Abstract
Galectin-3 (Gal-3) is a multifunctional lectin, unique to galectins by the presence of a long N-terminal tail (NT) off of its carbohydrate recognition domain (CRD). Many previous studies have investigated binding of small carbohydrates to its CRD. Here, we used nuclear magnetic resonance spectroscopy (15N–1H heteronuclear single quantum coherence data) to assess binding of 15N-Gal-3 (and truncated 15N-Gal-3 CRD) to several, relatively large polysaccharides, including eight varieties of galactomannans (GMs), as well as a β(1 → 4)-polymannan and an α-branched mannan. Overall, we found that these polysaccharides with a larger carbohydrate footprint interact primarily with a noncanonical carbohydrate-binding site on the F-face of the Gal-3 CRD β-sandwich, and to a less extent, if at all, with the canonical carbohydrate-binding site on the S-face. While there is no evidence for interaction with the NT itself, it does appear that the NT somehow mediates stronger interactions between the Gal-3 CRD and the GMs. Significant Gal-3 resonance broadening observed during polysaccharide titrations indicates that interactions occur in the intermediate exchange regime, and analysis of these data allows estimation of affinities and stoichiometries that range from 4 × 104 to 12 × 104 M−1 per site and multiple sites per polysaccharide, respectively. We also found that lactose can still bind to the CRD S-face of GM-bound Gal-3, with the binding of one ligand attenuating affinity of the other. These data are compared with previous results on Gal-1, revealing differences and similarities. They also provide research direction to the development of these polysaccharides as galectin-targeting therapeutics in the clinic.
Keywords: galactose, glycan, lectin, NMR, protein
Introduction
Glycans can be considered biochemical platforms that store information, which is translated into cellular effects by interactions with their lectin receptors (Gabius 2009). A number of lectin folds have been identified, demonstrating the structural breadth of carbohydrate–lectin recognition patterns (Gabius et al. 2011). The galectins (named for their affinity toward β-galactosides; Barondes 2008) are potent effectors of diverse physiological processes, e.g., mediating cell adhesion/ attachment and glycoprotein routing or controlling growth or chemo/cytokine production (Klyosov et al. 2008; Smetana et al. 2013). All galectins share a β-sandwich-folded carbohydrate recognition domain (CRD) with a key Trp moiety that mediates C–H/π-interactions to galactose. Members of the galectin family are classified by structural organization into three groups: proto-type (homodimeric CRDs), tandem-repeat-type (two covalently linked CRDs) and chimera-type (CRD with a relatively long N-terminal tail (NT) containing phosphorylation sites and collagen-like repeats) (Nesmelova, Dings, et al. 2008). This diversification is phylogenetically maintained, with variations in number of the first two groups (Kaltner and Gabius 2012). However, all known animal genomes harbor the sequence of chimera-type galectin-3 (Gal-3), apparently due to it being biologically indispensible, and thus heightening interest in their interactions with various carbohydrate-binding partners.
Gal-3 can accommodate various β-galactosides in its canonical-binding site, with high affinity histo-blood groups epitopes, ganglioside GM1-pentasaccharide and clustered core 1 O-glycans, as well as immunologically (immune defense) relevant epitopes like xenoantigen, Galβ1,3Gal termini on Leishmania (and also Galβ1,4Gal) or LacdiNAc on parasites (Nesmelova, Dings, et al. 2008; Kopitz et al. 2010; Krzeminski et al. 2011; Gunning et al. 2013). In immune defense, Gal-3 is also known to interact with core components of bacterial lipopolysaccharide in both carbohydrate-dependent and -independent ways, the latter with participation of non-CRD elements (Mey et al. 1996). Curiously, Gal-3 also interacts in an unknown fashion with β1,2-linked oligomannosides on the surface of Candida albicans (Kohatsu et al. 2006). Both the binding with mannosides and lack of structural insight into that binding process prompted us to use nuclear magnetic resonance (NMR) spectroscopy to explore binding of Gal-3 to a panel of mannans, including galactomannans (GMs) with α1,6-galactose branched to a β1,4-mannose backbone and to define their contact region with Gal-3.
To this end, our present study investigates interaction of Gal-3 with GMs, a β(1 → 4)-linked polymannan (the backbone of the GMs) and an α-branched polymannan. GMs from various plant sources are reported to have a ratio of Man/Gal between about 1.1 and 4.0 (Mestechkina and Shcherbukhin 1991; Shcherbukhin 1992; Daas et al. 2000; Ilyina et al. 2006). The α-galactose unit can apparently be active as a (ga)lectin ligand, as reported for a galactogluco-mannan and for hydroxypropyl guar galactomannan (with α1-6-linked galactose branches on a β1-4 polymannan), purportedly interacting with Gal-3 at the canonical site (Woodward et al. 2012). We previously reported that galectin-1 (Gal-1) can bind α-galactosides at the canonical site (Miller et al. 2011), as well as to GMs at a relatively large surface area on the F-face of the CRD β-sandwich opposite to its canonical S-face carbohydrate-binding site (Miller, Klyosov, et al. 2009). However, the GM binding region on Gal-1 crosses this lectin's dimer interface, raising the question as to whether the dimer structure is required for GM binding. This study with Gal-3, which does not dimerize under the conditions of our experiments, also addresses the question of whether GMs can still bind to its F-face in the context of a native galectin monomer.
Overall, this work expands our understanding of how Gal-3 interacts with relatively large, complex polysaccharides, including cell surface glycans that are composed to a large extent of β-galactose (23% of all terminal monosaccharides), as well as, e.g., α-galactose (2.3%) and mannose (18.9% in various anomeric states) (Werz et al. 2007). In addition, these studies may provide research direction to the development of these polysaccharides as galectin-targeting therapeutics in the clinic (Klyosov and Traber 2012).
Results
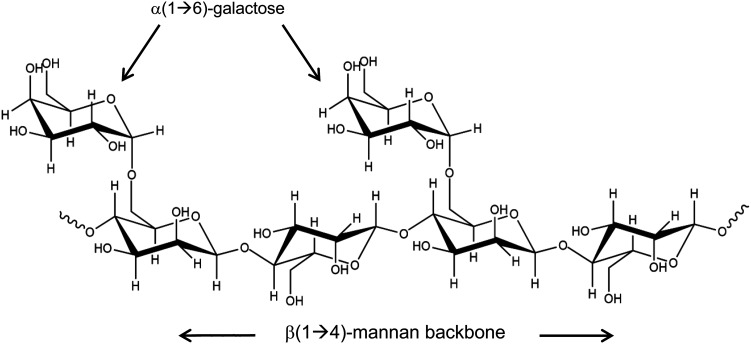
Heteronuclear single quantum coherence (HSQC) spectra of 15N-labeled Gal-3 (15N-Gal-3) were measured as a function of the concentration (0.01, 0.02, 0.04, 0.07, 0.12, 0.18, 0.3, 0.6 and 1.2 mg/mL) of eight α-GMs (i.e., (1 → 6)-α-d-galacto-(1 → 4)-β-d-mannans) that vary in their weight-averaged molecular weight and mannose/galactose (Man/Gal) ratio, i.e., 1.1, 1.7, 1.8, 2, 2.2, 2.4, 3 and 4. Figure 1 illustrates a possible structural unit of these GMs. However, bear in mind that GMs are heterogeneous in that their α(1 → 6)-galactose groups are randomly attached throughout the mannan backbone. For ease of discussion, we refer to them by their Man/Gal ratio (e.g., GM 1.1). To assess the role of the α-(1 → 6)-linked galactose branch in GMs, we also investigated a β(1 → 4)-mannan which lacks the branch, yet maintains the same backbone structure. Because Gal-3 is secreted from activated macrophages in host defense, we also assessed its potential to target yeast α-mannan.
Fig. 1.
One possible repeat unit of a galactomannan structure is illustrated. Note that the backbone is made up of (1 → 4)-linked β-d-mannopyranosyl units to which α-d-galactopyranosyl residues are attached via (1 → 6)-linkages. Only one such possible chemical structure is shown because galactose side chains are randomly distributed along the mannan backbone.
