Abstract
Tumor-specific mutations can be identified in circulating, cell-free DNA in plasma or serum and may serve as a clinically relevant alternative to biopsy. Detection of tumor-specific mutations in the plasma, however, is technically challenging. First, mutant allele fractions are typically low in a large background of wild-type circulating, cell-free DNA. Second, the amount of circulating, cell-free DNA acquired from plasma is also low. Even when using digital PCR (dPCR), rare mutation detection is challenging because there is not enough circulating, cell-free DNA to run technical replicates and assay or instrument noise does not easily allow for mutation detection <0.1%. This study was undertaken to improve on the robustness of dPCR for mutation detection. A multiplexed, preamplification step using a high-fidelity polymerase before dPCR was developed to increase total DNA and the number of targets and technical replicates that can be assayed from a single sample. We were able to detect multiple cancer-relevant mutations within tumor-derived samples down to 0.01%. Importantly, the signal/noise ratio was improved for all preamplified targets, allowing for easier discrimination of low-abundance mutations against false-positive signal. Furthermore, we used this protocol on clinical samples to detect known, tumor-specific mutations in patient sera. This study provides a protocol for robust, sensitive detection of circulating tumor DNA for future clinical applications.
Carcinogenesis occurs primarily through the acquisition of somatic mutations in the form of base substitutions, insertions, deletions, and chromosomal duplications, and these changes may potentially serve as biomarkers for diagnosis, monitoring tumor progression, response to therapy, and drug resistance.1, 2 Cancer diagnosis and identification of mutations present in an individual tumor often rely on analysis of biopsy or cytology samples. For mutation detection, however, this approach has significant disadvantages. Tissue biopsy or cytology generally samples only a fraction of the tumor and, therefore, may not provide a complete representation of tumor heterogeneity.3 Furthermore, tumor biopsy represents a single time point. Repeat sampling to assess tumor response, clonal evolution, and development of resistance mutations is both costly and invasive and may not be ethical because of inherent risk of certain biopsy procedures. Therefore, the development of a less-invasive, more cost-effective alternative to tissue biopsy is wanted to improve on current clinical practices for cancer patients.
It has been recognized for many years that cells, including cancer cells, release cell-free nucleic acids during processes, including apoptosis, necrosis, and active secretion (exocytosis),4, 5, 6 and that these nucleic acids can be found in bodily fluids (plasma/serum, urine, and sputum).7 Spurred by recent technological advances, there is now a growing interest in detection of cancer-specific mutations in bodily fluids, and, in particular, in plasma/serum as a possible way to overcome the limitations of repeated biopsy or cytology collection procedures. It has been demonstrated that tumor-associated mutations can be detected in plasma of patients with many different types of cancer8; circulating, cell-free, tumor DNA (ctDNA) can be used to monitor response to therapy or evolution of therapeutic resistance; and ctDNA may provide a more complete picture of mutation heterogeneity.9 Therefore, assessing ctDNA in the blood by liquid biopsy as a means to detect and monitor cancer may serve as a less invasive and clinically relevant alternative to tumor biopsy.
Detection of tumor-specific biomarkers in the blood, however, does not come without some significant challenges. First, the total amount of circulating, cell-free DNA (ccfDNA) isolated from plasma or serum is low (typically <20 ng/mL of plasma or approximately 6000 genome equivalents/mL).10, 11 Second, the fraction of ccfDNA that is derived from the tumor can be <0.1% for early-stage tumors (fewer than six mutant copies/mL of plasma).8 This combination of low mutant allele fraction and low absolute mutant copy numbers is beyond the capability of most conventional methods for robust, reliable mutation detection and quantification.
One recently developed technology, digital PCR (dPCR), has the potential to meet these requirements. dPCR operates as a standard, probe-based, allelic discrimination reaction that is partitioned into thousands or millions of independent reactions using either physical separation or an oil-based emulsion of droplets.12 Partitioning greatly reduces the number of template molecules per reaction (potentially as low as one template per partition) and, as a result, the fraction of mutant alleles per positive partition is effectively increased to facilitate detection. When pushed to the point where most partitions contain zero or only one template molecule, Poisson's distribution can be used for accurate quantification of absolute copy number of wild-type or mutant alleles, thereby facilitating precise calculation of mutant allele frequency. With enough partitions, and assuming adequate DNA, it is possible to detect mutations with allele frequencies <1:180,000 when starting with 3.3 μg of genomic DNA (gDNA).13 However, in our experience with esophageal adenocarcinoma, 52% (60 of 115) of plasma samples yielded <150 ng ccfDNA when starting with 16- to 20-mL blood samples. Therefore, when dealing with ccfDNA, the low DNA yields and resulting low mutant allele copy numbers remain a challenge for several reasons. First, technical/assay or instrument noise can produce false-positive partitions, making it difficult to discriminate between true positive signal and background noise.14, 15 Second, technical replicates often cannot be performed because this requires dividing the limited sample into separate reactions. Similarly, analysis of more than one mutation per sample can only be performed if the assays are multiplexed, which requires substantial optimization and potentially increases background noise.15, 16 Finally, potentially positive results cannot be verified using an alternative method because of lack of remaining sample. Thus, the current study was undertaken to overcome these limitations and to improve on the sensitivity and robustness of dPCR for mutation detection in the plasma of cancer patients.
