Abstract
Many investigators recognize the importance of data sharing, however they lack the capability to share data. Research efforts could be vastly expanded if Alzheimer's disease data from around the world was linked by a global infrastructure that would enable scientists to access and utilize a secure network of data with thousands of study participants at risk for or already suffering from the disease. We discuss the benefits of data sharing, impediments today and solutions to achieving this on a global scale. We introduce the Global Alzheimer's Association Interactive Network (GAAIN), a novel approach to create a global network of Alzheimer's disease data, researchers, analytical tools and computational resources to better our understanding of this debilitating condition. GAAIN has addressed the key impediments to Alzheimer's disease data sharing with its model and approach. It presents practical, promising, yet data owner sensitive data sharing solutions.
Keywords: Alzheimer's disease data sharing, Data Integration
1. Introduction
Good, reliable, curated and complete data is at the core of meaningful scientific research today. In recent years the advancement of technology has empowered us to generate, collect and manage massive amounts of data in the field of Alzheimer's disease research. As imaging, clinical, biological and genetic data acquisition becomes more widespread and in aggregate becomes “big data”, scientific research becomes increasingly data-driven and computationally intensive in terms of analysis, maintenance and storage. In today's world, data are not only the end result of research studies but also the beginning of new hypotheses and opportunities for innovation that may never have been otherwise explored. The possibilities can further multiply considerably if we can successfully share and integrate these “big data” across organizations, groups and countries. There have been major strides in data sharing in the field of Alzheimer's disease research, including the Alzheimer's Disease Neuroimaging Initiative25 (ADNI) in the United States, the Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing11 (AIBL) in Australia, neuGRID for You or “N4U”28 in Europe, the French National Alzheimer's Information System and Databank3 the National Institute of Aging Genetics of Alzheimer's Disease Data Storage Site or NIAGADS33 and SveDem – the Swedish Dementia Registry35 to name a few. ADNI was launched in 2004 to collect longitudinal data from 58 sites around the United States for clinical, imaging and genetic data types from elderly participants with normal cognition and participants diagnosed with mild cognitive impairment or Alzheimer's disease. The data are freely accessible to any researcher, resulting in an unprecedented number of research articles using these data29. These efforts and systems have succeeded in providing models and frameworks that groups can adopt to share their data. However there remains open the challenge of being able to share Alzheimer's disease data across any set of organizations across the globe, regardless of how they maintain their own individual archives. Table 1 provides a more comprehensive list of Alzheimer's disease research and data organizations around the globe.
Table 1. Alzheimer's Disease Data.
| Alzheimer's Disease Data Center, Core or Consortium | Host Institution for Data | Database or Data Characteristics |
|---|---|---|
| National Institute of Aging Genetics of Alzheimer's Disease Data Storage Site (NIAGADS)/Alzheimer's Disease Genetics Consortium (ADGC) | National Institute of Aging and the University of Pennsylvania | A national genetics data repository in order to facilitate access by qualified investigators to genotypic data for the study of the genetics of late-onset Alzheimer's disease. |
| Alzheimer's Disease Neuroimaging Initiative | University of California San Francisco | Comprised of neurocognitive, imaging, genetic and demographic longitudinal subject data from 58 sites in North America since 2004. |
| French National Alzheimer's Database (BNA) | Nice University Hospital in France | Registers all medical acts performed by memory units and independent specialists where subject data is collected at several hundred memory centers all over France |
| Integrated NeuroDegenerative Disease database (INDD) | University of Pennsylvania | Integrated database of multiple, aging related neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis, and frontotemporal lobar degeneration |
| AlzPharm and BrainPharm | Yale University | Database(s) of drugs for the treatment of different neurological disorders. AlzPharm is the “semantic” representation of BrainPharm i.e., it actually represents knowledge and relationships in the data. |
| COMET-AD | Indian University and the State of Indiana | Indiana state-wide registry of adverse events with Alzheimer's disease drugs. |
| Layton Aging and Alzheimer's Disease Center(LAADC) | Oregon Health and Sciences University | Longitudinal research database of over 3000 subjects with clinical, neuroimaging, biomarkers, and neuropathology data |
| neuGRID for you (N4U) | European consortium of multiple partners | Grid network of European Alzheimer's disease data |
| Dominantly Inherited Alzheimer's Network (DIAN) | Washington University at St. Louis | Focused on Autosomal Dominant Alzheimer's Disease (ADAD) drug trials |
| National Alzheimer's Coordinating Center (NACC) | University of Washington | Data is collected from the 27 NIA-funded Alzheimer's Disease Centers (ADCs) across the United States, NACC has developed and maintains a large relational database of standardized clinical and neuropathological research data. |
| Australian Imaging Biomarkers and Lifestyle Flagship Study of Ageing (AIBL) | Commonwealth Scientific and Industrial Research Organization (CSIRO) Australia and partner universities | 4.5+year prospective longitudinal study of cognition. Large-scale cohort study: 1000+ participants (minimum age 60 years). Patients with Alzheimer's disease (AD), mild cognitive impairment (MCI) and healthy volunteers. All data is collected at two centers (40% subjects from Perth in Western Australia, 60% from Melbourne, Victoria). |
