Abstract
The progressive loss of neurons causes neurodegenerative diseases. Because the accumulation of DNA breaks results in neuronal apoptosis, the lack of a variety of DNA damage repair-related proteins contributes to neurodegeneration. The ubiquitin ligase RNF8 plays an important role in DNA double-strand break repair via histone ubiquitination. However, the function of RNF8 in terminally differentiated neurons remains unknown. This study aimed to determine whether RNF8 is involved in the DNA damage response in neurons and contributes to neurodegeneration. Here, we present evidence suggesting that RNF8 deficiency results in DNA damage accumulation and neuronal apoptosis. RNF8−/−mice exhibit neuronal degeneration and reactive astrocytosis. Neurons from RNF8−/− mice appear to be more susceptible to X-ray-induced DNA damage. These changes were consistent with the behavioral performances of the RNF8-deficient mice, which included impaired performances in the open-field test and step-down avoidance task. Overall, these findings show that RNF8 is required for DNA damage repair in neurons. RNF8 deficiency is sufficient to cause neuronal pathology and cognitive decline, and the loss of RNF8 results in neuron degeneration.
Keywords: ubiquitin ligase, RNF8, DNA damage repair, double-strand break, neurodegeneration
1. Introduction
Mammalian cells constantly experience DNA damage resulting from exogenous and endogenous damage-inducing elements. Each cell in the human body receives tens of thousands of DNA lesions per day[1]. Because of their toxicity and the difficulty in repairing these lesions, DNA double-strand breaks (DSBs) are the most deleterious lesions caused by DNA-damaging agents[1, 2]. In response to DSBs, hundreds of proteins are involved in an elaborate network that signals and repairs the DNA damage[3, 4]. The DNA damage response (DDR) is involved in the surveillance and repair of DNA damage to maintain genome stability[1, 5].
DSB repair defects lead to genomic instability, apoptosis, and cell death. Moreover, the loss of genomic integrity promotes cancer, neurodegenerative disease and immunodeficiency[6-8]. Mature neurons are post-mitotic cells; therefore, these neurons cannot be replenished after severe damage. To ensure the longevity and functionality of neurons that may encounter DNA damaging threats, neurons possess elaborate mechanisms to preserve their genetic integrity[6]. Defects in DSB repair have been detected in the cells of several human neurodegenerative syndromes, such as ataxia-telangiectasia (A-T)[9].
The protein ubiquitination cascade induced by DSBs has emerged as a crucial posttranslational modification that activates cell cycle checkpoints and DNA repair to maintain the genomic integrity[2, 10-16]. One important component of DSB repair is the E3 ubiquitin ligase RNF8, which is a 485-residue nuclear polypeptide with an N-terminal forkhead-associated (FHA) domain and a C-terminal really interesting new gene (RING) domain[1]. Histone H2AX phosphorylation (γH2AX) is among the initial DSB-induced histone modifications and is a marker of DNA damage. In response to DSBs, RNF8 interacts with phosphorylated MDC1 bound to γ-H2AX, and this complex is rapidly recruited to DNA damage sites through the FHA domain of RNF8. The RING domain of RNF8 interacts with the E2 ubiquitin-conjugating enzyme Ubc13 to ubiquitinate histones, catalyze the ubiquitination cascade, and recruit 53BP1 and BRCA1 to the DNA lesions[17]. Our recent studies and others have demonstrated that RNF8 deficiency may induce male infertility, class switching and meiotic defects in mice[13, 17]. However, in terminally differentiated neurons, whether RNF8 plays important roles in the DSB response and whether RNF8 deficiency in mice results in neuronal loss remain unknown. We investigated the role of RNF8 in neuronal DSBs and the effect of RNF8 deficiency on neuronal survival in RNF8-deficient mice.
2. Materials and Methods
Animals and X-ray Irradiation
The RNF8-deficient mice were provided by Xiaochun Yu and have been previously described[18]. The gene trap embryonic stem (ES) cell clone RRR260 was used to generate the RNF8-deficient mice. RNF8+/− mice were intercrossed to generate RNF8−/− mice. In total, 60 RNF8−/−mice were randomly divided into 2 groups (RNF8−/− and RNF8−/−IR [irradiated]). The homologous WT littermates were used as controls and were designated as WT and WT IR accordingly. Each group consisted of 30 mice. To minimize the number of animals, 24 mice in each group were used for the morphological and behavioral experiments, and 6 mice were used for Western blot analysis and the comet assay. Three-month-old WT IR and RNF8−/− IR mice were treated with 2 Gy X-ray irradiation using a 130 kV X-ray machine (Faxitron RX-650, Tucson, AZ, USA) at a dose rate of 50 cGy/min. These mice were used for Cresyl Violet (CV) staining and behavioral experiments. All animals were maintained in the SPF laboratory at the Lanzhou University Animal Center. The animals were housed under a 12:12 h light:dark cycle at a constant temperature in a pathogen-free environment and were given free access to food and water. The experimental protocols were approved by the ethics committee of Lanzhou University.
Frozen Coronary Sections
After IR (2 Gy) treatment and further recovery for relevant amounts of time, mice were sacrificed and then perfused and fixed in 4% paraformaldehyde. The brains were dehydrated with 4% paraformaldehyde containing 30% sucrose. Frozen coronary sections (20 μm) were generated using freezing microtome (Leica CM1850, Germany).
Neuron Isolation
Neuron isolation assays were performed as described by Lee Y et al. [19] with some minor modification. Briefly, the neurons were purified from the 6-month-old cerebral cortex and hippocampus. The neurons were treated with 2-Gy IR and then subjected to Western blot analysis and comet assay.
Immunostaining
For immunostaining, sections from 6-month-old mice were stained for RNF8, MDC1, 53BP1, BRCA1 and NeuN. Sections were boiled in 0.01 M sodium citrate (pH 6.0) for 10 min to retrieve the antigens. Standard immunostaining procedures were used. The following antibodies were used: anti-γH2AX [phosphor S139] (ab26350, Abcam, 1:400 dilution), rabbit anti-MDC1 (ABC155, Millipore, 1:500 dilution), anti-53BP1 (ab36823, Abcam), anti BRCA1 (H-100) (sc-7867, Santa Cruz, 1:200 dilution), and anti-NeuN (clone A60, MAB377, Millipore, 1:500 dilution). For secondary detection, goat anti-mouse IgG-FITC (sc-2010, Santa Cruz, 1:400 dilution) and goat anti-rabbit IgG-Cy3 (A22220, abbkine, 1:400 dilution) were used. Then, the neuronal nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI; D9542, Sigma). After a final wash with PBS, sections were mounted in anti-fading medium (glycerin containing p-phenylenediamine). All images were obtained with an Olympus fluorescence microscope (BX53).