Binding of GMs to Gal-3
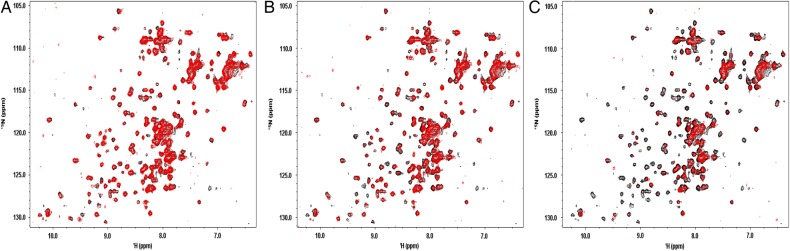
As each GM is titrated into the Gal-3 solution, resonances from both NT and CRD of the lectin are differentially decreased in intensity (broadened). In fact, some cross-peaks become so broad that they fall into the noise, while others are only partially reduced in intensity or remain mostly unchanged. These observations are exemplified in Figure 2, which shows 15N–1H HSQC spectra of 15N-Gal-3 in the absence (cross-peaks in black) and presence (cross-peaks in red) of increasing concentrations of GM 2.4. Similar effects are found with all of the other GMs, as exemplified in Supplementary data, Figure S1 that shows HSQC spectra of 15N-Gal-3 in the absence (cross-peaks in black) and presence (cross-peaks in red) of increasing concentrations of GM 1.1 (A, B and C) and GM 4 (D, E and F).
Fig. 2.
1H–15N HSQC spectra are shown for 15N-enriched Gal-3 alone (cross-peaks in black) and in the presence of GM 2.4 (cross-peaks in red) at 0.01 mg/mL (A), 0.07 mg/mL (B) and 0.6 mg/mL (C). Solution conditions were 20 mM potassium phosphate, pH 6.9. HSQC spectra (32 scans per transient) were acquired at 700 MHz (proton frequency) at 30°C with a sweep width of 16 ppm in the 1H dimension (2k points) and 30 ppm in the 15N dimension (256 points).
Although GM-induced broadening of Gal-3 resonances is consistent with exchange effects from Gal-3 binding to GMs, there are other possibilities. One of them is a polysaccharide-induced increase in solution viscosity. To address this possibility, we acquired HSQC spectra with Gal-3 in the presence of 25% (w/w) sucrose or 50% (w/w) glycerol and found that Gal-3 resonances were globally broadened (not differentially broadened), but not to the extreme extent as observed with GMs. Another possibility is Gal-3 oligomerization. In solution, Gal-3 is primarily monomeric (Birdsall et al. 2001) in equilibrium with a small fraction of oligomers (Morris et al. 2004) and can presumably oligomerize via its NT segment in the presence of multivalent oligosaccharides (Hernandez and Baum 2002; Ahmad et al. 2004; Leffler et al. 2004). However, we observed minimal resonance broadening as a function of Gal-3 (0.02–0.4 mM) or NT peptide (0.05–1 mM) concentration, and GM- and mannan-induced changes in Gal-3 chemical shifts and resonance intensities occur equally for CRD and NT resonances, suggesting that NT-mediated Gal-3 oligomerization plays no significant role in explaining GM-induced resonance broadening. In fact, when the polysaccharide–Gal-3 molar ratio is relatively low at the start of the titration, resonances of many Gal-3 resonances (CRD and NT) are decreased, and not increased, in line width. Thus, we concluded that the observed resonance broadening results from interactions between Gal-3 and these GMs that occur on the intermediate exchange regime on the chemical shift time scale. Our HSQC data also suggest that the overall β-sandwich/sheet fold of the CRD is not significantly perturbed by GM binding, because the dispersion of NH resonances that can be followed during the titration is essentially the same as that in the absence of the polysaccharides.
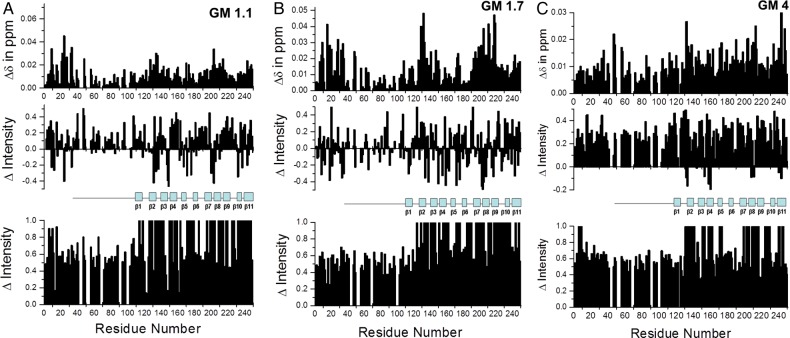
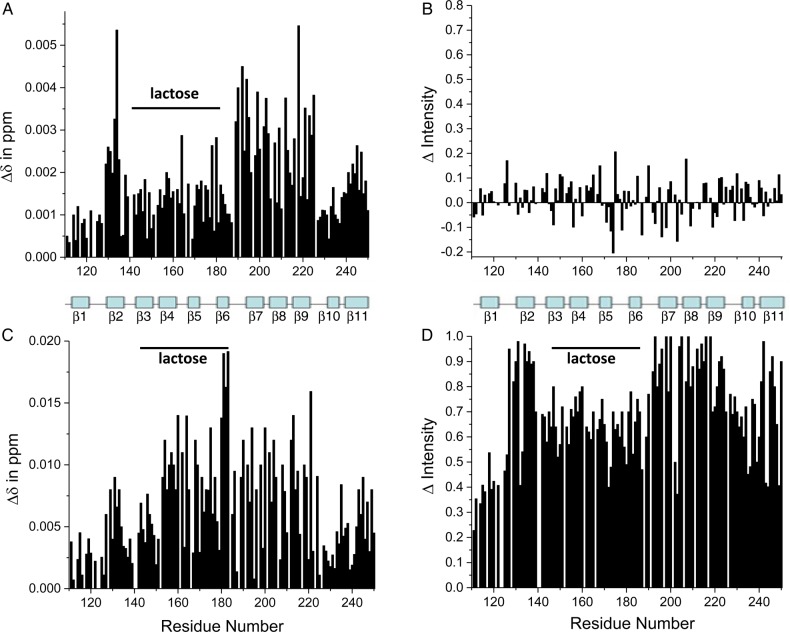
Insights into how GMs interact with Gal-3 were gained by observing changes in Gal-3 resonance intensities and chemical shifts during the titrations. Because binding of Gal-3 to these GMs falls in the intermediate exchange regime, binding-induced 15N-Gal-3 chemical shift changes, Δδ, are small. Nevertheless, they are useful to assess which residues of the lectin are most affected in their chemical environments during the binding process, especially when inferred at a relatively low polysaccharide/Gal-3 ratio where most resonances are still observable. At relatively high polysaccharide/Gal-3 ratios, resonance broadening increases and many resonances become unobservable. There are several features observed in all HSQC titrations that suggest common modes of interaction between Gal-3 and these GMs. Because resonance broadening and chemical shift changes are similar (albeit not identical) for all GMs, we only show representative results for GM 1.1, GM 1.7 and GM 4 (Figure 3). For each GM, chemical shift changes at a relatively low GM/Gal-3 ratio are shown at the top, and changes in intensities at low and high GM/Gal-3 ratios are shown in the middle and bottom panels, respectively. In each case, parameters are plotted vs. the sequence of Gal-3.
Fig. 3.
HSQC chemical shift and resonance broadening maps are shown for the binding of GM 1.1 (A), GM 1.7 (B) and GM 4 (C) to 15N-enriched Gal-3. Chemical shift changes, Δδ, are plotted at the lowest GM concentration vs. the sequence of Gal-3 in the top panel of each figure. At this relatively low glycan concentration, the molar ratio of Gal-3-to-GM is relatively high, such that resonances, although broadened to some extent, remain in the spectrum allowing most Δδ values to be determined. Fractional changes in Gal-3 resonance intensities observed for Gal-3 in the presence of the lowest (middle panels) and highest (bottom panels) GM concentration are plotted vs. the amino acid sequence of Gal-3. Changes in resonance intensity are calculated as fractional changes by taking the intensity of a given HSQC cross-peak at each titration point divided by that in ligand-free Gal-3 at the same protein concentration, and subtracting it from one. In the middle and bottom panels, a value of 1 indicates that the resonance associated with that particular residue is no longer apparent, and a value of 0 indicates no change in resonance intensity. In the middle panel, negative values indicate an increase in resonance intensity at a particular site; this may be accounted for by ligand binding-induced increased internal motions, more rapid conformational exchange or reduction in the NH proton exchange rate with solvent. Approximate positions of the 11 β-strands in the CRD are indicated, and a solid line above part of the sequence indicates the canonical lactose-binding site on the S-face of the lectin. This is figure available in black and white in print and in color at glycobiology online.
