Abstract
The extracellular matrix (ECM) microenvironment plays a central role in cell migration by providing physiochemical information that influences overall cell behavior. Much of this external information is accessed by direct interaction of the cell with ECM ligands and structures via integrin-based adhesions that are hypothesized to act as mechanosensors for testing the surrounding microenvironment. Our current understanding of these mechanical complexes is derived primarily from studies of cellular adhesions formed on two-dimensional (2D) substrates in vitro. Yet the rules of cell/ECM engagement and mechanosensing in three-dimensional (3D) microenvironments are invariably more complex under both in vitro and in vivo conditions. Here we review the current understanding of how cellular mechanosensing occurs through adhesion complexes within 3D microenvironments and discuss how these mechanisms can vary and differ from interactions on 2D substrates.
Keywords: Adhesion, mechanotransduction, collagen, 3D migration, contractility, dynamics
Introduction
Cell migration is the foundation of many crucial physiological events that begin in embryonic development with gastrulation and later during the formation of epithelial organs and the migration of cells from the neural crest to generate craniofacial structures. During adult life, cell migration is vital for wound healing and immune function. While the chemical composition of the extracellular milieu in the form of soluble growth factors, chemokines, and ECM molecules can initiate global cellular responses, the physical microenvironment plays an equally important role in controlling cell migration and other important processes. The contributions of the physical microenvironment to the regulation of cell fate, gene and protein expression, and signal transduction suggest that cells can “feel” the physical attributes of their surroundings1, 2. This ability to feel or sense the microenvironment occurs at the cell/ECM interface, particularly through integrin ligation of ECM proteins such as collagen and fibronectin. Integrins bridge the cell membrane and interact with numerous cytoskeletal and signaling proteins that accumulate in a force-dependent manner into focal adhesions and other types of cell-matrix adhesions. These adhesions in turn interact with the actomyosin cytoskeleton to provide the mechanical link through which cellular forces are transmitted to and from the extracellular environment.
This process of mechanosensing, where cells respond to the physical properties of the external environment, has been characterized for a variety of cell-matrix interactions. For example, Weiss characterized bidirectional effects of cells on matrix and vice versa, including the process of contact guidance of cell migration along matrix fibers3. More recently, Lo et al.4 demonstrated that fibroblasts prefer rigid 2D substrates over soft, and that they migrate towards tension locally applied with a microneedle to elastic 2D polyacrylamide gels.. Although 2D substrates with uniform stiffness have been used as the primary model for studying cellular mechanosensing, especially at focal adhesions, 3D models composed of single or multiple ECM proteins in vitro or native 3D environments in vivo present unique physical features of the ECM. These more-complex elements can alter cellular responses and have revealed important dimension-and architecture-dependent differences. In this review, we will focus mainly on how recent research and modeling of adhesion-based mechanosensing can differ in 3D microenvironments, describe new conceptual insights, and suggest future directions in which studies of cell interactions with 3D environments can help to answer important mechanobiology questions.
Differences in 3D ECM structure at multiple levels can affect cell mechanotransduction
Before we explore how adhesions and mechanosensing in 3D differ from their 2D counterparts, we will first describe how the 3D microenvironment can change the rules of mechanosensing. The obvious difference between 2D and 3D environments is dimensionality, with the simplest version of 3D consisting of two or more ECM surfaces in contact with a cell. Beningo et al.5 demonstrated that fibroblasts acquire a spindle-like, linearized cytoskeletal morphology reminiscent of cells migrating within a 3D ECM6 by simply “sandwiching” fibroblasts between two 2D soft polyacrylamide gels. The sandwich also promoted dorsal and ventral adhesion anchorage while reducing the number and size of focal adhesions, as well as reducing cell migration rate. These major effects of this simplest of 3D environments suggest that dimensionality alone can alter cellular responses.