Western Blot Analysis
For Western blot analyses, histone protein extraction assays were performed as described [18]. The protein concentrations were quantified using the Bradford assay. The histone proteins were subjected to electrophoresis and Western blotting and were then probed with the following antibodies: anti-ubiquityl-histone H2A [clone E6C5] (05-678), anti-ubiquityl-histone H2B [clone 56] (05-1312), anti-H2A (ABE327), anti-H2B (AB1623), and anti-H4 (07-108) (Millipore, Milford, MA, USA).
Neutral Single-Cell Gel Electrophoresis (Comet Assay)
The neurons were isolated from WT and RNF8−/− mice. After X-ray IR (2 Gy) treatment and a recovery period of 2 or 24 h, the cell suspension was mixed with 0.6% low melting point agarose and was then spread onto a microscope slide that had been treated with normal melting point agarose. The slide was incubated at 4 °C for 15 min. Then, the coverslip was gently removed, and the slide was submerged in lysis solution (2.5 M NaCl, 100 mM EDTA, 10 mM Tris-HCl, 1% sodium lauroyl sarcosine, 10% dimethyl sulfoxide, pH 10.0, and 1% Triton X-100, which was added just before use) overnight at 4°C. Next, the slide was placed in electrophoresis solution (300 mM NaOH and 1 mM EDTA, pH>13) for 20 min to allow for DNA unwinding, and electrophoresis was conducted for 20 min at 25V and 300 mA. After electrophoresis, the slide was drained, neutralized with neutralization buffer (0.4 M Tris-HCl, pH 7.5) and stained with ethidium bromide (EB, 5 μg/ml). The slide was covered with a coverslip and analyzed under an Olympus fluorescence microscope (BX53). Images of 50 nuclei were randomly selected for analysis with Casp1.2.3 software (Institute of Theoretical Physics, University of Wroclaw, Wroclaw, Poland).
Immunohistochemistry
Sections from six-month-old mice were incubated with 3% H2O2 for 30 min at room temperature. The following antibodies were used for immunohistochemistry: anti-GFAP (AB5804, Millipore, 1:400 dilution); anti-caspase-3 [cleaved Asp175] (ab52293, Abcam, 1:400 dilution); goat anti-rabbit IgG (H&L); biotin-conjugated, affinity-purified antibody (AP132B, Millipore, 1:500 dilution); and peroxidase-conjugated streptavidin (SA202, Millipore, 1:500 dilution). A standard avidin-biotin-immunoperoxidase complex method was used.
Fluoro-Jade C and TUNEL Assay
Slices from six-month-old mice were immersed in an 80% alcohol solution that contained 1% NaOH for 5 min, in 70% ethanol for 2 min and in distilled water for 2 min. Next, the slices were immersed in 0.06% potassium permanganate solution for 10 min on a shaker to suppress the background noise. The slices were then washed with distilled water for 2 min. Next, the slices were stained in dye liquor (Fluoro-Jade C, Chemicon, Millipore) and washed three times with distilled water for 1 min. The slices were completely dried and then immersed in xylene for at least 1 min and covered with neutral gum. An apoptosis analysis was performed using the DeadEnd™ Fluorometric terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) System (Promega).
CV staining
The slices from 6-, 9- and 12-month-old mice were fixed on glass slides coated with 2% gelatin and then dried in the oven (50°C) for at least half an hour. Slices were immersed into 75%, 85%, 95%, 100% and 100% alcohol for 8 minutes each to dehydrate and were then rehydrated by being immersed into 95%, 85%, 75% and 50% alcohol for 3 minutes each. Slices were stained with CV for 20 minutes. Next, slices were immersed into 85%, 95%, 100% and 100% alcohol for 30 seconds separately each. The slides were immersed into xylene and 100% alcohol solution at a ratio of 1:10 (v/v). Then, the slides were then immersed into xylene two times for 10 minutes and covered with neutral gum.
Silver staining
The slides from 12-month-old mice were placed in 20% silver nitrate under dark conditions at 20 °C for 2 h. The sections were placed in 10% formaldehyde for 5 min until the sections became light brown. Next, the slides were placed in ammonium silver solution and stained at 37 °C for 5 min. The slides were placed directly in 8% formaldehyde until the sections became dark brown (approximately 1 min or less). After washing three times, the sections were toned in 0.2% gold chloride for 5 min and then washed three times. Finally, the slides were placed in 5% sodium thiosulfate for 1 min and washed three times. Dehydrating and clearing were promoted using 95% ethyl alcohol, absolute alcohol and xylene, and the slides were covered with neutral gum.
Behavioral tests
For the open-field test, the mice were placed in a square open-field box (625 mm [L]×740 mm [W]×510 mm [H]) equipped with an infrared sensor (TM-Vision behavioral assay system, OFT-100) and attached to a computer to record the following parameters: the total distance, center distance, total move time, center time, corner time and side time. Each test session lasted 30 min, and the data were collected at 5 min intervals.
For the step-down avoidance test, a YLS-3TB instrument with five boxes (120×120×180 mm for each box) was used and attached to a computer to record the latency and error times. After training for 3 min, the mice were placed on a platform. During the training sessions, the mice received a 0.10-mA, 36-V foot shock for 2 s immediately upon stepping down. Each test session lasted 8 min, and the data were collected at 5-min intervals.
Statistical analysis
One-way analysis of variance (ANOVA) with the Student-Newman-Keuls test was used for the statistical analyses. The means and standard deviations were plotted. A P<0.05 was considered significant.
