA, February 2–7, 2016; abstract #8 (February 4, 2016)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Catalano PM, Ehrenberg HM. The short- and long-term implications of maternal obesity on the mother and her offspring. BJOG. 2006;113:1126–33. doi: 10.1111/j.1471-0528.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 2.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA. 2012;307:491–7. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- 3.Tanda R, Salsberry PJ, Reagan PB, Fang MZ. The Impact of Prepregnancy Obesity on Children’s Cognitive Test Scores. Matern Child Health J. 2013;7:222–9. doi: 10.1007/s10995-012-0964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krakowiak P, Walker CK, Bremer AA, et al. Maternal metabolic conditions and risk for autism and other neurodevelopmental disorders. Pediatrics. 2012;129:e1121–8. doi: 10.1542/peds.2011-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez A, Miettunen J, Henriksen TB, et al. Maternal adiposity prior to pregnancy is associated with ADHD symptoms in offspring: evidence from three prospective pregnancy cohorts. Int J Obes (Lond) 2008;32:550–7. doi: 10.1038/sj.ijo.0803741. [DOI] [PubMed] [Google Scholar]
- 6.Edlow AG, Vora NL, Hui L, Wick HC, Cowan JM, Bianchi DW. Maternal obesity affects fetal neurodevelopmental and metabolic gene expression: a pilot study. PLoS One. 2014;9:e88661. doi: 10.1371/journal.pone.0088661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edlow AG, Hui L, Wick HC, Fried I, Bianchi DW. Assessing the fetal effects of maternal obesity via transcriptomic analysis of cord blood: a prospective case-control study. BJOG. 2016;123:180–189. doi: 10.1111/1471-0528.13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang SS, Kurti A, Fair DA, Fryer JD. Dietary intervention rescues maternal obesity induced behavior deficits and neuroinflammation in offspring. Journal of neuroinflammation. 2014;11:156. doi: 10.1186/s12974-014-0156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ovilo C, Gonzalez-Bulnes A, Benitez R, et al. Prenatal programming in an obese swine model: sex-related effects of maternal energy restriction on morphology, metabolism and hypothalamic gene expression. Br J Nutr. 2014;111:735–46. doi: 10.1017/S0007114513002948. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan EL, Grayson B, Takahashi D, et al. Chronic consumption of a high-fat diet during pregnancy causes perturbations in the serotonergic system and increased anxiety-like behavior in nonhuman primate offspring. J Neurosci. 2010;30:3826–30. doi: 10.1523/JNEUROSCI.5560-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilbo SD, Tsang V. Enduring consequences of maternal obesity for brain inflammation and behavior of offspring. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:2104–15. doi: 10.1096/fj.09-144014. [DOI] [PubMed] [Google Scholar]
- 12.Gallou-Kabani C, Vige A, Gross MS, et al. C57BL/6J and A/J mice fed a high-fat diet delineate components of metabolic syndrome. Obesity (Silver Spring) 2007;15:1996–2005. doi: 10.1038/oby.2007.238. [DOI] [PubMed] [Google Scholar]
- 13.Heerwagen MJ, Stewart MS, de la Houssaye BA, Janssen RC, Friedman JE. Transgenic increase in N-3/n-6 Fatty Acid ratio reduces maternal obesity-associated inflammation and limits adverse developmental programming in mice. PLoS One. 2013;8:e67791. doi: 10.1371/journal.pone.0067791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eijssen LM, Jaillard M, Adriaens ME, et al. User-friendly solutions for microarray quality control and pre-processing on ArrayAnalysis.org. Nucleic acids research. 2013;41:W71–6. doi: 10.1093/nar/gkt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B (Methodological) 1995;57:289–300. [Google Scholar]
- 17.Ingenuity Systems. [Accessed November 18, 2015];Calculating and interpreting the p-values for functions, pathways, and lists in IPA. 2013 Available at: http://www.ingenuity.com/wp-content/themes/ingenuity-qiagen/pdf/ipa/functions-pathways-pval-whitepaper.pdf.
- 18.Ingenuity Systems. [Accessed November 18, 2015];Ingenuity Upstream Regulator Analysis whitepaper: identify the cascade of upstream transcriptional regulators and explain gene expression changes using IPA. Available at: http://pages.ingenuity.com/IngenuityUpstreamRegulatorAnalysisWhitepaper.html.