In this study, we use the RainDance dPCR platform (RainDance Technologies, Inc., Billerica, MA) and describe the development of a multiplexed preamplification PCR step to increase the total amount of starting material and increase to the number of targets and technical replicates that can be assayed from a single sample. Specifically, we used a multiplexed preamplification PCR step composed of three different primer pairs targeting cancer-relevant genes SMAD4, TP53, and KRAS using a high-fidelity polymerase to reduce PCR-induced errors. We demonstrate that multiplexed preamplification allowed for easy detection of 0.05% mutant fraction for all three targets of interest in a single, 50-ng, tumor-derived sample mix compared with non-preamplified sample where mutations were not detectable. For the SMAD4 point mutation, we were able to further increase sensitivity to detect 0.01% mutant frequency (one mutant allele in a background of 10,000 wild-type alleles). More important, background noise did not proportionately increase in the preamplified samples. Furthermore, we were also able to use this protocol for clinical samples to detect mutations in the serum of patients with known tumor-specific mutations. Overall, this proof-of-principle study provides a robust approach for assessment of ctDNA (liquid biopsy) using dPCR for future clinical application.
Materials and Methods
Patients and Sample Collection
Research on human specimens was performed with approval of Institutional Review Boards at all participating institutions, and all patients provided informed consent for use of their tissues and clinical data in research studies. Fresh-frozen esophageal adenocarcinoma tissue specimens and blood were obtained from patients treated at the University of Pittsburgh Medical Center (Pittsburgh, PA) between 2002 and 2008. Tumor tissues were frozen immediately in liquid nitrogen and then stored at −80°C until use. Blood samples were collected in red-top vacutainers (BD, Franklin Lakes, NJ), allowed to clot for 1 hour, and then centrifuged at 3750 rpm (2531 × g) for 5 minutes. Serum was aliquoted for storage at −80°C.
Three patient samples were selected for this study on the basis of the presence of established cancer-associated, single-nucleotide variants identified in previous studies using whole exome sequencing and/or targeted resequencing.17 The mutations were as follows: a TP53 R273C mutation (patient MS310), a SMAD4 R361G mutation (patient 4873), and a KRAS G12S mutation (patient MS1079).
gDNA Preparation
gDNA was extracted from the Barrett's esophageal cell line, CP-A, and from fresh-frozen tissue samples using the QiaAmp DNA Mini Kit (Qiagen, Valencia, CA). DNA concentrations were quantified with the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) and stored at −20°C.
Tumor DNA Dilution
According to our AmpliSeq sequencing data (data not shown), CP-A does not contain the mutations of interest assayed in this study, and was therefore used as a source of wild-type DNA for diluting mutations present in the tumor DNA samples. Confirmation of our sequencing data on tumor DNA with specific mutations of interest and allele frequencies were validated through dPCR analysis. Mutant percentages determined through dPCR were used to calculate dilutions. To dilute the tumor DNA mutations, wild-type gDNA from CP-A cells was used to reduce mutation frequency to 3% for each patient sample, with a final concentration of 25 ng/μL. All three samples were then combined at an equal ratio to yield a 1% mutation frequency for all three mutations. Lower mutant frequencies were prepared directly from the 1% mutant stock by diluting with 25 ng/μL wild-type CP-A gDNA.
Serum/Plasma DNA Extraction
Extraction of ccfDNA from pooled normal human plasma (wild-type control; Innovative Research, Novi, MI) and patient serum (1 to 3 mL per patient) was performed using the QIAamp Circulating Nucleic Acid Kit (Qiagen), according to the manufacturer's protocol, and ccfDNA was eluted in 45 μL of TE buffer, pH 8.0. ccfDNA concentrations were quantified with the Qubit 2.0 Fluorometer and stored at −20°C.
Multiplexed Preamplification
Multiplexed preamplification of gDNA mixtures was performed in 10-μL reactions using 50 ng of gDNA, 1× Q5 Hot Start High-Fidelity Master Mix (New England Biolabs, Ipswich, MA) or 1× TaqMan Genotyping Master Mix (Life Technologies), and 50 nmol/L each of forward and reverse primers for KRAS [5′-CTGAAAATGACTGAATATAAACTTGTGG-3′ (forward) and 5′-TAGCTGTATCGTCAAGGCACTC-3′ (reverse)], TP53 [(5′-CTGCCTCTTGCTTCTCTTT-3′ (forward) and 5′-GAGATTCTCTTCCTCTGTGC-3′ (reverse)], and SMAD4 [5′-CAAGCTGCCCTATTGTTACT-3′ (forward) and 5′-GCTCTCTCAATGGCTTCTG-3′ (reverse)] (final reaction concentrations). In addition, reactions without DNA were included as no template controls (NTCs). Reactions were preamplified in a GeneTouch Thermal Cycler (Bioer Technology, Hangzhou, China) with a temperature profile of 98°C for 3 minutes, followed by nine cycles of amplification (98°C for 10 seconds, 63°C for 3 minutes, and 72°C for 30 seconds), and a final 72°C extension for 2 minutes.18 Preamplified reactions were immediately placed on ice after the final extension and diluted with 90 μL TE buffer, pH 8.0, to inactivate the Q5 polymerase.