3. Results
RNF8 participates in neuronal DSB repair
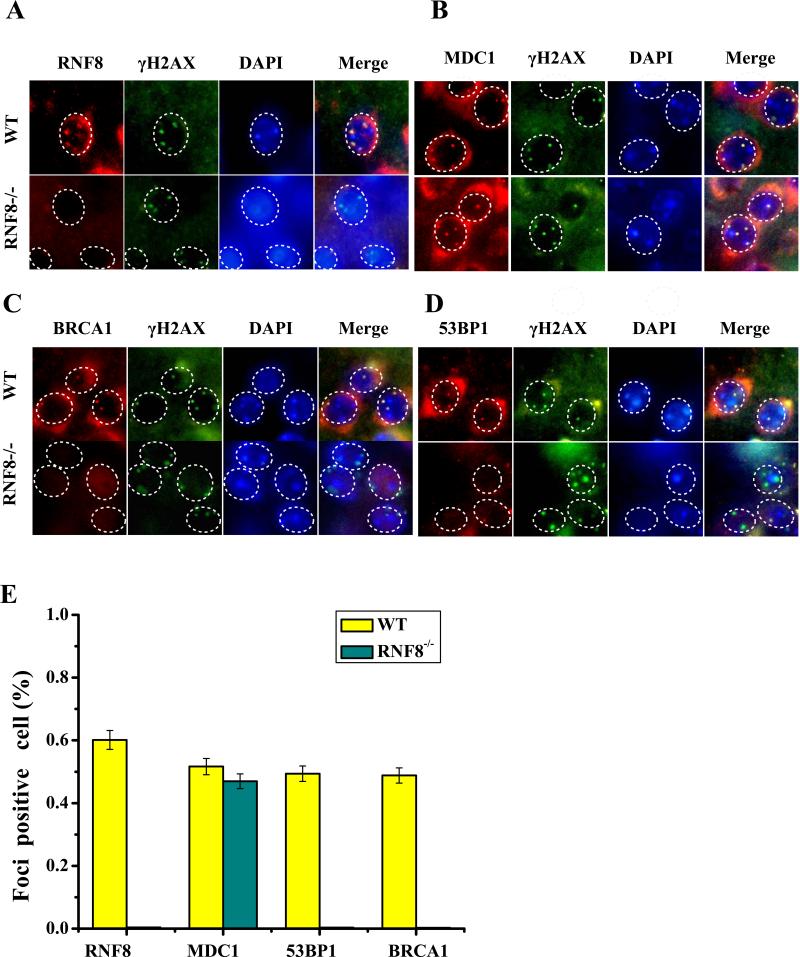
The RNF8−/− mice appeared to have a slightly lower body weight compared with their WT littermates (Figure S2B, S2D). The RNF8−/− mice also demonstrated less and slower spontaneous locomotor activity. To determine the roles of RNF8 in neuronal integrity and survival, we examined the DNA DSB repair pathway in neurons from RNF8-deficient mice (Figure 1) and in HT22 cells, which are an immortalized mouse hippocampal neuronal cell line. If RNF8 is involved in protein ubiquitination at the DNA damage sites in neurons, then RNF8 deficiency may abrogate the ionizing radiation-induced focus (IRIF) of the downstream DDR elements at the DNA damage site. We examined the IRIF formation of γ-H2AX, RNF8, MDC1, 53BP1 and BRCA1 after X-ray irradiation (2 Gy). As shown in Figure 1A, RNF8 IRIF was not observed in the neurons from RNF8−/− mice. MDC1 (Figure 1B) foci overlapping with the DNA damage marker γ-H2AX were observed after exposure to IR (2 Gy) in neurons from WT and RNF8−/− mice. In contrast, 53BP1 and BRCA1, which act downstream of RNF8, were not observed in the neurons from RNF8−/− mice (Figure 1C). siRNF8-treated HT22 cells demonstrated results that were similar to those observed due to RNF8 deficiency, such as impaired BRCA1 and 53BP1 IRIF formation, but the MDC1 and γ-H2AX IRIF formation remained intact (Figure S3). These results support the hypothesis that RNF8 participates in DSB repair in neurons.
Figure 1. RNF8 is required for DNA damage repair.
The WT mice (RNF8+/+) and RNF8 knockout mice (RNF8−/−) were divided into groups with or without IR treatment. In the IR-treated mice, DNA damage was induced by X-ray at 3 months of age, and the mice were allowed to recover for 6 h. Transverse brain slices were processed for immunofluorescence staining. Sections were stained with polyclonal anti-RNF8, MDC1, 53BP1 and BRCA1 antibodies and with monoclonal γ-H2AX antibody. (A) RNF8 can form clear foci at DNA damage sites in WT mice but failed to form foci in the neurons from RNF8−/− mice (B, C). (E) The percentages of foci-positive neurons in panels A, B, C, and D are summarized in the histogram (means±SD). RNF8 deficiency impaired BRCA1 and 53BP1 foci formation, whereas the MDC1 and γ-H2AX IRIF formation remained intact.
RNF8 is involved in the neuronal DDR via the ubiquitination of H2A and H2B
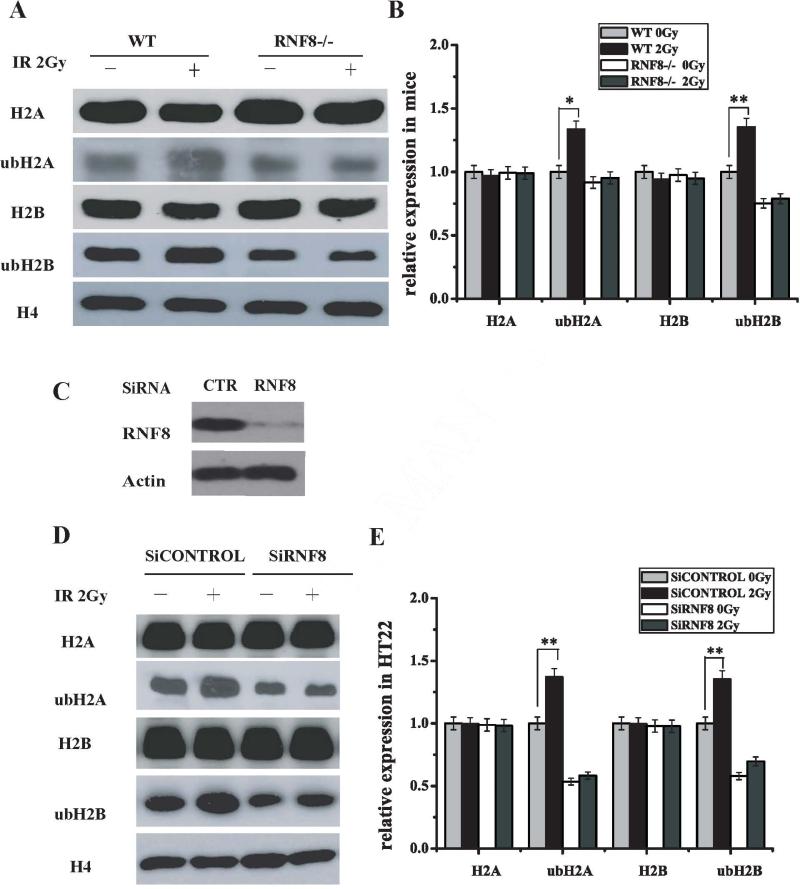
To verify the substrate of RNF8 as an ubiquitin ligase during the DDR in neurons, we performed Western blot analysis to evaluate the capacity of RNF8 to catalyze histone ubiquitination in neurons. Because H2A and H2B ubiquitination is a downstream event of initial IR-induced protein ubiquitination catalyzed by the RNF8/UBC13 complex, ubiquitination of H2A, H2AX, and H2B is regulated by RNF8 at DNA damage sites. The ubH2A and ubH2B levels decreased in the neurons from the brain tissues of the RNF8-deficient mice compared with those from the WT mice (Figure 2A and B). IR-induced increases in the H2A and H2B ubiquitination levels were observed in the neurons from WT mice; however, IR-induced increases in ubH2A and ubH2B were not observed in neurons from RNF8-deficient mice (Figure 2A and B). The same results were observed in the RNF8 siRNA-treated HT22 cells (Figure 2D and E). Meanwhile, as shown in Figure 2A, B, D and E, the H2A and H2B levels in the neurons from WT and RNF8-deficient mice (treated or not with 2-Gy IR) did not display obvious changes. These results suggest that RNF8 is involved in the neuronal DDR via H2A and H2B ubiquitination.
