Abstract
CRISPR-Cas9 nucleases are widely used for genome editing but can induce unwanted off-target mutations. Existing strategies for reducing genome-wide off-targets of the broadly used Streptococcus pyogenes Cas9 (SpCas9) are imperfect, possessing only partial or unproven efficacies and other limitations that constrain their use. Here we describe SpCas9-HF1, a high-fidelity variant harboring alterations designed to reduce non-specific DNA contacts. SpCas9-HF1 retains on-target activities comparable to wild-type SpCas9 with >85% of single-guide RNAs (sgRNAs) tested in human cells. Strikingly, with sgRNAs targeted to standard non-repetitive sequences, SpCas9-HF1 rendered all or nearly all off-target events undetectable by genome-wide break capture and targeted sequencing methods. Even for atypical, repetitive target sites, the vast majority of off-targets induced by SpCas9-HF1 were not detected. With its exceptional precision, SpCas9-HF1 provides an alternative to wild-type SpCas9 for research and therapeutic applications. More broadly, our results suggest a general strategy for optimizing genome-wide specificities of other RNA-guided nucleases.
CRISPR-Cas9 nucleases enable highly efficient genome editing in a wide variety of organisms1–3 but can also cause unwanted mutations at off-target sites that resemble the on-target sequence4–13. These off-target effects can confound research experiments and also have potential implications for therapeutic uses of the technology. Various strategies have been described to reduce genome-wide off-target mutations of the commonly used SpCas9 nuclease, including: truncated sgRNAs bearing shortened regions of target site complementarity8, 14, SpCas9 mutants such as the recently described D1135E variant15, paired SpCas9 nickases16, 17, and dimeric fusions of catalytically inactive SpCas9 (dSpCas9) to a non-specific FokI nuclease18–20. However, these