For multiplexed, preamplification of serum-derived ccfDNA, 15 μL (7.5 to 31.5 ng) of wild-type ccfDNA and patient ccfDNA was used in 35 μL reactions using the same protocol outlined above, except the reactions were diluted with 140 μL TE buffer, pH 8.0. Samples were either used immediately for dPCR or stored at −20°C.
dPCR Data
Emulsion-based dPCR was performed using the RainDrop Digital PCR system from RainDance Technologies, Inc. For samples without preamplification, 50 ng of DNA (or equivalent volume of water for NTCs) was loaded into 25-μL dPCRs. For preamplified samples, a maximum volume of 8 μL of the diluted, preamplified sample was added to each 25-μL dPCR. dPCRs contained 1× TaqMan Genotyping Master Mix (Life Technologies), 0.5 μmol/L target primers, 0.2 μmol/L (KRAS) or 0.4 μmol/L (TP53 and SMAD4) wild-type target probe, and 0.2 μmol/L mutant target probe (Table 1). In addition, 1× proprietary hydrofluorinated droplet stabilizer (RainDance Technologies, Inc.) was added to aid in emulsion formation (final reaction concentrations).
Table 1.
Sequences and Final Concentrations of Probes Used in Digital PCR Assays
| Target mutation | Wild-type probe sequence | Final, μmol/L | Mutant probe sequence | Final, μmol/L |
|---|---|---|---|---|
| KRAS G12S | 5′-VICTTGGAGCTGGTGGCGTAMGBNFQ-3′ | 0.2 | 5′-6FAMTTGGAGCTAGTGGCGTAMGBNFQ-3′ | 0.2 |
| TP53 R273C | 5′-TETAGGTGCGTGZENTTIABKFQ-3′ | 0.4 | 5′-6FAMAGGTGTGTGZENTTIABKFQ-3′ | 0.2 |
| SMAD4 R361G | 5′-TETAGGAGATCGZENCTTTIABKFQ-3′ | 0.4 | 5′-6FAMAGGAGATGGZENCTTTIABKFQ-3′ | 0.2 |
KRAS probes were designed by RainDance Technologies, Inc. (Billerica, MA), and generated by Life Technologies (Carlsbad, CA), and TP53 and SMAD4 probes were designed and generated by Integrated DNA Technologies. Wild-type probes were labeled with either VIC or TET fluorophore, and mutant probes were labeled with 6-FAM fluorophore. Mutations of interest are in bold, and bases with locked nucleic acids are underlined.
For 0.01% mutant assay and ccfDNA from serum/plasma samples, the same protocol was followed, but 50-μL reaction volumes were used to maximize the amount of input (20 μL) that could be analyzed. Preamplified NTCs with TE buffer alone and 20 μL of ccfDNA from normal plasma served as negative and wild-type alone controls, respectively, to assess false-positive signal/background noise. Three to six replicates were run for all assays.
Emulsions of each reaction were prepared on the RainDrop Source instrument (RainDance Technologies, Inc.) to produce 5 to 10 million, 5-pL-volume droplets per 25- or 50-μL reaction. Thereafter, the emulsions were placed on a thermal cycler to amplify the target and generate signal. The temperature profile for amplification was an activation step of 95°C for 10 minutes, followed by 40 cycles of amplification [95°C for 15 seconds, 56°C (TP53 and SMAD4 assays) or 64°C (KRAS assay) for 15 seconds, and 60°C for 45 seconds], and a polymerase inactivation step of 98°C for 10 minutes. Reactions were allowed to cool to at least 50°C before placing them on the RainDrop Sense instrument (RainDance Technologies, Inc.) for signal detection.
dPCR Analysis and Statistical Analysis
RainDrop Analyst (RainDance Technologies, Inc.) was used to determine positive signal for each allele type, either wild type (VIC/TET) on the y axis or mutant (FAM) on the x axis. Gates were applied to regions of clustered droplets to define positive hits for each allele, according to the manufacturer's instructions. Gating within each experiment was applied to NTC and wild-type alone controls to determine false-positive rates and inherent background noise from the assay and instrument. Data were exported to Microsoft Excel (Redmond, WA) to calculate averages for mutant signal and significance values via t-test.
Results
Preamplification for Detection of Low-Frequency Alleles Requires the Use of a High-Fidelity DNA Polymerase to Minimize PCR-Induced Errors
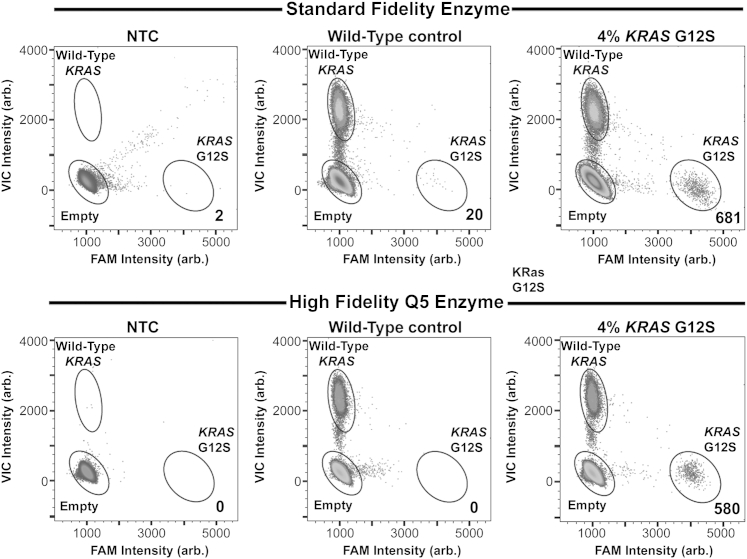
During initial optimization of preamplification conditions using 1× TaqMan Genotyping Master Mix and multiple dPCR assays, we consistently observed a small, yet significant, number (mean = 26; n = 10) of false-positive droplets in the wild-type control (Figure 1) that were not present when gDNA was analyzed without preamplification (data not shown). After ruling out the presence of low levels of the variant alleles either in the gDNA or because of contamination, we hypothesized that the false-positive background was originating from Taq-induced errors during the nine-cycle preamplification step. Therefore, we tested a high-fidelity DNA polymerase, Q5 Hot Start, in the preamplification step to determine whether we could reduce or eliminate false-positive signal observed in reactions containing TaqMan Genotyping mix. Using KRAS primers, we preamplified wild-type gDNA, gDNA with 4% KRAS G12S allele, or water as NTC with both enzymes and used our KRAS G12S assay for dPCR. Using gating parameters for the positive control (4% KRAS G12S), we observed that, although the NTCs for both enzymes were free of false-positive background noise, the wild-type control for the standard fidelity enzyme had 20 KRAS G12S mutant droplets, whereas the high-fidelity enzyme yielded no false-positive signal in the wild-type control (Figure 1). Furthermore, it appeared that using a high-fidelity enzyme tightened the empty, wild-type, and mutant droplet populations and produced less spray from those clusters.