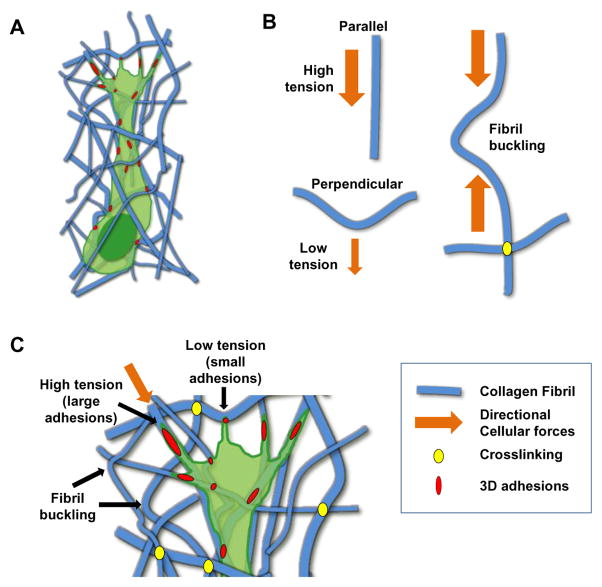
2D mechanosensing is highly dependent on distinguishing between different levels of ECM stiffness. Cells are able to detect stiffness gradients over the length of a single cell and migrate up these gradients in the process of durotaxis4, 7. Yet in 3D microenvironments, ECM stiffness can vary immensely depending on the experimental conditions. These variations are in part due to other ECM-dependent factors, such as ECM ligand density, fibril alignment, ECM pore size, and intra- and extra-fibril crosslinking that can influence matrix stiffness. For example, in collagen type I gels, ECM concentration directly affects both matrix elasticity and the ECM pore size8, 9, while changing the temperature at which the collagen is polymerized alters its ECM fibril size, pore size, overall architecture, and local fibril stiffness10. An interesting conundrum in characterizations of 3D fibrillar matrix involves macro/gel stiffness versus fiber stiffness: individual fibers of collagen and fibrin can have a Young’s modulus in the MPa range, yet a gel can be multiple orders of magnitude softer11, 12 (Fig. 1). This discrepancy can occur because the bulk mechanical properties of a gel are strongly dependent on fiber architecture and organization rather than on fiber strength/stiffness13, 14. In nonaligned hydrogels (collagen and fibrin), application of shear force in one direction results in fiber alignment along the axis of force to develop tension, while the remaining fiber population undergoes varying degrees of compression or buckling, which reduces the macroscopic or “bulk” mechanical properties of the gel15. Because of these mechanical characteristics, it is possible for an individual cell to locally sense part of a matrix as stiff or soft depending on whether tension is generated parallel (stiff) or perpendicular (soft) to a particular fiber. Kubow et al.16 demonstrated that alignment of a cell adhesion with a fiber correlates with adhesion size in collagen gels as well as on electro-spun fibers, suggesting a cellular mechanical response to the stiffness of a fiber along its length. In fact, a recent study established that mechanical pre-alignment of collagen fibrils increases gel stiffness, demonstrating the importance matrix architecture in gel stiffness17. Tumor cells can align collagen fibrils and then subsequently migrate outward along these aligned fibrils18. These findings support the notion that aligned matrices and alignment of cellular forces along a fiber can be perceived by the cell as being stiffer and result in altered cellular responses.
Figure 1. Differential mechanosensing of tension in 3D.
(A) Schematic representation of the effects of direction of force application in cell sensing of 3D microarchitecture. (B) For forces applied parallel or perpendicular to a fibril, the cell will perceive high or low tension, respectively. Fibril buckling and crosslinking may also play important roles in cell perceptions of tension from the surrounding ECM. (C) Within many 3D matrices, cells can interact locally with both parallel and perpendicular fibrils, which can affect adhesion size and possibly adhesion dynamics due to local perceived differences in tension.
Matrix type matters
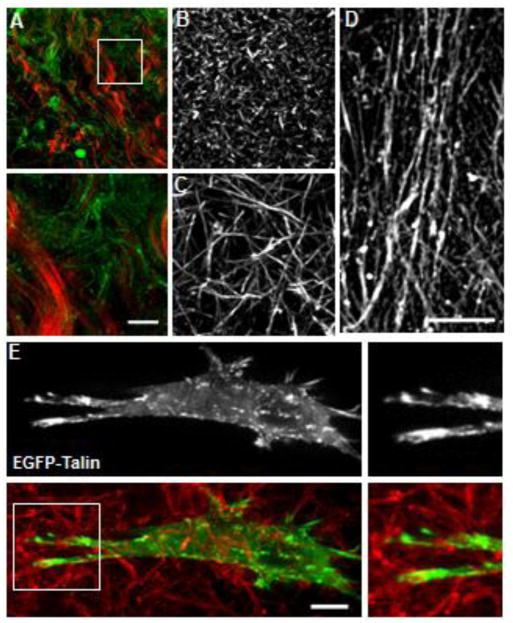
The type of 3D ECM is important, since ECMs can differ greatly in composition and architecture. Although the truest of 3D environments for understanding mechanosensing is found in tissues in vivo, imaging and other studies of this type of 3D environment are by far the most complicated, time consuming, and least controllable of current systems (Fig. 2A). In vitro “polymerizable” 3D ECMs that include collagen type I (from bovine and rat sources, Fig. 2B–C), fibrin, and Matrigel, and in vivo-like 3D cell-derived matrices (3D CDMs) all are good alternatives, but they differ greatly in their physical characteristics. 3D CDMs are considered to be the closest to in vivo conditions during embryonic development19; they contain mostly fibronectin, but also collagens I and IV, perlecan, hyaluronic acid, and other proteins20, while consisting of highly organized, frequently aligned, robust ECM fibers that promote directional cell migration and the formation of 3D matrix adhesions (Fig. 2D)19, 21–25. In comparison, collagen and fibrin often consist of a single protein that polymerizes in a random 3D ECM topography, while Matrigel forms a gel without discernable fibers at the cellular level such that fibroblasts fail to spread or generate adhesions 24.