Figure 2. RNF8 participates in H2A and H2B ubiquitination in neurons.
(A) The H2A, ubH2A, H2B and ubH2B expression levels in neurons from WT and RNF8−/− mice with or without 2-Gy IR treatment were detected using Western blot analysis. Neurons were harvested 6 h post-IR. The histones were prepared, separated by SDS-PAGE, and blotted with ubH2A, ubH2B and H4 antibodies. (B) The relative expression levels of H2A, ubH2A, H2B and ubH2B in Figure A are summarized in the histogram (means±SD). Error bars indicate standard deviations. (C) HT22 cells were transiently transfected with RNF8 siRNA. Immunoblotting was performed using anti-RNF8 and β-actin antibodies. (D) HT22 cells were transfected with control siRNA or RNF8 siRNA and subjected to IR treatment. The H2A, ubH2A, H2B and ubH2B expression levels were detected by Western blot analysis. (E) Bar graphs showing the density of the blots from Figure D. ** represents P<0.01 vs WT 0 Gy.
RNF8 deficiency results in DNA damage accumulation and neuron apoptosis
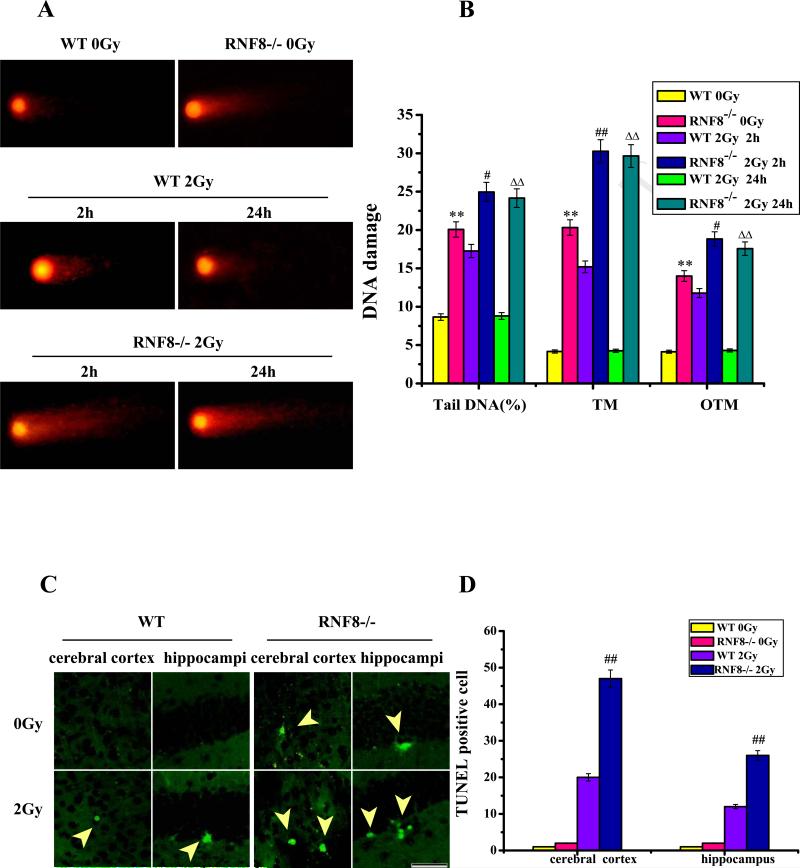
We utilized the comet assay (single-cell gel electrophoresis) to assess the DNA damage. If DNA head (DNAH) is the sum of intensities of all points of the head, and DNA tail (DNAT) is the sum of intensities of all points of the tail, then percent tail DNA (%DNAT), and %DNAT = 100DNAT/(DNAH + DNAT). If tail moment (TM) is the product of the tail length and %DNAT, then TM = TL × (%DNAT). If olive tail moment (OTM) is the product of the distance (in × direction) between the center of gravity of the head (CGH) and the center of gravity of the tail (CGT) and %DNAT, then OTM = (CGTx − CGHx) /%DNAT. Based on the results of the neutral comet assay, we detected more damage in the 6-month-old RNF8−/− mice compared with the WT littermates. The %DNAT, TM and OTM values were higher in the neurons from the RNF8−/− mice and the WT mice 2 h after 2-Gy IR compared with the control mice (Figure 3A and B). Sharp increases in these parameters were observed in the RNF8−/− mice 2 and 24 h after 2-Gy IR, whereas there were no significant increases in the WT littermates after 24 h IR treatment (Figure 3A and B). These data highlight the importance of RNF8 and its role in neuronal DSB repair. These results were also consistent with the results from the TUNEL assay. IR treatment induced a greater number of TUNEL-positive cells in the brain tissues from the RNF8−/− mice compared with those from the WT littermates (Figure 3D and E).
Figure 3. DNA damage and apoptosis assay in neurons after IR treatment (2 Gy or 0 Gy).
(A) Plot of the comet assay for neurons from the hippocampi of WT and RNF8−/− mice 2 and 24 h after IR treatment. (B) The amounts of DNA damage were measured as the means of the %DNAT, TM and OTM. Error bars indicate standard deviations. (C) Plot of the TUNEL staining of cells from 6-month-old WT and RNF8−/− mice with or without 2-Gy IR treatment. Scale bar=50 μm. (D) Bar graph showing the TUNEL staining in the cerebral cortices and hippocampi of WT and RNF8−/− mice. The values are the means±SD of 6 mice, and per mouse is based on the analysis of 6 sections. Error bars indicate standard deviations. ** represents P<0.01 vs WT 0Gy. ## represents P<0.01 vs WT 2 Gy 2 h. △△ represents P<0.01 vs WT 2 Gy 24 h.