approaches are only partially effective, have as-yet unproven efficacies on a genome-wide scale, and/or possess the potential to create more new off-target sites. Furthermore, some require expression of multiple sgRNAs and/or fusion of additional functional domains to Cas9, which can reduce targeting range and create challenges for delivery with viral vectors that have limits on nucleic acid payload size. Thus, a major challenge for the field remains the development of a robust and easily employed strategy that eliminates off-target mutations on a genome-wide scale.
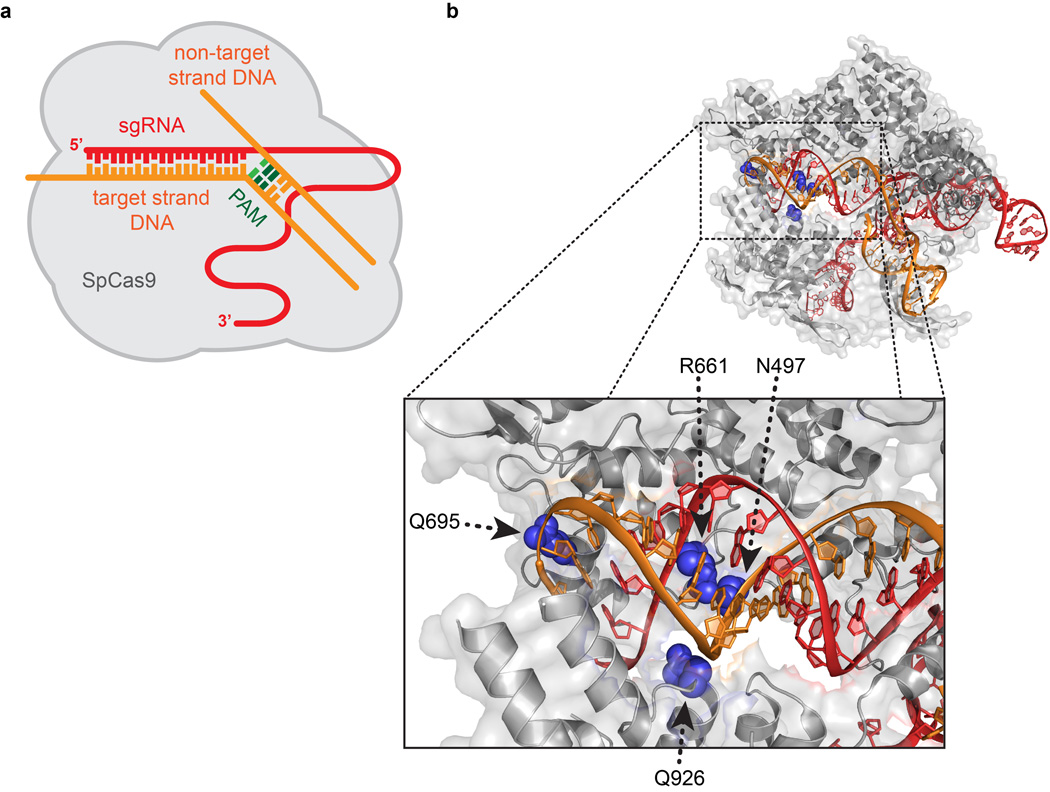
We initially hypothesized that off-target effects of SpCas9 might be minimized by decreasing non-specific interactions with its target DNA site. SpCas9-sgRNA complexes cleave target sites composed of an NGG PAM sequence (recognized by SpCas9)21–24 and an adjacent 20 bp protospacer sequence (which is complementary to the 5’ end of the sgRNA)22, 25–27. We previously theorized that the SpCas9-sgRNA complex might possess more energy than is needed for optimal recognition of its intended target DNA site, thereby enabling cleavage of mismatched off-target sites14. Structural studies have suggested that the SpCas9-sgRNA-target DNA complex includes several SpCas9-mediated DNA contacts, including direct hydrogen bonds made by four SpCas9 residues (N497, R661, Q695, Q926) to the phosphate backbone of the target DNA strand28, 29 (Fig. 1a and Extended Data Figs. 1a and 1b). We envisioned that disruption of one or more of these contacts might alter the energetics of the SpCas9-sgRNA complex so that it might retain enough for robust on-target activity but have a diminished ability to cleave mismatched off-target sites.
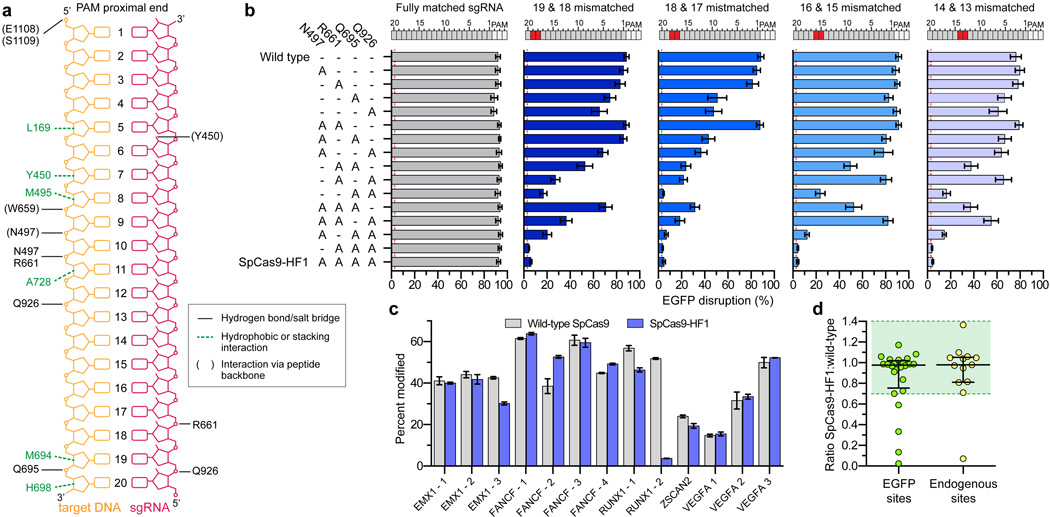
Figure 1. Identification and characterization of SpCas9 variants bearing substitutions in residues that form non-specific DNA contacts.