Figure 2. 3D ECMs are not all the same.
(A) Second harmonic generation (SHG: red) imaging together with immuno-labeling for mouse collagen type I (green) showing the diverse collagen microarchitectures found within a mouse ear. Inset (below) shows thick bundled fibers next to thinner loose fibrils (green) that are not detected by SHG imaging. (B and C) Collagen polymerized at 37°C (B) and 16°C (C) demonstrate different microarchitectures. Concentrations for both are 3 mg/ml. (D) An example of an aligned 3D CDM directly labeled with fluorescent dye. (E) Human dermal fibroblast transiently transfected with EGFP-talin (top and green below) migrating through a collagen gel polymerized at 16°C. Insets to the right show robust 3D adhesions at the leading edge. Scale bars: 10 μm.
Other 3D matrix-specific physical factors include whether the environment is linearly elastic (3D CDM, mouse ear explants22) or not (fibrin26, collagen22). The nonlinear elasticity of fibrin gels has been reported to permit fibroblast communication via long-distance contractile forces, but only when cell density is high26. It has also been established that the type of elastic behavior can regulate the mode of 3D cell migration; nonlinear elastic collagen results in primarily lamellipodia-based migration, whereas linear elastic 3D CDMs promote pressure-based lobopodia to drive migration25. Recently, the use of non-biological hydrogels consisting of polyethylene glycol (PEG) that incorporate RGD integrin-binding sites27, 28 and electrospun fibers of hyaluronic acid or polycaprolactone (PCL) coated with collagen have been used to measure 3D traction forces28, promote chondrogenesis in human mesenchymal stem cells29, and test the effect of fiber size and orientation on cell adhesion size16, respectively. These highly controllable 3D environments are helpful for establishing which specific characteristics of more complex systems are important for proper mechanosensing in vivo.
3D matrix adhesions
Cell matrix adhesions in 3D environments show many similarities and a few key differences with respect to the proteins involved from adhesions formed on 2D substrates that may be relevant to mechanosensing in specific 3D ECMs. Cukierman et al.19 originally demonstrated in 3D CDMs that 3D matrix adhesions associated with fibronectin-based fibrils differ in composition from adhesions forming on 2D fibronectin; the α5 integrin is absent from paxillin-positive 2D focal adhesions, yet shows nearly complete colocalization in 3D adhesions. Dimensionality was key for the divergence, since mechanical compression of 3D CDMs to form a 2D CDM led to fibroblast development of 2D-like adhesions. This greater association of α5 integrin with 3D CDMs may suggest that 3D adhesions are under high tension, since the α5β1 integrin binds directly to the synergy site of fibronectin in an interaction associated with stabilization of 2D adhesions that are under tension30. These features may be responsible for the high longevity of 3D matrix adhesions compared to 2D focal adhesions21. Although 3D matrix adhesions and 2D focal adhesions are comprised of many of the same proteins, integrin β3 and α5 integrin are absent from these 3D and 2D focal adhesions (but not fibrillar adhesions), respectively. For other 3D environments, the composition of cell adhesions is less defined, although many proteins have been identified; β1 integrin (activated and total), vinculin, and paxillin are found in adhesions in fibrin gels31, while for collagen gels, these proteins as well as zyxin have been confirmed31, 32. We have also recently found tensin-1 and talin associated with 3D adhesions in cells migrating through collagen10 (Fig. 2E). In general, the molecular players that comprise 3D adhesions appear to be similar to those of 2D substrates, but the phosphorylation of several proteins can differ. Focal adhesion kinase (FAK) is highly phosphorylated at autophosphorylation site tyrosine 397 on 2D glass surfaces33, but shows little phosphorylation in 3D matrix adhesions found in 3D CDM19 or within collagen34. However, cells plated on top of collagen gels and assayed 4 hours after plating show FAK tyrosine 397 phosphorylation similar to 2D35, demonstrating the complexity of such pathways; they may depend on experimental conditions, including the type of 3D matrix. Kubow et al.16 showed that overall tyrosine phosphorylation was similar in 2D compared to 3D adhesions. The amounts of vinculin and zyxin, but not paxillin, increase with 3D adhesion size, indicating that some characteristics of focal adhesions in 2D can be found in 3D environments.