RNF8−/−mice display neuronal degeneration and reactive astrocytosis
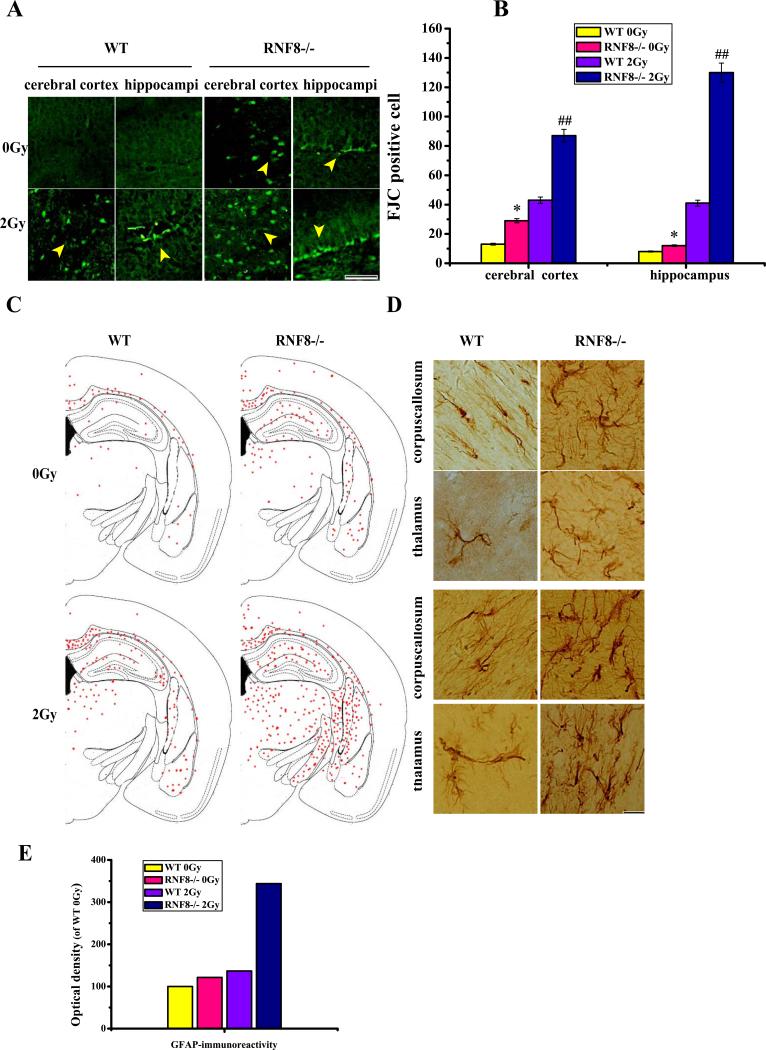
To further evaluate the degenerating neurons, we stained for Fluoro-Jade C (FJC). FJC selectively stains degenerating neurons and may be used as a biomarker of neurodegeneration in brain tissues. Neuronal degeneration was observed in the brain tissues of the RNF8−/− mice, and treatment with 2-Gy IR resulted in a dramatic increase in neuron degeneration in the RNF8−/− mice. In contrast, the WT mice barely exhibited any neuronal degeneration, and the WT mice treated with 2-Gy IR exhibited only sporadic degenerative neurons (Figure 4A and B). The RNF8 deficiency resulted in DDR failure; therefore, the DDR pathways may be insufficient for coping with these lesions and resulting in neurodegeneration. The upregulation in glial fibrillary acidic protein (GFAP) and the highly branched GFAP-positive cells are indicative of reactive astrocytes, which are known to be markers of neuronal injury and degeneration. Consistent with the FJC staining, the brain tissues from the RNF8−/− mice exhibited increased GFAP immunostaining compared with those from the WT mice. The 2-Gy IR treatment induced a dramatic increase in the GFAP staining in the brains of RNF8−/− mice and induced only a slight increase in the brains of WT mice (Figure 4C and D). These results indicate that the IR treatment results in greater activation of astrocytosis in the RNF8−/− mice.
Figure 4. Neurodegenerative changes in RNF8 deficient mice.
(A) Plot of FJC staining in cells from 6-month-old WT and RNF8−/− mice before and after 2 Gy IR treatment. Scale bar=50 μm. (B) Bar graph showing the FJC-positive cells in the cerebral cortices and hippocampi of WT and RNF8−/− mice. The values are the means±SD of 6 mice, and per mouse is based on the analysis of 6 sections. ** represents P<0.01 vs WT 0 Gy. ## represents P<0.01 vs WT 2 Gy. (C) Brain slices processed for GFAP immunohistochemistry exhibited the normal gross histoarchitecture of the dorsal hippocampi and surrounding brain structures in 6-month-old WT mice and had increased GFAP staining throughout the brain tissues of RNF8−/− mice. GFAP staining in the hippocampus and thalamus was also increased. (D) Plot of GFAP-positive cells in selected transverse brain sections of WT and RNF8−/− mice, illustrating the widespread distribution of activated astrocytosis. Scale bar=20 μm. (E) Bar graphs showing the optical density of WT 0 Gy for GFAP.