a, Schematic depicting wild-type SpCas9 interactions with the target DNA:sgRNA duplex, based on PDB 4OO8 and 4UN3 (adapted from refs. 28 and 29, respectively). b, Characterization of SpCas9 variants that contain alanine substitutions in positions that form hydrogen bonds with the DNA backbone. Wild-type SpCas9 and variants were assessed using the human cell EGFP disruption assay when programmed with a perfectly matched sgRNA or partially mismatched sgRNAs. Error bars represent s.e.m. for n = 3; mean level of background EGFP loss represented by red dashed line. c, On-target activities of wild-type SpCas9 and SpCas9-HF1 across 13 endogenous sites measured by T7E1 assay. Error bars represent s.e.m. for n = 3. d, Ratio of on-target activity of SpCas9-HF1 to wild-type SpCas9. The median and interquartile range are shown; the interval with >70% of wild-type activity is highlighted in green.
Alteration of SpCas9 DNA contacts
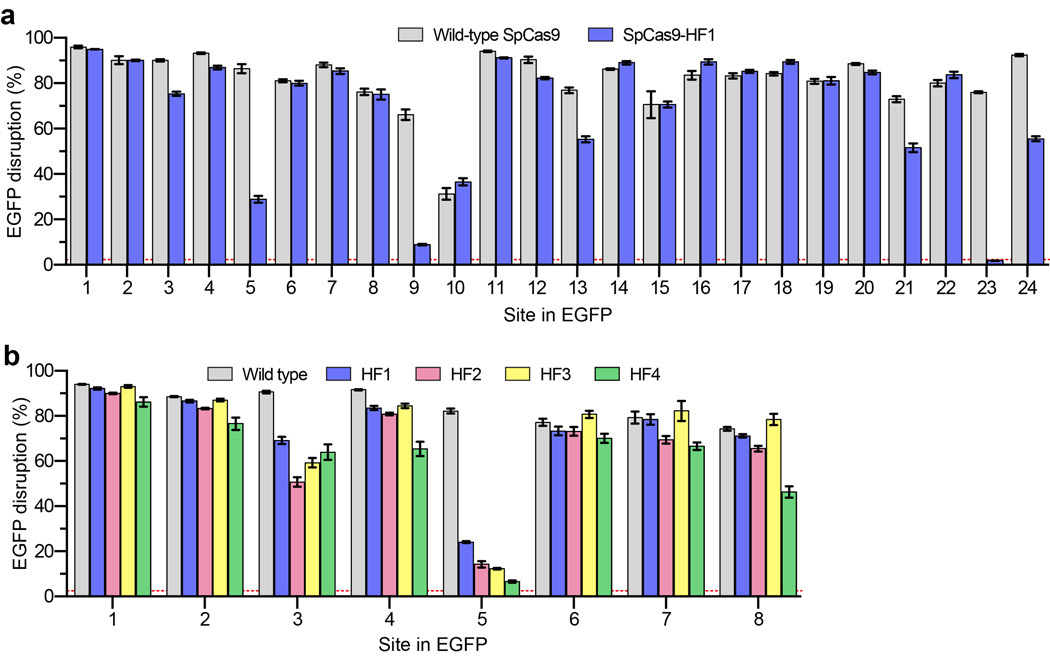
Guided by this excess energy hypothesis, we first constructed 15 different SpCas9 variants bearing all possible single, double, triple and quadruple combinations of N497A, R661A, Q695A, and Q926A substitutions to test whether contacts made by these residues might be dispensable for on-target activity (Fig. 1b). For these experiments, we used a previously described human cell-based EGFP-disruption assay30. Using an EGFP-targeted sgRNA, which we have previously shown can efficiently induce insertion or deletion mutations (indels) in an EGFP reporter gene when paired with wild-type SpCas9 (ref. 4), we found that all 15 SpCas9 variants possessed activities comparable to that of wild-type SpCas9 (Fig. 1b, grey bars). Thus, alanine substitution of one or all of these residues did not reduce on-target cleavage efficiency of SpCas9 with this EGFP-targeted sgRNA.