Cell phenotype and cell migration: indirect information about 3D mechanosensing
Potential roles of specific proteins in mechanosensing may be found in knockout or knockdown studies. For example, even though genetic ablation of vinculin in mouse embryonic fibroblasts (MEFs) has no significant effect on cell morphology on rigid 2D ECMs, and in fact increases the rate of cell migration, these same cells in 3D collagen show a round cell morphology after vinculin ablation rather than their usual spindle shape36. Moreover, loss of vinculin reduces cellular traction force generation and stability of cell protrusions, both of which are indispensable for persistent 3D migration and potentially for mechanosensing37. A similar phenotypic effect in 3D collagen can be observed after loss of NEDD938. 3D ECMs can also influence the binding partners of an adhesion protein. In 3D CDMs, vinculin was shown to bind preferentially to Hic-5, a paxillin family member, over paxillin39, opposite to findings on 2D substrates. This interaction depends on whether vinculin is in its open or active conformation, which is spatially regulated by Rac and RhoA activity. Moreover, Hic-5 knockdown in cancer cells induces an amoeboid phenotype in both 3D CDMs and collagen gels that is associated with an increase in RhoA activity, yet no phenotype is observed on 2D substrates40.
Kutys and Yamada41 discovered an extracellular-matrix-specific dependent interaction of the guanine exchange factor (GEF) βPix that regulates cell migration in 3D collagen but not in 3D CDMs. Unlike on 2D fibronectin, βPix fails to localize to focal adhesions on fibrillar collagen. Knockdown of βPix induces a hyper-contractile cellular phenotype analogous to that reported by Deakin et al.39, 40, but only in association with native fibrillar collagen; no effect was found on 2D globular collagen or when cells interacted with 2D/3D matrices enriched in fibronectin rather than collagen. Mechanistically, βPix plays an important role in both activating Cdc42 and inhibiting srGAP1-induced RhoA activation, but solely when cells associate with native collagen. Studies such as these that compare cellular functions in 2D versus 3D environments containing different ECM molecules suggest that specific proteins and/or signaling pathways can be important for proper adhesion functioning and mechanosensing depending on both the composition and dimensionality of a 3D matrix.
As for cell morphological and contractile phenotypes, cell motility can be affected by adhesion function because normal mesenchymal migration involves adhesion to the surrounding environment. One of the greatest paradoxes in cell migration is the contradictory roles of contractility in migration for 2D and 3D ECMs: contractility is dispensable for 2D mesenchymal cell migration42 yet is required for migration in 3D environments10, 16, 18, 21, 23. Some of this functional divergence in different dimensions can be attributed to ECM pore size in 3D; smaller pores can impede the translocation of the largest cellular organelle, the nucleus8, 43. The lack of such physical obstructions in 2D environments permits lamellipodial-driven migration that is independent of contractile force. However, pore size obstruction alone cannot explain the fact that in 1D fibrillar migration, where cells migrate rapidly on single 1.5 micron wide micropatterns, nuclear movement and migration speed are also reduced after loss of myosin II contractility, even without physical obstruction21. For human dermal fibroblasts, the requirement for contractility is partly explained by a need to break adhesions resulting from high levels of integrin activation in order to migrate; furthermore, in 3D collagen gels, unlike 2D substrates, integrin activation is not affected by inhibiting contractility with blebbistatin (25 μM)10.
Cell motility often requires effective 3D mechanosensing. Requirements for many proteins in the cell adhesions used for mechanosensing have been identified in gene knockdown or ablation models. Knockdown or knockout of p130Cas, α-actinin, talin, paxillin, FAK, NEDD9, and VASP all show reduced 3D migration32, 38, with vinculin yielding different results depending on the publication36, 44. Conversely, zyxin knockout increases cell migration in single tumor cells and mesenchymal fibroblasts within 3D collagen gels32, 45, 46. Another indirect indicator of 3D mechanosensing associated with 3D adhesions is the cell-induced alignment of collagen fibrils in isotropic gels18, 47. Homogenous collagen gels can permit visualization of tension/force-induced matrix reorganization that provides insight into cell/ECM interactions in 3D. For example, inhibition of cellular contractility or loss of vinculin inhibits cell-mediated collagen fibril alignment in 3D collagen36, indicating a requirement for tension at cell adhesions for matrix organization. While these indirect indicators associated with adhesion function can provide useful hints, direct measurement of adhesion kinetics can provide better insight into mechanisms of 3D mechanosensing through cell adhesions.