Figure 1.
KRAS G12S assay sensitivity using either standard (top panels) or high-fidelity (bottom panels) polymerase enzyme during preamplification of 10 ng of gDNA. Preamplification was performed on water alone [no template control (NTC)], wild-type genomic DNA (gDNA), and gDNA containing 4% KRAS G12S allele frequency. After preamplification, digital PCR was performed on each sample. The numbers shown in the bottom right of each plot indicate the number of KRAS G12S mutants detected for each condition. Arb., arbitrary.
Evaluating the Sensitivity and Specificity of Multiplexed Preamplification
For initial testing of protocol feasibility, we assessed the sensitivity and specificity of multiplexed preamplification in a DNA sample with predefined mutant allele fractions for three genes. Specifically, DNAs from three independent tumor samples (tumors MS1079, MS310, and 4873) with three cancer-relevant mutations (KRAS G12S, TP53 R273C, and SMAD4 R361G, respectively) were mixed into a single pool such that each mutant allele was present at approximately 0.05%. DNA (50 ng; approximately 7.5 mutant copies) was either thermocycled through the multiplexed preamplification protocol or remained untreated (no preamplification) before dPCR.
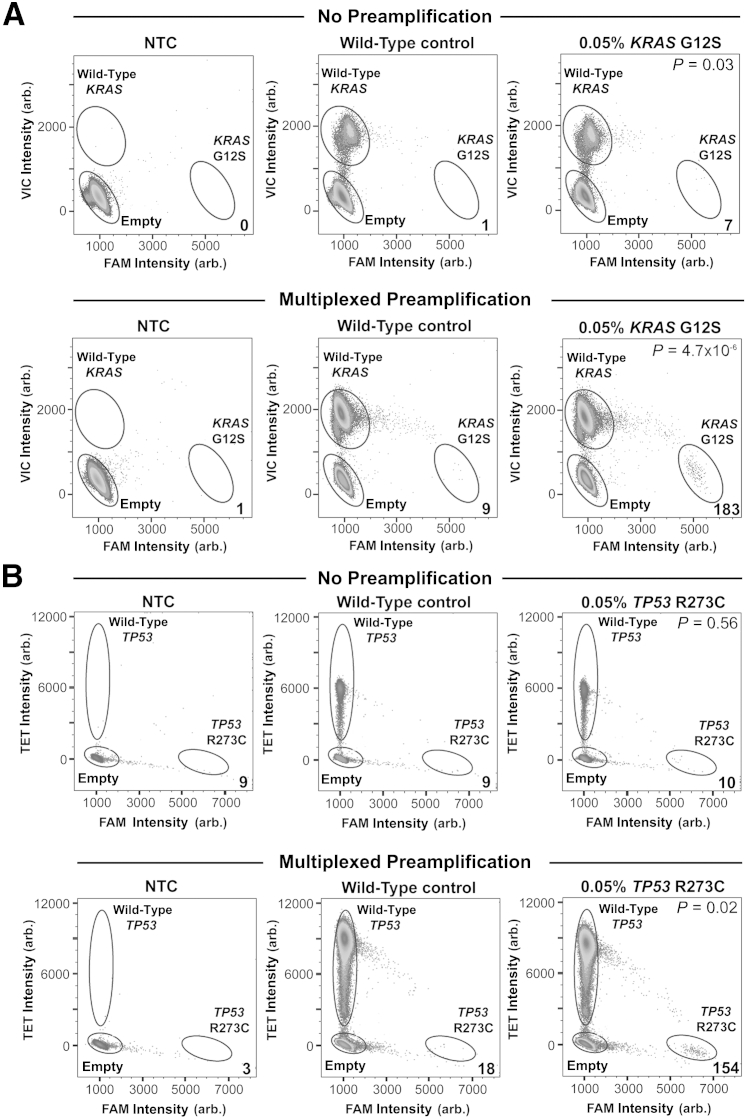
For the KRAS G12S assay, an average of seven positive mutant droplets (n = 3) were detected in the non-preamplified 0.05% mutant sample, with one false-positive droplet detectable on average in the wild-type control (n = 3, P = 0.03) (Figure 2A). In the multiplexed preamplified sample, an average of 183 positive KRAS G12S droplets (n = 3) were detected in the 0.05% mutant sample (Figure 2A) compared with nine false-positive droplets on average in the wild-type control sample (n = 3, P = 4.7 × 10−6). Notably, although there was a 27-fold increase in detectable mutant droplets between the non-preamplified and the preamplified 0.05% mutant sample, false-positive noise did not increase proportionately (ninefold). For the TP53 R273C assay, we detected an average of 10 positive mutant droplets (n = 3) in the non-preamplified 0.05% mutant sample and nine false-positive droplets in the wild-type control (n = 3), consequently yielding statistically insignificant results (P = 0.56), thus failing detection (Figure 2B). In the multiplexed preamplified 0.05% mutant sample, an average of 154 positive mutant droplets (n = 3) were detected against an average background of 18 false-positive mutant droplets in the wild-type control (n = 3), resulting in statistical significance (P = 0.02) (Figure 2B). Again, the fold increase in positive mutant signal in the 0.05% mutant sample (15.4-fold) was much greater than the increase in false-positive signal observed in the wild-type control (twofold).