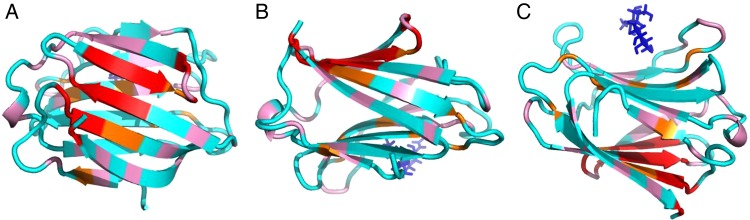
With the binding of any GM, small yet significant chemical shift changes, Δδ, are noted for residues within Gal-3. In general, the most shifted resonances arise from residues within β-strands 2, 7, 8, 9 and 11 that comprise the F-face β-sheet of the CRD β-sandwich that is opposite to the canonical carbohydrate-binding site on the S-face. This is illustrated in Figure 4 which shows three views of the structure of the Gal-3 CRD (pdb access code: 1A3K; Seetharaman et al. 1998) in which residues displaying the largest Δδ values (composite of shift changes from all GMs) are highlighted. These data support the proposal that sites within GMs bind primarily to the F-face of the CRD. Nevertheless, we cannot exclude the possibility of some interactions at the canonical carbohydrate-binding site, because some Δδ values of residues on the S-face of the CRD, while smaller, are still significant, as indicated in Figure 3 by the line and label over the primary lactose-binding sequence.
Fig. 4.
The primary GM binding surface on the CRD of Gal-3 is shown. Segments containing residues that are most affected by binding to these GMs (as a composite) are highlighted in red, orange, pink and blue on the structure of Gal-3, as discussed in the text. The X-ray structure of lactose-loaded Gal-3 CRD was used in this figure (pdb access code: 1A3K, Seetharaman et al. 1998). Residues that are most chemically shifted or broadened by binding to GMs are highlighted in red (>2SD above the average Δδ value), orange (between 1SD to 2SD), pink (between the average and 1 SD) and light blue (below the average). Different orientations of the Gal-3 CRD structure are shown: (A) the F-face is oriented toward the reader and the S-face is at the back; (B) the F-face is oriented at the top after a 90° rotation relative to the orientation shown in (A) and (C) the S-face and canonical lactose-binding site are at the top. In all instances, a molecule of lactose (stick model) bound to the S-face canonical site is shown in dark blue.
Analyzing changes in resonance intensities provides another dimension to assess Gal-3 binding to GMs (Miller, Klyosov, et al. 2009; Miller, Nesmelova, et al. 2009; Miller et al. 2012). Differential resonance broadening indicates that not all NH sites in Gal-3 are affected equally by interactions with GMs. The extent of broadening at a particular 15NH depends on a combination of factors. These include direct interaction of that residue with the polysaccharide, lifetime of Gal-3 in the glycan-loaded state (binding kinetics), binding-induced changes to internal motions, conformational exchange, potential oligomer exchange as well as Gal-3 exchange between multiple binding sites within the polysaccharides. And because the extent of broadening at any given site depends on a combination of factors, intensity changes are not necessarily correlated with chemical shift changes. Therefore, we should be cautious in our interpretation of resonance broadening effects.
Surprisingly, some Gal-3 resonances are initially increased in intensity (reduction in line width) at the start of the titration when the GM:Gal-3 molar ratio is relatively low (see middle panels in Figure 3A–C). An increase in intensity at a particular site could be accounted for by ligand binding-induced increased internal motions, more rapid conformational exchange or reduction in the NH proton exchange rate with solvent. There is precedence for increased internal motions with lactose binding to Gal-1 (Nesmelova et al. 2010), Gal-7 (Ermakova et al. 2013) and Gal-3 CRD (Diehl et al. 2010), where the ligand binding-induced increase in flexibility was associated with an increase in conformational entropy that contributed favorably to the free energy of ligand binding. The most narrowed resonances are generally located within and throughout the CRD and are associated mostly with hydrophobic residues within the core and on both faces of the β-sandwich. Thus GM binding on the surface of the CRD would seem to impact on the core of the sandwich and could be related to allosteric signaling between S- and F-faces of the CRD.
As the titration continues and the GM:Gal-3 molar ratio is increased, all Gal-3 resonances eventually become significantly, yet differentially, broadened (decreased in intensity). The reason that binding of some GMs increases the intensity of only a few Gal-3 resonances at the start of the titration (e.g., GM 4, Figure 3C) is likely due to greater overall ligand binding. As the concentration of any GM is increased, so is the population of GM-loaded Gal-3, and resonance broadening dominates even for these initially more narrowed resonances. At 0.6 or 1.2 mg/mL of any GM, most Gal-3 resonances within the CRD have reached maximal observable broadening, including those within the binding site on the F-face and around the canonical carbohydrate-binding site on the S-face. During most of the titration and prior to the end, the most broadened resonances are generally (although not always for reasons noted above) the same as the most chemically shifted ones highlighted on the folded structure of the Gal-3 CRD in Figure 4.
Less can be inferred about the NT, primarily because it is reported to be unstructured, highly flexible and engaged in transient interactions with the CRD (Kopitz et al. 2003; Morris et al. 2004; Ippel et al. 2015; Berbís et al. 2014; Halimi et al. 2014). However, our HSQC data do indicate that GM binding to Gal-3 also perturbs residues within the NT. For example, NT residues S6, H8, D9, A10, G15, A33, S40, G47, G65 and A88 are all chemically shifted and maximally broadened by the end of the titration. There are several possibilities to explain GM-induced effects at the NT. For one, GMs may interact independently with the CRD and the NT, block transient interactions between the NT and CRD or simultaneously interact with both when the NT is in its CRD “bound” state. Any of these scenarios could explain the observed NT chemical shift and intensity changes. For insight into this question, we performed titrations with two GMs (GM 1.7 and GM 1.1) on the truncated 15N-labeled Gal-3 CRD and on two synthetic peptides derived from the Gal-3 NT (one having residues 1–50 and the other having residues 51–107). Because the NT-derived peptides were not isotopically enriched, we followed the titrations (up to 1.2 mg/mL GM) using NOESY and TOCSY spectra.
Based on the absence of GM-induced chemical shift or intensity changes with either of the two synthetic NT-derived peptides, we concluded that these GMs do not bind to the NT independent of the CRD. On the other hand, GMs still bind to the F-face of the truncated Gal-3 CRD, but with some differences relative to full-length Gal-3. For one, a higher GM concentration is required to attain the same degree of broadening with the truncated CRD vis-à-vis the CRD in full-length Gal-3. This is exemplified in Supplementary data, Figure S2 that shows three HSQC overlays acquired during the titration with GM 1.7. Even at 1.2 mg/mL (Supplementary data, Figure S2C), GM-induced resonance broadening with the truncated CRD is not as great as that observed with full-length Gal-3 and any of the GMs. Figure 5 shows chemical shift and intensity changes vs. the Gal-3 CRD sequence. At a relatively low concentration of GM 1.7, Δδ values (Figure 5A) are greatest for residues within β-strands 2, 7, 8, 9 and 11 that define the CRD F-face, and resonance intensities are either decreased or increased (Figure 5B). This is essentially what was observed at the start of the titration with full-length Gal-3. By the end of the titration, however, clear differences are noted. At 1.2 mg/mL (Figure 5D), broadening is greatest for resonances from the F-face of the CRD (albeit not as large as with the CRD in full-length Gal-3 at the same GM concentration), whereas the largest Δδ values (Figure 5C) are observed for both resonances within the F-face and those within the canonical carbohydrate-binding site. This was surprising. One plausible explanation is that both the F-face and canonical S-face sites in truncated CRD have an equal potential to interact with sites (likely different ones) in these GMs. In full-length Gal-3, GM interactions with the S-face are apparently attenuated relative to those with the F-face. However, with the truncated CRD, it appears that the S-face can bind to its GM sites to an equal extent as the F-face can bind to its GM-binding epitope(s). Note that Δδ values of lactose-binding residues are relatively similar in value in both Gal-3 and truncated Gal-3, whereas those at the F-face are considerably lower in value in truncated Gal-3 vis-à-vis full-length Gal-3 (Figure 3). This suggests two conclusions: (1) binding affinity to the F-face, but not to the S-face, is reduced, and relatedly (2) that the NT in full-length Gal-3 plays a role in GM-binding affinity to the CRD F-face.
Fig. 5.
HSQC chemical shift and resonance broadening maps are shown for the binding of GM 1.7 to 15N-enriched truncated Gal-3 CRD (0.07 mM). Changes in chemical shift, Δδ, and intensity, Δ Intensity, are plotted vs. the sequence of the truncated Gal-3 CRD using residue numbers as in full-length Gal-3. Results are shown at GM 1.7 concentrations of 0.1 mg/mL (A and B) and 1.2 mg/mL (C and D). Changes in resonance intensity are calculated as fractional changes by taking the intensity of a given HSQC cross-peak at each titration point divided by that in ligand-free truncated Gal-3 CRD at the same protein concentration, and subtracting it from one. A value of 1 indicates that the resonance associated with that particular residue is no longer apparent, and a value of 0 indicates no change in resonance intensity. Approximate positions of the 11 β-strands in the CRD are indicated, and a solid line above part of the sequence indicates the canonical lactose-binding site on the S-face of the lectin. This is figure available in black and white in print and in color at glycobiology online.