Adhesion kinetics in 3D ECMs
Adhesion assembly and disassembly rates, adhesion longevity, and adhesion protein kinetics determined by fluorescence recovery after photobleaching (FRAP) have been characterized using rapid imaging techniques48. An obstacle impeding analyses of 3D mechanosensing, particularly adhesion dynamics, involves the technical difficulties associated with live-cell 3D and 4D (3D plus time) imaging. 3D CDMs provide a relatively thin matrix (10–30 μm) and are a good alternative to thicker collagen and fibrin gels; e.g., collagen has high autofluorescence at 500–530 nm that can hamper visualization of EGFP-tagged adhesion proteins49. Another factor is the expression level of fluorescently tagged adhesion proteins, where excess protein can accumulate in the cytoplasm and hinder visualization of the cytoskeletal fraction16, 49. This problem contributed to initial confusion about whether cell adhesions actually exist in 3D31, 49. Choosing “low-expressor” cells or using a weakened promoter can alleviate this type of background fluorescence issue. With the current growth of rapid and high-resolution 3D imaging techniques, such as lightsheet, structured illumination, and two-photon resonant-scanning confocal microscopy, it is now possible to easily observe adhesion kinetics in complex 3D and even in vivo environments. Several reviews provide more information about such imaging techniques50, 51.
For 2D adhesions, there is a known progression of adhesion maturation and adhesion dynamics: Force-independent nascent adhesions can mature into force-dependent focal adhesions if proper linkages are established to the actin cytoskeleton, and focal adhesions can in turn develop into fibrillar adhesions found primarily beneath the cell body52, 53. This process of force-dependent adhesion maturation is less understood in 3D conditions, where rapid live-cell imaging in 3D is more difficult. However, we recently demonstrated the presence of nascent adhesions containing paxillin in 3D collagen, with adhesion lifetimes less than 2 minutes 10. The relative sizes of populations of nascent versus maturing adhesions was shown to depend on ECM stiffness, and is important for overall cell migration rate. Broussard et al.54 recently automatically tracked several adhesion metrics, including adhesion size, adhesion lifetime, and assembly and disassembly rate constants for adhesions containing paxillin and vinculin in 3D collagen gels. They found a strong correlation between adhesion size and lifetime, a relationship recently established in 2D associated with vinculin-mediated tension55.
2D/3D mechanosensing mechanism
What are the mechanisms of mechanosensing? The complete answer to this question is currently unknown, even for 2D substrates, but the proposed mechanisms are intriguing. Although it would be simplest for a single protein to serve as the linchpin that triggers mechanosensing within an adhesion, none has yet been identified. Instead, it is likely that adhesions containing several hundred proteins act as a cellular machine to sense extra- and intracellular stiffness and tension. Several key adhesion proteins including talin, vinculin, and p130cas are known to either stretch or unfold under applied force and are vital to overall function of cell adhesions56–58. Moreover, both integrins and myosins can form catch bonds, where force application increases bond longevity; catch bond formation may be part of cell tension-sensing mechanisms59–61. Interestingly, both integrins and myosin II engagement with their ligands are essential for force sensing, which may suggest that an adhesion-based tension sensor may require one or two sets of catch bonds possibly working against each other to function.
Current research on mechanosensing on 2D surfaces suggests it involves the dynamic interplay of proteins within adhesion sites62. Focal adhesions are thought to act like a molecular clutch, binding and slowing down the fast retrograde movement of actin, and integrating contractile force with relatively immobile integrins. Through single particle tracking and FRAP of fluorescently-tagged proteins, it is now known that adhesion proteins (talin, paxillin, vinculin, etc.) demonstrate an intermediate dynamic rate of movement when compared to fast-moving actin and slow-moving integrins21, 48, 62, 63. This movement reflects their dissociation rate within an adhesion, is force-dependent, and varies depending on environmental conditions48. It has been speculated that the relative rates of protein movement or the stretching rate of proteins like talin and vinculin may be important for tension sensing and providing a variable sensor64, 65. Several reviews provide details about the proposed molecular clutch and mechanosensing66, 67.
There is no evidence at present that mechanosensing in 3D requires major differences in mechanisms compared to 2D substrates, with the caveat that 2D and 3D have microenvironmental and architectural differences. Recently, we demonstrated that a molecular clutch does exist for adhesions in 3D collagen. FRAP analysis of adhesions containing eGFP-zyxin indicate that 3D ECM architecture, including the local stiffness of fibrils, alters adhesion protein dynamics that can be up to 3-fold faster than in 2D conditions10. This increased turnover coincided with a decrease in adhesion stability for softer collagen gels, with increased adhesion retraction due to adhesion slipping. Retraction was due to a force imbalance between the cell and the local ECM stiffness: in soft matrices, adhesions slipped, while in stiffer matrices adhesions gripped. This balance could be shifted by changing integrin ligation, attenuating contractility, or changing ECM stiffness. These findings suggest that the two-spring model of 2D mechanosensing theorized by Bell and Schwarz et al. may exist in 3D68, 69. Chiu et al.70 revealed that both actin and paxillin demonstrate different diffusion rates inside and outside of adhesion sites in 3D collagen consistent with a clutch-like mechanism. For vinculin, however, the half-life FRAP recovery rate in porcine dermis is similar to 2D rates70, 71, suggesting protein-to-protein differences in regulation and response to different types of ECM. Clearly, how adhesion dynamics respond to differences in ECMs remains to be fully characterized.