Figure 2.
Detection of 0.05% mutant allele frequency with and without multiplexed preamplification before digital PCR. Water alone [no template control (NTC)], 50 ng wild-type DNA, and 50 ng tumor DNA containing 0.05% mutant allele frequency were either analyzed by digital PCR alone (no preamplification) or preamplified before digital PCR (multiplexed preamplification). Non-preamplified samples (top panels) and samples subjected to multiplexed preamplification (bottom panels) were analyzed for the presence of KRAS G12S (A) or TP53 R73C (B) mutation. The numbers shown in the bottom right of each plot indicate the number of mutant alleles detected for each condition. P values are denoted in the top right of each plot. Arb., arbitrary.
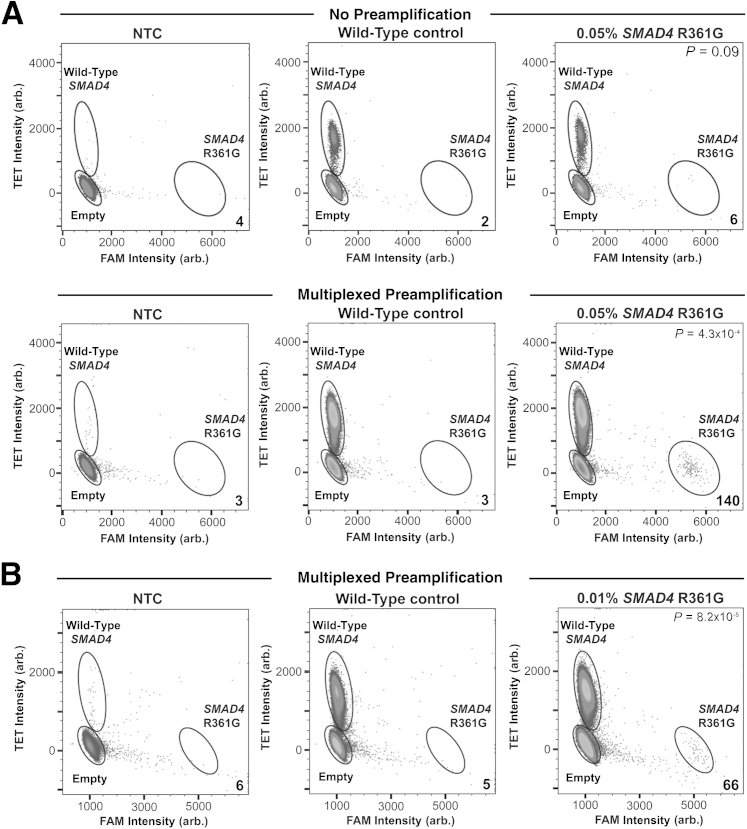
For the SMAD4 R361G assay, non-preamplified 0.05% mutant sample yielded, on average, six positive mutant droplets (n = 3) versus two false-positive droplets in the wild-type control (n = 3, P = 0.09) (Figure 3A). In the multiplexed, preamplified, 0.05% mutant sample, an average of 140 positive mutant droplets (n = 3) were detected, with an average of three false-positive droplets in the wild-type control (n = 3), thereby yielding highly significant results (P = 4.3 × 10−4) (Figure 3A). Overall, a 22-fold increase in positive mutant signal was observed in the 0.05% mutant sample compared with the 1.5-fold increase in false-positive signal observed in the wild-type control. Furthermore, in an effort to push the limit of detection for this assay, we further diluted the SMAD4 R361G mutant to 0.01%, to determine whether this low allelic frequency was still detectable. Using a 50-μL reaction and 10 million droplets, we were able to detect, on average, 66 positive droplets (n = 3) in the preamplified 0.01% mutant sample against a false-positive background average of five droplets in the wild-type control (n = 3), again yielding statistically significant results (P = 8.2 × 10−5) (Figure 3B).
Figure 3.
