Abstract
Objective
We comprehensively examined the rare variants in the IPO11-HTR1A region to explore their roles in neuropsychiatric disorders.
Method
Five hundred seventy-three to 1,181 rare SNPs in subjects of European descent and 1,234-2,529 SNPs in subjects of African descent (0 < minor allele frequency (MAF) < 0.05) were analyzed in a total of 49,268 subjects in 21 independent cohorts with 11 different neuropsychiatric disorders. Associations between rare variant constellations and diseases and associations between individual rare variants and diseases were tested. RNA expression changes of this region were also explored.
Results
We identified a rare variant constellation across the entire IPO11-HTR1A region that was associated with attention deficit hyperactivity disorder (ADHD) in Caucasians (T5: p=7.9×10−31; Fp: p=1.3×10−32), but not with any other disorder examined; association signals mainly came from IPO11 (T5: p=3.6×10−10; Fp: p=3.2×10−10) and the intergenic region between IPO11 and HTR1A (T5: p=4.1×10−30; Fp: p=5.4×10−32). One association between ADHD and an intergenic rare variant, i.e., rs10042956, exhibited region- and cohort-wide significance (p=5.2×10−6) and survived correction for false discovery rate (q=0.006). Cis-eQTL analysis showed that, 29 among the 41 SNPs within or around IPO11 had replicable significant regulatory effects on IPO11 exon expression (1.5×10−17≤p<0.002) in human brain or peripheral blood mononuclear cell tissues.
Conclusion
We concluded that IPO11-HTR1A was a significant risk gene region for ADHD in Caucasians.
Keywords: IPO11, HTR1A, ADHD, rare variant constellations, non-coding RNA
Introduction
5-hydroxytryptamine (serotonin) receptor 1A gene (HTR1A) encodes the 5-HT1A receptor that binds the endogenous neurotransmitter serotonin. This receptor is a G protein-coupled receptor (GPCR). GPCR is coupled to Gi/Go and mediates inhibitory neurotransmission. In the human central nervous system, 5-HT1A receptors have been found in the cerebral cortex, hippocampus, amygdala, septum and raphe nucleus in high densities. The activation of 5-HT1A receptor may increase dopamine release in the medial prefrontal cortex, striatum, and hippocampus. This dopamine release may inhibit the release of glutamate and acetylcholine in various areas of the brain, and thus may impair cognition, learning, and memory. This release may also increase impulsivity and inhibition of human behaviors. Therefore, the activation of 5-HT1A receptor is likely to be related to the development of neuropsychiatric diseases. Using the candidate gene approach, HTR1A at 5q11.2-q13 has been associated with numerous neuropsychiatric disorders and related traits in human, including antidepressant response (citalopram, fluvoxamine, fluoxetine, sertraline and paroxetine) [Arias et al., 2005; Lemonde et al., 2004; Serretti et al., 2004; Suzuki et al., 2004; Villafuerte et al., 2009; Yevtushenko et al., 2010; Yu et al., 2006], antipsychotic drug response [Reynolds et al., 2006], anxiety- and depression-related personality traits [Schmitz et al., 2009; Strobel et al., 2003], impulsivity [Benko et al., 2010], depression [Anttila et al., 2007; Chen et al., 2004; Haenisch et al., 2009; Kraus et al., 2007], schizophrenia, substance use disorder, panic attack [Huang et al., 2004], alcoholism [Lee et al., 2009; Wojnar et al., 2006], and migraineurs [Marziniak et al., 2007]. However, HTR1A is a small gene (1,269bp) with only one exon. Only 110 variants have been detected within the open reading frame (ORF) of this gene so far (see NCBI dbSNP), which leads to a hypothesis that its associations with the neuropsychiatric disorders might be driven by the variants from the flanking regions.
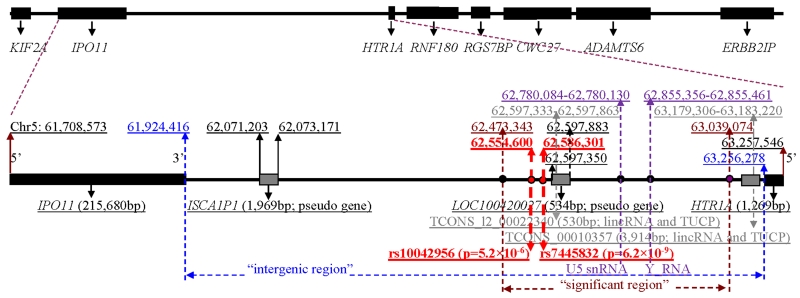
In a recent genome-wide association study (GWAS), we found a unique replicable intergenic risk region between importin 11 gene (IPO11) and HTR1A (called “significant region” in the context; 0.5Mb wide; Figure 1) that was most significantly associated with alcohol and nicotine co-dependence (AD+ND) (peak SNP rs7445832: p=6.2×10−9) at genome-wide significance level in subjects of European descent [Zuo et al., 2013a]. This “significant region” was enriched with numerous common risk variants [minor allele frequency (MAF) > 0.05] for AD+ND in European-Americans and European-Australians. Many of these variants had significant cis-acting regulatory effects. Common variants in this intergenic region were neither significantly associated with any non-alcoholism neuropsychiatric disorder, nor with AD+ND in African-Americans. We speculated that this region might harbor causal variant(s) for AD+ND in subjects of European descent [Zuo et al., 2013a].
Figure 1. Structure of IPO11-HTR1A region.
[The numbers in the middle track are chromosome positions (Build 37); “significant region” is a risk region for alcohol and nicotine co-dependence identified by a previous study; lincRNAs, large intergenic non-coding RNAs; TUCPs, transcripts of uncertain coding potential; rs10042956 (p=5.2×10−6) is the peak association marker for ADHD; rs7445832 (p=6.2×10−9) is the peak association marker for AD+ND]
This recent GWAS used the common variants as markers, as did the aforementioned candidate gene studies. However, in recent years, an increasing number of human diseases appear to be caused by constellations of multiple rare, regionally concentrated, variants, rather than by common variants, and the synthetic effects of region-wide rare variant constellations on diseases might be more significant than individual rare variants in some cases. So far, the hypothesis that rare variants in this intergenic region, in the entire IPO11-HTR1A region (including IPO11, intergenic region and HTR1A) or even in the extended flanking regions might be associated with neuropsychiatric disorders has never been tested. In the present study, we aimed to test the associations between rare variants (MAF < 0.05) across the entire IPO11-HTR1A region and 11 neuropsychiatric disorders including attention deficit hyperactivity disorder (ADHD), schizophrenia, AD+ND, autism, major depression, bipolar disorder, Alzheimer’s disease, amyotrophic lateral sclerosis (ALS), early onset stroke, ischemic stroke, and Parkinson’s disease. These disorders were all hypothesized to be related to serotoninergic system, and the data on these disorders were all of those with neuropsychiatric disorders available for our analysis from the dbGaP database at the moment of analysis (http://www.ncbi.nlm.nih.gov/gap/). Furthermore, after the specific disorder(s) that was associated with this region was identified, we also extended this region to a larger flanking region to explore the associations of rare variants with that specific disorder(s).
Materials and Methods
Subjects
A total of 49,268 subjects in 21 independent cohorts with 11 different neuropsychiatric disorders were analyzed (Table I). These 21 cohorts included case-control and family-based samples, genotyped on Illumina, Affymetrix or PERLEGEN microarray platforms (Table I). More detailed demographic information for these samples has been published elsewhere [Zuo et al., 2013b].