Insight into GM-binding affinity and stoichiometry
The extent of resonance broadening is related to binding affinity and stoichiometry, in addition to any binding-induced changes in internal motions and conformational exchange, as discussed in the previous section. Therefore, one might look at this parameter as binding avidity, or the net ability of a GM to bind molecules of Gal-3. However, because Gal-3 binding to GMs falls in the intermediate exchange regime, one cannot accurately determine binding affinity (or stoichiometry), other than to say that the equilibrium dissociation constant, Kd, falls in the ∼0.002 to ∼0.05 mM range. Nevertheless, if we assume that binding-induced changes in internal motions and conformational exchange are, for the most part, the same for each Gal-3 molecule regardless of the GM, the extent of broadening should primarily and comparatively reflect the overall fraction of bound Gal-3.
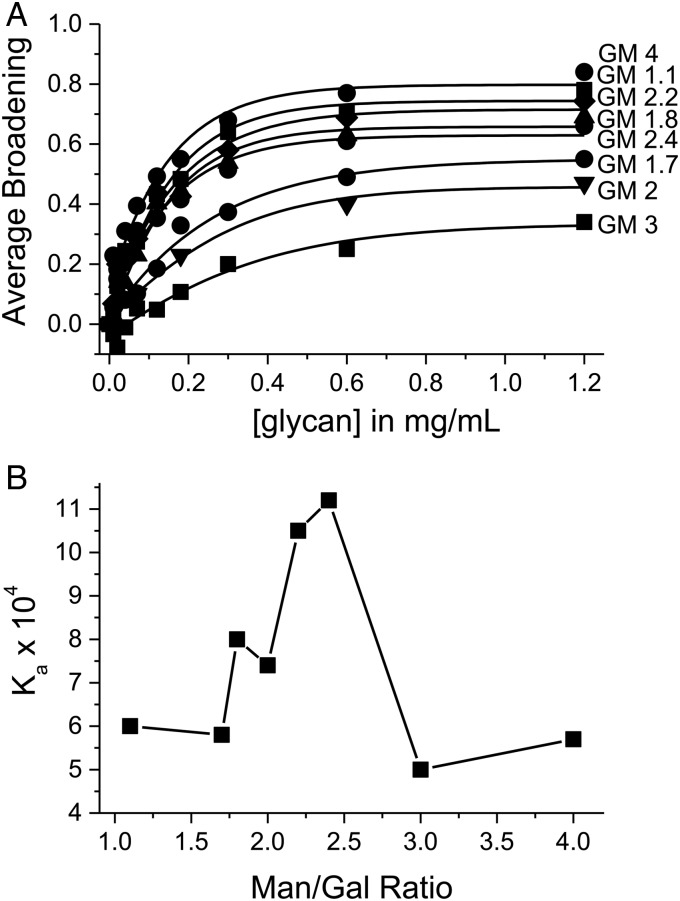
Under this assumption, we estimated binding parameters by sigmoidal or Gaussian curve fitting plots of Δ intensity averaged over all Gal-3 resonances vs. the concentration of each GM (Figure 6A). At 1.2 mg/mL, each GM has apparently reached 90% “saturation” with Gal-3. A Δ intensity value of 1.0 is not always reached, because the apparent level of “saturation” is a composite of average binding affinity per site (i.e., lifetime of the bound state(s) or exchange dynamics) and binding stoichiometry. If we take the concentration of any GM at this “saturation” point relative to the concentration of Gal-3 used in any given titration (0.093–0.124 mM), we can estimate binding stoichiometry. For GM 1.7 (59 kDa), e.g., 1.2 mg/mL corresponds to a concentration of 0.02 mM. Therefore, with 0.124 mM Gal-3 (used for this titration) divided by 0.02 mM GM 1.7, binding stoichiometry is ∼6 molecules of Gal-3 per one molecule of GM 1.7. The Kd is estimated by taking the GM concentration at 50% ligand-loaded Gal-3 (0.0028 mM) and multiplying it by the stoichiometry of 6, to yield an average Kd per binding site of ∼0.017 mM. Binding parameters for GMs calculated in this way are listed in Table I. When we perform the same analysis for the binding of truncated Gal-3 CRD to GM 1.7 (Table I), we find that the overall binding avidity has decreased, along with the number of bound lectin molecules and the average Ka per site. Once again, removal of the NT from Gal-3 has a detrimental effect on GM binding. As a cautionary note, these values should be taken cum grano salis, because the binding process does fall within the intermediate exchange regime, and these are not actual Kd values. Nevertheless, this range is expected for exchange events within the intermediate exchange regime and should reflect relative differences in binding over the set of GMs.
Fig. 6.
(A) The figure plots resonance broadening averaged over all Gal-3 resonances vs. the concentration of each GM. Resonance broadening was calculated as the fractional change in resonance intensity as discussed in the text. A value of 1 indicates that a resonance is no longer apparent, and a value of 0 indicates no change in resonance intensity. The average fraction of resonance broadening from 15N-enriched Gal-3 HSQC resonance intensity changes vs. the concentration (mg/mL) of GMs is shown. Solid lines are fits to the data using a Gaussian/Boltzmann function. (B) Plot of the average equilibrium association binding constants vs. the Man/Gal ratio for each GM glycan investigated.
Table I.
Binding parameters from HSQC titrations
| MW (kDa) | [∼90% sat.] (10−6 M) | [Gal-3] (10−6 M) | Number of sites | [50% sat.] (10−6 M) | Ave. Kd/site (10−6 M) | Ave. Ka/site (104 M−1) | |
|---|---|---|---|---|---|---|---|
| Full-length Gal-3 | |||||||
| GM 1.1 | 83 | 14.4 | 93 | 6.5 | 2.4 | 15 | 6.4 |
| GM 1.7 | 59 | 20.3 | 124 | 6.1 | 2.8 | 17 | 5.9 |
| GM 1.8 | 100 | 12 | 121 | 10 | 1.4 | 14 | 7.1 |
| GM 2 | 690 | 1.7 | 121 | 71 | 0.2 | 14 | 7.1 |
| GM 2.2 | 215 | 5.5 | 121 | 22 | 0.4 | 8.8 | 11.3 |
| GM 2.4 | 790 | 1.5 | 121 | 81 | 0.1 | 8.1 | 12.3 |
| GM 3 | 685 | 1.7 | 121 | 71 | 0.4 | 28 | 3.6 |
| GM 4 | ∼1000 | 1.2 | 93 | 78 | 0.2 | 16 | 6.2 |
| α-mannan | 50 | 12 | 117 | 5 | 1.2 | 6 | 16.7 |
| β-mannan | 5 | 240 | 121 | 0.5 | 60 | 30 | 3.3 |
| Truncated Gal-3 CRD | |||||||
| GM 1.1 | 83 | 26 | 80 | 3.1 | 8 | 25 | 4 |
| GM 1.7 | 59 | 42 | 80 | 2 | 10 | 20 | 5 |
For full-length Gal-3, 90% saturation was assumed at 1.2 mg/mL of each glycan, and the concentration given is calculated accordingly. For truncated Gal-3 CRD, 90% saturation was assumed at 2.5 mg/mL of each glycan, and the concentration given is calculated accordingly. The number of glycan sites bound (# sites) was calculated by dividing the concentration at 90% saturation into the lectin concentration used in the HSQC experiments. The 50% saturation was taken at that point on binding curves with 50% lectin is bound. The average Kd/site is calculated by multiplying the 50% saturation concentration by the number of binding sites. For each glycan, the MW listed is the weight-averaged MW.
Based on this analysis, the number of Gal-3 molecules bound per GM ranges from about 5 to 81. Certainly, the binding of 81 (or even 71) Gal-3 molecules per GM molecule is unrealistic and underscores the inaccuracies of this approach. Therefore, we use these values only to compare one GM to another. Figure 6B, in which average Ka value/site is plotted vs. the Man/Gal ratio, indicates that GM 2.2 and GM 2.4 bind Gal-3 most strongly. In this regard, a GM molecular signature should define optimal Gal-3 binding, and this should be related to the distribution of Man and Gal residues which will be nonhomogeneous and aperiodic. With GM 1.1, most mannose units will have one α(1 → 6)-linked galactose, whereas GM 4 will have numerous Gal-free mannan segments. The distribution will be more even, albeit random, in GM 2.2 and GM 2.4 which were also the most optimal Gal-1 binders (Miller, Klyosov, et al. 2009). With Gal-1, we reported that the optimal-binding signature was α-d-galactopyranosyl doublets flanked by regions of Gal-free mannan (Miller, Klyosov, et al. 2009). Nevertheless, because Gal-3 apparently has multiple glycan binding sites, each estimated “Ka” is some weighted average with unknown weighting factors for each site.