Other important considerations in 3D microenvironments
Although most recent studies of cellular mechanosensing have focused on the integrin-based focal adhesions that link the contractile actin cytoskeleton to the external environment, recent evidence suggests that other cellular proteins/organelles/systems could indirectly influence either mechanical interactions with the ECM or cellular responses. For example, interstitial fluid stresses can initiate integrin activation and formation of focal adhesions in 3D collagen, leading to tumor cell polarization and directional migration72. Petrie et al.25 discovered that directional migration of fibroblasts in 3D CDMs is dependent upon compartmentalization of intracellular pressure in front of the nucleus, which is driven by actomyosin contraction. Contraction pulls the nucleus forward via a myosin IIA/vimentin/nesprin 3 mechanical axis; this mechanism might alter local tension on anterior adhesions and alter their mechanosensing. Microchannels have been used by several investigators to confine cells to mimic a 3D environment in which cells must “chimney” through small pore-like channels to migrate73–75. Under these conditions, force-dependent focal adhesions are dispensable, and other factors such as intracellular pressure may be more important than mechanosensing in processes that may also be highly cell-type specific.
Besides integrins, other transmembrane proteins could play an important role in 3D mechanosensing; Discoidin domain receptors (DDR) 1 and 2, as well as syndecans, have been implicated in mechanotransduction in 2D and some 3D models. Over-expression of DDR1 stimulates β1 integrin binding to collagen, enhances 3D collagen reorganization, and increases 2D integrin activation and the formation of adhesions containing paxillin, talin, and vinculin, all of which could contribute to mechanosensing76. Genetic ablation of syndecan-4 reduces directional migration in 3D CDMs in a Rac1-dependent manner, and more recently syndecan-4 has been implicated in control of integrin recycling and 2D focal adhesion dynamics77, 78. The cellular glycocalyx could also play a role. Overexpression of mucin 1 is known to occur in many cancer cells and may help to drive integrin clustering and mechanical loading on 2D substrates79, but it remains unknown whether or how this mechanism functions in 3D fibrillar environments. These and other “nontraditional” pathways may indirectly influence cellular mechanosensing through integrins, and their roles remain to be tested.
Future directions in 3D mechanosensing
The study of 3D mechanobiology, especially as it pertains to 3D matrix adhesions, is clearly in its infancy. As we highlighted throughout this review, the context of the 3D microenvironment in terms of composition and physical properties is of great importance to understanding mechanosensing processes and future research directions. Obvious next steps will include the confirmation (or not) of current knowledge about 2D mechanosensing in diverse 3D environments, including 3D CDMs, collagen gels, and fibrin, as well as dermal explants and ultimately in vivo for determining the effects of composition, dimensions, and local architectural and other physical properties at the single-cell scale. These comparative assessments across multiple in vitro and in vivo 3D models will be crucial for a full understanding of how different model systems can affect our scientific interpretation of this mechanism, in order to avoid narrow or incomplete views of 3D mechanosensing.
Further exploration of 3D mechanosensing by and between cells undergoing collective migration, as well as understanding these phenomena in developing organs, needs to be explored, because a majority of cells in vivo interact with other cells as well as with ECM. The recent availability of rapid imaging and super-resolution technologies will help examine multiple aspects of mechanotransduction at cell, cell adhesion, and even molecular levels once thought impossible to explore. They should also make it possible to directly observe multiple aspects of the molecular clutch in 3D. Nevertheless, 2D mechanisms should still be explored. In fact, research on 1D or 2D fibrillar ECMs (2D native collagen fibrils, fibronectin fibers) could provide simpler systems to help elucidate how topography and local physical properties influence adhesion kinetics, providing good alternatives to complex 3D systems.