RNF8−/−mice exhibit neuronal loss in the brain
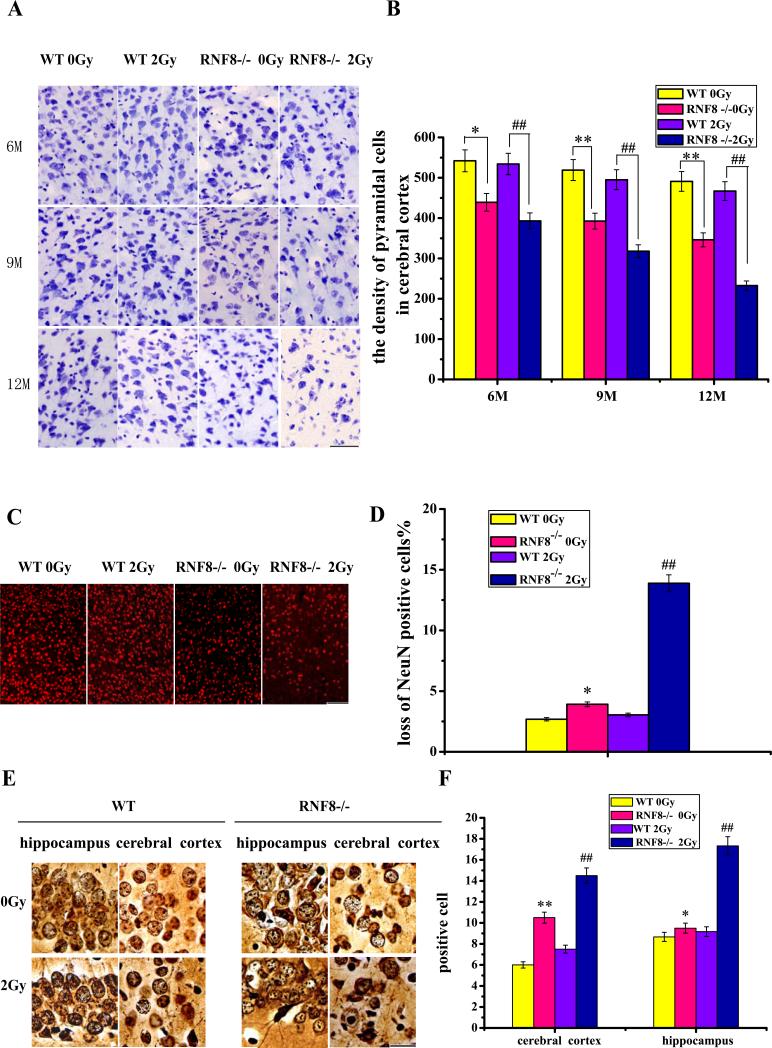
Analysis of CV-stained sections revealed a prominent decrease in the number of pyramidal cells in the cerebral cortices of RNF8−/− mice compared with the WT mice from 6 to 12 months. The 2-Gy IR-treated RNF8−/− mice also exhibited a dramatic decrease in the number of pyramidal cells in the cerebral cortex compared with the RNF8−/− mice without IR. However, in the WT mice, there was no obvious difference between IR-treated mice and the mice without IR treatment (Figure 5A and B). After IR treatment, increased numbers of sporadic cells with pyknotic nuclei, karyopyknosis, vacuoles, nuclear concentrations in the margin and perineural phenomena were observed in the cortices and hippocampi of the RNF8−/− mice (Figure S4A), indicating that the neurons underwent cell death. The number of NeuN-positive neurons significantly decreased in the cortices of RNF8−/− mice compared with the WT mice (Figure 5C). The IR treatment induced a loss of cortex neurons in the RNF8−/− mice, whereas the treatment did not induce neuron loss in the WT mice. Approximately 13.88% of the neurons were lost in the IR-treated RNF8−/− mice compared with only a 3.03% loss in the IR-treated WT mice. These results suggest that the absence of RNF8 results in neuron loss in mice and that the RNF8−/− mice are more vulnerable to DNA damage.
Figure 5. Neuron lost in the RNF8-deficient mice.
(A) Plot of pyramidal cells in the cerebral cortices of 6-, 9- and 12-month-old WT and RNF8−/− mice. Transverse brain slices stained with CV. Scale bar, 50 μm. (B) Bar graph shows the density of pyramidal cells in the cerebral cortices of WT and RNF8−/− mice with or without 2-Gy IR treatment. (C) Sections from 12-month-old WT and RNF8−/− mice were stained with NeuN to quantify the neuronal density. Scale bar, 100 μm. (D) The percentages of neuronal loss in the cortices of 12-month-old WT and RNF8−/− mice were calculated. (E) Plot of the silver-stained cells in the cerebral cortex and hippocampus. Scale bar, 20 μm. (F) Bar graph showing the density of abnormal cells in the cerebral cortex and hippocampus. The values are the means±SD of 6 mice. ** represents P<0.01 vs WT 0 Gy. ## represents P<0.01 vs WT 2 Gy. * represents P<0.05 vs WT 0 Gy.
Direct evidence for neuronal degeneration was obtained by silver staining. Viable neurons were light brown with pale nuclei containing dark nucleoli, whereas the degenerated neurons exhibited dark staining of nuclei and irregularly shaped cells. The neurons and the nerve fibers analyzed via silver staining appeared normal in the WT mice. The RNF8−/− mice exhibited sporadic degenerated neurons with dark and irregularly shaped cells. After IR treatment, increases in the amount of degenerated neurons were observed in the cortices and hippocampi of the RNF8−/− mice, and the axons were spirally twisted, swollen and even ruptured in the RNF8−/− mice (Figure S4B d, e, and f). Some argyrophilic plaque-like deposits were observed in the RNF8−/− mice; these deposits require further identification (Figure S4B a).
RNF8−/− mice display memory impairment and reduced exploratory behavior in the open-field test
We used an open-field exploratory test to analyze the spontaneous locomotor activity of the mice. Three-month-old mice were treated with IR (0 Gy for controls and 2 Gy for the treatment groups). Behavior tests were performed 2, 4 and 6 months after IR treatment. The horizontal activity of the RNF8−/− mice decreased significantly. The IR-treated RNF8−/− mice also demonstrated decreased activity compared with the RNF8−/− mice without IR treatment. The WT mice with 2-Gy IR treatment demonstrated no obvious difference in activity compared with that of the WT mice without IR treatment. Additionally, step-down avoidance tests were performed to measure the memory retention of foot shock. The incidence of errors increased in the RNF8−/− mice compared with that in the WT mice at 6 months of age. The 2-Gy IR treatment on 3-month-old RNF8−/−mice increased the incidence of errors compared with that of the RNF8−/− mice without IR treatment (3 months after IR, 6 months of age), P<0.05; 6 months after IR, P<0.01). The step-down latency (SDL) decreased in the 2-Gy IR-treated RNF8−/− mice compared with the RNF8−/− mice without IR treatment (P<0.05 at 3 months after IR [6 months of age] and P<0.01 at 6 months after IR [9 months of age]). Regarding the WT groups, the 2-Gy IR-treated WT mice demonstrated no difference in the incidence of errors or the SDL compared with the WT mice without IR treatment (Figure 6E). These data suggest that RNF8, which is one of the DSB repair factors, maintains brain function and that RNF8 deficiency causes deterioration in short-term memory and reduces the spontaneous locomotor activity in mice.
Figure 6. RNF8−/− mice demonstrate memory impairment and reduced exploratory behavior in the open-field test.