Binding of mannan to Gal-3
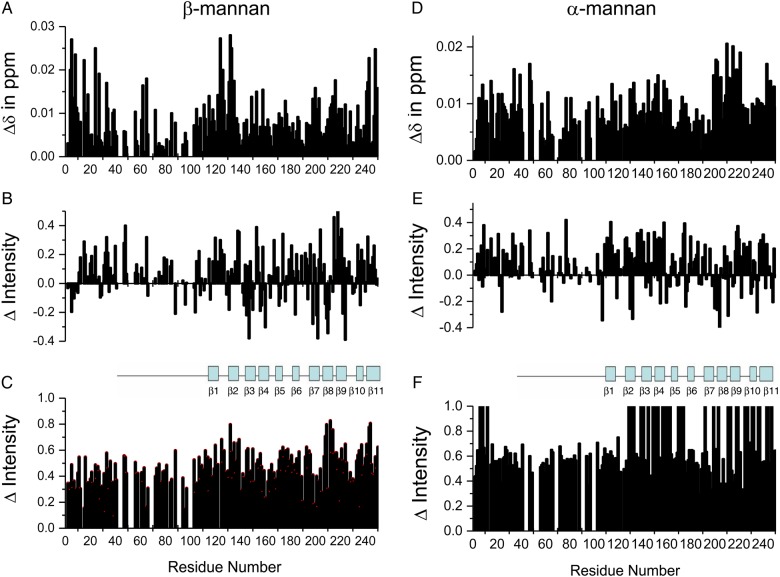
To assess whether Gal-3 can bind to the GM mannan backbone absent galactose residues, we performed a HSQC titration with a linear β(1 → 4)-linked mannan. As shown in Figure 7, this β-mannan binds Gal-3 somewhat differently from the GMs. The most significant chemical shift changes within the CRD occur at β-strands 2 and 11 that form the lower part of the β-sandwich F-face (Figure 7A). With the GMs, it appears that binding is more centered at the upper part of the F-face within β-strands 2, 7, 8 and 9. A number of resonances from the N-terminal part of the NT are also significantly shifted, as observed with GMs above. Similar effects are noted with intensity changes at both low and high mannan/Gal-3 molar ratios (Figure 7B and C, respectively). In fact, initial resonance broadening is significant for NT residues G32AG34, S40 and G47Q48 (Figure 7B). At the higher β-mannan concentration (Figure 7C), resonance broadening at the CRD is on average higher than that at the NT, something that could be accounted for by inherently greater flexibility of the NT.
Fig. 7.
HSQC chemical shift and resonance broadening changes are shown for the binding of a β-mannan (A–C) and an α-mannan (D–F) to 15N-enriched Gal-3, as discussed in the text. Chemical shift changes, Δδ, are plotted at the lowest glycan concentration vs. the sequence of Gal-3 in panels (A) and (D). Fractional changes in Gal-3 resonance intensities observed for Gal-3 in the presence of the lowest (panels (B) and (E)) and highest (panels (C) and (F)) mannan concentrations are plotted vs. the amino acid sequence of Gal-3. As above, a value of 1 indicates that the resonance associated with that particular residue is no longer apparent, and a value of 0 indicates no change in resonance intensity. This is figure available in black and white in print and in color at glycobiology online.
An α-linked mannan induces similar HSQC spectral effects on 15N-Gal-3 (Figure 7). Within the CRD, the most significant chemical shift changes are noted at β-strands 7, 8 and 9 that form the upper part of the β-sandwich F-face, suggesting interactions of the α-mannan at this site (Figure 7D). However, residues within the canonical site on the S-face are also perturbed, suggesting at least some interactions here. There are also a number of significant NH shifts at sites within the N-terminal part of the NT. Similar effects are noted with intensity changes at both low and high mannan/Gal-3 molar ratios. In fact, initial changes in resonance broadening are highly significant for NT residues L7, D9, G15, G21, G24, A33, G35, S40, Y41, A62, G65 and G77 (Figure 7E). At the higher α-mannan concentration (Figure 7F), resonance broadening at the CRD is generally greater than that observed at the NT; this could be due to inherently greater flexibility of the NT and/or greater avidity of the α-mannan for Gal-3.
As with the GMs, neither of these mannans interacts with the free NT peptides. Estimated affinities and stiochiometries for Gal-3 binding to these mannans are shown in Table I. With β(1 → 4)-mannan (5 kDa), the 90% saturation point is ∼0.240 mM, and binding stoichiometry is about 0.3 molecules of Gal-3 (0.121 mM) per one molecule of β-mannan. At 50% Gal-3 bound (0.06 mM), the average Kd per binding site is about 0.03 mM. With the α-mannan (50 kDa), 90% saturation is ∼0.024 mM, yielding a binding stoichiometry of about five molecules Gal-3 per one of this α-mannan. At 50% Gal-3 bound (0.0012 mM), the average Kd per binding site is ∼0.006 mM. Interestingly, this α-mannan does appear to bind Gal-3 better than does the β-mannan.
Effect of lactose on GM-bound Gal-3
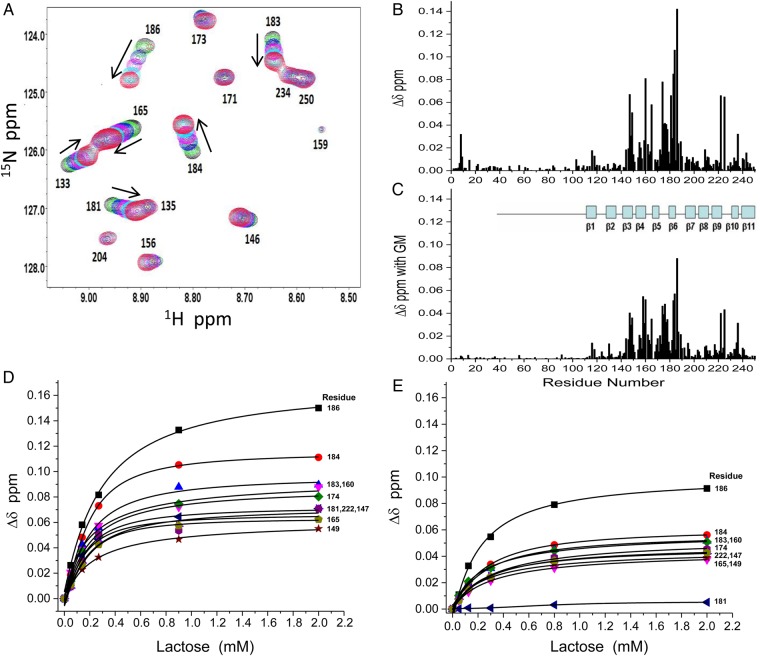
Figure 8 shows the effect that lactose has on Gal-3 bound to GM 1.7. Because GM 1.7 at 1.2 mg/mL causes many Gal-3 resonances to be highly broadened with many being unobservable, we performed these studies with GM 1.7 at 0.3 mg/mL where most Gal-3 resonances are still observed. 15N-Gal-3 HSQC spectral expansions are overlaid in Figure 8A for GM-bound Gal-3 alone (black cross-peaks) and GM-bound Gal-3 in the presence of increasing concentrations of lactose (0.05–2 mM; multicolored cross-peaks). Note that whereas some Gal-3 cross-peaks are highly shifted, others are not. Arrows indicate how some Gal-3 resonances are shifted during the titration. Resonances that are most shifted are associated with residues located within the canonical lactose-binding site.
Fig. 8.
Lactose binds Gal-3 in the presence of GM 1.7 (Davanat). (A) Overlay of expansions from HSQC spectra of 15N-enriched Gal-3 in the presence of GM 1.7 (0.3 mg/mL) (black cross-peaks) and with the addition of increasing concentrations of lactose from 0.05 to 2 mM. (B and C) Lactose (2 mM) induced chemical shift changes (Δδ) for Gal-3 alone (B) and Gal-3 in the presence of GM 1.7 (0.3 mg/mL) (C) are shown vs. the amino acid sequence of Gal-3. (D) 15N–1H weighted chemical shift differences (Δδ) of Gal-3 in the absence of GM 1.7 vs. the total concentration of lactose added. The 10 most shifted resonances are shown. The solid lines represent sigmoidal fits to the data. (E) 15N–1H weighted chemical shift differences (Δδ) of Gal-3 in the presence of GM 1.7 (0.3 mg/mL) vs. the total concentration of lactose added. The 10 most shifted resonances are shown. The solid lines represent sigmoidal fits to the data.