| European Medical Information Framework (EMIF) | Consortium of European universities, research organizations, pharmaceutical companies, public bodies and non-profit groups | EMIF-AD is the Alzheimer's Disease focused thrust of this effort, where the goals of EMIF-AD include (1) Setting up a large data repository of patient data to allow biomarker discovery studies within the EMIF. (2) Linking data from research cohorts to electronic health registry data. (3) Identifying new potential targets for AD drug development using genomics and proteomics approaches in presymptomatic and prodromal AD. |
| Framingham Heart Study (FHS) | Boston University | Database of genetic, phenotypic and biomarkers data from the Framingham Heart Study. |
| Brain Health Registry (BHR) | University of California San Francisco and partners | Expected to soon make available an Investigator Portal that provides Alzheimer's Disease researchers with access to de-identified brain health registry data. |
| Texas Alzheimer's Research Care Consortium (TARCC) | University of Texas San Antonio Health Sciences Center and partners. | Database of longitudinal study data from over 2000 subjects in Texas. |
| Aged Brain Sys Bio (ABSB) | French National Institute of Health | Expected to generate novel resources for the European ageing research scientific community, including a novel open-access database |
| Alzheimer's Preventative Initiative/Banner Alzheimer's Institute (API/BAI) | Banner Alzheimer's Institute in Phoenix, Arizona | Data from the Alzheimer's Prevention Initiative including the ‘Columbia Study’ |
| Women's Healthy Aging Project (WHAP) | University of Melbourne | A prospective, longitudinal, epidemiological study of 438 Australian women that has spanned two decades |
| The Three City Study (3C) | The consortium of the three cities of Bordeaux, Dijon and Montpellier, France. | A population-based longitudinal study of the relation between vascular diseases and dementia in persons aged 65 years and older. A total of 9,294 participants (3,649 men and 5,645 women) were recruited from three French cities. |
| Swedish Dementia Registry | The registry database, SveDem, is maintained at the Uppsala Clinical Research Center in Sweden | Includes dementia and demographic information of 28, 742 followed-up as of October 2014. 95% of all memory clinics in Sweden are currently participating in SveDem. |
| OPTIMA | University of Oxford | Database of neuropsychological assessments, brain scans, blood samples, cerebrospinal fluid samples (CSF), physical examination data and histopathological information following brain donation |
| Wisconsin Registry for Alzheimer's Prevention (WRAP) | University of Wisconsin | Includes Over 1500 participants |
| Dallas Lifespan Brain Study (DLBS) | University of Texas at Dallas | Focuses on the study of healthy adults and is playing a significant role in understanding the aging mind. There are 350 adults, 50 from each decade from 20 to 89 tested thoroughly for characterize cognition, brain structure and function across the adult lifespan |
| European Alzheimer's Disease Initiative (EADI) | Central repository of data across seven academic sites of the European Alzheimer's Disease Consortium (EADC) | |
| Neuroanatomical Database of Normal Japanese Brains | Tohuko University | A dataset on 1547 normal subjects between the ages of 16 and 79 years has been collected. |
| Canadian Longitudinal Study on Aging | Consortium of 26 universities across Canada | Consists of a national, stratified, random sample of 50,000 Canadian women and men aged 45 to 85 years at the time of recruitment. Participants will undergo repeated waves of data collection at three-year intervals and will be followed for at least 20 years. |
| AlzGene | Alzforum, operated by the Biomedical Research Forum (BRF) LLC (in Cambridge, MA) | The goal of the AlzGene database is to serve as a comprehensive, unbiased, publicly available and regularly updated field-synopsis of published genetic association studies performed on AD phenotypes. The database includes data from over 1600 AD gene studies. Over 300 AD gene candidates have been systematically subjected to meta-analysis and the results showing over 40 AD-associated genes are publicly available on the AlzGene.org website. |
Technology, or rather the lack of it, is not the factor that can explain why data sharing is not universal in Alzheimer's disease research. The more significant impediments today are often sociological. A major concern for investigators is data “ownership”. The collection and integration of clinical, genetic and/or imaging data requires significant resources, including time, money and expertise. Providing access to one's data too soon may feel similar to giving away work “for free” or also raise concerns that a competing researcher may “find the answer first”. A related issue is unauthorized use and/or redistribution of data. Many groups require users to agree to a ‘Data Use Agreement.’ However it is practically impossible to ensure that rules are followed, and most policing methods are inappropriate. Subject data privacy and unauthorized access to data are also key concerns for potential data providers.
The remainder of this paper is organized as follows. The next section describes our approach. We first describe various data sharing models and explain the rationale for the GAAIN design. We describe the GAAIN approach with early experimental indications, and provide a conclusion summarizing our experience as well as directions for the future.
2. Methods
Data can be shared and integrated in multiple ways using different models. The term model encompasses the nature of technology and software as well as the processes employed for data sharing10. There are a few fundamental dimensions for classifying data sharing models that we must consider in designing the approach best suited for GAAIN. One dimension is that of flexibility and extensibility, which is how well (or not) does a data sharing model lend itself to a dynamic and evolving data sharing environment. A second dimension is around the sensitivity of the data and relates to how well (or not) data ownership and privacy concerns are addressed by the data sharing model. Data sharing itself, and data ownership and privacy are mutually competing and must be balanced. The presidential bioethics advisory committee report17 also emphasizes the benefits of sharing and the development of solutions that achieve data sharing but without compromising subject data privacy.
