Abstract
Molecular and genomic analysis of microscopic quantities of tumor from formalin-fixed, paraffin-embedded biopsy specimens has many unique challenges. Herein, we evaluated the feasibility of obtaining transcriptome-wide RNA expression to measure prognostic classifiers in diagnostic prostate needle core biopsy specimens. One-hundred fifty-eight samples from diagnostic needle core biopsy specimens (BX) and radical prostatectomies (RPs) were collected from 33 patients at three hospitals; each patient provided up to six tumor and benign samples. Genome-wide transcriptomic profiles were generated using Affymetrix Human Exon arrays for comparison of gene expression alterations and prognostic signatures between the BX and RP samples. A sufficient amount of RNA (>100 ng) was obtained from all RP specimens (n = 77) and from 72 of 81 of BX specimens. Of transcriptomic features detected in RP, 95% were detectable in BX tissues and demonstrated a high correlation (r = 0.96). Likewise, an expression signature pattern validated on RPs (Decipher prognostic test) showed correlation between BX and RP (r = 0.70). Of matched BX and RP pairs, 25% showed discordant molecular subtypes. Genome-wide exon arrays yielded data of comparable quality from biopsy and RP tissues. The high concordance of tumor-associated gene expression changes between BX and RP samples provides evidence for the adequate performance of the assay platform with samples from prostate needle biopsy specimens with limited tumor volume.
Prostate cancer is the second leading cause of cancer-related mortality in US men.1 The American Cancer Society estimates that >220,000 new cases of prostate cancer will be recorded in 2015, accounting for >25% of all cancers in men.2 Prostatic needle core biopsy is currently the most reliable standard for diagnosis of prostate cancer, and it is estimated that >800,000 patients undergo prostate biopsy annually in the United States.3 However, in addition to the recognized concerns about tumor heterogeneity and sampling errors associated with biopsy, the pathological findings and tumor grade do not always accurately predict tumor behavior and patient outcome. In addition, tumor grading has poor interobserver reproducibility in some cases, which can lead to uncertainty in grade assignment and subsequent misclassification of disease severity.4 Therefore, developing more sensitive and accurate biomarkers and prognostic tools is of critical clinical need to better risk-stratify patients when cancer is first diagnosed at biopsy, and will allow patients to make the most informed treatment management decisions possible.
Before molecular tests can be accepted into standard clinical practice, there is a need to demonstrate their analytical feasibility and clinical utility. When using formalin-fixed, paraffin-embedded (FFPE) tissue specimens as starting material, the FFPE processing causes degradation of RNA that generates challenges in using expression patterns as a clinical biomarker; the oxygen and hydroxyl radicals in formalin crosslink RNA, and the high temperatures of the wax involved in embedding the sample cause irreversible damage to RNA, with fragmentation into 150 to 200 bases long oligonucleotides.5 Run-to-run variations in processing parameters, as well as processing and storage variations from one institution to the next, can also affect RNA levels and degradation rates in FFPE tissue specimens. Tumor heterogeneity and the limited amount of tumor in biopsy material further affect expression analyses, introducing multiple, sometimes discordant, expression signatures. In addition, the multifocal and heterogeneous nature of prostate cancers poses even more challenges.6, 7, 8 More studies that use biopsy tumor samples and address the opportunities for biomarker and molecular signature evaluation studies are needed to improve patient management from the time of diagnosis.
Herein, we demonstrate the feasibility of using transcriptome-wide oligonucleotide microarray technology [Human Exon 1.0 ST GeneChips (Affymetrix Inc., Santa Clara, CA) with 1.4 million probe selection regions (PSRs)] optimized for use with RNA extracted from FFPE tissue specimens, and this protocol is performed in a Clinical Laboratories Improvement Amendment–certified reference laboratory, allowing it to be used to generate tumor expression data for clinical use in prognostic assays.9 For example, the Decipher test score—performed on prostate tumor tissue—is designed to predict metastatic prostate cancer risk after radical prostatectomy (RP), and is based on the expression of 22 markers from the 1.4 million PSRs on the chip. These markers relate to cell proliferation, differentiation, androgen signaling, motility, and immune modulation10, 11 and have been validated to predict metastatic progression after RP in several independent cohorts from multiple institutions.12, 13, 14 This genomic assay is currently covered by Medicare for helping to guide postoperative therapy decision making in patients with adverse pathological features.15 Use of this expression array protocol also allows for evaluation of various combinations of PSRs, and thus permits one to simultaneously assess other expression marker panels and data sets,10, 16, 17 as well as evaluate new ones.
To explore the transcriptomic differences between prostate biopsy and matched RP samples, and evaluate the effects of heterogeneity, we compared transcriptomic data generated using Human Exon arrays obtained from 158 different prostate tissue samples from 33 patients seen at three different institutions. This cohort provides, for the first time, an opportunity to compare the whole transcriptome array-based expression profiles obtained from matched biopsy and RP specimens from multiple institutional sources. It is particularly important as different procurement, processing, and sampling conditions are represented in this cohort for a more thorough evaluation of expression-based genomic classifiers such as Decipher test.18, 19, 20 Finally, we use the data to explore tumor heterogeneity and the assignments of recently described molecular subtypes of prostate cancers by comparing and contrasting expression patterns in this specimen cohort.20, 21
Materials and Methods
Patients and Samples
A total of 158 FFPE samples from 33 patients with matching biopsy and RP were collected from three institutions: University of California San Francisco (UCSF; n = 13), Cedars Sinai Medical Center (CSMC; Los Angeles, CA; n = 11), and the University Health Network (Toronto, ON, Canada; n = 9) (Supplemental Figure S1). Each institution's institutional review board committees gave approval of this study. These 158 samples comprised 64 tumor samples (33 from biopsy and 31 from RP), 47 benign adjacent to tumor tissue samples (24 from biopsy and 23 from RP), and 47 benign contralateral tissue samples (24 from biopsy and 23 from RP) (Table 1). For 23 patients from UCSF and CSMC, six prostate tissue samples were obtained from each patient: tumor biopsy, tumor RP, benign adjacent biopsy, benign adjacent RP, benign contralateral biopsy, and benign contralateral RP (Table 1). Tumor grade, tumor content, and stromal content were assigned by expert uropathologists (J.P.S., B.S.K., T.v.d.K.) on hematoxylin and eosin review for each biopsy core and RP tumor section used, with the area to be sampled for RNA extraction marked on each slide (Table 1). Except for cases analyzed before 2006 (two cases), all were categorized as robotic prostatectomies, which have similar warm and cold ischemia times. These cases were all fixed within 1 hour of resection by formalin injection technique. Biopsy specimens are all fixed immediately on removal of the specimen.
Table 1.