Acknowledgments
We would like to thank Brian DuChez, Will Daley, Josh Collins, and Dan Sykora for critical reading of this review. This work was supported by the NIDCR Division of Intramural Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 3.Weiss P, Garber B. Shape and Movement of Mesenchyme Cells as Functions of the Physical Structure of the Medium: Contributions to a Quantitative Morphology. Proc Natl Acad Sci U S A. 1952;38:264–280. doi: 10.1073/pnas.38.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beningo KA, Dembo M, Wang YL. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc Natl Acad Sci U S A. 2004;101:18024–18029. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003;13:264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- 7.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf K, et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roeder BA, Kokini K, Voytik-Harbin SL. Fibril microstructure affects strain transmission within collagen extracellular matrices. J Biomech Eng. 2009;131:031004. doi: 10.1115/1.3005331. [DOI] [PubMed] [Google Scholar]
- 10.Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat Commun. 2015 doi: 10.1038/ncomms9720. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guthold M, et al. A comparison of the mechanical and structural properties of fibrin fibers with other protein fibers. Cell Biochem Biophys. 2007;49:165–181. doi: 10.1007/s12013-007-9001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An KN, Sun YL, Luo ZP. Flexibility of type I collagen and mechanical property of connective tissue. Biorheology. 2004;41:239–246. [PubMed] [Google Scholar]
- 13.Pedersen JA, Swartz MA. Mechanobiology in the third dimension. Ann Biomed Eng. 2005;33:1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- 14.Lee B, et al. A three-dimensional computational model of collagen network mechanics. PLoS One. 2014;9:e111896. doi: 10.1371/journal.pone.0111896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munster S, et al. Strain history dependence of the nonlinear stress response of fibrin and collagen networks. Proc Natl Acad Sci U S A. 2013;110:12197–12202. doi: 10.1073/pnas.1222787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubow KE, Conrad SK, Horwitz AR. Matrix microarchitecture and myosin II determine adhesion in 3D matrices. Curr Biol. 2013;23:1607–1619. doi: 10.1016/j.cub.2013.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riching KM, et al. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys J. 2014;107:2546–2558. doi: 10.1016/j.bpj.2014.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Provenzano PP, Inman DR, Eliceiri KW, Trier SM, Keely PJ. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J. 2008;95:5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 20.Kutys ML, Doyle AD, Yamada KM. Regulation of cell adhesion and migration by cell-derived matrices. Exp Cell Res. 2013;319:2434–2439. doi: 10.1016/j.yexcr.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle AD, et al. Micro-environmental control of cell migration--myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J Cell Sci. 2012;125:2244–2256. doi: 10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J Cell Biol. 2012;197:439–455. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng Part A. 2011;17:713–724. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062–1065. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winer JP, Oake S, Janmey PA. Non-linear elasticity of extracellular matrices enables contractile cells to communicate local position and orientation. PLoS One. 2009;4:e6382. doi: 10.1371/journal.pone.0006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JS, et al. Bioactive hydrogels made from step-growth derived PEG-peptide macromers. Biomaterials. 2010;31:3736–3743. doi: 10.1016/j.biomaterials.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legant WR, et al. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim IL, Khetan S, Baker BM, Chen CS, Burdick JA. Fibrous hyaluronic acid hydrogels that direct MSC chondrogenesis through mechanical and adhesive cues. Biomaterials. 2013;34:5571–5580. doi: 10.1016/j.biomaterials.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 31.Harunaga JS, Yamada KM. Cell-matrix adhesions in 3D. Matrix Biol. 2011;30:363–368. doi: 10.1016/j.matbio.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraley SI, et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Miron-Mendoza M, Seemann J, Grinnell F. The differential regulation of cell motile activity through matrix stiffness and porosity in three dimensional collagen matrices. Biomaterials. 2010;31:6425–6435. doi: 10.1016/j.biomaterials.2010.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thievessen I, et al. Vinculin is required for cell polarization, migration, and extracellular matrix remodeling in 3D collagen. FASEB J. 2015 doi: 10.1096/fj.14-268235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldmann WH, Auernheimer V, Thievessen I, Fabry B. Vinculin, cell mechanics and tumour cell invasion. Cell Biol Int. 2013;37:397–405. doi: 10.1002/cbin.10064. [DOI] [PubMed] [Google Scholar]