Table I. Associations between rare IPO11-HTR1A variants and different psychiatric or neurological disorders.
| Human Diseases | Dataset name | Ethnicity | Design | SNP # (total) |
SNP # (p<0.05) |
SNP # (P<α) |
SNP # (q<0.05) |
Minimal p value |
Most sig. rare SNP |
Gene | Affected |
Unaffected |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | MAF | N | MAF | |||||||||||
| ADHD | IMAGE | CA | Fam | 1143 | 64 | 1 | 1 | 5.2×10−6 | rs10042956 | Sig. Region | 924 | 0.020 | 924 | 0.024 |
| Schizophrenia | MGS_nonGAIN | EA | CC | 735 | 29 | 0 | 0 | 3.2×10−5 | rs13178180 | Intergenic | 1437 | 0.004 | 1347 | 0.033 |
| Schizophrenia | GAIN | EA | CC | 760 | 15 | 0 | 0 | 3.8×10−3 | rs32186 | Intergenic | 1351 | 0.022 | 1378 | 0.044 |
| Schizophrenia | GAIN | AA | CC | 1774 | 31 | 0 | 0 | 9.5×10−4 | rs346405 | Intergenic | 1195 | 0.026 | 954 | 0.044 |
| AD+ND | SAGE+COGA | EA | CC | 1016 | 75 | 0 | 0 | 4.3×10−3 | rs35620767 | Intergenic | 818 | 0.029 | 1396 | 0.032 |
| AD+ND | OZ-ALC | EAu | Fam | 964 | 24 | 0 | 0 | 3.7×10−3 | rs77016362 | Intergenic | 907 | 0.003 | 907 | 0.0002 |
| AD+ND | SAGE+COGA | AA | CC | 2529 | 56 | 0 | 0 | 7.7×10−5 | rs7737739 | Intergenic | 449 | 0.003 | 480 | 0.027 |
| Autism | AGP | EA | Fam | 1145 | 82 | 0 | 0 | 3.9×10−3 | rs79567137 | Intergenic | 1330 | 0.0008 | 1330 | 0.002 |
| Major Depression | PRSC | CA | CC | 1110 | 11 | 0 | 0 | 8.2×10−3 | rs74881455 | Intergenic | 1805 | 0.030 | 1820 | 0.019 |
| Bipolar Disorder | BDO+GRU | EA | CC | 633 | 13 | 0 | 0 | 8.4×10−3 | rs32167 | Intergenic | 368 | 0.031 | 1034 | 0.008 |
| Bipolar Disorder | BARD+GRU | EA | CC | 706 | 19 | 0 | 0 | 3.0×10−3 | rs10057026 | Intergenic | 653 | 0.006 | 1034 | 0.021 |
| Bipolar Disorder | BARD+GRU | AA | CC | 1234 | 40 | 0 | 0 | 2.0×10−3 | rs1978467 | Intergenic | 141 | 0.162 | 671 | 0.049 |
| Alzheimer’s Disease | LOAD × 4 | CA | Fam | 1136 | 19 | 0 | 0 | 5.7×10−3 | rs7706026 | Intergenic | 2298 | 0.016 | 2298 | 0.014 |
| Alzheimer’s disease | GenADA | EA | CC | 573 | 6 | 0 | 0 | 0.027 | rs73760871 | Intergenic | 806 | 0.042 | 782 | 0.026 |
| ALS | GRU | CA | CC | 838 | 57 | 0 | 0 | 2.5×10−4 | rs78531402 | Intergenic | 261 | 0.002 | 246 | 0.038 |
| Early Onset Stroke | GEOS × 3 | EA | CC | 1000 | 38 | 0 | 0 | 1.7×10−3 | rs73760721 | Intergenic | 372 | 0.050 | 430 | 0.018 |
| Early Onset Stroke | GEOS × 3 | AA | CC | 2491 | 115 | 0 | 0 | 3.0×10−4 | rs74947586 | IPO11 | 309 | 0.027 | 290 | 0.002 |
| Ischemic Stroke | ISGS | CA | CC | 789 | 136 | 0 | 0 | 0.011 | rs17176034 | Intergenic | 219 | 0.063 | 266 | 0.029 |
| Parkinson’s Disease | NGRC | CA | CC | 1181 | 69 | 0 | 0 | 1.2×10−3 | rs2052493 | Intergenic | 2000 | 0.004 | 1986 | 0.0006 |
| Parkinson’s Disease | PDRD+GRU | CA | CC | 1142 | 36 | 0 | 0 | 4.3×10−3 | rs114119382 | Intergenic | 900 | 0.052 | 867 | 0.022 |
| Parkinson’s Disease | lng_coriell_pd | CA | CC | 1048 | 13 | 0 | 0 | 6.6×10−3 | rs260989 | Intergenic | 940 | 0.038 | 801 | 0.021 |
Only the most significant rare risk markers are listed. The significance level (α) is corrected for the numbers of effective genetic markers (calculated by SNPSpD) and the number of cohorts (i.e., 21). ADHD, Attention deficit hyperactivity disorder; AD+ND, alcohol and nicotine co-dependence; ALS, Amyotrophic Lateral Sclerosis. “Sig. Region”, a significant risk region for alcohol and nicotine co-dependence identified previously. N, sample size; MAF, minor allele frequency; AA, African-American; EA, European-American; EAu, European-Australian; CA, Caucasian; CC, case-control design; Fam, family-based design. Dataset names correspond to dbGaP. In family-based cohorts, N= sample size of offspring; “affected MAF”=“transmitted MAF”, “unaffected MAF”=“untransmitted MAF” in offspring.
In particular, the sample with ADHD was the same one used previously, whose demographic data have been described in details before [Brookes et al., 2006]. This sample was genotyped on the PERLEGEN platform (599,171 markers). In brief, 922 parent-child trios (totally 2,757 subjects with 924 ADHD children) from the International Multisite ADHD Genetics (IMAGE) project were included. Children in this study were between the ages of 6 and 17 years old. One or more sibling(s) in the same age range was included. Both parents or one parent plus two or more siblings were available to provide DNA samples. Each child’s IQ was above 70. They were free of single-gene disorders known to be associated with ADHD (e.g. fragile-X, phenylketonuria, hypercalcaemia, thyroid hormone resistance), and free of neurological disease and damage (e.g. hemiplegia and other cerebral palsies, epilepsy, hydrocephalus, post-encephalitic syndromes, psychosis, sensorimotor handicaps). They were diagnosed using DSM-IV criteria and did not meet the criteria for autism or Asperger’s syndrome.
Imputation
To make the genetic marker sets consistent across different cohorts, we imputed the untyped SNPs across the entire IPO11-HTR1A region using the same reference panels that included the rare variants from whole-genome sequencing data. This entire IPO11-HTR1A region started from the transcript start site (TSS) of IPO11 to the TSS of HTR1A at Chr5: 61,708,573-63,257,546 (Build 37), including the ORFs of IPO11 and HTR1A and the intergenic region between them. We used the following strategies to maximize the success rate and accuracy of imputation. (1) We used both 1,000 Genome Project and HapMap 3 genotype panels as references, and separated the European and African ethnicities during the imputation processes. Only the genotypes that were consistently imputed from these two independent reference panels were selected for analysis. (2) We used a Markov Chain Monte Carlo (MCMC) algorithm implemented in the program IMPUTE2 [Howie et al., 2009] to derive full posterior probabilities (i.e., not the “best-guess”) of the genotypes of each SNP to minimize the inference bias. (3) We set the imputation parameters at burnin=10,000, iteration=10,000, k=100, Ne=11,500 and confidence level=0.99 when using IMPUTE2 [Howie et al., 2009]; that is, the uncertainty rate of inference was less than 1%. (4) Within the same ethnicity, we merged the following types of datasets during imputation process, in order to increase sample sizes and marker density for imputation: a) the cases and controls were merged if they were paired within the same study; b) the different panels of array data were merged if they were genotyped in the same subjects; and c) the separate samples were merged if they had the same phenotype and were genotyped on the same microarray platform. (5) Because the imputation process using IMPUTE2 did not incorporate the family relationship information, Mendelian errors might occur in the imputed data. Thus, the families with at least one individual who had more than 0.5% Mendel errors (considering all SNPs tested) and the SNPs with more than 0.5% Mendel errors (considering all individuals tested) were excluded. Meanwhile, we also used the program BEAGLE [Browning and Browning 2009] to impute genotypes independently. The imputation process using BEAGLE does incorporate the family relationship information. Only the genotypes that were consistently imputed by both IMPUTE2 and BEAGLE were selected for analysis. And (6) we stringently cleaned the imputed genotype data after imputation (see below). Furthermore, only the SNPs that had similar minor allele frequencies (with frequency difference < 0.2%) in the healthy controls across different cohorts and HapMap database (within the same ethnicity) were selected for analysis. After this strict selection, we were highly confident with the quality of these imputed genotype data. Finally, for SNPs that were directly genotyped, we used the direct genotypes rather than the imputed. To prevent the loss of the originally-genotyped SNPs during the process of imputation, which might happen sometimes, we also performed regular association analysis on the original unimputed but cleaned genotype data, and then we merged these results back into those generated after imputation (this step was missed in a previous GWAS using the same samples [Zuo et al., 2013a]).
Data cleaning
We stringently cleaned the phenotype and genotype data within each ethnicity before association analysis (detailed previously [Zuo et al., 2012]). Subjects with poor genotypic data, allele discordance, sample relatedness, missing race, non-European and non-African ethnicity, a mismatch between self-identified and genetically-inferred ethnicity, or a missing genotype call rate ≥2% across all SNPs were filtered out. Furthermore, we excluded monomorphic SNPs and SNPs with allele discordance, Mendelian errors (in family samples), or an overall missing genotype call rate ≥2%. We also filtered out the SNPs with MAF differences ≥ 2% or missing rate differences ≥ 2% between two cohorts that had the same ethnicity, phenotype and microarray platform. The SNPs with MAF=0 in either cases or controls were excluded, because it could not be determined if they were missed during the imputation process or truly non-polymorphic in nature in some disease groups. Finally, only a total of 573-1181 (in subjects of European descent) and 1234-2529 (in subjects of African descent) SNPs with 0<MAF<0.05 in either cases or controls were extracted for association analysis. The diagnoses, dataset names, ethnicities, study designs, cleaned sample sizes, and cleaned SNP numbers of all cohorts are shown in Table I.