In Figure 1 we have enumerated and placed different data sharing models along the (two) dimensions of flexibility, and data ownership sensitivity. It is important to understand various existing models in designing any new approach (GAAIN) to network them all together. The shared data repository approach stores data from multiple places in a single location. Such a model applies when providers can more readily share and distribute their data. It is also better suited for environments where evolution of the data sharing network i.e., new data providers or datasets is relatively less dynamic. Examples are the Integrated Neurodegenerative Disease Database or INDD36 described in Table1:R3 (row 3) and AlzPharm21 Table 1:R4 which is another neurodegenerative integrated repository for Alzheimer's. INDD is an integrated database of Alzheimer's research data developed by a consortium of University of Pennsylvania investigators that includes data related to multiple neurodegenerative disorders such as Alzheimer's disease, Parkinson's disease, ALS (amyotrophic lateral sclerosis), and FTLD (frontotemporal lobar degeneration). Using INDD as a research tool, investigators are able to obtain data across several disease groups and conduct comparative studies to elucidate distinct and common features and mechanisms of these disorders. AlzPharm is also a general purpose integrated data repository for Alzheimer's disease data that has integrated databases such as BrainPharm21 and knowledge resources such as SWAN14. AlzPharm is based on the “semantic-web” framework which we discuss shortly. The Indiana Network for Patient Care Comparative Effectiveness Research Trial of Alzheimer's Disease Drugs or COMET-AD23 Table 1:R5, is an integrated repository of data from hospitals and payers statewide for Indiana. Entities participating in COMET-AD submit patient registration records, laboratory test results, diagnoses, procedure codes, and other data for various types of healthcare encounters. The COMET-AD project is using data to monitor healthcare processes and outcomes and to build systems to monitor patients for adverse (Alzheimer's) drug events. The French National Alzheimer's Databank, called ‘BNA’ (Table 1:R2) has Alzheimer's subject data from over 300 memory centers in France3. This data is transmitted by individual memory centers at hospitals and is maintained in a centralized data bank.
Figure 1. Data Sharing Modalities.
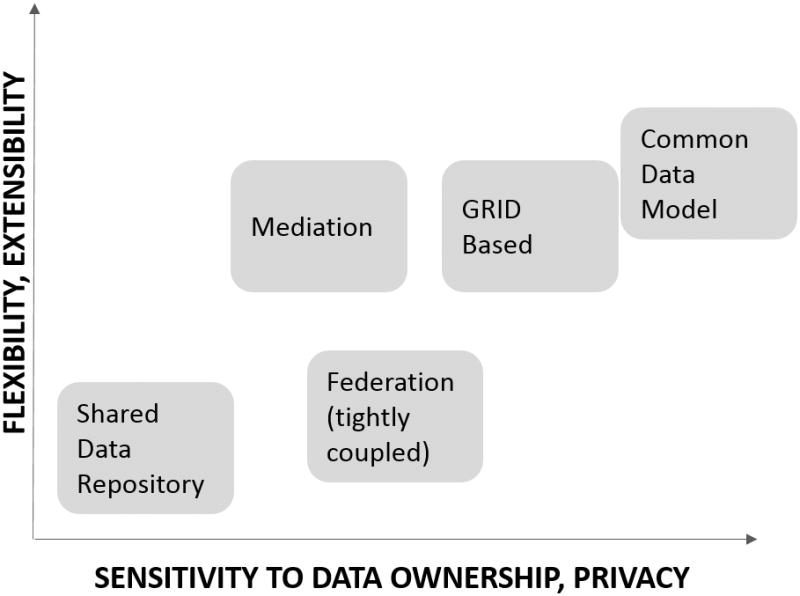
A (tightly coupled) federation is a model that is more flexible and also more sensitive to data ownership. In a federated model, a set of organizations come together to form a federation (group) and commit that their databases would all use a single, agreed upon data model and schema. Data resides with the individual data organizations but can be brought together when users require it. The Human Imaging Database HID27 is an example. HID is an extensible database management system developed to handle the increasingly large and diverse datasets collected as part of the MBIRN and FBIRN collaboratories and throughout clinical imaging communities at large. Certain research groups have adopted the HID data schema for storing their subject data27 thus becoming part of the HID federation. We must mention that the notion of a “federated database” in the original sense was that of what we are referring to as a tightly coupled federation i.e., where multiple databases agree to use the same schema. Over the years however the biomedical community has taken a looser interpretation of the term federation, using it to refer to any non-warehousing based data integration approach, including mediation. In the rest of this paper we will also assume the latter convention of a more expansive interpretation of the term ‘federation’.
The mediation approach is significantly more flexible compared to the (tightly coupled) federated approach. The approach is based on the idea of an information broker10 that harmonizes data from independent, heterogeneous and distributed data repositories. The term “mediate” refers to the process of taking user requests, translating them to individual requests to individual data sources, harmonizing the results from the individual data sources and returning the integrated results to the user. As opposed to a tightly-coupled federation, no agreement and enforcement of a common data model is required across data partners and data model harmonization is left to the mediator information broker. The Biomedical Informatics Research Network or BIRN5 the Extensible Neuroimaging Archive Toolkit or XNAT22 and the Neuro-informatics Information Framework or “NIF” mediator15 have employed mediation approaches in the neuro-informatics context. The mediation approach is less appropriate in environments where data ownership concerns are high, as it typically requires direct access to any data provider's databases, other storage systems or “APIs” (application programming interfaces). Grid-based approaches13 are similar to mediation but are more partner data sensitive, with data access control and security capabilities supported by the grid infrastructure. There are also hybrid approaches that draw from multiple modalities. For instance the European Medical Information Framework – EMIF12 in Table 1:R7 and NeuGRID (Table 1:R12) initiatives have elements of both the shared data repository and mediation models.