Line broadening and chemical shifts of Gal-3 resonances are also affected by the presence of lactose. Figure 8 shows chemical shift changes (Δδ) of resonances vs. residue number for Gal-3 upon addition of 2 mM lactose in the absence (Figure 8B) or presence (Figure 8C) of GM 1.7. Addition of lactose to a sample of Gal-3 bound to GM 1.7 partially reverses the intensity loss caused by GM binding (data not shown), indicating that binding of lactose at the canonical site attenuates the fraction of Gal-3 bound to GM. However, even though lactose binding apparently reduces the avidity of GM 1.7 for the lectin, the GM remains bound to Gal-3 as indicated by sustained resonance broadening. In fact, broadening is least reduced in resonances associated with residues at the CRD F-face (β-strands 2, 7–9 and 11), supporting the idea that the GM primarily interacts with residues at the F-face and that lactose and GM can bind Gal-3 simultaneously. Note also that in the absence of GM 1.7, lactose binding to Gal-3 induces chemical shift changes of resonances within the NT (Figure 8B), suggesting that binding of this ligand to the Gal-3 canonical site affects interactions between the NT and CRD. However, when GM is bound to Gal-3, lactose binding-induced chemical shift changes within the NT are considerably less (Figure 8C) than those when lactose binds in the absence of GM. This supports the proposal that GM binding to the F-face displaces or inhibits NT interactions with the CRD.
Moreover, whereas lactose binding at the S-face weakens affinity of GM 1.7 at the F-face, binding of GM 1.7 also reduces the affinity of Gal-3 for lactose. Lactose-induced chemical shift changes for the 10 most perturbed backbone NH resonances of Gal-3 residues are shown in Figure 8D. The side chains of most of these residues (R186, E184, R183, W181, N174, E165 and N160) make crucial contacts (mostly H-bonds) with the disaccharide. Figure 8E shows how the same 10 resonances from the Gal-3/GM 1.7 complex respond to the addition of lactose. Note that throughout the lactose titration with Gal-3 in the presence of GM 1.7 (Figure 8E), Δδ values are more attenuated. In fact, the backbone NH of W181 is barely shifted. W181 is an important residue within the canonical-binding site, because its aromatic side chain makes key hydrophobic interactions with the galactose ring of the disaccharide. Reduced Δδ values reflect relatively weaker lactose binding. Using the data in Figure 8D and E, we quantified this by determining the free lactose concentration when the fraction of Gal-3 bound with ligand was 0.5. This concentration provides a measure of the equilibrium dissociation constant, Kd. In the absence of GM 1.7, the Kd value for lactose binding to Gal-3 is 0.13 mM. In the presence of GM 1.7, the Kd value is increased to 0.2 mM, indicating weaker affinity for lactose binding to the canonical site of Gal-3.
Discussion
When considering glycan associations, binding at the canonical β-galactoside site with its conserved galactose-Trp C-H/π-interaction has so far been the only carbohydrate-binding site reported for Gal-3. Here we report that this lectin also binds to GMs, as well as to α- and β-mannans. With these larger polysaccharides, the primary site for interaction is at the F-face of the CRD β-sandwich. Even though we cannot exclude some interactions at the canonical carbohydrate-binding site on the S-face of the CRD β-sandwich, we found that lactose can still bind at the canonical site when a GM is bound at the F-face. In fact, we also found that ligand occupation at one site attenuates ligand affinity at the other, an effect that is likely mediated via a conformational transition through the β-sandwich and inter-connecting loops.
The F-face binding region is composed of charged (R129, K139, K199, E205, D215, H223 and R224) and polar residues (T133, T137, Q201, N204, Q220 and N222,), as well as some solvent-exposed hydrophobic residues (L131, V138, L203, A212, A216, V225, Y247 and M249). Although this set of residues is essentially the same as that found for GM binding to the F-face of Gal-1 (Miller, Klyosov, et al. 2009), the GM-binding region on Gal-1 crosses the dimer interface, raising the question as to whether the galectin dimer structure is required for GM binding. This study with Gal-3, which does not dimerize under the conditions of our experiments, addressed this question in that GMs can still bind to its F-face. Nevertheless, the commonality in GM binding to Gal-3 and Gal-1 stands in contrast to the absence of GM binding of tandem-repeat-type galectins-8 and -9 (Werz et al. 2007).
Several reports have suggested that the NT of Gal-3 interacts with the CRD (Kopitz et al. 2003; Morris et al. 2004; Ippel et al. 2015; Berbís et al. 2014; Halimi et al. 2014). Because chemical shifts and narrow line widths of NT resonances indicate that this segment of Gal-3 is random coil and highly dynamic (Ippel et al. 2015), the NT-CRD interaction must be transient and therefore relatively weak. Here, we found that GMs bind to the CRD in Gal-3 relatively strongly, such that their binding to the CRD could inhibit interactions between the NT and CRD. Nevertheless, our results also indicate that even though GMs do not bind to the NT segment alone, the NT somehow plays a role in optimizing GM interactions with the CRD in the context of full-length Gal-3. We conclude this primarily because GM binding is reduced in truncated Gal-3 CRD, even though GMs can still interact with the F-face of the CRD in both full-length and truncated Gal-3.
Binding affinities (or avidities) of GM and mannan for Gal-3 are considerably greater than those for small saccharides like lactose and N-acetyllactosamine (Nesmelova, Dings, et al. 2008). Our crude estimates of average Kd values range from about 0.008 to 0.025 mM per Gal-3 binding site, indicating that the carbohydrate-binding footprint on Gal-3, and galectins in general, is larger for these polysaccharides than that defined by simple disaccharides. That having been said, it is important to realize that because these relatively large and complex polysaccharides generally have multiple binding sites (stiochiometries greater than one) for Gal-3, it is likely that Gal-3 interacts with different sites having different binding affinities. In this regard, our average Kd values should be taken cum grano salis.
Nevertheless, we found that our Gal-3 binding affinity to GMs is correlated with the Man/Gal ratio, thus providing insight into structure–activity relationships. GMs have a heterogeneous or aperiodic distribution of Man and Gal residues. With GM 1.1, we have few “naked” mannan segments, whereas with GM 4 we have numerous “naked” mannan segments and fewer segments of concentrated galactose residues. With the Man/Gal ratio of 2.2–2.4, there is a more even distribution, albeit random, of segments of naked mannan and of high galactose content. Gal-3 binding to GMs is greatest for Man/Gal ratios of 2–2.4, a finding that we reported earlier with Gal-1 binding to GMs (Miller et al. 2012). As with Gal-1, a statistical analysis using a model parameterized for the fraction of Gal/Man clusters, the fraction of “naked” Man residues, and the number of sequential Gal/Man residues surrounded by “naked” Man residues suggests that the optimal GM-binding signature is an α-d-galactopyranosyl doublet flanked by regions of “naked” mannan. However, regions of “naked” mannan by themselves do not provide a good binding epitope for Gal-3. We conclude this because even though β-mannan (the backbone of these GMs) interacts with the F-face of the Gal-3 CRD like the GMs, Gal-3 avidity to the β-mannan is significantly lower. In this regard, the α-(1 → 6)-linked galactose residues in these GMs are crucial to optimal binding. That having been said, it was surprising to find that the α-mannan bound Gal-3 with significantly greater avidity for Gal-3 than the GMs. In this instance, however, binding apparently occurs at both the F-face and S-face of the lectin. The reason for this is unclear and requires further investigation.
Because most of these polysaccharides are pectin derived, the question of physiological relevance in terms of binding to the Gal-3 F-face arises. Nevertheless, the larger and more relevant mannans also bind to the F-face, and we did find that lactose can bind to the canonical site even when a GM is bound at the F-face. Even though the highly conserved tryptophan within the canonical site is absent from the F-face, both ligand binding sites do share some compositional and structural features, as illustrated in Figure 4B and C. The sites both display overall concave shapes and contain numerous charged and polar residues that would be crucial for interacting with any polysaccharide. Uncertainty with physiological relevance notwithstanding, it should be noted that binding of the galectin to tissue sections, e.g., of tumors, has prognostic relevance (Plzák et al. 2004; Dawson et al. 2013), prompting the interest in defining in situ binding partners. In this sense, our study broadens the view on the interactive potential. Equally important, the comparable activity profiles of Gal-3 and Gal-1 would let functional competition become possible, as seen in growth control (Kopitz et al. 2001; Sanchez-Ruderisch et al. 2010). Overall, this work expands our understanding of how Gal-3 interacts with polysaccharides, opening the possibility for binding to polysaccharides in host defense.