Detection of 0.05% and 0.01% allele frequency of SMAD4 R361G mutation. A: Detection of 0.05% SMAD4 R361G mutation with and without multiplexed preamplification before digital PCR. Water alone [no template control (NTC)], 50 ng wild-type DNA, and 50 ng tumor DNA containing 0.05% mutant allele frequency were either analyzed by digital PCR alone (no preamplification) or preamplified before digital PCR (multiplexed preamplification). Non-preamplified samples (top panels) and samples subjected to multiplexed preamplification (bottom panels) were analyzed for the presence of SMAD4 R361G. B: Detection of 0.01% SMAD4 R361G mutation with multiplexed preamplification before digital PCR. The numbers shown in the bottom right of each plot indicate the number of mutant alleles detected for each condition. P values are denoted in the top right of each plot. Arb., arbitrary.
Detection of Low-Frequency Mutant Alleles in Serum-Derived ccfDNA from Cancer Patients Using Multiplexed Preamplification in dPCR
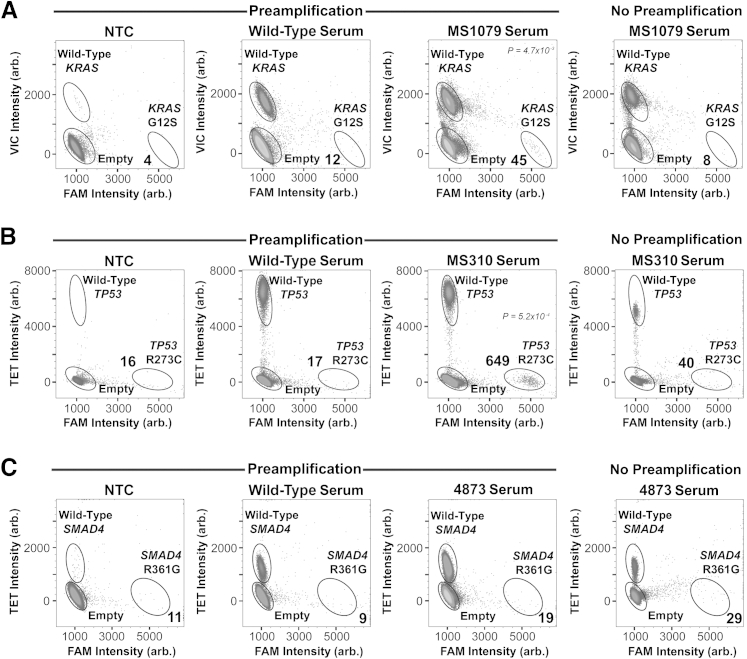
We next assessed the detection of rare mutant alleles in serum-derived ccfDNA from cancer patients (tumors MS1079, MS310, and 4873) both with and without multiplex preamplification. The total amount of ccfDNA available from these patients was 84, 20, and 64 ng, respectively. For the KRAS G12S assay (MS1079), a 37.5-ng ccfDNA input yielded eight positive mutant droplets in the non-preamplified reaction (Figure 4A) compared with zero in the NTC and two in the wild-type control (data not shown). Using the preamplification protocol starting with 31.3 ng of ccfDNA, we observed a clear positive mutant signal with an average of 45 positive droplets (n = 3). Four and 12 false-positive droplets, on average, were detected in the NTC and wild-type control samples, respectively (n = 6); therefore, results were significant (P = 4.7 × 10−3) (Figure 4A). For the TP53 R273C assay (MS310), a 9-ng ccfDNA input yielded 40 positive mutant droplets in the non-preamplified sample (Figure 4B) compared with 3 in the NTC and 11 in the wild-type control (data not shown). No statistical analysis could be performed because the MS310 sample could only be analyzed once because of limiting DNA. Preamplifying 7.5 ng ccfDNA, we observed a statistically significant positive mutant cluster of 649 droplets (n = 3) against a background average of 17 false-positive droplets in the wild-type control (n = 4, P = 5.2 × 10−4) (Figure 4B). Last, we assessed ccfDNA from patient 4873 for the SMAD4 R361G mutation and were unable to detect a positive mutant signal when starting with 28.8 ng of ccfDNA either with or without preamplification (Figure 4C).
Figure 4.
Detection of low-frequency mutant alleles in serum-derived, circulating, cell-free DNA (ccfDNA) from cancer patients using multiplexed preamplification in digital PCR. Water alone [no template control (NTC)], wild-type ccfDNA, and ccfDNA from three cancer patients with the KRAS (patient MS1079; A), TP53 (patient MS310; B), or SMAD4 (patient 4873; C) mutation were subjected to multiplexed preamplification and assessed for presence of the mutation using digital PCR. A single digital PCR was performed on each patient ccfDNA sample to assess presence of mutation without preamplification (right panels). The numbers shown in the bottom right of each plot indicate the number of mutant alleles detected for each condition. P values are denoted in the top right of each plot. Arb., arbitrary.
Discussion
With the development of new technologies for highly sensitive detection of cancer-associated DNA mutations, there has recently been much interest in assessing ctDNA fragments in plasma or serum as a liquid biopsy for detection and treatment monitoring of cancer. One such technology, dPCR, is capable of detecting and quantifying extremely low-frequency mutations, but difficulties remain when trying to detect ctDNA. The combination of low DNA abundance in plasma and the small fraction of DNA that is derived from the tumor cells means that the absolute number of mutant DNA copies can be extremely challenging to detect. In this scenario, splitting of the DNA sample is often not feasible and, as a result, technical replicates cannot be performed to generate statistically validated results and it is difficult to analyze multiple mutations in the same DNA sample. Herein, we have demonstrated that incorporation of a high-fidelity, multiplex preamplification step to increase DNA quantity before dPCR partitioning improves the robustness of rare mutant allele detection, enables technical replicates to be performed, and allows multiple mutations to be analyzed from the same starting DNA sample.