Association tests for region-wide rare variant constellations
Associations between rare variant constellations and diseases were tested using a score-type program, SCORE-Seq [Lin and Tang 2011]. The mutation information was aggregated by virtue of a weighted linear combination across all rare variants of the entire IPO11-HTR1A region or across each sub-region within IPO11-HTR1A region (i.e., IPO11, HTR1A and intergenic region), and then related to disease phenotypes using regression models. Sex, age, smoking and the first 10 principal components served as the covariates in the regression models. The principal component scores of our samples were derived from all autosomal SNPs across the genome using principal component analysis (PCA) implemented in the software package EIGENSTRAT [Price et al., 2006]. Each individual received scores on each principal component. These principal components reflected the population structure of our samples. The first principal component (PC1) separated the self-identified European-American and African-American subjects very well. Other principal components also accounted for small fractions of the total variance. The first 10 principal component scores accounted for >95% of variation. These PCs serving as covariates in the regression model can control for the population stratification and admixture effects on association analysis. For the regression analysis of those non-alcoholism disorders, alcohol drinking was also included as a covariate.
Two types of tests, i.e., T5 and Fp, were performed to derive the overall p values. (1) In the T5 test, the weight was fixed at 1. (2) In the Fp tests, the weight was 1/sqrt(p(1-p)) where p was the estimated MAF with pseudo counts in the pooled sample. Statistical significance was assessed by using one million times of permutation. All association analyses were performed within the same ethnicity.
Association tests for individual rare variants
For case-control samples, the allele frequencies of each SNP were compared between cases and controls using logistic regression analysis as implemented in PLINK [Purcell et al., 2007]. Diagnosis served as the dependent variable, alleles served as the independent variables, and sex, age, alcohol drinking (for non-alcoholism cohorts only), smoking and the first 10 principal components served as the covariates. For family samples, we tested the allele-disease associations using the program FBAT [Horvath et al., 2001], adjusting for covariates and assuming an additive genetic model under the null hypothesis of no linkage and no association, biallelic mode, minimum number of informative families of 10 for each analysis and offset of zero. These family-based association tests avoided confounding effects from population stratification or admixture [Laird et al., 2000]. Different cohorts were analyzed independently. The MAFs and p values of the most significant risk SNPs and the numbers of the nominally-significant risk SNPs (p<0.05) in all cohorts are shown in Table I.
Correction for multiple testing in single-point association tests
The experiment-wide significance levels (α) were corrected for the number of cohorts (i.e., n=21) and the numbers of effective markers that were calculated by the Bonferroni-type program SNPSpD [Li and Ji 2005] that takes the linkage disequilibrium (LD) structure into account. Approximately 200-300 effective SNPs captured most of the information content of all rare variants across the entire IPO11-HTR1A region in these cohorts. Thus, the corrected significance levels (α) for single-point association tests were set at 7.9×10−6-1.2×10−5. In particular, α was set at 8.4×10−6 for ADHD cohort and 1.1×10−5 for schizophrenia cohort (MGS_nonGAIN). The numbers of the statistically-significant (i.e., p<α) risk SNPs in all cohorts are shown in Table I. The false discovery rate (FDR) (q value) for each SNP was estimated from the p values within each cohort using the R package QVALUE [Storey and Tibshirani 2003].
Association tests in the extended KIF2A-ERBB2IP region
After the specific disorder (here ADHD) that was associated with the IPO11-HTR1A region was identified, we further extended the IPO11-HTR1A region toward both 3’ and 5’ ends to a larger region, i.e. the KIF2A-ERBB2IP region, to explore the roles of the SNPs at the IPO11-HTR1A flanking region in this disease. This extended region harbors KIF2A-DIMT1L-IPO11-HTR1A-RNF180-RGS7BP-SREK1IP1-CWC27-ADAMTS6-CENPK-PPWD1-TRIM23-C5of44-SGTB-NLN-ERBB2IP, starting from chr5:61,637,745 to 65,412,606 (Table II and Figure 1). The syntenic Kif2a-Erbb2ip region in mouse and rat extended the Ipo11-Htr1a region about 0.5Mb from both ends (see below and the Supplemental Table SI). The imputation, data cleaning, association tests for rare variant constellations, association tests for individual rare variants and correction for multiple testing in this extended region were the same as the above.
Table II. p values for associations between ADHD and rare SNPs in the extended IPO11-HTR1A region.
| Genomic region | Positions (Build 37) |
SNP #
Total |
SNP #
(p<0.05) |
SNP #
(p<10−4) * |
SNP #
(q<0.05) |
Rare variant constellation | |
|---|---|---|---|---|---|---|---|
| p-value (T5) | p-value (Fp) | ||||||
| KIF2A | 61601989-61682210 | 48 | 5 | 0 | 0 | 0.374 | 0.378 |
| DIMT1L | 61684351-61699728 | 6 | 0 | 0 | 0 | 0.624 | 0.501 |
| DIMT1L × IPO11 | 61699729-61708572 | 7 | 1 | 0 | 0 | 0.509 | 0.238 |
|
Entire IPO11-HTR1A
region |
61708573-63257546 | 1143 | 64 | 2 | 1 | 8.0×10−31 | 1.3×10−32 |
| IPO11 | 61708573-61924416 | 94 | 6 | 0 | 0 | 3.6×10−10 | 3.2×10−10 |
| IPO11 × HTR1A | 61924417-63256278 | 1048 | 54 | 2 | 1 | 4.1×10−30 | 5.4×10−32 |
| “Significant region” | 62473343-63039074 | 585 | 17 | 2 | 1 | 4.0×10−17 | 1.1×10−17 |
| ISCA1P1 | 62071203-62073171 | 2 | 0 | 0 | 0 | 0.955 | 0.896 |
| TCONS_l2_00022340 | 62597333-62597863 | 0 | 0 | 0 | 0 | - | - |
| LOC100420027 | 62597350-62597883 | 1 | 0 | 0 | 0 | 0.606 | 0.606 |
| U5 snRNA | 62780084-62780130 | 0 | 0 | 0 | 0 | - | - |
| Y_RNA | 62855356-62855461 | 1 | 0 | 0 | 0 | 0.419 | 0.419 |
| TCONS_00010357 | 63179306-63183220 | 3 | 0 | 0 | 0 | 0.828 | 0.596 |
| HTR1A | 63256278-63257546 | 1 | 0 | 0 | 0 | 0.564 | 0.564 |
| HTR1A × RNF180 | 63257547-63461670 | 105 | 2 | 0 | 0 | 0.164 | 0.056 |
| RNF180 | 63461671-63668696 | 86 | 4 | 0 | 0 | 0.034 | 8.8×10−3 |
| RNF180 × RGS7BP | 63668697-63802451 | 65 | 0 | 0 | 0 | 0.112 | 0.065 |
| RGS7BP | 63802452-63908126 | 56 | 4 | 0 | 0 | 0.203 | 0.084 |
|
RGS7BP ×
SREK1IP1 |
63908127-64013974 | 81 | 3 | 0 | 0 | 0.579 | 0.245 |
| SREK1IP1 | 64013975-64064496 | 42 | 3 | 0 | 0 | 0.682 | 0.374 |
| CWC27 | 64064755-64314590 | 153 | 8 | 0 | 0 | 0.600 | 0.534 |
|
CWC27 ×
ADAMTS6 |
64314589-64444562 | 124 | 9 | 1 | 1 | 0.946 | 0.991 |
| ADAMTS6 | 64444563-64777704 | 225 | 20 | 0 | 0 | 0.403 | 0.325 |
|
ADAMTS6 ×
CENPK |
64777705-64813592 | 14 | 2 | 0 | 0 | 0.725 | 0.405 |
| CENPK | 64813593-64858995 | 26 | 1 | 0 | 0 | 0.577 | 0.530 |
| PPWD1 | 64859131-64883373 | 18 | 0 | 0 | 0 | 0.433 | 0.277 |
| TRIM23 | 64885507-64920187 | 17 | 1 | 0 | 0 | 0.333 | 0.185 |
| C5of44 | 64920558-64961954 | 24 | 3 | 0 | 0 | 0.206 | 0.202 |
| SGTB | 64961755-65017941 | 19 | 0 | 0 | 0 | 0.285 | 0.153 |
| NLN | 65018023-65125111 | 70 | 3 | 0 | 0 | 0.217 | 0.125 |
| NLN × ERBB2IP | 65125112-65222383 | 69 | 5 | 0 | 0 | 0.545 | 0.530 |
| ERBB2IP | 65222384-65376850 | 62 | 2 | 0 | 0 | 0.575 | 0.605 |
| Total | 61601989-65376850 | 2460 | 140 | 3 | 2 | ||
T5 and Fp, association tests using SCORE-Seq. x, intergenic region between two genes; ISCA1P1 and LOC100420027, pseudo gene loci; TCONS_l2_00022340 and TCONS_00010357, long non-coding RNAs. “Significant region”, a significant risk region for alcohol and nicotine co-dependence reported previously.
details are shown in Table III.