(A) Representative example of open-field test plots. (B) Quantification of the total times the mice stayed in the center of the field, demonstrating the reduced ambulatory behavior of the RNF8−/− mice at relevant times before and after IR treatment in the center of the field. *represents P<0.05. (D) Bar graph illustrating the incidence of errors of WT and RNF8−/− mice with or without IR treatment (2 Gy) at 3 and 6 months after treatment. The data represent the means±SD. One-way ANOVA revealed a significant effect on the performance of RNF8−/− mice 3 and 6 months after IR treatment (P<0.05 and P<0.01, respectively). The incidence of errors of WT mice with IR treatment did not differ from that of the no-IR treatment controls.
4. Discussion
Appropriate signaling responses to DNA damage in neurons are required for homeostasis maintenance and organism survival. DNA DSBs induced by exogenous and endogenous agents initiate a signaling cascade to repair this damage. During embryonic development, embryonic cells sequentially undergo division, migration and differentiation[20]; however, an important distinction exists between developing and mature neurons. During the early proliferative phase, homologous recombination (HR) is required to repair transcription- and replication-associated DNA breaks and naturally occurring oxidative lesions[21]. HR deficiencies in repairing these breaks may lead to the accumulation of damage and cell death, resulting in embryo lethality[22]. As shown in Figure S2A, the percentage of viable RNF8-deficient mice was only 10.8%, and several stillbirths were observed among the neonatal mice (Figure S2C). During differentiation and migration, however, the non-homologous end-joining (NHEJ) pathway, which involves repair during all cell cycle phases, plays a more prominent role in DSB repair in mammals. NHEJ pathway deficiencies may result in the loss of neurons[21]. The NHEJ pathway functions in both proliferating and non-proliferating cells, whereas HR only functions in proliferating cells[23]. Therefore, NHEJ repair may be responsible for IR-induced neuronal DSBs. Failure to repair DSBs will frequently lead to a loss of genetic material, and NHEJ is thought to be the major mechanism of repair during the first 4-6 h following IR treatment and repairs ~80-85% of the induced DNA DSBs[24, 25].
Because of the complexity and heterogeneity of the nervous system, the challenges presented in this study involve biological changes to the DNA DSBs in neurons. We explored the DNA damage pathways in neurons using RNF8-deficient mice. Based on the neutral single-cell gel electrophoresis results, the neurons in RNF8−/− mice failed to repair the DSBs post-IR treatment (2 Gy). Generally, the alkaline assay detects both SSBs and DSBs, whereas the neutral assay is more specific for DSBs[26]. Cells with DNA damage demonstrate increased migration of DNA fragments from the nucleus toward the anode during electrophoresis[27], and the intensity of the comet tail relative to the head reflects the number of DNA breaks[28]. During the 24-h recovery period, the WT mice may have repaired the DSBs induced by the IR treatment. In contrast, because of the deficiency in the repair mechanism, the %DNAT, TM and OTM increased significantly for the neurons of the IR-treated RNF8−/− mice compared with the untreated RNF8−/− mice and WT mice. Additionally, the neurons from the WT and RNF8−/− mice without IR treatment exhibited slight DSB damage, which may be due to the various types of applied damaging agents and endogenous agents.
In response to the DDR, MDC1 directly binds to the phosphorylated S139 of γ-H2AX through its BRCT domains[22]. The FHA and RING domain-containing protein RNF8 has been reported to participate in the DDR by acting downstream of H2AX and MDC1[13, 29]. MDC1 is recruited to the DNA damage lesion through the ATM-dependent phosphorylation of a cluster of S/T-Q residues, which provide binding sites for RNF8. RNF8 participates in the DNA damage response by interacting with MDC1, which is directly recruited to DNA lesions through its interaction with γ-H2AX. H2AX and MDC1 mediate the recruitment of 53BP1 to the IRIFs[13]. If RNF8 is involved in the ubiquitination of proteins at the damaged sites, then the RNF8 deficiency may abrogate the accumulation of the downstream proteins at IRIF. Next, RNF8 catalyzes histone ubiquitination and enables the DSB-flanking chromatin to gather other DSB regulators, such as 53BP1 and BRCA1[13, 24, 29-32]. As shown in Figure 1 and Figure S3, 53BP1 and BRCA1 foci formation were abrogated in siRNF8 and RNF8-deficient neurons. In contrast, MDC1 and γ-H2AX recruitment to the IR-induced foci were not affected. Therefore, 53BP1 and BRCA1 act downstream of RNF8, and MDC1 acts upstream of RNF8. The absence of RNF8 increases the sensitivity of the mice to ionizing radiation; after IR treatment, the survival rate of the RNF8-deficient mice declined, as shown in Figure S2E.
The FHA domain of RNF8 is required for its recruitment to DNA lesions through its interaction with MDC1; its activity as an E3 ubiquitin ligase, which promotes histone ubiquitination at DNA damage sites, is required to recruit downstream DNA damage response factors[12, 29]. At the DNA lesions, RNF8 ubiquitinates histones, facilitating the recruitment of downstream DNA damage response factors[16, 29]. Histone ubiquitination may directly affect the electrical charge of histone octamers and may promote their affinity to DNA[33]. The type of histone ubiquitination on histone H2A at lysine 119 and H2B at lysine 120 is primarily mono-ubiquitination. H2A ubiquitination is highly enriched at the DNA damage site[11, 13, 15, 29, 34, 35], and H2B ubiquitination is induced by DNA damage[36]. The RNF8 deficiency decreased the overall IR-induced ubiquitination of H2A and H2B, even in non-irradiated neurons and siRNF8-treated neurons. RNF8 is involved in the histone ubiquitinationin response to DSBs (Figure 2). Our results demonstrate that RNF8 participates in DSB repair in neurons via H2A and H2B ubiquitination. However, whether RNF8 deficiency leads to neurodegeneration remains unknown. Non-repaired or mis-repaired neuronal DSBs lead to various changes in neuropathology[37]. In the brains of RNF8-deficient mice, a variety of neuronal abnormalities were detected. Based on the FJC staining, the brains of the RNF8−/− mice exhibited sporadic neuron degeneration, and more sporadic neurons were detected in the 2-Gy IR-treated RNF8−/− mice. These results further demonstrated that the neurons of the RNF8−/− mice underwent degeneration and that exogenous DNA damage accelerated the process. Additionally, we found that failure to repair the DNA damage may lead to neuronal apoptosis in the brain. The 2-Gy IR-treated RNF8−/− mice exhibited an increased number of apoptotic cells (Figure 3C and D). GFAP is an astrocyte activation marker that increases in expression following neuronal injury and degeneration[38]. We demonstrated that GFAP is upregulated in response to DNA damage and that the number of astrocytes increases. Hypertrophy, soma swelling, increased protrusion and elongation were also observed. We also found that the IR-treated RNF8−/− mice had more reactive astrocytes (Figure 4C).These data suggest that impaired DSB repair resulting from RNF8 deletion induces neurodegeneration.