Some recent studies have addressed the idea of amplifying small amounts of DNA before dPCR using either whole genome amplification19 or coamplification at lower denaturation temperature PCR.20 In addition, it has been previously established that multiplex preamplification can be used to increase total DNA from a limited sample and also increase sampling power by incorporating multiple target primers within the preamplification reaction.21 To our knowledge, however, this approach has not yet been described to overcome the limitations of ctDNA detection in plasma or serum using dPCR. In this setting, one concern is that the preamplification step would elevate background noise in addition to increasing the true positive signal; indeed, when using the standard dPCR DNA polymerase, TaqMan Genotyping master mix, we consistently detected a small amount of false-positive background in the wild-type gDNA controls (Figure 1). After careful cleanup, and testing multiple sources of wild-type gDNA, this observation continued, leading us to believe that errors introduced by Taq polymerase during the preamplification step were likely the source of these mutant alleles. This was confirmed by the use of a high fidelity enzyme with >100-fold lower error rate than Taq polymerase, which completely eliminated any additional false-positive background in wild-type gDNA controls compared with non-preamplified samples (Figure 1). This finding allowed us to proceed with testing the sensitivity of this protocol with little to no erroneous signal from the preamplification step impeding the dPCR readout.
In initial experiments, our objective was to determine the ability of multiplex preamplification to improve the reliability and robustness for identification of three different mutations at an allelic frequency of 0.05% when starting with a 50-ng DNA sample. This translates to an expected 7.5 positive mutant DNA molecules in a wild-type background of approximately 15,000. Because this was an artificial mixing experiment, we had sufficient DNA to run replicates with and without preamplification and therefore could run a direct comparison. We found that two of the three mutations (TP53 and SMAD4) were only detectable with statistical significance when the preamplification step was used. Furthermore, although the KRAS mutation was detectable without preamplification because of low assay noise, preamplification greatly increased the signal/noise ratio, resulting in a dramatically lower P value. In fact, this was the case for all assays and, overall, preamplification increased true-positive signal 15- to 27-fold over the signal from non-preamplified samples compared with only a 1.5- to 9-fold increase in false-positive signal. Therefore, the results clearly demonstrate that multiplex preamplification can increase detection sensitivity by increasing true signal without proportionately increasing background noise.
Translating the multiplex preamplification protocol to clinical samples, we next detected ctDNA in serum from three cancer patients. When analyzed without preamplification, all samples provided inconclusive results because of low mutant signal, high false-positive signal in wild-type controls, and lack of replicates for statistical analysis. However, we were able to successfully detect the presence of ctDNA in the serum of two of three cancer patients when using the preamplification step. For patient MS1079, the KRAS G12S mutation was detectable at 0.018% when starting with 31.3 ng of DNA, which corresponds to approximately 1.7 mutant alleles in a wild-type background of 95,000. For patient MS310, although only 7.5 ng of DNA was available for preamplification, we were able to easily distinguish a TP53 mutation population (2.7%) that had been inconclusive in the non-preamplified sample. Finally, the SMAD4 mutation assessed in patient 4873 was not detectable in the serum even when using preamplification. Interestingly, this patient had stage I disease, whereas the other two patients both had stage II disease. Therefore, this negative result may occur because of the lower disease burden. In all cases, perhaps the most important benefit from the preamplification step was the ability to perform dPCR replicates, which enabled statistical analysis to be performed. Furthermore, the mutant fraction does not actually need to be exceptionally low for preamplification to be of benefit. Although the TP53 mutant was present at 2.7% in patient MS310, the low total amount of DNA meant that the positive signal without preamplification was too close to call conclusively without replicates. With preamplification, however, the positive signal became abundantly clear and was statistically significant. Thus, preamplification provides a benefit when absolute mutant copy numbers are exceptionally low because of either a low mutant allele fraction or a low starting DNA input.
One potential concern with our approach is that multiplex preamplification may introduce measurement bias because targets may not be equally amplified during preamplification.22, 23 This could be a concern if the relative abundance of multiple mutations in a single sample is important for the clinical or research question being addressed. Similarly, if absolute quantification, or quantification and comparison of data from longitudinal samples, were required, it may be necessary to rigorously validate the quantification process and possibly include exogenous controls in the preamplification step. In this study, we are simply attempting to detect mutation and not quantify mutant allele fraction. However, we did review our data to determine whether an amplification bias was evident and found that all three targets, KRAS, TP53, and SMAD4, in the multiplexed, preamplified sample were amplified 29-, 32-, and 33-fold, respectively, compared with the non-preamplified samples (data not shown). This implies that all primer pairs in the multiplexed reaction have similar amplification efficiencies during preamplification and, thus, the limit of detection is expected to be similar for all three digital assays. Furthermore, the mutant fractions of all three mutant alleles assessed were not significantly different between the preamplified and non–preamplified samples (data not shown). Therefore, our data suggest that the mutant fraction detected in dPCR after preamplification is highly representative of the actual mutant allele frequency in the original sample. In summary, we have shown that multiplexed preamplification using a high-fidelity polymerase before dPCR can do the following: i) increase the total amount of starting material, ii) increase the number of targets and technical replicates that can be assayed from a single DNA sample, and iii) improve assay signal/noise ratio to easily detect low-frequency mutations. Last, integration of higher-order multiplexing (ie, incorporating commercially available cancer panels to increase the number of targets assayed) should prove to be uncomplicated, thereby providing a vast number of cancer-relevant regions for assessment by dPCR.