Cis-acting expression of quantitative trait locus (cis-eQTL) analysis on all available SNPs in the IPO11-HTR1A region in two primary human cells
To examine whether the SNPs in the IPO11-HTR1A region influence the gene expression of IPO11 and HTR1A, we tested the associations between the genotypes and the exon-level expression changes of these two genes in two European samples (Table III). Expression data of these two genes in 93 autopsy-collected frontal cortical brain tissue samples with no defined neuropsychiatric condition and 80 peripheral blood mononuclear cell (PBMC) samples collected from living healthy donors were evaluated [Heinzen et al., 2008]. The expression data were evaluated using Affymetrix Human ST 1.0 exon arrays and were confirmed by quantitative RT-PCR. Forty-one SNPs within or around IPO11 and 24 SNPs within or around HTR1A were genotyped in these samples. The SNP-expression associations were analyzed using a linear regression model by correcting for age, sex and source of tissues. The cis-eQTL analysis served as a validation for the SNP-disease association findings.
Table III. cis-regulatory effects on the exon-level expression of IPO11 in brain and PBMC tissues.
| SNP | Position (Build 37) |
Gene | p-values |
|
|---|---|---|---|---|
| Brain | PBMC | |||
| rs3822485 | 61605062 | 5′ flanking | 5.6×10−13 | 6.1×10−6 |
| rs10471545 | 61607267 | 5′ flanking | 9.1×10−11 | 7.0×10−5 |
| rs7734679 | 61607717 | 5′ flanking | 0.020 | 0.078 |
| rs7446543 | 61608378 | 5′ flanking | 0.001 | 0.001 |
| rs7718580 | 61611988 | 5′ flanking | 3.2×10−10 | 1.8×10−4 |
| rs264529 | 61626533 | 5′ flanking | 0.020 | 0.078 |
| rs264524 | 61630878 | 5′ flanking | 0.030 | 0.020 |
| rs959899 | 61651247 | 5′ flanking | 2.9×10−13 | 2.4×10−6 |
| rs153867 | 61679611 | 5′ flanking | 2.6×10−4 | 0.017 |
| rs35015 | 61687540 | 5′ flanking | 3.2×10−10 | 1.8×10−4 |
| rs2272290 | 61689519 | 5′ flanking | 0.016 | 0.025 |
| rs17467190 | 61717763 | IPO11 | 0.062 | 3.6×10−5 |
| rs152186 | 61719505 | IPO11 | 1.5×10−17 | 4.6×10−14 |
| rs247235 | 61739462 | IPO11 | 7.2×10−10 | 2.0×10−4 |
| rs247230 | 61750666 | IPO11 | 0.131 | 0.060 |
| rs26645 | 61788236 | IPO11 | 7.2×10−10 | 6.0×10−5 |
| rs3776633 | 61794163 | IPO11 | 0.062 | 5.6×10−6 |
| rs3776637 | 61825234 | IPO11 | 4.9×10−8 | 2.3×10−7 |
| rs32181 | 61825632 | IPO11 | 7.2×10−10 | 6.0×10−5 |
| rs32179 | 61826662 | IPO11 | 9.6×10−9 | 0.001 |
| rs7722692 | 61830977 | IPO11 | 4.7×10−11 | 7.9×10−6 |
| rs32163 | 61857576 | IPO11 | 1.9×10−12 | 5.5×10−8 |
| rs32162 | 61858162 | IPO11 | 0.001 | 1.2×10−4 |
| rs10058598 | 61871008 | IPO11 | 1.5×10−17 | 4.6×10−14 |
| rs1477358 | 61891621 | IPO11 | 9.6×10−17 | 2.2×10−13 |
| rs16890857 | 61895123 | IPO11 | 0.013 | 0.078 |
| rs7719851 | 61941042 | 3′ flanking | 2.3×10−13 | 2.0×10−12 |
| rs11750272 | 61948933 | 3′ flanking | 0.038 | 8.1×10−5 |
| rs4700505 | 61978988 | 3′ flanking | 8.3×10−9 | 6.8×10−6 |
| rs11955532 | 61980981 | 3′ flanking | 0.001 | 0.129 |
| rs1469095 | 61983847 | 3′ flanking | 0.003 | 0.029 |
| rs1423386 | 61984852 | 3′ flanking | 0.001 | 0.002 |
| rs4552552 | 61994696 | 3′ flanking | 0.001 | 0.046 |
| rs37764 | 62002139 | 3′ flanking | 0.025 | 0.137 |
| rs903421 | 62007686 | 3′ flanking | 0.025 | 0.135 |
| rs10434537 | 62010639 | 3′ flanking | 0.002 | 0.066 |
| rs7706346 | 62010846 | 3′ flanking | 0.121 | 0.142 |
| rs10514950 | 62020356 | 3′ flanking | 0.022 | 0.010 |
| rs13171600 | 62021805 | 3′ flanking | 3.4×10−4 | 0.039 |
| rs10461474 | 62022335 | 3′ flanking | 3.4×10−4 | 0.055 |
| rs1121882 | 62024478 | 3′ flanking | 0.111 | 0.018 |
All available SNPs in this dataset are listed. Rare variants are underlined. α=0.002 (=0.05/31 where “31” is the number of exons in IPO11)
Expression of the syntenic transcripts in mouse and rat brains and genome-wide eQTL analysis of these transcripts in mouse brain
We generated the expression data across the LXS and HXB/BXH RI panels in mouse using Affymetrix Exon Arrays and the expression data across the BN-Lx/CubPrin panels in rat using RNA-Seq technology, and then analyzed the transcripts syntenic to all genes within the human KIF2A-ERBB2IP genomic region. We identified 19 syntenic transcripts that coded protein (Table IV). For both mouse and rat, we collapsed the exon-level probes and reported results on the transcript level (i.e., the integration of exon-level probes was used to define a “core” transcript). We ascertained the level of expression in mouse and rat brains of these syntenic transcripts. We also identified the loci across the mouse genome that regulated the expression level of each syntenic transcript (eQTL analysis). A Lod score above 3 indicates a significant regulation. Furthermore, we analyzed the expression of non-coding RNAs (ncRNAs) within the syntenic Ipo11-Htr1a regions in rat. We reported the number of reads and FPKM (Reads Per Kilobase of exon model per Million mapped reads) values to reflect the level of transcript abundance of these ncRNAs. A FPKM value above 5 indicates that transcript is present in brain.
Table IV. RNA expression and eQTL analysis of the genes in mouse and rat brains syntenic to the human KIF2A-ERBB2IP region.