An approach that has gained success in recent years, especially in the clinical, health and medical data sharing domains, is the common data model approach. Data providers agree to adopt a common data model to which they would transform their data (or portions thereof), for sharing. This model is more data ownership and privacy sensitive as data providers control exactly what parts of their data they offer for transformation and sharing. The Clinical Data Integration Standards Consortium20 (CDSIC) has developed a set of common concepts or elements for the Alzheimer's disease domain. TAUG-Alzheimer's32 is the CDISC Alzheimer's Therapeutic Area User Guide which describes the most common research concepts relevant to studies of Alzheimer's disease and mild cognitive impairment, and the necessary metadata to represent such data consistent with CDISC standards.
An orthogonal issue is the choice of the semantic data model or framework5 for data sharing i.e., the formal representation of the data as it is shared. The relational model is appropriate for integrating structured “row and column” oriented data10 that is typically stored in database systems or spreadsheets. XML (the eXtensible Markup Language) based approaches are better suited for data that has a more hierarchical representation, such as in neuroimaging19,22. The Semantic-Web approach10, which is the vision of interconnected nodes of open internet resources such as Web pages has also been applied to biomedical data sharing. In recent years this vision has evolved into the “Linked Data” effort18 where content or data providers tag their data in terms of common or universally understood elements and these common element tags are used to link data in multiple sources together. The approach is directed towards freely available data that can be readily and openly shared on the internet. OpenPHACT16 an open linked data system for pharmaceutical data, is an example.
In designing the GAAIN approach we prioritized the following:
Concerns regarding data ownership and data privacy are paramount. Any practical solution must address these issues.
The scope of GAAIN is global and data may come from any relevant institution and group, and not a predetermined fixed set of groups or institutions.
The nature of the GAAIN network is dynamic where new data partners could be added frequently, also the data from existing partners can be updated.
The GAAIN data is highly structured, and relational. The data shared is primarily subject data that is organized by structured categories such as subject demographics, family history, various kinds of assessments, etc., and is typically stored in granular, well defined data elements or fields.
The expectation of GAAIN users is that of getting harmonized data from multiple providers, and the harmonization is complex given multiple factors such as syntactic and semantic heterogeneity30, where there are various representations of identical elements, data organizational differences and also data format differences across different datasets. As examples of semantic heterogeneity across datasets consider 1) The Italians' Logical Memory tests range from (0-15), instead of (0-25) as in the United States, 2) The LAADC (Table:R7) measures the size of the hippocampi differently than how ADNI does, and 3) INDD (Table 1:R4) measures CSF levels of total tau protein using a different antibody than ADNI.
Table 2 is a summary matrix of various data sharing models with the key data sharing concerns or features that are relevant to GAAIN. A ‘+’ denotes that that model is well suited to addressing a particular concern or feature and a ‘-’denotes that the model exacerbates the concern.
Table 2. Data sharing issues and models.
| Shared Repository | Federation | Mediation | Grid | Linked Data | Common Model | |
|---|---|---|---|---|---|---|
|
Data ownership Data provider sensitivity to data distributed externally, and control of data. |
- | - | - | + | ||
|
Global scope Ability to integrate any relevant dataset into network. |
- | + | + | + | + | |
|
Dynamic nature The network is dynamic, allowing for any relevant data provider to join, as opposed to a consortium of fixed members. |
- | - | + | + | ||
|
Data structure The data in the Alzheimer's disease domain is highly structured, and that must be preserved in sharing. |
+ | + | + | + | + | |
|
Harmonization Bring disparate datasets to a unified representation. |
+ | + | + | + | + | + |
We have adopted the common data model approach for GAAIN. As Table 2 summarizes, models such as a shared repository or mediation are disadvantageous for addressing data ownership concerns. The tightly-coupled federated model will not scale to the dynamic nature and global scope of GAAIN.
The GAAIN common model approach has the additional key aspects:
GAAIN utilizes an extensible common data model to accommodate data from any data partner.
GAAIN uses a ‘transform-and-cache-onsite’ approach where data is prepared for sharing at the partner site. GAAIN does not store or persist any of that data.
GAAIN access to data partner databases and other systems is non-intrusive and is controlled by the data partner.
The common model, at present, comprises of twenty four data elements about a subject taken over four years. We are initially focusing on the data elements that are most frequently used by investigators for cohort discovery and that are also (mostly) present in the datasets that we have integrated into GAAIN so far. These elements are listed in Table 4 and we are currently expanding the common model to hundreds of data elements.