Footnotes
Supported by NIH Exploratory/Developmental Research grant award (R21) 9500302322 (T.E.G.).
Disclosures: T.E.G. and J.B.J. received reimbursement from RainDance Technologies, Inc., for travel related to presentation of some of the data reported herein. These authors report no other relationships relevant to the work presented herein. KRAS probes were designed by RainDance Technologies, Inc., and generated by Life Technologies, and TP53 and SMAD4 probes were designed and generated by Integrated DNA Technologies.
References
- 1.Chin L., Hahn W.C., Getz G., Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aung K.L., Board R.E., Ellison G., Donald E., Ward T., Clack G., Ranson M., Hughes A., Newman W., Dive C. Current status and future potential of somatic mutation testing from circulating free DNA in patients with solid tumours. Hugo J. 2010;4:11–21. doi: 10.1007/s11568-011-9149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., Varela I., Phillimore B., Begum S., McDonald N.Q., Butler A., Jones D., Raine K., Latimer C., Santos C.R., Nohadani M., Eklund A.C., Spencer-Dene B., Clark G., Pickering L., Stamp G., Gore M., Szallasi Z., Downward J., Futreal P.A., Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stroun M., Lyautey J., Lederrey C., Olson-Sand A., Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–142. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 5.Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F.O., Hesch R.D., Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 6.Gahan P.B., Stroun M. The biology of circulating nucleic acids in plasma and serum (CNAPS). Extracellular Nucleic Acids. In: Rykova E., Kikuchi Y., editors. Springer; Berlin: 2010. pp. 167–189. [Google Scholar]
- 7.Gonzalez-Masia J.A., Garcia-Olmo D., Garcia-Olmo D.C. Circulating nucleic acids in plasma and serum (CNAPS): applications in oncology. Onco Targets Ther. 2013;6:819–832. doi: 10.2147/OTT.S44668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz L.A., Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamkovich S.N., Bryzgunova O.E., Rykova E.Y., Permyakova V.I., Vlassov V.V., Laktionov P.P. Circulating nucleic acids in blood of healthy male and female donors. Clin Chem. 2005;51:1317–1319. doi: 10.1373/clinchem.2004.045062. [DOI] [PubMed] [Google Scholar]
- 11.Devonshire A.S., Whale A.S., Gutteridge A., Jones G., Cowen S., Foy C.A., Huggett J.F. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406:6499–6512. doi: 10.1007/s00216-014-7835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B., Kinzler K.W. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milbury C.A., Zhong Q., Lin J., Williams M., Olson J., Link D.R., Hutchison B. Determining the lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Det Quant. 2014;1:8–22. doi: 10.1016/j.bdq.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker M. Digital PCR hits its stride. Nat Methods. 2012;9:541–544. [Google Scholar]
- 15.Taly V., Pekin D., Benhaim L., Kotsopoulos S.K., Le Corre D., Li X., Atochin I., Link D.R., Griffiths A.D., Pallier K., Blons H., Bouche O., Landi B., Hutchison J.B., Laurent-Puig P. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722–1731. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- 16.Zhong Q., Bhattacharya S., Kotsopoulos S., Olson J., Taly V., Griffiths A.D., Link D.R., Larson J.W. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab Chip. 2011;11:2167–2174. doi: 10.1039/c1lc20126c. [DOI] [PubMed] [Google Scholar]
- 17.Dulak A.M., Stojanov P., Peng S., Lawrence M.S., Fox C., Stewart C. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson D., Akrap N., Svec D., Godfrey T.E., Kubista M., Landberg G., Stahlberg A. Properties of targeted preamplification in DNA and cDNA quantification. Expert Rev Mol Diagn. 2015;15:1085–1100. doi: 10.1586/14737159.2015.1057124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid A.L., Freeman J.B., Miilward M., Ziman M., Gray E.S. Detection of BRAF-V600E and V600K in melanoma circulating tumor cells by droplet digital PCR. Clin Biochem. 2015;48:999–1002. doi: 10.1016/j.clinbiochem.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Castellanos-Rizaldos E., Paweletz C., Song C., Oxnard G.R., Mamon H., Jänne P.A., Makrigiorgos G.M. Enhanced ratio of signals enables digital mutation scanning for rare allele detection. J Mol Diagn. 2015;17:284–292. doi: 10.1016/j.jmoldx.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piggott M.P., Bellemain E., Taberlet P., Taylor A.C. A multiplex pre-amplification method that significantly improves microsatellite amplification and error rates for faecal DNA in limiting conditions. Conserv Genet. 2004;5:417–420. [Google Scholar]
- 22.Sanders R., Huggett J.F., Bushell C.A., Cowen S., Scott D.J., Foy C.A. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 2011;83:6474–6484. doi: 10.1021/ac103230c. [DOI] [PubMed] [Google Scholar]
- 23.Whale A.S., Cowen S., Foy C.A., Huggett J.F. Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS One. 2013;8:e58177. doi: 10.1371/journal.pone.0058177. [DOI] [PMC free article] [PubMed] [Google Scholar]