| Human |
Mouse Information (based on LXS RI panel) . |
Rat |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene Title | Gene Symbol | Location chr (Mb |
Transcript Cluster ID |
Location chr (Mb) |
eQTL Max Locus ID |
eQTL Location chr (Mb) |
LOD score (p-value) |
FPKM**(s) |
| kinesin family member 2A | Kif2a | chr5 (61.6) | 6815797 | 13 (107.7) | rs29631328 | 13 (104.5) | 2.7 (0.308) | 3.6 |
| DIM1 dimethyladenosine transferase 1 homolog (S. cerevisiae) | DIMT1 | chr5 (61.7) | 6915037 | 4 (84.6) | rs27901635 | 2 (48.8) | 2.8 (0.178) | 0.5 |
| importin 11 | Ipo11 | chr5 (61.7) | 6815792 | 13 (107.6) | rs33426574 | 17 (45.5) | 2.3 (0.544) | 2.4 |
| leucine rich repeat containing 70 | LRRC70 | chr5 (61.9) | Ensembl indicates that the “possible” mouse ortholog is Ipo11 | 0.2 | ||||
| 5-hydroxytryptamine (serotonin) receptor 1A | Htr1a | chr5 (63.3) | 6809880 | 13 (106.2) | rs47752022 | 17 (76.3) | 2.4 (0.433) | 3.3 |
| ring finger protein 180 | Rnf180 | chr5 (63.5) | 6815749 | 13 (105.9) | rs33717556 | 9 (60.9) | 2.0 (0.748) | 9.5 |
| RIKEN cDNA 4933425L06 gene | 4933425L06Rik | chr5 (63.7) | 6809876 | 13 (105.9) | rs4186276 | 16 (51.0) | 3.6 (0.050) | N/A |
| regulator of G-protein signalling 7 binding protein | Rgs7bp | chr5 (63.8) | 6815739 | 13 (105.7) | rs45852014 | 9 (58.3) | 2.0 (0.784) | 13.5 |
| family with sequence similarity 159, member B | FAM159B | chr5 (64.0) | 6815733 | 13 (105.6) | rs31760078 | 7 (29.5) | 3.5 (0.046) | 0.3 |
| SREK1-interacting protein 1 | SREK1IP1 | chr5 (64.0) | 6809851 | 13 (105.6) | rs29085977 | X (92.3) | 2.2 (0.521) | 6.8 |
| CWC27 spliceosome-associated protein homolog (S. cerevisiae) | CWC27 | chr5 (64.1) | 6815726 | 13 (105.4) | rs29070885 | X (90.0) | 1.9 (0.842) | 1.1 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 6 | ADAMTS6 | chr5 (64.4) | 6809817 | 13 (105.1) | rs29121906 | X (78.4) | 3.9 (0.019) | 0.4 |
| centromere protein K | CENPK | chr5 (64.8) | 6809812 | 13 (105.0) | rs28213218 | 11 (79.0) | 2.2 (0.595) | 0.2 |
| peptidylprolyl isomerase domain and WD repeat containing 1 | PPWD1 | chr5 (64.9) | 6815708 | 13 (105.0) | rs29631328 | 13 (104.5) | 23.9 (<0.001) | 1.7 |
| tripartite motif containing 23 | TRIM23 | chr5 (64.9) | 6809810 | 13 (105.0) | rs33426574 | 17 (45.5) | 2.6 (0.318) | 5.7 |
| chromosome 5 open reading frame 44 | C5orf44 | chr5 (64.9) | 6815702 | 13 (104.9) | rs29631328 | 13 (104.5) | 2.4 (0.401) | 7.7 |
| small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta | SGTB | chr5 (65.0) | 6809806 | 13 (104.9) | rs29631328 | 13 (104.5) | 8.0 (<0.001) | 16.6 |
| neurolysin (metallopeptidase M3 family) | NLN | chr5 (65.0) | 6815698 | 13 (104.8) | rs4186276 | 16 (51.0) | 2.6 (0.294) | 4.2 |
| Erbb2 interacting protein (in mouse: predicted gene 2590) | Erbb2ip (Gm2590) | chr5 (65.3) | 6815687 | 13 (104.6) | rs51663211 | 13 (107.2) | 4.0 (0.025) | 4.4 |
All of the genes listed in this table are expressed in mouse brain. Rat information is based on RNA-Seq of brain tissue from BN-Lx/CubPrin.
FPKM > 5 is considered expressed above background in rat brain. N/A, no RefSeq mRNA ID available in rat.
Results
Rare variant constellation across the entire IPO11-HTR1A region was associated with ADHD in Caucasians (T5: p=7.9×10−31; Fp: p=1.3×10−32). When testing the rare variant constellations within each sub-region, the variant constellations within IPO11 (T5: p=3.6×10−10; Fp: p=3.2×10−10), the intergenic region (T5: p=4.1×10−30; Fp: p=5.4×10−32) and the “significant region” (T5: p=4.0×10−17; Fp: p=1.1×10−17) were highly significantly associated with ADHD. Only one rare variant (rs6294) in HTR1A was studied and it was not significantly associated with ADHD (Table II). When extending to the KIF2A-ERBB2IP region, the rare variant constellation within RNF180 was modestly associated with ADHD (T5: p=0.034; Fp: p=8.8×10−3). No rare variant constellation across the IPO11-HTR1A region was significantly associated with any other disorder examined, e.g., schizophrenia (MGS_nonGAIN) in European-Americans (for entire region: T5: p=0.638; Fp: p=0.733; for IPO11: T5: p=0.559; Fp: p=0.824; for intergenic region: T5: p=0.658; Fp: p=0.738).
Single-point association analysis showed that among a total of 1,143 individual rare variants in Caucasians, 64 SNPs were nominally associated with ADHD (p<0.05). The association of rs10042956 with ADHD was significant (p=5.2×10−6) after region- and cohort-wide correction (α=8.4×10−6) or FDR correction (q=0.006) (Tables I and III). This intergenic marker is located in the middle of the “significant risk region” (Figure 1). When extending to the KIF2A-ERBB2IP region, an association of rs114984365 with ADHD was significant (p=1.6×10−5) after region-wide correction (α=1.7×10−4) or FDR correction (q=0.017) (Table V). This rs114984365 was located between CWC27 and ADAMTS6. We noted that minor alleles of rare risk variants were protective alleles for ADHD (OR<1) (Table V). No significant individual rare variant was associated with any other disease examined after correction. For example, among a total of 735 individual rare variants in European-Americans, 29 SNPs were nominally associated with schizophrenia (MGS_nonGAIN) (p<0.05). The associations of two variants (p=3.2×10−5 for rs13178180 and p=4.7×10−5 for rs66582641) with schizophrenia were suggestive but not significant after region- and cohort-wide correction (α=1.1×10−5) (Table I).
Table V. Top-ranked rare risk variants (p<10−4) in the extended IPO11-HTR1A region for ADHD.
| SNP | Position (Build 37) |
Genomic region |
Minor allele |
Minor allele frequency |
OR | p-value | q-value | |
|---|---|---|---|---|---|---|---|---|
| T | U | |||||||
| rs10042956 | 62554599 | IPO11 × HTR1A (“Sig. region”) | T | 0.020 | 0.060 | 0.328 | 5.2×10−6 | 6.3×10−3 |
| rs10057026 | 62642906 | IPO11 × HTR1A (“Sig. region”) | A | 0.005 | 0.093 | 0.056 | 9.6×10−5 | 8.1×10−2 |
| rs114984365 | 64316615 | CWC27 × ADAMTS6 | C | 0.009 | 0.104 | 0.083 | 1.6×10−5 | 1.7×10−2 |
MAF, minor allele frequency. T, transmitted; U, untransmitted. x, intergenic region between two genes. “Sig. region”, a significant risk region for AD+ND identified previously.
Cis-eQTL analysis in human cells showed that, among the 41 SNPs within or around IPO11, 39 had nominal regulatory effects on IPO11 exon expression (1.5×10−17≤p<0.05) in brain or PBMC; 29 had significant regulatory effects on IPO11 exon expression after correction for multiple testing (α=0.002=0.05/31 where “31” is the number of exons in IPO11). Most of these associations were well replicable between brain tissue and PBMC (Table III). However, no SNPs within or around HTR1A had significant regulatory effects on HTR1A mRNA expression (p>0.05; data not shown).
The range of the IPO11-HTR1A region in human, the ranges of its syntenic Ipo11-Htr1a regions in mouse and rat, and their extended ranges we explored are presented in the Supplemental Table SI. In these extended syntenic regions, we found 19 protein coding transcripts; all of them were expressed in mouse brain, and six of them showed detectable level of expression in rat brain, including Rnf180 (FPKM=9.5), Rgs7bp (FPKM=13.5), Srek1ip1 (FPKM=6.8), Trim23 (FPKM=5.7), C5orf44 (FPKM=7.7) and Sgtb (FPKM=16.6). Two loci on mouse chr13 significantly cis-controlled the expression of Ppwd1 (peak marker: rs29631328, LOD=23.9, p<0.001), Sgtb (peak marker: rs29631328, LOD=8.0, p<0.001) and Erbb2ip (peak marker: rs51663211, LOD=4.0, p=0.025), respectively, and three other loci significantly trans-controlled the expression of 4933425L06Rik (peak marker: rs4186276 on chr16, LOD=3.6, p=0.050), Fam159b (peak marker: rs31760078 on chr7, LOD=3.5, p=0.046) and Adamts6 (peak marker: rs29121906 on chrX, LOD=3.9, p=0.019), respectively (Table IV). Furthermore, the ncRNAs within the syntenic Ipo11-Htr1a regions we detected in rat brain are illustrated in the Supplemental Figure S1 and listed in details in the Supplemental Table SII. Ninteen known ncRNAs (Table SIIa) and 27 novel ncRNAs (Table SIIb) in this region were detected in the rat brain (FPKM>6), but nine other known ncRNAs (Table SIIc) were not.