Materials and methods
Gal-3 preparation
The expression plasmid for human Gal-3 and truncated Gal-3 CRD was constructed by inserting the appropriate cDNAs (1–250 amino acids of human Gal-3 or 111–250 amino acids of truncated human Gal-3 CRD) into the vector pET-22b(+) between the Ndel and BamHI cut sites. This expression vectors were kindly provided by Dr Tai. Uniformly 15N-labeled lectins were expressed in BL21 (DE3) competent cells (Novagen), grown in minimal media, purified over a lactose affinity column and fractionated on a gel filtration column, as described previously for production of Gal-1 (Nesmelova, Pang, et al. 2008). About 8 mg of purified protein were obtained from 1 L of cell culture. Protein purity was checked by using SDS PAGE and mass spectrometry.
Polysaccharide preparations
1,4-β-d-Galactomannan from Cyamopsis tetragonoloba (guar gum; Man/Gal = 1.7) of weight-averaged molecular weight 59 kDa (Miller, Klyosov, Platt, et al. 2009) was produced by Galectin Therapeutics Inc. (Newton, MA) under the commercial name Davanat® and described in US Pat. Nos. 6,642,205 and 7,893,252 (Platt et al. 2006; Miller, Klyosov, et al. 2009; Miller, Klyosov, Platt, et al. 2009). 1,4-β-d-Galactomannan from Ceratonia siliqua (locust bean; >1000 kDa; Man/Gal = 4.0) was obtained from Sigma (St. Louis, MO), catalog no. G-0753, Lot 109H0899. Other 1,4-β-d-galactomannans used here were kindly provided by Dr Vladimir D. Shcherbukhin, Institute of Biochemistry, Russian Academy of Sciences, Moscow, Russia (Mestechkina and Shcherbukhin 1991; Shcherbukhin 1992; Ilyina et al. 2006). These include the following, with weight-averaged molecular weights and Man/Gal ratios given in parentheses:
| GM from Medicago falcata (yellow lucerne) | (83 kDa; Man/Gal 1.1) |
| GM from Cyamopsis tetragonoloba (guar gum) | (100 kDa; Man/Gal 1.8) |
| GM from Lagonychium farctum (mimosa) | (690 kDa; Man/Gal 2) |
| GM from Gleditsia triacanthos (honey locust) | (215 kDa; Man/Gal 2.2) |
| GM from Gleditsia triacanthos (honey locust) | (790 kDa; Man/Gal 2.4) |
| GM from Gleditsia capsa (locust tree) | (685 kDa; Man/Gal 3) |
α-Branched mannan from Saccharomyces cerevisiae (weight-averaged molecular weight of 50 kDa) and β(1 → 4)-mannan (5 kDa) were purchased from Sigma Chemical (St. Louis, MO) and were used without further purification.
Heteronuclear NMR spectroscopy
Uniformly 15N-labeled Gal-3 or 15N-labeled truncated Gal-3 CRD was dissolved at concentrations of 0.08–0.124 mM in 20 mM potassium phosphate buffer at pH 6.9, made up using a 95% H2O/5% D2O mixture. 1H–15N HSQC NMR experiments were used to investigate binding of a series of polysaccharides. 1H and 15N resonance assignments for recombinant human Gal-3 were previously reported (Ippel et al. 2015) and were used here to analyze our HSQC data.
NMR experiments were carried out at 30°C on Bruker 600, 700 or 850 MHz spectrometers equipped with H/C/N triple-resonance probes and x/y/z triple-axis pulse field gradient units. A gradient sensitivity-enhanced version of the two-dimensional 1H–15N HSQC experiment (32 scans per transient) was applied with 256 (t1) × 2048 (t2) complex data points in 15N and 1H dimensions, respectively, and a sweep width of 16 ppm in the 1H dimension and 30 ppm in the 15N dimension. Raw data were converted and processed by using NMRPipe (Delaglio et al. 1995) and were analyzed by using NMRview (Johnson and Blevins 1994).
Resonance broadening of peaks in HSQC spectra was determined by automated peak picking and/or by manual assessment of resonance intensities. In instances where peaks were well resolved, the automated approach was as accurate as manual determination. In instances where peaks were in a crowded area or partially overlapped, intensities had to be determined by manual assessment. In some cases, resonances were integrated, and resulting relative volumes were found to compare favorably with determination of peak intensities. Changes in resonance intensities during any given titration were calculated as fractional changes by taking the intensity of a given HSQC cross-peak at each titration point divided by that in ligand-free Gal-3 at the same protein concentration, and subtracting it from one. Therefore, a value of 1 indicates that a resonance is no longer observed (i.e., highly broadened), and a value of 0 indicates no change in resonance intensity. In some instances, resonance intensities increased during titrations, and thus fractional values became negative as per the calculation and as discussed in the text.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Conflict of interest statement
KHM is a paid consultant for Galectin Therapeutics.
Abbreviations
CRD, carbohydrate recognition domain; Gal-3, galectin-3; Gal-7, galectin-7; GM, galactomannan; HSQC, heteronuclear single quantum coherence; NMR, nuclear magnetic resonance; NT, N-terminal tail.
Acknowledgments
We thank Prof. Tai for providing the expression vector for human Gal-3, and Prof. Gabius for reading the manuscript and making helpful additions to the text. Funding for NMR instrumentation at the University of Minnesota was provided by the Office of the Vice President for Research, the Medical School, the College of Biological Sciences, NIH, NSF, and the Minnesota Medical Foundation.
References
- Ahmad N, Gabius H-J, André S, Kaltner H, Sabesan S, Roy R, Liu B, Macaluso F, Brewer CF. 2004. Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. J Biol Chem. 279:10841–10847. [DOI] [PubMed] [Google Scholar]
- Barondes SH. 2008. Stumbling on galectins. In: Klyosov AA, Witczak ZJ, Platt D, editors, Galectins. Hoboken: Wiley, p. 1–8. [Google Scholar]
- Berbís MÁ, André S, Cañada FJ, Pipkorn R, Ippel H, Mayo KH, Kübler D, Gabius HJ, Jiménez-Barbero J. 2014. Peptides derived from human galectin-3 N-terminal tail interact with its carbohydrate recognition domain in a phosphorylation-dependent manner. Biochem Biophys Res Commun. 443:126–131. [DOI] [PubMed] [Google Scholar]
- Birdsall B, Feeney J, Burdett ID, Bawumia S, Barboni EA, Hughes RC. 2001. NMR solution studies of hamster galectin-3 and electron microscopic visualization of surface-adsorbed complexes: Evidence for interactions between the N- and C-terminal domains. Biochemistry. 40:4859–4866. [DOI] [PubMed] [Google Scholar]
- Daas PJH, Schols HA, de Jongh HHJ. 2000. On the galactosyl distribution of commercial galactomannans. Carbohyd Res. 329:609–619. [DOI] [PubMed] [Google Scholar]
- Dawson H, André S, Karamitopoulou E, Zlobec I, Gabius H-J. 2013. The growing galectin network in colon cancer and clinical relevance of cytoplasmic galectin-3 reactivity. Anticancer Res. 33:3053–3059. [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 6:277–293. [DOI] [PubMed] [Google Scholar]
- Diehl C, Engström O, Delaine T, Håkansson M, Genheden S, Modig K, Leffler H, Ryde U, Nilsson UJ, Akke M. 2010. Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3. J Am Chem Soc. 132:14577–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova E, Miller MC, Nesmelova IV, Lopez-Merino L, Berbis MA, Nesmelov Y, Tkachev YV, Lagartera L, Daragan VA, André S et al. 2013. Lactose binding to human galectin-7 (p53-induced gene 1) induces long-range effects through the protein resulting in increased dimer stability and evidence for positive cooperativity. Glycobiology. 23:508–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabius H-J, editor. 2009. The Sugar Code. Fundamentals of Glycosciences. Weinheim: (Germany: ): Wiley-VCH. [Google Scholar]
- Gabius H-J, André S, Jiménez-Barbero J, Romero A, Solís D. 2011. From lectin structure to functional glycomics: Principles of the sugar code. Trends Biochem Sci. 36:298–313. [DOI] [PubMed] [Google Scholar]