Clinicopathologic Variables and QC Characteristics of Samples and Tissue Types
| Variables/characteristics | Biopsy |
Radical prostatectomy |
Total | ||||
|---|---|---|---|---|---|---|---|
| Contralateral | Adjacent | Tumor | Contralateral | Adjacent | Tumor | ||
| Sample storage age, mean (SD), years | 3.29 (2.69) | 3.06 (2.69) | |||||
| Available tissue | 24 | 24 | 33 | 23 | 23 | 31 | 158 |
| Clinicopathological | |||||||
| Tumor content, % | |||||||
| Median | 0 | 0 | 70 | 0 | 0 | 80 | |
| Range | 0–0 | 0–0 | 10–90 | 0–0 | 0–0 | 50–90 | |
| Stromal content, % | |||||||
| Median | 60 | 60 | 25 | 60 | 60 | 20 | |
| Range | 40–80 | 40–80 | 10–80 | 10–80 | 40–80 | 5–50 | |
| Gleason score | |||||||
| 6 | 0 | 0 | 13 | 0 | 0 | 10 | 23 |
| 7 | 0 | 0 | 14 | 0 | 0 | 16 | 30 |
| 8 | 0 | 0 | 4 | 0 | 0 | 4 | 8 |
| 9 | 0 | 0 | 2 | 0 | 0 | 2 | 4 |
| Quality control | |||||||
| Successful RNA extraction | 23 | 22 | 27 | 23 | 23 | 31 | 149 |
| Failed RNA extraction | 1 | 2 | 6 | 0 | 0 | 0 | |
| Median RNA yield, ng | 463.28 | 283.74 | 278.25 | 2814.3 | 2319.04 | 2846.7 | |
| Successful cDNA amplification | 23 | 22 | 27 | 22 | 23 | 31 | 148 |
| Failed cDNA extraction | 0 | 0 | 0 | 1 | 0 | 0 | |
| Median cDNA yield, ng | 7474.68 | 6984.275 | 7066.44 | 6704.88 | 7011.2 | 6461.91 | |
| Array good QC | 23 | 22 | 26 | 22 | 23 | 31 | 147 |
| Median % present | 54.34 | 51.16 | 48.65 | 46.91 | 51.03 | 49.44 | |
The array quality is equivalent across tissue types and samples.
QC, quality control.
Tissue Selection and Sampling
At CSMC and UCSF, tumor tissues were sampled from FFPE biopsy cores by dissecting a portion of the tumor tissue directly from the blocks in the areas corresponding to marked areas on each hematoxylin and eosin–stained slide using either a 1-mm sterile biopsy punch tool (UCSF) or 0.6-mm cylindrical full-thickness cores using the Tissue microarrayer (Pathology Devices, Westminster, MD). Next, RNA was extracted as described below. Either one to two 0.6-mm punches from the center of the tumor (CSMC) or one to five superficial punches sampling half of the tumor in a single core (UCSF) were obtained. The histologically benign peripheral zone glandular tissue was sampled in an analogous manner. Benign tissues were defined as adjacent to tumor (benign adjacent) when they were within 1 to 5 mm of the tumor. Benign contralateral tissues were obtained from the side of the prostate opposite to the tumor, and as far away from any other tumor or high-grade prostatic intraepithelial neoplasia areas as possible. To minimize any effects because of tumor heterogeneity, the area matching the biopsy specimen was identified in the RP specimen and punched. Tissues were punched in the same manner at locations matching those where the biopsy punches were taken for each tissue type (tumor, benign adjacent to tumor, and benign contralateral to tumor). In the RP specimens, only a single punch was obtained for each tissue type. For example, if the biopsy specimen used for RNA extraction reportedly was from the right apex, then the tumor in that portion of the RP also was sampled for extraction. This matching was performed for all tissue types (tumor, benign adjacent, and benign contralateral). At University Health Network, both biopsy and RP specimens were divided into sections (4 μm thick) to generate unstained sections, and the areas of interest were macrodissected (scraped) from the slides for RNA extraction. Six unstained sections were used to isolate RNA from biopsy tissue, and four to isolate RNA from RP tissue. RP tissue was sampled in locations matching the location of tumor in the biopsy cores.
RNA Extraction, Quantification, and Quality Control
Total RNA was extracted and purified using the RNeasy FFPE kit (Qiagen, Valencia, CA). RNA was amplified and labeled using the Ovation WTA FFPE system (NuGen, San Carlos, CA) and hybridized to Human Exon 1.0 ST GeneChips (Affymetrix Inc.), according to the manufacturer's recommendations. Using this approach, the expression of >1.4 million PSRs was quantified. Quality and quantity of RNA extracted and cDNA amplified were measured with a NanoDrop 1000 (Thermo Scientific Inc., Wilmington, DE). RNA (50 to 100 ng) was required for cDNA amplification. The ratio of absorbance (260/280 nm) used to assess the purity of RNA and ratio values between 1.7 and 2.2 were considered of acceptable purity. Quality control for microarray data was performed with the Affymetrix Power Tools packages and with internally developed metrics, including percentage present—the percentage of probes detected above the background defined the detection level of background probes with similar GC content. The positive versus negative area under the curve (AUC) was used as an additional metric to assess microarray quality by measuring the signal between positive control probes, which measure the expression of housekeeping genes, and negative control probes, which measure antigenomic sequences and hence should exhibit background intensity levels.10 This metric can represent the quality of the RNA sample, with an AUC of 1 reflecting perfect separation that indicates no false positives are detected, whereas all true positives are measured.
Expression Data Processing Analysis
The expression of approximately 1.4 million PSRs was normalized and summarized using SCAN22 to the Affymetrix core transcript cluster level (approximately 22,000 genes). Expression data were uploaded to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, accession number GSE72220). To reduce measurement error because of laboratory variability, matching biopsy and RP samples were processed in the same batch, and batch correction was performed using ComBat (http://www.bu.edu/jlab/wp-assets/ComBat/Abstract.html, last accessed January 7, 2015)23 on the expression data before analysis. Differential expression analysis using paired median fold difference (MFD). (MFD, ) was used to identify discriminative features between tumor and benign tissues. Dimensionality reduction was performed using principal component analysis (PCA), and significant sources of experimental and biological signal associated with the genomic variance captured by each principal component were assessed using the Mann-Whitney U test. The Fisher's exact test was used to assess the significance of concordance between Decipher measured from biopsy and Decipher measured from RP samples.
Field Effect Assessment
Matching tumor, benign adjacent to tumor, and benign contralateral samples for each patient were used to assess a potential genomic prostate cancer field effect. The expression profiles for features on the array were evaluated between the tumor and the two benign samples using Pearson's correlation. One-tailed P values were computed and adjusted using the false discovery rate method. Features correlated between tumor and benign with a P < 0.05 were considered candidate field effect features. This assessment was performed on the RP and biopsy samples separately to minimize confounding.
Prostate Cancer Molecular Subtyping
Patients in this study were classified into four previously published molecular subtypes that are mutually exclusive of each other: ERG+, ETS+, SPINK1+, and triple negative.21 Outlier expression analysis of ERG, ETS (ETV1, ETV4, ETV5, and FLI1), and SPINK1 was used to assign each tumor sample to one of the subtypes. Patients exhibiting an outlier profile in either of genes were annotated with +, and − otherwise. Patients with high ERG expression profile (ERG+) and not exhibiting outlier profiles for the other genes were classified as ERG+ subtype, patients who were ETV1+, ETV4+, ETV5+, or FLI1+ and ERG− and SPINK1− were classified as ETS+ subtype, patients who were SPINK1+, ERG−, and ETS− were classified as SPINK1+, patients not exhibiting outlier profiles of any of these six genes were classified as the triple-negative subtype.
Results
Clinical and Pathological Characteristics of Patient Samples
Matched biopsy and RP specimens from three institutions using several sampling techniques were used to investigate the feasibility of obtaining high-quality and comprehensive whole transcriptome profiling data from FFPE tissue samples using Human Exon arrays. Patients had a median time between RP and biopsy of 86 days. The tumors in the samples covered a wide spectrum of Gleason scores (GSs) (Table 1) and in the RPs included GS = 3 + 3 (13 cases), GS = 3 + 4 (six cases), GS = 4 + 3 (eight cases), GS = 4 + 4 (four cases), and GS = 4 + 5 (two cases). The individual GSs in the biopsy specimens that were sampled matched those in the corresponding RP specimen, with the exception of two cases with GS 3 + 3 in the biopsy that were upgraded in the RP specimen. None of the cases were downgraded at RP.