Discussion
We found that rare IPO11-HTR1A variants conferred risk for ADHD in Caucasians, and the association signals mainly came from the intergenic region and IPO11, which was supported by both rare variant constellation analysis and individual rare variant analysis. The most significant risk variant for ADHD was located in the “significant region” that was previously identified as a significant risk region for AD+ND in subjects of European descent using common variant marker set [Zuo et al., 2013a]. The rare variants in this IPO11-HTR1A region had no significant association with any other neuropsychiatric disorder examined, although the same set of markers was explored. The variants in this region might influence the risk for diseases via regulating transcription of the causal variant(s), which was supported by our cis-eQTL findings.
Our study provided an additional example to support the hypothesis that the region-wide rare variant constellations could have synthetic effects on diseases. The synthetic effects of region-wide rare variant constellations across IPO11-HTR1A locus on ADHD were much more significant than the effects of individual rare variants (p=10−31 vs. 10−6), and the specificity of variant-disease associations to ADHD became much more apparent when using the synthetic effect analysis (p=10−31 for ADHD vs. p>0.05 for other diseases) than using the individual effect analysis (p=10−6 for ADHD vs. p=10−5 for schizophrenia). Thus, rare variant constellation analysis could play an important role in the association studies.
Common variants in the IPO11-HTR1A region were significantly associated with AD+ND at genome-wide significance level (α=5×10−8), but not with ADHD (minimal p=2.8×10−4 > α) [Zuo et al., 2013a]. Rare variant constellations were only significantly associated with ADHD, but not AD+ND. These findings might reflect the difference between these two diseases and the different roles of common variants and rare variants in diseases. However, both most significant risk SNPs [i.e., rare variant rs10042956 for ADHD (p=5.2×10−6) and common variant rs7445832 for AD+ND (p=6.2×10−9)] were closely located in the same “significant region” (Figure 1), which might suggest that ADHD and AD+ND share sources of genetic liability. This genetic commonality may underlie high rate of comorbidity of ADHD and AD+ND. It has been reported that 32% of patients with a substance use disorder met the criteria for ADHD [Clure et al., 1999; Ohlmeier et al., 2007], and 50% of individuals with continuing ADHD symptoms in adults showed symptoms of substance abuse [Ohlmeier et al., 2007; Sullivan and Rudnik-Levin 2001]. There is also a greater likelihood of adolescents with ADHD developing an addiction to cigarettes compared to adolescents without ADHD [Milberger et al., 1997; Ohlmeier et al., 2007; Pomerleau et al., 1995; Wilens 2004].
We found that the association signals of rare variants for ADHD mainly came from the intergenic region between IPO11 and HTR1A, which might be related to the roles of non-coding RNAs (ncRNAs) existing within this intergenic region. We detected tens of ncRNAs (mainly tRNAs) in this region in rat. Two large intergenic non-coding RNAs (lincRNAs), i.e., TCONS_l2_00022340 (530bp) and TCONS_00010357 (3,914bp), a U5 snRNA (46bp) and an Y_RNA (106bp) were also previously reported in this region in human (see UCSC genome browser). TCONS_l2_00022340, U5 snRNA and Y_RNA are all located in the “significant region”, close to the two peak association markers for ADHD and AD+ND (Figure 1). TCONS_00010357 is located closely to the 3’ of HTR1A (Figure 1). Recent evidence suggests that ncRNAs play an important and dynamic role in transcriptional regulation, epigenetic signaling, stress response, and plasticity in the nervous system. Dysregulation of ncRNAs are thought to contribute to many, and perhaps all, neuropsychiatric disorders, including ADHD and drug addiction [Sartor et al., 2012]. Additionally, both U5 snRNA and Y_RNA are a part of ribonucleoprotein (RNP) complexes, including spliceosome complexes, and they may be important in determining the translated isoforms of many protein coding transcripts in brain [Kershaw et al., 2009; Sim and Wolin 2011]. The Y-RNAs have also been recently identified as part of the quality control process for other ncRNAs, including snRNAs [Chen et al., 2003; Langley et al., 2010]. Variations in the U5 snRNA and/or Y_RNA may affect the expression of the isoforms.
The association signals of rare variants for ADHD also came from IPO11. IPO11 is a gene flanking to 3’ of HTR1A. It encodes the importin 11 that is a member of the karyopherin/importin-beta family of transport receptors. This receptor mediates the nuclear import of ubiquitin-conjugating enzyme E2E3 (UBE2E3) [Plafker and Macara 2000]. Ubiquitin-conjugating enzyme may ligate small ubiquitin-related modifier to target proteins in brain, resulting in changes of their localization, activity, or stability. In the present study, we found most markers within or around IPO11 had significant cis-acting regulatory effects on IPO11 mRNA expression both in the human brain and PBMC tissues. Ipo11 was also found to express in mouse brain. Thus, IPO11 might play important roles in neuropsychiatric disorders. Our study is the first time to detect the association between this gene and ADHD.
However, as expected, the association signals for ADHD did not come from HTR1A. HTR1A is a small gene with only one rare variant included in this study. We did not find significant or functional SNPs within or around human HTR1A in the present study, and the expression level of Htr1a in rat brain was found to be low (FPKM=3.3). Our findings may suggest that its associations with the neuropsychiatric disorders reported before might be driven by variants (1) in the flanking intergenic region, in which several ncRNAs existed, (2) in the flanking gene IPO11, in which the variant-disease association signals and the functional (i.e., cis-QTL) signals were much more significant than HTR1A, or (3) in other flanking genes like RNF180, CWC27 and ADAMTS6.
RNF180 encodes ring finger protein 180 and is a gene flanking to 5’ of HTR1A. A rare variant constellation across RNF180 was modestly associated with ADHD. This gene was also found to be expressed in human [Ogawa et al., 2008], mouse and rat brains (particularly in the ventricular layer of the cerebral cortex at embryonic stages [Ogawa et al., 2008]). RNF180 protein is a membrane-bound E3 ubiquitin-protein ligase. E3 ubiquitin-protein ligase may promote polyubiquitination and degradation of target proteins (e.g., dopamine D4 receptor in brain [Rondou et al., 2008]) by the proteasome pathway of ZIC2, and thus might play roles in neuropsychiatric disorders.
CWC27 is a spliceosome-associated protein homolog gene and is flanking to 3’ of ADAMTS6 (ADAM metallopeptidase with thrombospondin type 1 motif, 6). One rare variant between CWC27 and ADAMTS6 was significantly associated with ADHD. These two genes were also found to be expressed in human and mouse brains, and expression of Adamts6 was significantly trans-controlled by a locus on chrX. The pathophysiological mechanism of how these two genes are involved in ADHD warrants further investigation.
RGS7BP encodes regulator of G-protein signaling 7 binding protein and is a gene flanking to 3’ of RNF180. This gene was also found to be expressed in human, mouse and rat brains [Gold et al., 1997; Larminie et al., 2004]. RGS7BP protein is a regulator of G protein-coupled receptor (GPCR) signaling. It controls the proteolytic stability of R7 proteins, probably by protecting them from degradation. When it is palmitoylated, RGS7BP initiates the activation of GPCRs, e.g, dopamine, epinephrine, norepinephrine, histamine, and glutamate receptors, etc., which contribute to the development of neuropsychiatric disorders. However, we did not detect significant association between RGS7BP and ADHD in human in the present study.
Three other genes, including Erbb2 interacting protein gene (Erbb2ip), Sgtb and Ppwd1, showed differential expression in mouse brain, and their levels of expression were also significantly cis-controlled from the same region. These strong cis-eQTLs usually can be extrapolated across organs and species, as we confirmed that Sgtb was detected and Erbb2ip was near the detection limit in rat brain. These strong cis-eQTLs are indicators of the locations of the control sequences for transcription of their transcripts. In particular, Erbin is a receptor of neuregulin. It is a scaffolding protein recently shown to be important in clustering of nicotinic cholinergic receptors and, thus, influencing their signaling properties (level of depolarization, etc.) [Simeone et al., 2010]. These nicotinic cholinergic receptors have been related to ADHD [Potter et al., 2006] and nicotine/alcohol dependence. Nicotinic receptor agonists, in general, have been linked to attention and cognition in several mental disorders, including ADHD [Wilens and Decker 2007], schizophrenia, dementia, etc. However, we did not detect significant association between these three genes and ADHD in human in the present study. They warrant further studies in other independent samples.