- 38.Zhong J, et al. NEDD9 stabilizes focal adhesions, increases binding to the extra-cellular matrix and differentially effects 2D versus 3D cell migration. PLoS One. 2012;7:e35058. doi: 10.1371/journal.pone.0035058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deakin NO, Ballestrem C, Turner CE. Paxillin and Hic-5 interaction with vinculin is differentially regulated by Rac1 and RhoA. PLoS One. 2012;7:e37990. doi: 10.1371/journal.pone.0037990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deakin NO, Turner CE. Distinct roles for paxillin and Hic-5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol Biol Cell. 2011;22:327–341. doi: 10.1091/mbc.e10-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kutys ML, Yamada KM. An extracellular-matrix-specific GEF-GAP interaction regulates Rho GTPase crosstalk for 3D collagen migration. Nat Cell Biol. 2014;16:909–917. doi: 10.1038/ncb3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Even-Ram S, et al. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 43.Wolf K, Friedl P. Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol. 2011;21:736–744. doi: 10.1016/j.tcb.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Mierke CT, et al. Vinculin facilitates cell invasion into three-dimensional collagen matrices. J Biol Chem. 2010;285:13121–13130. doi: 10.1074/jbc.M109.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fraley SI, Feng Y, Giri A, Longmore GD, Wirtz D. Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nat Commun. 2012;3:719. doi: 10.1038/ncomms1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li N, Goodwin RL, Potts JD. Zyxin regulates cell migration and differentiation in EMT during chicken AV valve morphogenesis. Microsc Microanal. 2013;19:842–854. doi: 10.1017/S1431927613001633. [DOI] [PubMed] [Google Scholar]
- 47.Gjorevski N, Piotrowski AS, Varner VD, Nelson CM. Dynamic tensile forces drive collective cell migration through three-dimensional extracellular matrices. Sci Rep. 2015;5:11458. doi: 10.1038/srep11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 49.Kubow KE, Horwitz AR. Reducing background fluorescence reveals adhesions in 3D matrices. Nat Cell Biol. 2011;13:3–5. doi: 10.1038/ncb0111-3. author reply 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer RS, Wu Y, Kanchanawong P, Shroff H, Waterman CM. Microscopy in 3D: a biologist’s toolbox. Trends Cell Biol. 2011;21:682–691. doi: 10.1016/j.tcb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frigault MM, Lacoste J, Swift JL, Brown CM. Live-cell microscopy - tips and tools. J Cell Sci. 2009;122:753–767. doi: 10.1242/jcs.033837. [DOI] [PubMed] [Google Scholar]
- 52.Yamada KM, Pankov R, Cukierman E. Dimensions and dynamics in integrin function. Braz J Med Biol Res. 2003;36:959–966. doi: 10.1590/s0100-879x2003000800001. [DOI] [PubMed] [Google Scholar]
- 53.Zamir E, et al. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat Cell Biol. 2000;2:191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- 54.Broussard JA, et al. Automated analysis of cell-matrix adhesions in 2D and 3D environments. Sci Rep. 2015;5:8124. doi: 10.1038/srep08124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hernandez-Varas P, Berge U, Lock JG, Stromblad S. A plastic relationship between vinculin-mediated tension and adhesion complex area defines adhesion size and lifetime. Nat Commun. 2015;6:7524. doi: 10.1038/ncomms8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hotta K, et al. Biophysical properties of intrinsically disordered p130Cas substrate domain--implication in mechanosensing. PLoS Comput Biol. 2014;10:e1003532. doi: 10.1371/journal.pcbi.1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Margadant F, et al. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 2011;9:e1001223. doi: 10.1371/journal.pbio.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kong F, et al. Cyclic mechanical reinforcement of integrin-ligand interactions. Mol Cell. 2013;49:1060–1068. doi: 10.1016/j.molcel.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci U S A. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 63.Lele TP, et al. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J Cell Physiol. 2006;207:187–194. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- 64.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thievessen I, et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J Cell Biol. 2013;202:163–177. doi: 10.1083/jcb.201303129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirata H, Sokabe M, Lim CT. Molecular mechanisms underlying the force-dependent regulation of actin-to-ECM linkage at the focal adhesions. Prog Mol Biol Transl Sci. 2014;126:135–154. doi: 10.1016/B978-0-12-394624-9.00006-3. [DOI] [PubMed] [Google Scholar]
- 68.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 69.Schwarz US, Erdmann T, Bischofs IB. Focal adhesions as mechanosensors: the two-spring model. Biosystems. 2006;83:225–232. doi: 10.1016/j.biosystems.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 70.Chiu CL, et al. Nanoimaging of focal adhesion dynamics in 3D. PLoS One. 2014;9:e99896. doi: 10.1371/journal.pone.0099896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tolde O, Rosel D, Janostiak R, Vesely P, Brabek J. Dynamics and morphology of focal adhesions in complex 3D environment. Folia Biol (Praha) 2012;58:177–184. doi: 10.14712/fb2012058050177. [DOI] [PubMed] [Google Scholar]
- 72.Polacheck WJ, German AE, Mammoto A, Ingber DE, Kamm RD. Mechanotransduction of fluid stresses governs 3D cell migration. Proc Natl Acad Sci U S A. 2014;111:2447–2452. doi: 10.1073/pnas.1316848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balzer EM, et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bergert M, et al. Force transmission during adhesion-independent migration. Nat Cell Biol. 2015;17:524–529. doi: 10.1038/ncb3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stroka KM, et al. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014;157:611–623. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Staudinger LA, et al. Interactions between the discoidin domain receptor 1 and beta1 integrin regulate attachment to collagen. Biol Open. 2013;2:1148–1159. doi: 10.1242/bio.20135090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bass MD, et al. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J Cell Biol. 2007;177:527–538. doi: 10.1083/jcb.200610076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morgan MR, et al. Syndecan-4 phosphorylation is a control point for integrin recycling. Dev Cell. 2013;24:472–485. doi: 10.1016/j.devcel.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paszek MJ, et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]