Table 4. GAAIN Common Model Elements (First Version).
| Element | Description |
|---|---|
| Age | Subject age at baseline visit |
| Gender | Gender of the subject |
| Handedness | Handedness of the subject |
| Race | Race of the subject |
| Ethnic Group | Ethnic group of the subject |
| Country | Country the subject lived in during the study |
| MMSE | Mini-Mental State Examination (total score) |
| Global CDR | Clinical Dementia Rating (global) |
| Diagnosis | Diagnosis of the subject |
| LIMM | Logical Memory - Immediate Recall (total score) |
| LDEL | Logical Memory - Delayed Recall (total score) |
| Mother Has AD | Family history of Alzheimer's Disease - mother |
| Father Has AD | Family history of Alzheimer's Disease - father |
| APOE Genotype | APOE genotype of the subject |
| CSF T-Tau | CSF level of total tau protein pg/mL |
| CSF P-Tau | CSF level of phosphorylated tau protein |
| CSF Abeta42 | CSF level of beta-amyloid 42 pg/mL |
| Plasma Abeta1-40 | Plasma level of amyloid-beta peptide 1-40 |
| Plasma Abeta1-42 | Plasma level of amyloid-beta peptide 1-42 |
| Right HIP Glucose | Glucose metabolism in right hippocampus (pons normalized) |
| Left HIP Glucose | Glucose metabolism in left hippocampus (pons normalized) |
| Brain Volume | Whole brain volume |
| Right HIP Volume | Volume of the right hippocampus |
| Left HIP Volume | Volume of the left hippocampus |
2.2 The Global Alzheimer's Association Interactive Network
GAAIN is a data federation infrastructure coupled to computational resources. The GAAIN architecture is built so the data resides at the site of the Data Partners. Partner data is transformed to a GAAIN common model data “cache”. All software processing to transform the data partner's data is done within the partner's data and computing environment. Data is never saved on the GAAIN servers or other locations thus reducing the risk of unauthorized access. Links between partners of GAAIN are monitored by the partner and the partner can disable the links dynamically. Data Partner identity is unambiguous in GAAIN with clear data use requirements. Data transformation overhead on part of the Data Partner is minimal. Data Partners are provided with a completely packaged and pre-configured software to perform any data transformation tasks. This approach has been successful with most Data Partners. In the occasional case where the Data Partner may not have resources to support data transformation client, we can compute the transformation locally i.e., at the GAAIN server.
For simplifying data integration, GAAIN provides automated and semi-automated tools to transform new datasets to the GAAIN common model. The GAAIN Entity Mapper (GEM)9 aligns data elements in a Data Partner's data sets with corresponding elements in the GAAIN model. Another system, the GAAIN Automated Data Transformer6 provides a general-purpose data transformation capability to quickly configure a new dataset for GAAIN.
The harmonization aspect in GAAIN is achieved through its common data model. The GAAIN common model draws from elements of the Clinical Data Interchange Standards Consortium – CDISC20, an organization that aims to establish standards to support the acquisition, exchange, submission and archive of clinical research data and metadata. CDISC has identified a set of terms to describe a number of clinical data measures commonly used in Alzheimer's disease research.
The Role of Data Standards and Ontologies in Data Sharing
The very interpretation of data “standardization” and its role in data sharing and integration needs careful consideration. Expectations that data standards would lead to universal data sharing were not realized as autonomous organizations that create or maintain data could not be forced to conform their data to standards specified by a central body4,31. Today however, communities have evolved to a more pragmatic and successful model, with the notion of Common Data Models (CDMs)15.
As opposed to standards, CDMs are designed “bottom up” from individual databases and datasets to be integrated. CDMs are comprised of multiple common data elements (CDEs). A significant aspect of CDEs is that they are typically anchored to more widely used (and accepted) knowledge resources or ontologies – for instance “SNOMED”8 and the “UMLS”7. The CDM approach has seen significant success in domains such as clinical data integration in recent years, an example being OMOP - the Observable Medical Outcomes Partnership common data model31.
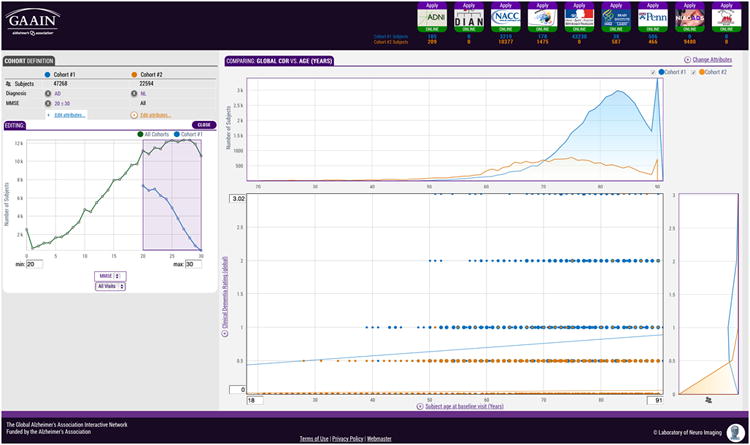
The GAAIN approach has successfully addressed the data sharing impediments discussed earlier. Table 3 summarizes the impediments along with the specific GAAIN technology and process elements that address them. From a user's point of view within GAAIN, data integration between multiple sources appears seamless. The GAAIN system invites exploration by presenting an interactive graphical interface tool for GAAIN data - the GAAIN Interrogator, through which data may be quickly and easily understood. The GAAIN Interrogator shows investigators how all of it fits together by graphing the data in interconnected views. Instead of typing numbers into text boxes, investigators search the data by interacting with graphs as illustrated in Figure 2. The data is “interrogated” by manipulating the graphs, which in turn change the search parameters and automatically update the entire display. Searching takes place by dynamically defining cohorts and visually exploring the differences between them. Every search result shows the number of subjects from each Data Partner and directs investigators how to obtain the data. Investigators can see which Data Partners have data ‘online’ and ‘offline’ on the GAAIN network and search for a study population, or cohort, across the online Data Partners.