This study has some limitations. The imputed genotypes were not directly observed from the molecular experiment, although we have high confidence with them after data cleaning. This warrants verification by direct sequencing of our samples in the future, which is out of the scope of the current study. Additionally, stringent cleaning process deleted many rare variants, so that not all rare variants within the candidate region were incorporated in the rare variant constellations. These rare variants can be filled in by direct sequencing in the future, which is out of the scope of the current study too. Finally, not all neuropsychiatric disorders and related traits were exhaustively examined in the present study; and thus, we do not completely exclude the possibility that other neuropsychiatric disorders not examined might share this risk genomic region with ADHD and AD+ND.
Supplemental Figure S1. Non-coding RNA expression within the syntenic Ipo11-Htr1a region in rat brain [This is the syntenic Ipo11-Htr1a region (chr2:36,434,518-38,109,822) in UCSC Genome Browser on Rat Nov. 2004 (Baylor 3.4/rn4) Assembly. The top track, Unannotated Expressed Regions track, indicates contiguous regions for which we showed RPKM > 6 and which were not explained by either known ncRNA or protein-coding exons. The middle track, Expressed ncRNA track, shows known ncRNA that had expression in rat brain. The bottom track, Known ncRNA Not Expressed track, indicates known ncRNAs for which no expression was detected in our samples]
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by National Institute on Drug Abuse (NIDA) grants K01 DA029643 and R01DA016750, National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants R21 AA021380, R21 AA020319, R21 AA023237 and R24 AA13162, National Alliance for Research on Schizophrenia and Depression (NARSAD) Award 17616 (L.Z.), ABMRF/The Foundation for Alcohol Research (L.Z.), National Foundation for Prevention of Chemical Dependency Disease Career Development Award and The Banbury Fund. We thank NIH GWAS Data Repository, the Contributing Investigator(s) who contributed the phenotype and genotype data from his/her original study (e.g., Drs. Faraone, Bierut, Edenberg, Heath, Singleton, Hardy, Foroud, Myers, Gejman, Sonuga-Barke, Sullivan, Nurnberger, Devlin, Monaco, etc.), and the primary funding organization that supported the contributing study. Funding and other supports for phenotype and genotype data were provided through the National Institutes of Health (NIH) Genes, Environment and Health Initiative (GEI) (U01HG004422, U01HG004436 and U01HG004438); the GENEVA Coordinating Center (U01HG004446); the NIAAA (U10AA008401, R01AA013320, P60AA011998); the NIDA (R01DA013423); the National Cancer Institute (P01 CA089392); the Division of Neuroscience, the NIA National Institute of Neurological Disorders and Stroke (NINDS); the NINDS Human Genetics Resource Center DNA and Cell Line Repository; the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C); the Center for Inherited Disease Research (CIDR); a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and Prevention; the NIH Office of Research on Women’s Health (ORWH) (R01NS45012); the Department of Veterans Affairs; the University of Maryland General Clinical Research Center (M01RR165001), the National Center for Research Resources, NIH; the National Institute of Mental Health (R01MH059160, R01MH59565, R01MH59566, R01MH59571, R01MH59586, R01MH59587, R01MH59588, R01MH60870, R01MH60879, R01MH61675, R01MH62873, R01MH081803, R01MH67257, R01MH81800, U01MH46276, U01MH46282, U01MH46289, U01MH46318, U01MH79469, U01MH79470 and R01MH67257); the NIMH Genetics Initiative for Bipolar Disorder; the Genetic Association Information Network (GAIN); the Genetic Consortium for Late Onset Alzheimer’s Disease; the Autism Genome Project, the MARC: Risk Mechanisms in Alcoholism and Comorbidity; the Molecular Genetics of Schizophrenia Collaboration; the Medical Research Council (G0601030) and the Wellcome Trust (075491/Z/04), University of Oxford; the Netherlands Scientific Organization (904-61-090, 904-61-193, 480-04-004, 400-05-717, NWO Genomics, SPI 56-464-1419) the Centre for Neurogenomics and Cognitive Research (CNCR-VU); Netherlands Study of Depression and Anxiety (NESDA) and the Netherlands Twin Register (NTR); and the European Union (EU/WLRT-2001-01254), ZonMW (geestkracht program, 10-000-1002). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the Genetic Consortium for Late Onset Alzheimer’s Disease, the GENEVA Coordinating Center (U01 HG004446), and the National Center for Biotechnology Information. Genotyping was performed at the Johns Hopkins University Center for Inherited Disease Research, and GlaxoSmithKline, R&D Limited. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap.
Footnotes
Conflict of Interest: The authors have no conflict of interest to declare.
References
- Anttila S, Huuhka K, Huuhka M, Rontu R, Hurme M, Leinonen E, Lehtimaki T. Interaction between 5-HT1A and BDNF genotypes increases the risk of treatment-resistant depression. J Neural Transm. 2007;114(8):1065–1068. doi: 10.1007/s00702-007-0705-9. [DOI] [PubMed] [Google Scholar]
- Arias B, Catalan R, Gasto C, Gutierrez B, Fananas L. Evidence for a combined genetic effect of the 5-HT(1A) receptor and serotonin transporter genes in the clinical outcome of major depressive patients treated with citalopram. Journal of psychopharmacology (Oxford, England) 2005;19(2):166–172. doi: 10.1177/0269881105049037. [DOI] [PubMed] [Google Scholar]
- Benko A, Lazary J, Molnar E, Gonda X, Tothfalusi L, Pap D, Mirnics Z, Kurimay T, Chase D, Juhasz G, Anderson IM, Deakin JF, Bagdy G. Significant association between the C(-1019)G functional polymorphism of the HTR1A gene and impulsivity. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):592–599. doi: 10.1002/ajmg.b.31025. [DOI] [PubMed] [Google Scholar]
- Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N, Anney R, Franke B, Gill M, Ebstein R, Buitelaar J, Sham P, Campbell D, Knight J, Andreou P, Altink M, Arnold R, Boer F, Buschgens C, Butler L, Christiansen H, Feldman L, Fleischman K, Fliers E, Howe-Forbes R, Goldfarb A, Heise A, Gabriels I, Korn-Lubetzki I, Johansson L, Marco R, Medad S, Minderaa R, Mulas F, Muller U, Mulligan A, Rabin K, Rommelse N, Sethna V, Sorohan J, Uebel H, Psychogiou L, Weeks A, Barrett R, Craig I, Banaschewski T, Sonuga-Barke E, Eisenberg J, Kuntsi J, Manor I, McGuffin P, Miranda A, Oades RD, Plomin R, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Taylor E, Thompson M, Faraone SV, Asherson P. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Molecular psychiatry. 2006;11(10):934–953. doi: 10.1038/sj.mp.4001869. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. American journal of human genetics. 2009;84(2):210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TJ, Yu YW, Hong CJ, Chen MC, Tsai SJ. Association analysis for the C-1019G promoter variant of the 5-HT1A receptor gene with auditory evoked potentials in major depression. Neuropsychobiology. 2004;50(4):292–295. doi: 10.1159/000080955. [DOI] [PubMed] [Google Scholar]
- Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Current biology : CB. 2003;13(24):2206–2211. doi: 10.1016/j.cub.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Clure C, Brady KT, Saladin ME, Johnson D, Waid R, Rittenbury M. Attention-deficit/hyperactivity disorder and substance use: symptom pattern and drug choice. The American journal of drug and alcohol abuse. 1999;25(3):441–448. doi: 10.1081/ada-100101871. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17(20):8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenisch B, Linsel K, Bruss M, Gilsbach R, Propping P, Nothen MM, Rietschel M, Fimmers R, Maier W, Zobel A, Hofels S, Guttenthaler V, Gothert M, Bonisch H. Association of major depression with rare functional variants in norepinephrine transporter and serotonin1A receptor genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(7):1013–1016. doi: 10.1002/ajmg.b.30912. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS biology. 2008;6(12):e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9(4):301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YY, Battistuzzi C, Oquendo MA, Harkavy-Friedman J, Greenhill L, Zalsman G, Brodsky B, Arango V, Brent DA, Mann JJ. Human 5-HT1A receptor C(-1019)G polymorphism and psychopathology. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2004;7(4):441–451. doi: 10.1017/S1461145704004663. [DOI] [PubMed] [Google Scholar]