Tissue Characteristics of Biopsy versus RP Samples
Samples were stored for an average of 3.1 years (σ = 2.6 years) before processing (Supplemental Table S1). The percentage of tumor involvement in the biopsy cores ranged from 5% to 70%, and the percentage tumor in the cylindrical punches of these cores ranged from 10% to 90%. The percentage tumor involvement of punches from RP specimens was higher and ranged from 50% to 90%. The stromal content in the punches of the biopsy cores ranged from 10% to 80%, whereas the percentage of benign epithelial content was <5% (Table 1). The amount of tissue that was provided for RNA extraction also varied between the three sites because of differences in the diameter of the punches (ie, 0.6 versus 1.0 mm) and sampling method (ie, a single cylindrical core using a biopsy punch versus multiple cores versus scraping from unstained sections). When the tumor occupied only 5% (0.5 mm) of the length of the biopsy specimen, the sample did not yield enough RNA (n = 2, 100%). However, at 1-mm tumor length and at least 35% tumor content in the punch area, the amount of RNA was sufficient to pass the quality threshold and generated high-quality data from the assay.
Transcriptome Data Quality from Biopsy versus RP Samples
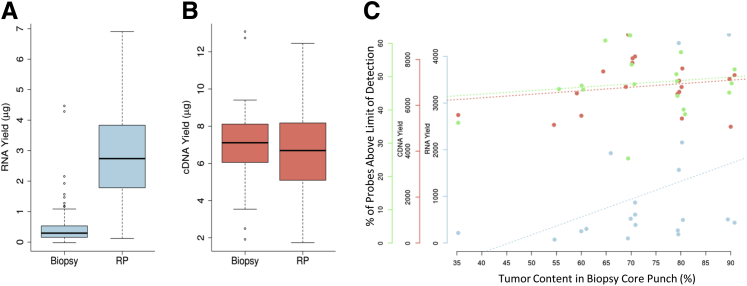
RNA was extracted from 0.6-mm (CSMC) and 1-mm (USCF) cylindrical punches and from macrodissected tumor from unstained sections (4 μm thick; University Health Network) from FFPE blocks. For each patient, the same procedure was used at each site for collecting samples from both biopsy and RP specimens. All 77 RP samples, but only 72 (89%) of 81 biopsy samples yielded sufficient RNA for cDNA amplification (Table 1). The lower yield of RNA from biopsy punches is explained by the smaller depth of tissue in biopsy specimens (at most, 1-mm core diameters) compared with RP (at least 2-mm thick tissue slice). In addition, RNA yield from unstained sections was, in general, lower than from punches (data not shown). Although the RNA yield from RP samples was approximately 10-fold greater than the RNA yield from biopsy samples (Figure 1A), when using 100 ng of RNA as a starting material, comparable amounts of cDNA were amplified from both sample sources (Figure 1B). All samples passed cDNA amplification, except one biopsy sample. A positive correlation was observed between RNA yield from punches of the biopsy cores and amount of tumor tissue in the punches. Overall cDNA yield remained relatively consistent between sample sources (Figure 1C).
Figure 1.
Quality control (QC) characteristics of biopsy and radical prostatectomy (RP) samples. A: Total RNA yield in RP samples is greater than in biopsy samples. B: An equal amount of extracted RNA was converted to cDNA, yielding similar amounts of cDNA in biopsy specimens and RPs. C: The three QC parameters (RNA yield, cDNA yield, and microarray probe signal intensity) are not affected by tumor content in the punch of the biopsy core samples. n = 77 (A, RP samples); n = 81 (A, biopsy samples).
Quality assessment of expression data generated from the assay was performed by assessing the sensitivity and specificity of the microarray. Sensitivity is assessed using the percentage of probe sets that provide a signal higher than the level of detection (LOD), whereas specificity is measured by the discrimination of positive and negative control probes, as calculated by the AUC. Biopsy and RP samples had similar medians of probe sets higher than the LOD, 51.3% for biopsy and 48.6% for RP samples; however, biopsy samples possessed greater AUCs than RP samples, with median AUCs of 0.76 versus 0.70 (P < 0.01), respectively. This suggests that the RNA quality of biopsy samples is higher than that of RP samples. Significantly, the percentage of probes higher than the LOD did not correlate with the tumor content present in the biopsy cores or punches evaluated (Figure 1C).
Transcriptome-Wide Expression Analysis in FFPE Tissues Obtained Through Biopsy
Having demonstrated that RNA extracted from needle core biopsy FFPE tissue samples was sufficient to generate transcriptome data using the assay and of comparable quality to that obtained from RP-derived samples, we next investigated the correlations of expression between biopsy specimens and RPs. Approximately 50% of the 1.4 million probe sets on the array provided signal intensities higher than the LOD (Table 1). Of these probe sets, 70% (490K probe sets) were detected in all RP samples, and of those, 95% were also detected in all of the biopsy samples, supporting the analytical feasibility of applying genome-wide exon arrays to prostate biopsy specimens.
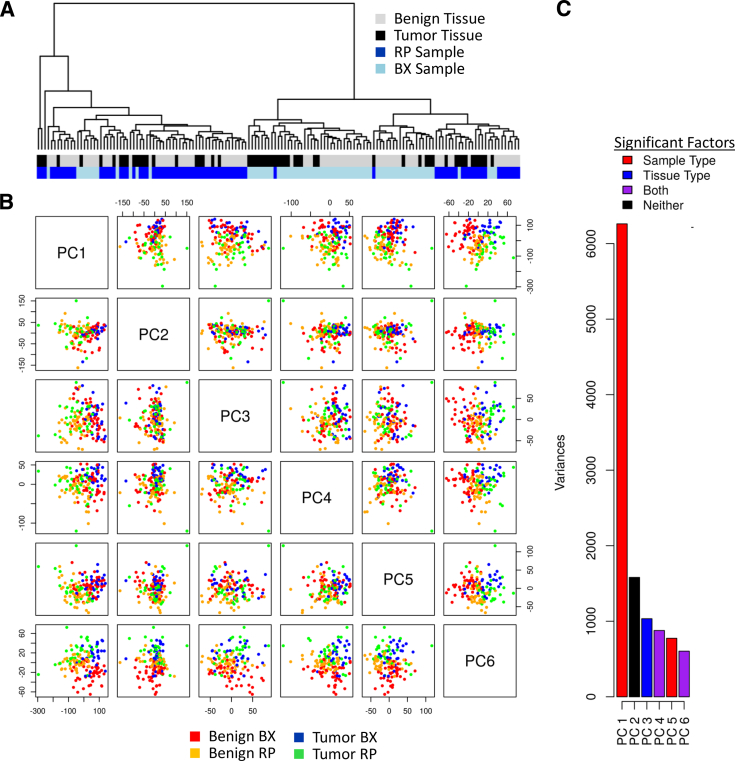
To examine if tumor-associated signals could be detected in the biopsy and RP samples, we first performed an unsupervised hierarchical clustering and PCA on the microarray expression data. The hierarchical clustering analysis revealed that biopsy and RP samples formed two large clusters (Figure 2A) and that the origin of the sample (ie, biopsy versus RP) was the main responsible determinant. Within each cluster, moderate sub-clustering of tumor and benign samples was also observed. The PCA confirmed the clustering results, demonstrating that the sample origin (biopsy versus RP) was the biggest source of variation at the global level. This was observed mainly in principle component 1 (PC1). Furthermore, with the exception of PC2, the first six PCs all associated with sample origin (biopsy versus RP) and tissue type (benign versus cancer). In addition to the origin of the sample, it became evident from the PCA that the tissue type (ie, tumor versus benign) was another main source of variance in the data set. As seen most clearly in PC6, samples from benign tissue in both biopsy and RP clustered together (Figure 2, B and C). Despite the limiting amount of tissue and sources of technical variability (eg, differences in time to fixation between biopsy and RP), tumor-associated signals were clearly identified in biopsy tissues using unsupervised and unbiased transcriptome-wide data analysis methods.