Table 3. GAAIN Elements for Practical Data Sharing.
| Challenge | GAAIN Technology and Process |
|---|---|
| Sociological impediments |
|
| Resource considerations |
|
| Data privacy and access controlconcerns |
|
Figure 2. GAAIN Interrogator.
For an initial stage of cohort discovery we have developed the GAAIN Scoreboard which provides an aggregated “snapshot” of the space of data available at that point via GAAIN. The GAAIN Scoreboard is a graphical interface tool that provides information such as counts of subjects in various GAAIN data sources that are online and active. This summary information is useful and informative to investigators for cohort discovery and also does not provide any actual instance level data from any data partner. Following cohort discovery, data access is determined by the Data Partner's data use provisions. By sharing meta-data, investigators retain ownership and access control while avoiding privacy policy violations.
3. Results
Although GAAIN technology development is still in its early stages, experimental evaluations evidence the promise of our approach, particularly towards simplifying data sharing. We have conducted experimental evaluations of the time and effort to transform Alzheimer's disease datasets from various data partners using the GAAIN data transformation tools, and demonstrated that the use of these tools leads to a significant reduction in developer time and effort to transform a dataset as compared to existing approaches.
The GAAIN Automated Data Transformer introduced more powerful mechanisms for specifying how a particular dataset should be transformed into the GAAIN common model. It also introduced a paradigm where the data from any data provider has a uniform canonical representation within the Automated Data Transformer system. The implication of this is that the specifications for data transformation, which are essentially a set of logical data transformation rules, “look alike” across different datasets. The structure and set of transformation rules is almost identical across different datasets, though they must account for the fact that different datasets could have different names and terminology for the same data elements.
This feature makes it easy to adapt and reuse a set of data transformation rules for one dataset for a different dataset but with the necessary modifications to work with the terminology of the new dataset. The GAAIN Automated Data Transformation tool provides a 75% reduction in developer effort in transforming a new dataset6. We have also optimized the actual execution time required to conduct a complete data transformation. While prior approaches result in an execution time of several hours to transform a single dataset, our approach reduces this to just a few minutes in a computing environment with identical configuration6. We have been able to achieve this by incorporating recent features in the underlying database technology which is part of the Automated Data Transformer.
Another key challenge for developers is data mapping i.e., the task of mapping data elements in a new (partner) dataset to corresponding data elements in the GAAIN common data model. Historically this is done manually, involving several weeks of effort per dataset. The GEM system is a tool that provides automated assistance for this task. GEM is built to leverage the documentation of data, particularly that in data dictionaries associated with the data, to propose mappings of elements across two partner datasets or from a dataset to the GAAIN common model. GEM has been demonstrated to provide highly accurate mappings, with precision and recall around 85% each, for actual GAAIN datasets8. We have also validated that the mapping accuracy of GEM is significantly higher (by about 15% for each of precision and recall) than that achieved by two (generic) schema-matching software tools with the same datasets. These other tools are Harmony24 and Coma++1. The GEM system was designed with a focus on the Alzheimer's disease and other medical domain, and is able to maximally leverage information in the data documentation that other generic matching tools cannot.
4. Discussion
The vision for GAAIN is to create and maintain a network of AD resources (data, tools and infrastructure) to establish and sustain the first global database for Alzheimer's disease research. GAAIN's innovative approach not only enables more accessible Alzheimer's disease research but also sets a benchmark for how big-data, distributed initiatives for other complex diseases could function in the future.
It is useful to represent GAAIN data with multiple levels of abstraction. This helps support a wider spectrum of information query. For instance, an investigator may be initially interested in just identifying available data sources and datasets, or they may be interested in obtaining an aggregate summary of data rather than the actual data. Data partners can easily provide metadata immediately but often take longer to provide the source subject data several weeks or months later. With the metadata available however, the process of integrating a dataset with transformations to the GAAIN common model can be initiated.
GAAIN's success is dependent on Data Partners joining and sharing their data through the network. We are currently in various phases of recruitment with over fifty five data sources in North America, Europe, Asia and Australia with an average of three to four new data sources every week. During the recruitment process, we receive consistent feedback about data sharing from all of our prospective data partners. All Data Partners agree that data sharing, in an approach like GAAIN, is the next logical step in big data and Alzheimer's disease research. Though potential partners see the benefit of GAAIN, the process of on-boarding can be a lengthy process, averaging at least three months. Partners often require approvals from an executive committee sometimes necessitating a process ranging from written proposals to web conference meetings or legal review. We ask each of our Data Partners to sign a non-legally binding Memorandum of Understanding (MOU) as a formal agreement to join the project. The MOU explains the GAAIN model and assures our Data Partners that GAAIN will not violate any Data Partner policies. This is a simple document with a one-time sign on requirement and does not require a more extensive proposal from a data partner. To date, there have been no objections to the MOU. It is our hope that as GAAIN continues to grow and the field of big data research, specifically in Alzheimer's disease, moves forward, research groups will continue to regard GAAIN as an attractive solution and appreciate the effect it can have on global science.