We observed the changes in the mouse nervous system via morphological analysis. Based on the CV staining, the IR-treated RNF8−/− mice exhibited a noticeable decrease in the number of pyramidal cells in the cerebral cortex. Furthermore, the RNF8−/− mice exhibited sporadic cells with pyknotic nuclei, karyopyknosis, vacuoles, nuclear concentrations in the margin and perineural phenomena in the cortex and hippocampus (Figure S4A). In addition to the changes in the soma, the neuronal fibers also exhibited corresponding changes. Based on the silver staining, abnormal argyrophilic fibers were found in the brains of RNF8−/− mice (Figure S4B d, e, and f). These results suggest that the neurons of the RNF8−/− mice experienced increased degeneration and neuronal loss after IR treatment.
Defects in DSB repair lead to complex neurology phenotypes; ATM deficiency causes progressive cerebellar atrophy, behavioral abnormalities and other neurological defects[39]. Our data indicate that RNF8 defects result in abnormal performances in behavioral tests. The horizontal activity of the RNF8−/− mice decreased significantly; IR-treated RNF8−/− mice demonstrated decreased activity compared with that of the RNF8−/− mice without IR treatment (Figure 6B and C). The step-down avoidance memory test had similar results (Figure 6D and E). These results suggest that RNF8, one of the DSB repair factors, maintains brain function and that RNF8 deficiency causes the deterioration of short-term memory and reduces the spontaneous locomotor activity of the mice. The RNF8−/− mice demonstrated increased sensitivity to IR treatment. The IR treatment resulted in further deterioration of the short-term memory and further reduced the spontaneous locomotor activity in the RNF8−/−mice. However, these results from WT mice do not correlate with the results shown in Figures 4 and 5. The reason for these discrepancies might be that such a loss of neurons could not cause the brain to function ineffectively.
In summary, our results indicate that RNF8 plays an important role in DSB repair in neurons. RNF8 is important for maintaining neuronal integrity. An absence of RNF8 is sufficient for causing neuronal pathology and cognitive decline. Inducing DNA damage with 2-Gy X-ray irradiation accelerated the process.
Supplementary Material
RNF8 is required for DNA damage repair in neurons.
RNF8 deficiency results in DNA damage accumulation and neuronal apoptosis in neuron.
Neurons from RNF8−/− mice appear to be more susceptible to X-ray-induced DNA damage.
RNF8 deficiency is sufficient to cause neuronal pathology and cognitive decline.
Acknowledgments
Funding
This work was supported by the National Natural Science Foundation of China [Grant No. 81171954 and 81472541 to D.W.] and by the Fundamental Research Funds for the Central Universities of Lanzhou University (Grant No.lzujbky-2012-145).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu C, Wang D, Wu J, Keller J, Ma T, Yu X. RNF168 forms a functional complex with RAD6 during the DNA damage response. J Cell Sci. 2013;126:2042–2051. doi: 10.1242/jcs.122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Englander EW. DNA damage response in peripheral nervous system: coping with cancer therapy-induced DNA lesions. DNA Repair (Amst) 2013;12:685–690. doi: 10.1016/j.dnarep.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barzilai A. DNA damage, neuronal and glial cell death and neurodegeneration. Apoptosis. 2010;15:1371–1381. doi: 10.1007/s10495-010-0501-0. [DOI] [PubMed] [Google Scholar]
- 7.Bohgaki T, Bohgaki M, Cardoso R, Panier S, Zeegers D, Li L, et al. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 2011;7:e1001381. doi: 10.1371/journal.pgen.1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaarsma D, van der Pluijm I, de Waard MC, Haasdijk ED, Brandt R, Vermeij M, et al. Age-related neuronal degeneration: complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. PLoS Genet. 2011;7:e1002405. doi: 10.1371/journal.pgen.1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reynolds JJ, Stewart GS. A nervous predisposition to unrepaired DNA double strand breaks. DNA Repair (Amst) 2013;12:588–599. doi: 10.1016/j.dnarep.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Alpi AF, Patel KJ. Monoubiquitylation in the Fanconi anemia DNA damage response pathway. DNA Repair (Amst) 2009;8:430–435. doi: 10.1016/j.dnarep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 12.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 14.Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 2009;8:436–443. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos MA, Huen MS, Jankovic M, Chen HT, Lopez-Contreras AJ, Klein IA, et al. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med. 2010;207:973–981. doi: 10.1084/jem.20092308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell. 2010;18:371–384. doi: 10.1016/j.devcel.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, Katyal S, Li Y, El-Khamisy SF, Russell HR, Caldecott KW, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain Research. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- 21.Rulten SL, Caldecott KW. DNA strand break repair and neurodegeneration. DNA Repair (Amst) 2013;12:558–567. doi: 10.1016/j.dnarep.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci U S A. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyama T, Wilson DM., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst) 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeggo PA, Geuting V, Lobrich M. The role of homologous recombination in radiation-induced double-strand break repair. Radiother Oncol. 2011;101:7–12. doi: 10.1016/j.radonc.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 25.Löbrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, et al. γH2AX foci analysis for monitoring DNA double-strand break repair: Strengths, limitations and optimization. Cell Cycle. 2014;9:662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 26.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 27.Speit G, Hartmann A. The Comet Assay. In: Henderson D, editor. DNA Repair Protocols. Humana Press; 2006. pp. 275–286. [Google Scholar]
- 28.Collins AR, Dobson VL, Dušinská M, Kennedy G, Štětina R. The comet assay: what can it really tell us? Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1997;375:183–193. doi: 10.1016/s0027-5107(97)00013-4. [DOI] [PubMed] [Google Scholar]
- 29.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, et al. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 34.Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, et al. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol Cell Biol. 2009;29:849–860. doi: 10.1128/MCB.01302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y, McKinnon PJ. Responding to DNA double strand breaks in the nervous system. Neuroscience. 2007;145:1365–1374. doi: 10.1016/j.neuroscience.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 38.Borgesius NZ, de Waard MC, van der Pluijm I, Omrani A, Zondag GC, van der Horst GT, et al. Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. J Neurosci. 2011;31:12543–12553. doi: 10.1523/JNEUROSCI.1589-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.