Figure 2.
Sources of variance in radical prostatectomies (RPs) and biopsy (BX) samples. A: Hierarchical clustering of samples from tumor and benign tissues from BX and RP origins. The two main clusters are generated on the basis of the origin (BX versus RP), containing both benign and tumor samples. Additional clusters are obtained on the basis of the tissue type (tumor versus benign). B and C: Principal component analysis (PCA) shows that sample origin (BX versus RP) contributes the biggest variation and that tissue type (tumor versus benign) contributes less to the total variation.
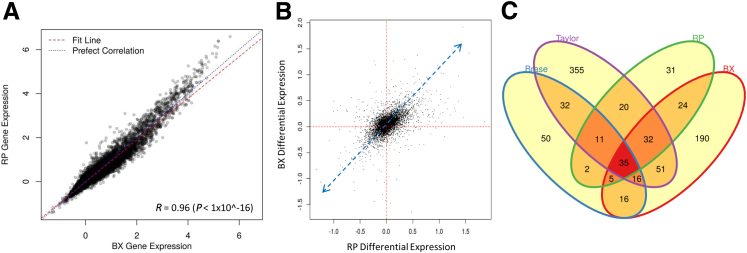
A comparison of overall gene expression levels between biopsy and RP samples demonstrated a highly positive correlation between expression values (r = 0.96) (Figure 3A and Supplemental Table S2). This suggested that, by comparison to expression levels generated from RP tissues, the assay could reliably quantify RNA expression levels in biopsy samples and, furthermore, that the expression profiles in biopsy and RP samples were highly analogous. A reduction in dynamic range in the biopsy samples was observed, but the effect was small. To further illustrate the within-patient RP-biopsy variability, correlation plots were generated for four randomly selected patients, which showed that there is strong consistency between RP and biopsy sample expression, even for individual patients (Supplemental Figure S2).
Figure 3.
Expression analysis between radical prostatectomies (RPs) and biopsy (BX). A: High concordance of gene expression levels between BX and RP (r = 0.96). B: Differential expression analysis using median fold difference (MFD) shows that the MFD between RP tumor and RP benign contralateral, and BX tumor and BX benign contralateral, is consistent in terms of directionality. C: The genes differentially expressed between tumor and benign contralateral in BX and RP are overlapping with genes differentially expressed between tumor and benign contralateral in external public data sets. Statistical significance was assessed via bootstrapping (P < 0.05).
To compare changes in gene expression levels that are specific to cancer, we determined the differential expression for each gene between cancer and contralateral benign tissue for both the biopsy and RP specimens separately. Benign adjacent samples were removed because of possible tumor contamination and field effect, which may confound the analysis, and to allow for a pairwise analysis. Differential expression analysis using paired MFD demonstrated the same directionality of expression changes in biopsy and RP and, in addition, similar magnitudes of expression changes in both biopsy and RP samples (Figure 3B). Although the magnitude of the MFD is affected by the difference in tumor content and efficiency of RNA extraction between biopsy and RP, the results nevertheless demonstrate that the assay faithfully captured the biological signal in both specimen types. An assessment of individual patients revealed similar trends, where benign-tumor gene expression differences in the biopsy and RP samples had significant, positive correlations (Supplemental Figure S3). In addition, prostate cancer–related genes that were found to be differentially expressed in analysis of two public data sets (Taylor et al24 and Brase et al25) were confirmed in our samples, independent of their origin from biopsy or RP (Figure 3C). Bootstrapping analysis revealed the observed overlap between these sets to be statistically significant (P < 0.05). Together, these data demonstrate that relevant and consistent biological prostate cancer–specific signals exist in data generated from both biopsy and RP specimen types.
Prostate Cancer Prognostic Signatures (Cuzick, Klein, Penney, and Decipher) in Prostate Needle Core Biopsy Specimens
Having demonstrated the feasibility of using the assay for evaluation of FFPE needle core biopsy specimens, we next assessed the robustness of four prostate cancer prognostic signatures. In addition to Decipher scores, we evaluated the expression levels of genes used in other published molecular signatures used for prostate cancer risk stratification, including expression signatures from Cuzick et al,19 Klein et al,18 and Penney et al,20 as previously described.5 For comparison, the RNA expression of the individual genes comprising these signatures was evaluated in both biopsy and RP samples. More than 94% of the features were higher than the LOD in all biopsy and RP samples, providing evidence of the ability of the assay to capture the biological signal of multiple prognostic signatures.
The Decipher scores showed a positive correlation (r = 0.70, P < 0.001) between biopsy and tumor RP samples (Figure 4A). Similarly, the Penney et al20 signature also showed a positive correlation (r = 0.65, P < 0.001) (Figure 4B). To show that this result is robust, we progressively removed one and then two points driving the correlation from the analysis and reevaluated the model's correlations (Supplemental Figure S4). We did not observe major changes in the correlations, except for Penney et al,20 which was found to have a borderline significant correlation after removing two of the driving points. Technical variables, such as percentage of tumor in punch, RNA yield, cDNA yield, and percentage of probes higher than the LOD, did not affect the Decipher scores in matched pairs of RP and biopsy tissues. Using validated cut points, Decipher patient risk categories between biopsy and RP were concordant in 75% of cases (Figure 4C). Most of the discordant cases were at the border between categories of low and intermediate risk of metastatic development. Using Fisher's exact test, the concordance in Decipher BX and RP scores trends toward significance (P = 0.08). This failure to reach significance is most likely because of the small sample size and the few patients classified as high risk. It is important to note that Decipher scores were independent of tumor content, demonstrating that the Decipher test is robust, despite limitations posed by formalin fixation and small amounts of cancer tissue in biopsy specimens.
Figure 4.
Robustness of the Decipher assay between biopsy (BX) and radical prostatectomies (RPs). A: Correlation of Decipher scores from BX and RP samples (r = 0.7). B: Correlation of scores for the Penney et al20 signature from BX and RP samples (r = 0.65). The blue dashed line represents the line of best fit, whereas the dotted blue lines represent the 95% CI. C: Concordance of Decipher category between cancers in RP and BX (Fisher's exact test P = 0.08). Int., intermediate.