The key design elements in the GAAIN approach, namely 1) partner sensitive data sharing, 2) simplifying data integration processes by automation, and 3) the common data model all contribute to achieving a global data federation. Our current efforts are focused on 1) The recruitment of and integration of additional key Alzheimer's disease data providers into GAAIN, 2) The further development of technology and infrastructure elements described above, and 3) Integration of additional analytic tools so that data and analyses are more tightly coupled and provided over the GAAIN network.
Acknowledgments
Funding: This research was supported by the GAAIN project which is funded by the Alzheimer's Association under grant number 003278-0001 and by the Laboratory of Neuro Imaging Resource (LONIR) NIH grant number P41EB015922. Additional research supported was provided by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award number U54EB020406.
Contributor Information
Arthur W. Toga, Email: toga@loni.usc.edu.
Priya Bhatt, Email: pbhatt@loni.usc.edu.
References
- 1.Algergawy A, Massmann S, Rahm E. Advances in Databases and Information Systems. Springer Berlin; Heidelberg: 2011. A clustering-based approach for large-scale ontology matching; pp. 415–428. [Google Scholar]
- 2.Anderson P. Contemporary Outcomes After the Fontan Procedure: A Pediatric Heart Network Multicenter Study. Journal of the American College of Cardiology. 2008;52:85–98. doi: 10.1016/j.jacc.2008.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony S, Pradier C, Chevrier R, Festraëts J, Tifratene K, Robert P. The French National Alzheimer database: a fast growing database for researchers clinicians. Dementia and Geriatric Cognitive Disorders. 2014;38(5-6):271–80. doi: 10.1159/000360281. [DOI] [PubMed] [Google Scholar]
- 4.Ashish N, Bamman MM, Cerny FJ, Cooper DM, D'Hemecourt P, Eisenmann JC, Ericson D, Fahey J, Falk B, Gabriel D, Kahn MG, Kemper HC, Leu SY, Liem RI, McMurray R, Nixon PA, Olin JT, Pianosi PT, Purucker M, Radom-Aizik S, Taylor A. The Clinical Translation Gap in Child Health Exercise Research: A Call for Disruptive Innovation. Clinical and Translational Science. 2014 Aug 11; doi: 10.1111/cts.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashish N, Ambite JL, Muslea M, Turner JA. Neuroscience data integration through mediation: an (F)BIRN case study. Frontiers in Neuroinformatics. 2010;(4):118. doi: 10.3389/fninf.2010.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashish N, Dewan P, Toga AW. Medical Data Transformation Using Query Rewriting; American Medical Informatics Association (AMIA) Annual Symposium; Washington, DC. Nov 2014. [Google Scholar]
- 7.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornet R, de Keizer N. Forty years of SNOMED: a literature review. BMC medical informatics and decision making. 2008;8(Suppl 1):S2. doi: 10.1186/1472-6947-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewan P, Ashish N, Toga AWA. A Schema-Mapping Tool for Mapping Alzheimer's Disease Datasets. 5th ACM Conference on Bioinformatics, Computational Biology and Health Informatics, 2014; Newport Beach. Sep 2014. [Google Scholar]
- 10.Doan A, Halevy A, Ives Z. Principles of data integration. Elsevier; 2012. [Google Scholar]
- 11.Ellis KA, Bush AI, Darby D, De Fazio D, Foster J, Hudson P, Ames D. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer's disease. International Psychogeriatrics. 2009;21(04):672–687. doi: 10.1017/S1041610209009405. [DOI] [PubMed] [Google Scholar]
- 12.EMIF: European Medical Information Framework. 2014 Web: http://www.emif.eu.