- Gunning AP, Pin C, Morris VJ. 2013. Galectin 3-β-galactobiose interactions. Carbohydr Polym. 92:529–533. [DOI] [PubMed] [Google Scholar]
- Halimi H, Rigato A, Byrne D, Ferracci G, Sebban-Kreuzer C, El Antak L, Guerlesquin F. 2014. Glycan dependence of Galectin-3 self-association properties. PLoS One. 9:e111836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JD, Baum LG. 2002. Ah, sweet mystery of death! Galectins and control of cell fate. Glycobiology. 12:127–136. [DOI] [PubMed] [Google Scholar]
- Ilyina AV, Mestechkina NM, Shcherbukhin VD, Varlamov VP. 2006. Depolymerization of legume seed galactomannan by Celloviridin G20x. Prikladnaya Biokhimiya I Mikrobiologiya. 42:580–586. [PubMed] [Google Scholar]
- Ippel H, Miller MC, Berbís MA, Suylen D, André S, Hackeng TM, Cañada FJ, Weber C, Gabius H-J, Jiménez-Barbero J, Mayo KH. 2015. 1H, 13C, and 15N backbone and side-chain chemical shift assignments for the 36 proline-containing, full length 29 kDa human chimera-type galectin-3. Biomol NMR Assign. 9:59–63. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. 1994. NMR view: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 4:603–614. [DOI] [PubMed] [Google Scholar]
- Kaltner H, Gabius H-J. 2012. A toolbox of lectins for translating the sugar code: The galectin network in phylogenesis and tumors. Histol Histopathol. 27:397–416. [DOI] [PubMed] [Google Scholar]
- Klyosov AA, Traber PG.. 2012. Galectins in disease and potential therapeutic approaches. In: Klyosov AA, Traber PG, editors. Galectins and Disease: Implications for Targeted Therapeutics. Washington, DC: American Chemical Society; p. 3–43. [Google Scholar]
- Klyosov AA, Witczak ZJ, Platt D, editors. 2008. Galectins. Hoboken, New Jersey: John Wiley & Sons. [Google Scholar]
- Kohatsu L, Hsu DK, Jegalian AG, Liu FT, Baum LG. 2006. Galectin-3 induces death of Candida species expressing specific β-1,2-linked mannans. J Immunol. 177:4718–4726. [DOI] [PubMed] [Google Scholar]
- Kopitz J, André S, von Reitzenstein C, Versluis K, Kaltner H, Pieters RJ, Wasano K, Kuwabara I, Liu F-T, Cantz M et al. 2003. Homodimeric galectin-7 (p53-induced gene 1) is a negative growth regulator for human neuroblastoma cells. Oncogene. 22:6277–6288. [DOI] [PubMed] [Google Scholar]
- Kopitz J, Bergmann M, Gabius H-J. 2010. How adhesion/growth-regulatory galectins-1 and -3 attain cell specificity: Case study defining their target on neuroblastoma cells (SK-N-MC) and marked affinity regulation by affecting microdomain organization of the membrane. IUBMB Life. 62:624–628. [DOI] [PubMed] [Google Scholar]
- Kopitz J, von Reitzenstein C, André S, Kaltner H, Uhl J, Ehemann V, Cantz M, Gabius H-J. 2001. Negative regulation of neuroblastoma cell growth by carbohydrate-dependent surface binding of galectin-1 and functional divergence from galectin-3. J Biol Chem. 276:35917–35923. [DOI] [PubMed] [Google Scholar]
- Krzeminski M, Singh T, André S, Lensch M, Wu AM, Bonvin AM, Gabius HJ. 2011. Human galectin-3 (Mac-2 antigen): Defining molecular switches of affinity to natural glycoproteins, structural and dynamic aspects of glycan binding by flexible ligand docking and putative regulatory sequences in the proximal promoter region. Biochim Biophys Acta. 1810:150–161. [DOI] [PubMed] [Google Scholar]
- Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. 2004. Introduction to galectins. Glyconj J. 19:433–440. [DOI] [PubMed] [Google Scholar]
- Mestechkina NM, Shcherbukhin VD. 1991. Galactomannan from Galega orientalis Lam. seeds. Appl Biochem Microbiol. 26:648–651. [Google Scholar]
- Mey A, Leffler H, Hmama Z, Normier G, Revillard J-P. 1996. The animal lectin Galectin-3 interacts with bacterial lipopolysaccharides via two independent sites. J Immunol. 156:1572–1577. [PubMed] [Google Scholar]
- Miller MC, Klyosov AA, Mayo KH. 2009. The α-galactomannan Davanat binds galectin-1 at a site different from the conventional galectin carbohydrate binding domain. Glycobiology. 19:1034–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Klyosov AA, Mayo KH. 2012. Structural features for α-galactomannan binding to galectin-1. Glycobiology. 22:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Klyosov A, Platt D, Mayo KH. 2009. Using pulse field gradient NMR diffusion measurements to define molecular weight distributions in glycan preparations. Carbohydr Res. 344:1205–1212. [DOI] [PubMed] [Google Scholar]
- Miller MC, Nesmelova IV, Platt D, Klyosov AA, Mayo KH. 2009. Carbohydrate binding domain on galectin-1 is more extensive for a complex glycan than for simple saccharides: Implications for galectin–glycan interactions at the cell surface. Biochem J. 421:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Ribeiro JP, Roldos V, Martín-Santamaría S, Cañada FJ, Nesmelova IA, André S, Pang M, Klyosov AA, Baum LG. 2011. Structural aspects of binding of α-linked digalactosides to human galectin-1. Glycobiology. 21:1627–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S, Ahmad N, André S, Kaltner H, Gabius H-J, Brenowitz M, Brewer CF. 2004. Quaternary solution structures of galectins-1, -3, and -7. Glycobiology. 14:293–300. [DOI] [PubMed] [Google Scholar]
- Nesmelova IV, Dings RPM, Mayo KH. 2008. Understanding galectin structure-function relationships to design effective antagonists. In: Klyosov A. editor. Galectins. New York: Oxford University Press. [Google Scholar]
- Nesmelova IV, Ermakova E, Daragan VA, Pang M, Menendez M, Lagartera L, Solis D, Baum LG, Mayo KH. 2010. Lactose binding to galectin-1 modulates structural dynamics, increases conformational entropy, and occurs with apparent negative cooperativity. J Mol Biol. 397:1209–1230. [DOI] [PubMed] [Google Scholar]
- Nesmelova IV, Pang M, Baum LG, Mayo KH. 2008. 1H, 13C, and 15N backbone and side-chain chemical shift assignments for the 29 kDa human galectin-1 protein dimer. Biomol NMR Assign. 2:203–205. [DOI] [PubMed] [Google Scholar]
- Platt D, Klyosov AA, Zomer E. 2006. In: Klyosov AA, Witczak ZJ, Platt D, editors. Carbohydrate Drug Design. American Chem. Soc., ACS Symposium Series 932. Washington, DC: Academic Press p. 49–66. [Google Scholar]
- Plzák J, Betka J, Smetana K Jr, Chovanec M, Kaltner H, André S, Kodet R, Gabius HJ. 2004. Galectin-3 - an emerging prognostic indicator in advanced head and neck carcinoma. Eur J Cancer. 40:2324–2330. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruderisch H, Fischer C, Detjen KM, Welzel M, Wimmel A, Manning JC, André S, Gabius H-J. 2010. Tumor suppressor p16INK4a: Downregulation of galectin-3, an endogenous competitor of the pro-anoikis effector galectin-1, in a pancreatic carcinoma model. FEBS J. 277:3552–3563. [DOI] [PubMed] [Google Scholar]
- Seetharaman J, Kanigsberg A, Slaaby R, Leffler H, Barondes SH, Rini JM. 1998. X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution. J Biol Chem. 273:13047–13052. [DOI] [PubMed] [Google Scholar]
- Shcherbukhin VD. 1992. Galactomannans from seeds of the leguminous plants found in the Soviet Union. Food Hydrocolloids. 6:3–7. [Google Scholar]
- Smetana K Jr, André S, Kaltner H, Kopitz J, Gabius HJ. 2013. Context-dependent multifunctionality of galectin-1: A challenge for defining the lectin as therapeutic target. Expert Opin Ther Targets. 17:379–392. [DOI] [PubMed] [Google Scholar]
- Werz DB, Ranzinger R, Herget S, Adibekian A, von der Lieth CW, Seeberger PH. 2007. Exploring the structural diversity of mammalian carbohydrates (“glycospace”) by statistical databank analysis. ACS Chem Biol. 2:685–691. [DOI] [PubMed] [Google Scholar]
- Woodward AM, Senchyna M, Williams R, Argueso P. 2012. Characterization of the interaction between hydroxypropyl guar galactomannan and galectin-3. Biochem Biophys Res Commun. 424:12–17. [DOI] [PubMed] [Google Scholar]