Transcriptome Expression Assessment with Respect to Field Effect
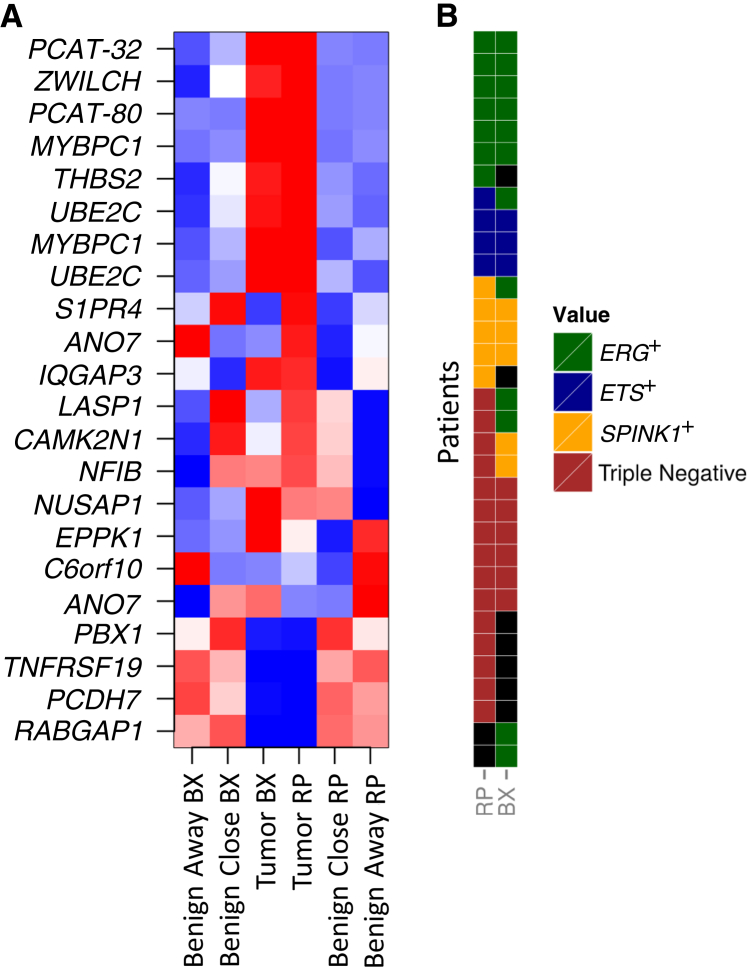
The 22 Decipher features were measured across the three tissue sources in the study: tumor, benign adjacent to tumor, and benign contralateral to tumor (Figure 5A). We observed 15 (68%) of the 22 markers displaying the same pattern of gene expression in both biopsy and RP across the three tissue types. Overall, the 22 features showed highly concordant expression patterns between matched tumor samples from biopsy and RP, with generally higher expression of these genes in tumor compared with matched benign samples.
Figure 5.
A: The expression of Decipher features is consistent across tissue types between radical prostatectomies (RPs) and biopsy (BX). Most features are overexpressed in tumor compared with benign in RP and BX. A group of features (LASP1, CAMK2N1, and NFIB) expressed at levels similar to the tumor specimens in benign adjacent to tumor (benign close to tumor) but not in benign contralateral to tumor (benign away from tumor). B: The concordance of prostate cancer molecular subtypes between BX and RP cancer samples.
Next, we explored molecular heterogeneity by comparing expression of prostate cancer lineage and subtype markers (ERG, ETV1, ETV4, ETV5, and SPINK1) for each RP–biopsy pair for tumor and benign samples. Samples were grouped into four mutually exclusive molecular subtypes, ERG+, ETS+, SPINK1+, and triple negative, as described in Materials and Methods.26 As expected, these prostate cancer subtype markers were found mostly in tumor tissues; however, a few benign samples had outlier expression of these genes, suggesting contamination of some tumor cells in the histologically benign tissue (Supplemental Figure S5). In biopsy samples, 12, 3, 5, and 6 of 26 were assigned to ERG+, ETS+, SPINK1+, and triple negative, respectively. In RP samples, 7, 4, 5, and 15 of 31 were assigned to ERG+, ETS+, SPINK1+, and triple negative, respectively. In matched biopsy and RP sample pairs, overall 18 (75%) of 24 had concordant subtypes (Figure 5B). In RP samples, four cases of the 23 adjacent benign sample demonstrated outlier expression of ERG, ETV5, and SPINK1 genes. Six of 24 matched biopsy and RP sample pairs showed different molecular subtypes: two were SPINK1+ in biopsy and triple negative in RP, two were ERG+ in biopsy and triple negative in RP, one was ERG+ in biopsy and SPINK1+ in RP, and, finally, one was ERG+ in biopsy and ETS+ in RP. The data herein indicate, for the first time, implementing molecular subtypes in prognostic assays to improve the currently available prognostic test for evaluation in prostate needle biopsy specimens and shed light on the, yet unmet, clinical need for integrating molecular subtypes and prognostic assays.
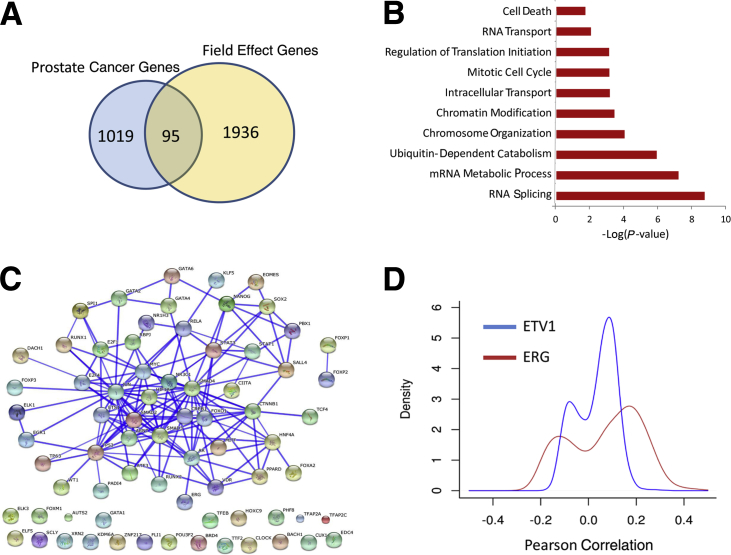
Finally, we assessed the extent of the prostate cancer field effect by examining the transcriptome-wide correlation between the tumor and matching benign samples within RP. We identified the genomic features on the microarray with significantly correlated expression (P < 0.05, after false discovery rate P value adjustment) between the tumor and the two types of benign samples. As expected, there were 7168 correlated features between RP tumor and benign adjacent samples compared with only 291 correlated features between RP tumor and benign contralateral samples (Supplemental Figure S6). Of 7168 features, 225 were identified in all three samples. As expected, the results show an overall higher correlation between tumor and benign adjacent samples compared with tumor and benign contralateral samples (Supplemental Figures S6 and S7 and Supplemental Tables S3 and S4). To shed further biological insights into the field effect genes, we used a subset (n = 2031) of the most correlated genes between paired tumor and adjacent benign samples (Pearson correlation >0.6 and P < 0.001) and compared it with a list of prostate cancer genes (n = 1114) that are differentially expressed between tumor and benign tissues in Memorial Sloan Kettering Cancer Center24 and German Cancer Research Center25 public data sets. Comparative analysis showed that field effect genes are distinct from prostate cancer genes, with only 95 genes overlapping (Figure 6A). Field effect genes are highly enriched with RNA splicing, chromosome organization, and intracellular transport biological processes (Figure 6B), and possess binding sites of key prostate cancer transcription factors, including AR, TP53, ETS1, JUN, CREB1, and FOXO1, on the basis of enrichment analysis of chromatin immunoprecipitation data sets using the ChEA tool27 (Figure 6C). To assess if field effect genes are correlated with ERG and ETV1 genomic rearrangements, a Pearson correlation coefficient between field effect genes and ERG and ETV1 expression was determined and revealed only a poor correlation (Figure 6D).