- 13.Foster I, Kesselman C, editors. The Grid 2: Blueprint for a new computing infrastructure. Elsevier; 2003. [Google Scholar]
- 14.Gao Y, Kinoshita J, Wu E, Miller E, Lee R, Seaborne A, Cayzer S, Clark T. SWAN: A Distributed Knowledge Infrastructure for Alzheimer Disease Research. Journal of Web Semantics. 2006;4(3) [Google Scholar]
- 15.Gardner D, Akil H, Ascoli GA, Bowden DM, Bug W, Donohue DE, Goldberg DH, Grafstein B, Grethe JS, Gupta A, Halavi M, Kennedy DN, Marenco L, Martone ME, Miller PL, Müller HM, Robert A, Shepherd GM, Sternberg PW, Van Essen DC, Williams RW. The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008 Sep;6(3):149–60. doi: 10.1007/s12021-008-9024-z. Epub 2008 Oct 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray AJ, Groth P, Loizou A, Askjaer S, Brenninkmeijer C, Burger K, Williams AJ. Applying linked data approaches to pharmacology: Architectural decisions and implementation. Semantic Web. 2014;5(2):101–113. [Google Scholar]
- 17.Guttman A. Privacy and Progress in Whole Genome Sequencing. Presidential Committee for the Study of Bioethical Issues. 2012 [Google Scholar]
- 18.Jain P, Hitzler P, Sheth AP, Verma K, Yeh PZ. Ontology Alignment for Linked Open Data. International Semantic Web Conference. 2010:402–417. [Google Scholar]
- 19.Keator DB, Helmer K, Steffener J, Turner JA, Van Erp TGM, Gadde S, Ashish N, Burns GA, Nichols BN. Towards structured sharing of raw and derived neuroimaging data across existing resources. Neuroimage. 2013 Nov 15;82:647–61. doi: 10.1016/j.neuroimage.2013.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuchinke W, Aerts J, Semler SC, Ohmann C. CDISC standard-based electronic archiving of clinical trials. Methods of information in medicine. 2009;48(5):408. doi: 10.3414/ME9236. [DOI] [PubMed] [Google Scholar]
- 21.Lam HY, Marenco L, Clark T, Gao Y, Kinoshita J, Shepherd G, Miller P, Wu E, Wong GT, Liu N, Crasto C, Morse T, Stephens S, Cheung KH. AlzPharm: integration of neurodegeneration data using RDF. BMC Bioinformatics. 2007 May 9;8(Suppl 3):S4. doi: 10.1186/1471-2105-8-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcus DS, Olsen T, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit (XNAT): An informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 23.McDonald CJ, Overhage JM, Barnes M, Schadow G, Blevins L, Dexter PR, Mamlin B INPC Management Committee. The Indiana network for patient care: a working local health information infrastructure. An example of a working infrastructure collaboration that links data from five health systems and hundreds of millions of entries. Health Aff (Millwood) 2005;24:1214–20. doi: 10.1377/hlthaff.24.5.1214. [DOI] [PubMed] [Google Scholar]
- 24.Mork P, Seligman L, Rosenthal A, Korb J, Wolf C. Journal on Data Semantics. XI. Springer Berlin; Heidelberg: 2008. The harmony integration workbench; pp. 65–93. [Google Scholar]
- 25.Mueller SG, Weiner MW, Thal LJ, Peterson RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Alzheimer's Disease Neuroimaging Initiative. Neuroimaging Clinic of North America. 2008;15(4) doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NDAR: National Database of Autism Research. 2014 Web: http://ndar.nih.gov.
- 27.Ozyurt IB, Keator DB, Wei D, Fennema-Notestine C, Pease KR, Bockholt J, Grethe JS. Federated web-accessible clinical data management within an extensible neuroimaging database. Neuroinformatics. 2010;8(4):231–249. doi: 10.1007/s12021-010-9078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redolfi A, McClatchey R, Anjum A, Zijdenbos A, Manset D, Barkhof F, Frisoni GB. Grid infrastructures for computational neuroscience: the neuGRID example. Future Neurology. 2009;4(6):703–722. [Google Scholar]
- 29.Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, Green RC, Hu X, Huentelman MJ, Kim S, Kauwe JS, Li Q, Liu E, Macciardi F, Moore JH, Munsie L, Nho K, Ramanan VK, Risacher SL, Stone DJ, Swaminathan S, Toga AW, Weiner MW, Saykin AJ. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging and Behaviour. 2014 Jun;8(2):183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheth AP. Interoperating geographic information systems. Springer; US: 1999. Changing focus on interoperability in information systems: from system, syntax, structure to semantics; pp. 5–29. [Google Scholar]
- 31.Stang P, Ryan P, Hartzema AG, Madigan D, Overhage JM, Welebob E, Reich CG, Scarnecchia T. Development and evaluation of infrastructure and analytic methods for systematic drug safety surveillance: Lessons and resources from the Observational Medical Outcomes Partnership. In: Andrews EB, Moore Nicholas, editors. Mann's Pharmacovigilance. 3rd. Chapter 28. Sussex, England: Wiley-Blackwell; 2014. [Google Scholar]
- 32.TAUG. 2014 Web: http://www.cdisc.org/therapeutic.
- 33.Wang L, Valladares O, Childress DM, Partch A, Laufer D, Iodice J, Lawrence C, Hu T, Malamon J, Tang M, Lin C, Arnold S, Stoeckert CJ, Schellenberg GD. Nia Genetics of Alzheimer'S Disease Data Storage Site (Niagads): 2014 Update. Alzheimer's and Dementia 2014. 2014 Jul;10(4) Supplement:634–635. [Google Scholar]
- 34.Wiley JC, Prattipati M, Lin CP, Ladiges W. Comparative Mouse Genomics Centers Consortium: the Mouse Genotype Database. Mutation Research. 2006 Mar 20;595(1-2):137–44. doi: 10.1016/j.mrfmmm.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Wimo A, Religa D, Spångberg K, Edlund AK, Winblad B, Eriksdotter M. Costs of diagnosing dementia: results from SveDem, the Swedish Dementia Registry. Int J Geriatr Psychiatry. 2013 Oct;28(10):1039–44. doi: 10.1002/gps.3925. [DOI] [PubMed] [Google Scholar]
- 36.Xie SX, Baek Y, Grossman M, Arnold SE, Karlawish J, Siderowf A, Hurtig H, Elman L, McCluskey L, Van Deerlin V, Lee VM, Trojanowski JQ. Building an integrated neurodegenerative disease database at an academic health center. Alzheimer's and Dementia. 2011;7:e84–93. doi: 10.1016/j.jalz.2010.08. [DOI] [PMC free article] [PubMed] [Google Scholar]