- 19.Hui L, Wick HC, Moise KJ, Jr, et al. Global gene expression analysis of amniotic fluid cell-free RNA from recipient twins with twin-twin transfusion syndrome. Prenatal diagnosis. 2013;33:873–83. doi: 10.1002/pd.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wick HC, Drabkin H, Ngu H, et al. DFLAT: functional annotation for human development. BMC bioinformatics. 2014;15:45. doi: 10.1186/1471-2105-15-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tozuka Y, Kumon M, Wada E, Onodera M, Mochizuki H, Wada K. Maternal obesity impairs hippocampal BDNF production and spatial learning performance in young mouse offspring. Neurochem Int. 2010;57:235–47. doi: 10.1016/j.neuint.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Niculescu MD, Lupu DS. High fat diet-induced maternal obesity alters fetal hippocampal development. Int J Dev Neurosci. 2009;27:627–33. doi: 10.1016/j.ijdevneu.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tozuka Y, Wada E, Wada K. Diet-induced obesity in female mice leads to peroxidized lipid accumulations and impairment of hippocampal neurogenesis during the early life of their offspring. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:1920–34. doi: 10.1096/fj.08-124784. [DOI] [PubMed] [Google Scholar]
- 25.Stachowiak EK, Oommen S, Vasu VT, et al. Maternal obesity affects gene expression and cellular development in fetal brains. Nutr Neurosci. 2013;16:96–103. doi: 10.1179/1476830512Y.0000000035. [DOI] [PubMed] [Google Scholar]
- 26.Gabory A, Ferry L, Fajardy I, et al. Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. PLoS One. 2012;7:e47986. doi: 10.1371/journal.pone.0047986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raz S, Goldstein R, Hopkins TL, et al. Sex differences in early vulnerability to cerebral injury and their neurodevelopmental implications. Psychobiology. 1994;22:244–53. [Google Scholar]
- 28.Smith AL, Alexander M, Rosenkrantz TS, Sadek ML, Fitch RH. Sex differences in behavioral outcome following neonatal hypoxia ischemia: insights from a clinical meta-analysis and a rodent model of induced hypoxic ischemic brain injury. Experimental neurology. 2014;254:54–67. doi: 10.1016/j.expneurol.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 29.McMillen MM. Differential mortality by sex in fetal and neonatal deaths. Science. 1979;204:89–91. doi: 10.1126/science.571144. [DOI] [PubMed] [Google Scholar]
- 30.Stevenson DK, Verter J, Fanaroff AA, et al. Sex differences in outcomes of very low birthweight infants: the newborn male disadvantage. Archives of disease in childhood -fetal and neonatal edition. 2000;83:F182–5. doi: 10.1136/fn.83.3.F182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hintz SR, Kendrick DE, Vohr BR, Kenneth Poole W, Higgins RD NICHD Neonatal Research Network. Gender differences in neurodevelopmental outcomes among extremely preterm, extremely-low-birthweight infants. Acta paediatrica. 2006;95:1239–48. doi: 10.1080/08035250600599727. [DOI] [PubMed] [Google Scholar]
- 32.Nunez JL, McCarthy MM. Androgens predispose males to GABAA-mediated excitotoxicity in the developing hippocampus. Experimental neurology. 2008;210:699–708. doi: 10.1016/j.expneurol.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos-Galindo M, Acaz-Fonseca E, Bellini MJ, Garcia-Segura LM. Sex differences in the inflammatory response of primary astrocytes to lipopolysaccharide. Biology of sex differences. 2011;2:7. doi: 10.1186/2042-6410-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanamsagar R, Bilbo SD. Sex differences in neurodevelopmental and neurodegenerative disorders: Focus on microglial function and neuroinflammation during development. The Journal of steroid biochemistry and molecular biology. 2015 doi: 10.1016/j.jsbmb.2015.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dada T, Rosenzweig JM, Al Shammary M, et al. Mouse model of intrauterine inflammation: sex-specific differences in long-term neurologic and immune sequelae. Brain, behavior, and immunity. 2014;38:142–50. doi: 10.1016/j.bbi.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edlow AG, Slonim DK, Wick HC, Hui L, Bianchi DW. The Pathway Not Taken: Understanding ‘Omics Data in the Perinatal Context. Am J Obstet Gynecol. 2015 doi: 10.1016/j.ajog.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guedj F, Pennings JL, Ferres MA, et al. The fetal brain transcriptome and neonatal behavioral phenotype in the Ts1Cje mouse model of Down syndrome. American journal of medical genetics Part A. 2015;167A:1993–2008. doi: 10.1002/ajmg.a.37156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Composition of maternal diets
Significantly differentially regulated genes in female embryonic brains for each diet group comparison
Significantly differentially regulated genes in male embryonic brains for each diet group comparison
Working files uploaded to Ingenuity Pathways Analysis for males and females, all diet group comparisons
Significantly differentially regulated microRNAs and putative dysregulated target genes and pathways
Significantly dysregulated functional annotations identified by Ingenuity Pathways Analysis for males and females by diet group comparison
Significantly dysregulated terms identified by GSEA/DFLAT for males and females by diet group comparison