Figure 6.
Functional analysis of the field effect signature. A: The field effect signature is distinct from prostate cancer genes differentially expressed between tumor and benign tissues (Memorial Sloan Kettering Cancer Center and German Cancer Research Center).24, 25B: Functional enrichment analysis reveals that the field effect signature is highly enriched with gene categories of RNA splicing, ubiquitin-dependent catabolism, epigenetics, and cellular transport. C: Functional interaction networks generated by STRING of 75 transcription factors whose targets are enriched in the field effect signature on the basis of a chromatin immunoprecipitation enrichment analysis tool.27D: Density plots of Pearson's correlation between field effect signature and ERG and ETV1 genes, suggesting that field effect is independent of ERG and ETS.
Discussion
Accurate pretreatment risk assessment using prostatic needle biopsy specimens, although challenging, is essential to proper prostate cancer patient management. When prostate cancer is first diagnosed, it is necessary to determine the best individualized treatment plan for each patient. Accurate prognostication at the time of diagnosis is challenging because of several reasons. First, the standard 12 core prostate needle biopsy only samples a small fraction of the prostate and, therefore, may not be a representative sampling of the most significant tumor, and may not provide sufficient material for deep expression profiling. Second, the accuracy of Gleason grading can be compromised by the sampling error of the biopsy process; the GS is increased at RP in approximately 20% to 50% of cases.28 Thus, the severity of the cancer is often underestimated at biopsy. In addition, the subjective nature of tumor grading and assigning GSs complicates prognostication. The unpredictability of disease progression is further affected by the genetic heterogeneity and multiclonality of tumors that can appear histologically identical or, surprisingly, even lower grade.6, 7 All of these challenges highlight the need for more sensitive and robust genomic-based risk stratification methods that are applicable to prostate biopsy specimens so that men can more confidently choose a proper cancer management strategy. The small amount of tumor usually present in FFPE tissue specimens poses a barrier that is particularly difficult to overcome in the analysis of tissue biomarkers.
Characterizing the RNA expression of the tumor in biopsy tissues may provide informative clinical insights into the true aggressiveness of the tumor. Herein, we took advantage of Human Exon 1.0 ST arrays, a high-density oligonucleotide microarray that measures 1.4 million transcriptome-wide PSRs representing all known genes and many noncoding RNAs. Because of well-characterized assay characteristics and excellent performance in RP FFPE tissues, we decided to test the workflow with its quality control standards in biopsy specimens using the matched RP specimens as a reference.
Several studies have been performed demonstrating the generation of high-quality gene expression data from FFPE specimens. Bibikova et al29 and Frank et al30 assessed the reproducibility of FFPE samples profiled with oligonucleotide arrays and found high concordance between replicate samples. Likewise, high correlations (r ≥ 0.83) were observed between array data from FFPE and snap-frozen tissues.30 Pillai et al31 profiled 462 FFPE metastatic tumor biopsy specimens with Pathchip arrays for a tumor of origin test and found high-quality data in 80% of cases. Our study builds on this foundation by focusing on the comparison between FFPE biopsy specimens and FFPE surgical samples from prostate cancer patients. Prostate biopsy specimens present a unique challenge because of issues of heterogeneity and significant stromal contamination. Likewise and unlike previously cited studies,29, 30, 31 which were all preformed in research laboratories, all samples in this study were assayed in a Clinical Laboratories Improvement Amendment–certified laboratory. Finally, unlike the cited feasibility studies with FFPE tissues, which used gene expression arrays, this study uses a transcriptome-wide, high-density, exon array to profile samples.
To assess the technical feasibility of running transcriptome-wide arrays on biopsy samples, we took advantage of a multi-institutional cohort representing various tissue sampling methods. The way the samples were obtained (direct biopsy of the FFPE block versus scraped area of unstained sections) and the variability in institution processing did not hinder the ability to generate genome-wide transcriptome data. The generated data demonstrate that a sufficient amount of RNA of suitable quality for molecular genomic analysis can be consistently and successfully derived from the limited tumor tissue in biopsy cores, even with minuscule tumor content (1 mm length of tumor in a biopsy core with at least 40% concentration of tumor cells). Optimally, we found the direct biopsy method of the block using a punch tool to give the highest yields, and with a single 1-mm diameter punch, almost all biopsy specimens could be investigated without depleting the block from cancer, as is often the case when biopsy specimens are divided into sections serially for hematoxylin and eosin diagnosis, immunohistochemistry, and genomic assays. This is important for many pathology laboratories, which do not want to deplete FFPE blocks of tumor cells for medicolegal reasons or want to preserve tumor for future clinical uses. Furthermore, not only have we demonstrated that biopsy-derived RNA is equivalent to that of RNA obtained from surgical specimens, it can be considered in some regards to be superior. This observation might not be surprising given that biopsy cores are typically fixed in formalin immediately after they are obtained from the patient, whereas surgical specimens experience a prolonged hypoxic period before they are immersed in the formalin fixative, and it takes more time for the fixative to penetrate and diffuse into the larger tissue volume if rapid fixation techniques, such as fixative injection, are not used. The delay could potentially lead to RNA degradation and perturb RNA expression profiles. However, we observed a high correlation overall between gene expression data generated from biopsy and RP samples (r = 0.96), suggesting that most genes do not change in abundance despite ischemic and/or slower fixation conditions.
We further examined the expression of prostate cancer genes from other prostate cancer prognostic tests (ie, Cuzick et al,19 Klein et al,18 and Penney et al20), which can be measured using Human Exon arrays. More than 94% of the genes from Cuzick et al,19 Klein et al,18 and Penney et al20 had signal higher than the LOD. The expression of the 22 features in the Decipher test and the Decipher scores were in general concordant between RP and biopsy. A patient-per-patient pairwise agreement between biopsy and RP samples using the Decipher risk category between biopsy and RP tumor samples showed good overall concordance (75%). This result provides preliminary evidence that the Decipher score may be predictive of disease progression in diagnostic prostate needle biopsy specimens and warrants a larger, adequately powered study to validate this observation.
To provide additional molecular insights, we examined whether matched tissues harbor similar molecular subtypes of prostate cancer. Overall, there is strong agreement in molecular subtypes between cancers in RP and biopsy specimens. In addition, the molecular analysis revealed the coexistence of several clones within the region of cancer that was analyzed. These results demonstrate that, in addition to the Decipher score, the molecular subtypes in the biopsy are representative of the cancer in the RP. Taken together, these observations provide additional evidence as to the potential clinical utility of high-resolution expression arrays in the biopsy setting for use in molecular classification, risk stratification, and prognosis.
Further characterization of the transcriptome data from tissues adjacent or contralateral to the tumor revealed that there is higher concordance in gene expression profiles between tumors and adjacent benign compared with contralateral benign samples, and that the expression of many Decipher features is higher in the tumor compared with the benign tissues. Although, in the RP specimens, we could clearly detect a field effect, with a higher proportion of correlated expressed genes between tumor and adjacent compared with contralateral histologically nonneoplastic tissue, this was not detected in the biopsy specimens. Functional characterization of the field effect genes revealed that they are related to key biological processes involved in tumor progression and key prostate cancer transcription factors, including AR, JUN, HIFA1, and TP53. On the other hand, they are not correlated with genomic rearrangements, suggesting that adjacent benign tissues harbor a distinct biology. Furthermore, the detection of tumor-specific subtype markers, such as ERG+, ETS+, and SPINK1+, in the benign specimens suggests the presence of tumor cells in histologically nonneoplastic tissue may be a confounder for the measurement of a field effect. Overall, the molecular subtyping results show that for 25% of patients analyzed, tumor heterogeneity could be detected between the biopsy and RP specimens.