- Kershaw CJ, Barrass JD, Beggs JD, O’Keefe RT. Mutations in the U5 snRNA result in altered splicing of subsets of pre-mRNAs and reduced stability of Prp8. RNA. 2009;15(7):1292–1304. doi: 10.1261/rna.1347409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus MR, Al-Taie O, Schafer A, Pfersdorff M, Lesch KP, Scheurlen M. Serotonin-1A receptor gene HTR1A variation predicts interferon-induced depression in chronic hepatitis C. Gastroenterology. 2007;132(4):1279–1286. doi: 10.1053/j.gastro.2007.02.053. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genetic epidemiology. 2000;19(Suppl 1):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Langley AR, Chambers H, Christov CP, Krude T. Ribonucleoprotein particles containing non-coding Y RNAs, Ro60, La and nucleolin are not required for Y RNA function in DNA replication. PloS one. 2010;5(10):e13673. doi: 10.1371/journal.pone.0013673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Brain research. 2004;122(1):24–34. doi: 10.1016/j.molbrainres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Lee YS, Choi SW, Han DH, Kim DJ, Joe KH. Clinical manifestation of alcohol withdrawal symptoms related to genetic polymorphisms of two serotonin receptors and serotonin transporter. European addiction research. 2009;15(1):39–46. doi: 10.1159/000173008. [DOI] [PubMed] [Google Scholar]
- Lemonde S, Du L, Bakish D, Hrdina P, Albert PR. Association of the C(-1019)G 5-HT1A functional promoter polymorphism with antidepressant response. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2004;7(4):501–506. doi: 10.1017/S1461145704004699. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Lin DY, Tang ZZ. A general framework for detecting disease associations with rare variants in sequencing studies. American journal of human genetics. 2011;89(3):354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marziniak M, Mossner R, Kienzler C, Riederer P, Lesch KP, Sommer C. Functional polymorphisms of the 5-HT1A and 5-HT1B receptor are associated with clinical symptoms in migraineurs. J Neural Transm. 2007;114(9):1227–1232. doi: 10.1007/s00702-007-0713-9. [DOI] [PubMed] [Google Scholar]
- Milberger S, Biederman J, Faraone SV, Chen L, Jones J. ADHD is associated with early initiation of cigarette smoking in children and adolescents. Journal of the American Academy of Child and Adolescent Psychiatry. 1997;36(1):37–44. doi: 10.1097/00004583-199701000-00015. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Mizugishi K, Ishiguro A, Koyabu Y, Imai Y, Takahashi R, Mikoshiba K, Aruga J. Rines/RNF180, a novel RING finger gene-encoded product, is a membrane-bound ubiquitin ligase. Genes Cells. 2008;13(4):397–409. doi: 10.1111/j.1365-2443.2008.01169.x. [DOI] [PubMed] [Google Scholar]
- Ohlmeier MD, Peters K, Kordon A, Seifert J, Wildt BT, Wiese B, Ziegenbein M, Emrich HM, Schneider U. Nicotine and alcohol dependence in patients with comorbid attention-deficit/hyperactivity disorder (ADHD) Alcohol and alcoholism (Oxford, Oxfordshire) 2007;42(6):539–543. doi: 10.1093/alcalc/agm069. [DOI] [PubMed] [Google Scholar]
- Plafker SM, Macara IG. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. The EMBO journal. 2000;19(20):5502–5513. doi: 10.1093/emboj/19.20.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerleau OF, Downey KK, Stelson FW, Pomerleau CS. Cigarette smoking in adult patients diagnosed with attention deficit hyperactivity disorder. Journal of substance abuse. 1995;7(3):373–378. doi: 10.1016/0899-3289(95)90030-6. [DOI] [PubMed] [Google Scholar]
- Potter AS, Newhouse PA, Bucci DJ. Central nicotinic cholinergic systems: a role in the cognitive dysfunction in attention-deficit/hyperactivity disorder? Behavioural brain research. 2006;175(2):201–211. doi: 10.1016/j.bbr.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds GP, Arranz B, Templeman LA, Fertuzinhos S, San L. Effect of 5-HT1A receptor gene polymorphism on negative and depressive symptom response to antipsychotic treatment of drug-naive psychotic patients. The American journal of psychiatry. 2006;163(10):1826–1829. doi: 10.1176/ajp.2006.163.10.1826. [DOI] [PubMed] [Google Scholar]
- Rondou P, Haegeman G, Vanhoenacker P, Van Craenenbroeck K. BTB Protein KLHL12 targets the dopamine D4 receptor for ubiquitination by a Cul3-based E3 ligase. The Journal of biological chemistry. 2008;283(17):11083–11096. doi: 10.1074/jbc.M708473200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, St Laurent G, 3rd, Wahlestedt C. The Emerging Role of Non-Coding RNAs in Drug Addiction. Frontiers in genetics. 2012;3:106. doi: 10.3389/fgene.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A, Kirsch P, Reuter M, Alexander N, Kozyra E, Kuepper Y, Osinsky R, Hennig J. The 5-HT1A C(-1019)G polymorphism, personality and electrodermal reactivity in a reward/punishment paradigm. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2009;12(3):383–392. doi: 10.1017/S1461145708009401. [DOI] [PubMed] [Google Scholar]
- Serretti A, Artioli P, Lorenzi C, Pirovano A, Tubazio V, Zanardi R. The C(-1019)G polymorphism of the 5-HT1A gene promoter and antidepressant response in mood disorders: preliminary findings. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2004;7(4):453–460. doi: 10.1017/S1461145704004687. [DOI] [PubMed] [Google Scholar]
- Sim S, Wolin SL. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley interdisciplinary reviews RNA. 2011;2(5):686–699. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone L, Straubinger M, Khan MA, Nalleweg N, Cheusova T, Hashemolhosseini S. Identification of Erbin interlinking MuSK and ErbB2 and its impact on acetylcholine receptor aggregation at the neuromuscular junction. J Neurosci. 2010;30(19):6620–6634. doi: 10.1523/JNEUROSCI.5778-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel A, Gutknecht L, Rothe C, Reif A, Mossner R, Zeng Y, Brocke B, Lesch KP. Allelic variation in 5-HT1A receptor expression is associated with anxiety- and depression-related personality traits. J Neural Transm. 2003;110(12):1445–1453. doi: 10.1007/s00702-003-0072-0. [DOI] [PubMed] [Google Scholar]
- Sullivan MA, Rudnik-Levin F. Attention deficit/hyperactivity disorder and substance abuse. Diagnostic and therapeutic considerations. Annals of the New York Academy of Sciences. 2001;931:251–270. doi: 10.1111/j.1749-6632.2001.tb05783.x. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Sawamura K, Someya T. The effects of a 5-hydroxytryptamine 1A receptor gene polymorphism on the clinical response to fluvoxamine in depressed patients. The pharmacogenomics journal. 2004;4(4):283–286. doi: 10.1038/sj.tpj.6500256. [DOI] [PubMed] [Google Scholar]
- Villafuerte SM, Vallabhaneni K, Sliwerska E, McMahon FJ, Young EA, Burmeister M. SSRI response in depression may be influenced by SNPs in HTR1B and HTR1A. Psychiatric genetics. 2009;19(6):281–291. doi: 10.1097/YPG.0b013e32832a506e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilens TE. Attention-deficit/hyperactivity disorder and the substance use disorders: the nature of the relationship, subtypes at risk, and treatment issues. The Psychiatric clinics of North America. 2004;27(2):283–301. doi: 10.1016/S0193-953X(03)00113-8. [DOI] [PubMed] [Google Scholar]
- Wilens TE, Decker MW. Neuronal nicotinic receptor agonists for the treatment of attention-deficit/hyperactivity disorder: focus on cognition. Biochemical pharmacology. 2007;74(8):1212–1223. doi: 10.1016/j.bcp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojnar M, Brower KJ, Jakubczyk A, Zmigrodzka I, Burmeister M, Matsumoto H, Wozny E, Sliwerska E, Hegedus AM, Husar A, Slufarska A, Lipinski M, Zucker RA. [Influence of impulsivity, suicidality and serotonin genes on treatment outcomes in alcohol dependence] Psychiatria polska. 2006;40(5):985–994. [PubMed] [Google Scholar]
- Yevtushenko OO, Oros MM, Reynolds GP. Early response to selective serotonin reuptake inhibitors in panic disorder is associated with a functional 5-HT1A receptor gene polymorphism. Journal of affective disorders. 2010;123(1-3):308–311. doi: 10.1016/j.jad.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Yu YW, Tsai SJ, Liou YJ, Hong CJ, Chen TJ. Association study of two serotonin 1A receptor gene polymorphisms and fluoxetine treatment response in Chinese major depressive disorders. Eur Neuropsychopharmacol. 2006;16(7):498–503. doi: 10.1016/j.euroneuro.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CR, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology. 2012;37(2):557–566. doi: 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CSR, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine co-dependence. Alcoholism, clinical and experimental research. 2013a;37(5):730–739. doi: 10.1111/acer.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang H, Malison RT, Li CSR, Zhang XY, Wang F, Lu L, Lu L, Wang X, Krystal JH, Zhang F, Deng H, Luo X. Rare ADH variant constellations are specific for alcohol dependence. Alcohol and alcoholism. 2013b;48:9–14. doi: 10.1093/alcalc/ags104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.