Previous reports of investigating multifocality in prostate RP specimens have shown between 41% and 67% discordance rates between ERG status within the same patient.26 Other studies have evaluated single prostates and detected numerous tumors with different clonal type and origin.6 To our knowledge, no previous study has reported the rate of subtype discordance between matched biopsy-RP specimens using six molecular markers (ERG, ETV1, ETV4, ETV5, FLI1, and SPINK1).
Although the conclusions drawn from this study might be limited by the small sample size, the data provide, for the first time, evidence of concordance of a genomic classifier across matched tumor samples from biopsy and RP in a multi-institutional cohort. Altogether, the data demonstrate the feasibility of measuring RNA expression in FFPE prostate needle biopsy specimens with small amounts of tumor using high-density expression arrays. These data suggest that a high-density array run on prostate biopsy specimens also provides potentially useful information on tumor heterogeneity, which can be combined with currently validated RNA expression–based tests to improve prediction of cancer progression.
Footnotes
Supported by GenomeDx Biosciences Inc. grant NIH/NCI R01CA131255-01A1 (B.S.K.), Spielberg Discovery Fund in Prostate Research (B.S.K.), Cedars Sinai institutional support grant NIH/NCI R01CA182438-01A1 (H.L.K.), and Department of Defense: Transformative Impact award W81XWH-13-2-0074 (J.M.C.).
Disclosures: N.E., H.S., M.A., L.L.C.L., and E.D. are employees of GenomeDx Biosciences Inc. P.R.C. has a research relationship with Genomic Health Inc. J.M.C.'s spouse is a full time employee of Myriad Genetics Labs, receiving salary, work travel reimbursements, and some stock shares. J.M.C. is a shared owner of a noncommercialized patent for using nutrient and germline genetic data for predicting aggressive prostate cancer. The University of California San Francisco is paid consulting fees by GenomeDx and Genomic Health on the basis of work performed by J.P.S., and J.P.S. receives unrestricted research funds from Myriad Genetics and Genomic Health.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2015.12.006.
Supplemental Data
A: Flow chart diagramming the composition of the study. University of California San Francisco (UCSF), Cedars Sinai Medical Center (Cedar Sinai; Los Angeles, CA), and University Health Network (UHN; Toronto, ON, Canada) each supplied patients for the study. For each patient from UCSF and Cedar Sinai, six samples were provided, whereas UHN provided two samples for each patient. Of the 162 possible samples, only 158 were available for this study. B: Diagram of the tissues sampled for each patient. In the case of UCSF and Cedar Sinai, a set of radical prostatectomies and a set of biopsy samples were taken from the tumor, benign adjacent, and benign contralateral tissue. UHN only provided tumor tissue. Image from Urosketch 3D.
Correlation plots showing the consistency between gene expressions measured from match biopsy (BX) and radical prostatectomy (RP) samples for four randomly selected patients. In general, these patients have relatively correlated gene expression.
Correlation plots showing the consistency between differential gene expression in the radical prostatectomies (RPs) and biopsy samples between tumor and benign tissue. The blue line shows the line of best fit.
Correlation plots for Decipher and Penney scores between radical prostatectomy (RP) and biopsy (BX) samples. In the first column, all of the patients are present; in the second column, one of the patients driving the correlation is removed; and in the third column, two of the patients driving the correlation are removed. For Decipher, even with removing two points driving the correlation of the significance, the correlation between BX and RP Decipher scores is preserved. This significance is also preserved for the Penney model when one patient is removed, but when two patients are removed only a borderline significance is observed. This demonstrates the robustness of the RP and BX correlations.
The expression distribution for each outlier gene used to define molecular subtype (ERG, SPINK1, ETV1, ETV4, ETV5, and FLI1) in each of the sample types. The horizontal bars indicate the cutoffs used to define outlier expression. BX, biopsy; RP, radical prostatectomy.
Venn diagram of the number of genes correlated between tumor and benign adjacent tissue and tumor and benign contralateral tissue. As expected, more genes were observed correlated between adjacent benign and tumor tissues than contralateral benign and tumor tissues. Of all of the genes significantly correlated in both sets, 225 significantly correlate in both.
Heat map of row-wise z-scored median expression of the 500 most correlated genes between tumor and benign tissue.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Wojno K., Hornberger J., Schellhammer P., Dai M., Morgan T. The clinical and economic implications of specimen provenance complications in diagnostic prostate biopsies. J Urol. 2015;193:1170–1177. doi: 10.1016/j.juro.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Chang A.J., Autio K.A., Roach M., Scher H.I. High-risk prostate cancer: classification and therapy. Nat Rev Clin Oncol. 2014;11:308–323. doi: 10.1038/nrclinonc.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie R., Chung J.-Y., Ylaya K., Williams R.L., Guerrero N., Nakatsuka N., Badie C., Hewitt S.M. Factors influencing the degradation of archival formalin-fixed paraffin-embedded tissue sections. J Histochem Cytochem. 2011;59:356–365. doi: 10.1369/0022155411398488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haffner M.C., Mosbruger T., Esopi D.M., Fedor H., Heaphy C.M., Walker D.A., Adejola N., Gürel M., Hicks J., Meeker A.K., Halushka M.K., Simons J.W., Isaacs W.B., De Marzo A.M., Nelson W.G., Yegnasubramanian S. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123:4918–4922. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindberg J., Kristiansen A., Wiklund P., Grönberg H., Egevad L. Tracking the origin of metastatic prostate cancer. Eur Urol. 2015;67:819–822. doi: 10.1016/j.eururo.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Boutros P.C., Fraser M., Harding N.J., de Borja R., Trudel D., Lalonde E. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 9.Abdueva D., Wing M., Schaub B., Triche T., Davicioni E. Quantitative expression profiling in formalin-fixed paraffin-embedded samples by affymetrix microarrays. J Mol Diagn. 2010;12:409–417. doi: 10.2353/jmoldx.2010.090155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erho N., Crisan A., Vergara I.A., Mitra A.P., Ghadessi M., Buerki C., Bergstralh E.J., Kollmeyer T., Fink S., Haddad Z., Sierocinski T., Ballman K.V., Triche T.J., Black P.C., Karnes R.J., Klee G., Davicioni E., Jenkins R.B. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alshalalfa M., Schliekelman M., Shin H., Erho N., Davicioni E. Evolving transcriptomic fingerprint based on genome-wide data as prognostic tools in prostate cancer. Biol Cell. 2015;107:232–244. doi: 10.1111/boc.201400097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karnes R., Bergstralh E., Davicioni E., Ghadessi M., Buerki C., Mitra A., Crisan A., Erho N., Lam L., Carlson R., Haddad Z., Triche T., Kollmeyer T., Ballman K., Black P., Klee G., Jenkins R. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein E.A., Yousefi K., Haddad Z., Choeurng V., Buerki C., Stephenson A.J., Li J., Kattan M.W., Magi-Galluzzi C., Davicioni E. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol. 2014;67:778–786. doi: 10.1016/j.eururo.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 14.Ross A.E., Feng F.Y., Ghadessi M., Erho N., Crisan A., Buerki C., Sundi D., Mitra A.P., Vergara I.A., Thompson D.J., Triche T.J., Davicioni E., Bergstralh E.J., Jenkins R.B., Karnes R.J., Schaeffer E.M. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 2014;17:64–69. doi: 10.1038/pcan.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Den R.B., Yousefi K., Trabulsi E.J., Firas A., Voleak C., Feng F.Y., Dicker A.P., Lallas C.D., Gomella L.G., Davicioni E., Karnes R.J. Genomic classifier identifies men with adverse pathology after radical prostatectomy who benefit from adjuvant radiation therapy. J Clin Oncol. 2015;33:944–951. doi: 10.1200/JCO.2014.59.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prensner J.R., Shuang Z., Erho N., Schipper M., Lyer M.K., Dhanasekaran S.M., Magi-Galluzzi C. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalonde E., Ishkanian A., Sykes J., Fraser M., Ross-Adams H., Erho N., Dunning M. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 2014;15:1521–1532. doi: 10.1016/S1470-2045(14)71021-6. [DOI] [PubMed] [Google Scholar]
- 18.Klein E.A., Cooperberg M.R., Magi-Galluzzi C., Simko J.P., Falzarano S.M., Maddala T., Chan J.M., Li J., Pelham R.J., Tenggara-Hunter I., Baehner F.L., Knezevic D., Febbo P.G., Shak S., Kattan M.W., Lee M., Carroll P.R. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66:550–560. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Cuzick J., Swanson G.P., Fisher G., Brothman A.R., Berney D.M., Reid J.E., Mesher D., Speights V.O., Stankiewicz E., Scardino P., Younus A., Flake D.D., Wagner S., Gutin A., Lanchbury J.S., Stone S. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penney K.L., Sinnott J.A., Fall K., Pawitan Y., Hoshida Y., Kraft P., Stark J.R., Fiorentino M., Setlur S., Johansson J.-E., Adami H.-O., Rubin M.A., Loda M., Golub T.R., Andrén O., Stampfer M.J., Mucci L.A. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J Clin Oncol. 2011;29:2391–2396. doi: 10.1200/JCO.2010.32.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomlins S.A., Alshalalfa M., Davicioni E., Erho N., Yousefi K., Zhao S., Haddad Z., Den R.B., Dicker A.P., Trock B.J., DeMarzo A., Ross A., Schaeffer E.M., Klein E.A., Magi-Galluzzi C., Karnes R.J., Jenkins R.B., Feng F.Y. Characterization of 1,577 primary prostate cancers reveals novel biological and clinicopathological insights into molecular subtypes. Eur Urol. 2015;68:555–567. doi: 10.1016/j.eururo.2015.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piccolo S.R., Sun Y., Campbell J.D., Lenburg M.E., Bild A.H., Johnson W.E. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012;100:337–344. doi: 10.1016/j.ygeno.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 24.Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B., Wilson M., Heguy A., Eastham J.A., Scardino P.T., Sander C., Sawyers C.L., Gerald W.L. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brase J.C., Johannes M., Mannsperger H., Falth M., Metzger J., Kacprzyk L.A., Andrasiuk T., Gade S., Meister M., Sirma H., Sauter G., Simon R., Schlomm T., Beissbarth T., Korf U., Kuner R., Sultmann H. TMPRSS2-ERG-specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11:507. doi: 10.1186/1471-2407-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlins S.A., Bjartell A., Chinnaiyan A.M., Jenster G., Nam R.K., Rubin M.A., Schalken J.A. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 27.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corcoran N.M., Hong M.K., Casey R.G., Hurtado-Coll A., Peters J., Harewood L., Goldenberg S.L., Hovens C.M., Costello A.J., Gleave M.E. Upgrade in Gleason score between prostate biopsies and pathology following radical prostatectomy significantly impacts upon the risk of biochemical recurrence. BJU Int. 2011;108(8 Pt 2):E202–E210. doi: 10.1111/j.1464-410X.2011.10119.x. [DOI] [PubMed] [Google Scholar]
- 29.Bibikova M., Yeakley J.M., Chudin E., Chen J., Wickham E., Wang-Rodriguez J., Fan J.-B. Gene expression profiles in formalin-fixed, paraffin-embedded tissues obtained with a novel assay for microarray analysis. Clin Chem. 2004;50:2384–2386. doi: 10.1373/clinchem.2004.037432. [DOI] [PubMed] [Google Scholar]
- 30.Frank M., Döring C., Metzler D., Eckerle S., Hansmann M.-L. Global gene expression profiling of formalin-fixed paraffin-embedded tumor samples: a comparison to snap-frozen material using oligonucleotide microarrays. Virchows Arch. 2007;450:699–711. doi: 10.1007/s00428-007-0412-9. [DOI] [PubMed] [Google Scholar]
- 31.Pillai R., Deeter R., Rigl C.T. Validation and reproducibility of a microarray-based gene expression test for tumor identification in formalin-fixed, paraffin-embedded specimens. J Mol Diagn. 2011;13:48–56. doi: 10.1016/j.jmoldx.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Flow chart diagramming the composition of the study. University of California San Francisco (UCSF), Cedars Sinai Medical Center (Cedar Sinai; Los Angeles, CA), and University Health Network (UHN; Toronto, ON, Canada) each supplied patients for the study. For each patient from UCSF and Cedar Sinai, six samples were provided, whereas UHN provided two samples for each patient. Of the 162 possible samples, only 158 were available for this study. B: Diagram of the tissues sampled for each patient. In the case of UCSF and Cedar Sinai, a set of radical prostatectomies and a set of biopsy samples were taken from the tumor, benign adjacent, and benign contralateral tissue. UHN only provided tumor tissue. Image from Urosketch 3D.
Correlation plots showing the consistency between gene expressions measured from match biopsy (BX) and radical prostatectomy (RP) samples for four randomly selected patients. In general, these patients have relatively correlated gene expression.
Correlation plots showing the consistency between differential gene expression in the radical prostatectomies (RPs) and biopsy samples between tumor and benign tissue. The blue line shows the line of best fit.
Correlation plots for Decipher and Penney scores between radical prostatectomy (RP) and biopsy (BX) samples. In the first column, all of the patients are present; in the second column, one of the patients driving the correlation is removed; and in the third column, two of the patients driving the correlation are removed. For Decipher, even with removing two points driving the correlation of the significance, the correlation between BX and RP Decipher scores is preserved. This significance is also preserved for the Penney model when one patient is removed, but when two patients are removed only a borderline significance is observed. This demonstrates the robustness of the RP and BX correlations.
The expression distribution for each outlier gene used to define molecular subtype (ERG, SPINK1, ETV1, ETV4, ETV5, and FLI1) in each of the sample types. The horizontal bars indicate the cutoffs used to define outlier expression. BX, biopsy; RP, radical prostatectomy.
Venn diagram of the number of genes correlated between tumor and benign adjacent tissue and tumor and benign contralateral tissue. As expected, more genes were observed correlated between adjacent benign and tumor tissues than contralateral benign and tumor tissues. Of all of the genes significantly correlated in both sets, 225 significantly correlate in both.
Heat map of row-wise z-scored median expression of the 500 most correlated genes between tumor and benign tissue.