Next, we sought to assess the relative activities of all 15 SpCas9 variants at mismatched target sites. To do this, we repeated the EGFP-disruption assay with derivatives of the EGFP-targeted sgRNA used in the previous experiment that contain pairs of substituted bases at positions ranging from 13 to 19 (numbering starting with 1 for the most PAM-proximal base and ending with 20 for the most PAM-distal base; Fig. 1b). This analysis revealed that one of the triply substituted variants (R661A/Q695A/Q926A) and the quadruple substitution variant (N497A/R661A/Q695A/Q926A) both showed minimal EGFP disruption at near-background levels with all four of the mismatched sgRNAs (Fig. 1b, colored bars). Based on these results, we chose the quadruple substitution variant (hereafter referred to as SpCas9-HF1 for High-Fidelity variant #1) for further analysis.
SpCas9-HF1 retains high on-target activities
To determine how robustly SpCas9-HF1 functions at a larger number of on-target sites, we performed direct comparisons between this variant and wild-type SpCas9 using additional sgRNAs. In total, we tested 37 different sgRNAs: 24 targeted to EGFP and 13 targeted to endogenous human gene targets. For 20 of the 24 sgRNAs tested using the EGFP disruption assay (Extended Data Fig. 2a) and 12 of the 13 sgRNAs tested using a T7 Endonuclease I (T7EI) mismatch assay (Fig. 1c), we found SpCas9-HF1 exhibited on-target activities that were at least 70% of what was observed with wild-type SpCas9 (Fig. 1d). Indeed, SpCas9-HF1 showed highly comparable activities (90–140%) to wild-type SpCas9 with the vast majority of sgRNAs (Fig. 1d). Three of the 37 sgRNAs tested showed essentially no activity with SpCas9-HF1 (EGFP sites 9 and 23, and RUNX1 site 2), and examination of these target sites did not suggest any obvious differences in the characteristics of these sequences compared to those for which we saw high activities (Supplementary Table 1). Overall, SpCas9-HF1 possesses comparable activities (greater than 70% of wild-type SpCas9 activities) for 86% (32/37) of the sgRNAs we tested.
Genome-wide specificity of SpCas9-HF1
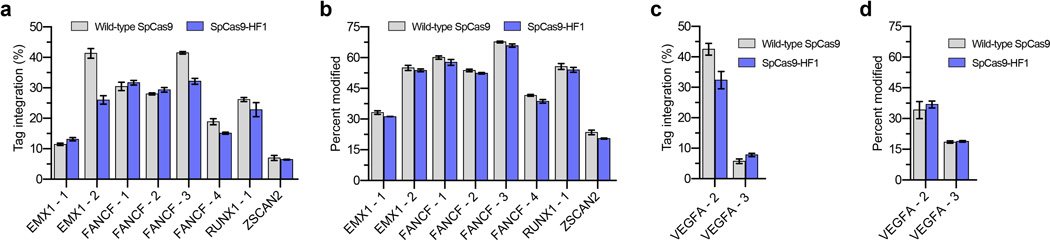
To test whether SpCas9-HF1 exhibits reduced off-target effects in human cells, we used the genome-wide unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq) method8 to assess eight different sgRNAs targeted to sites in the endogenous human EMX1, FANCF, RUNX1, and ZSCAN2 genes. The sequences targeted by these sgRNAs have variable numbers of predicted mismatched sites in the reference human genome (Extended Data Table 1). Assessment of on-target double-stranded oligodeoxynucleotide (dsODN) tag integration (by restriction fragment length polymorphism (RFLP) assay) and indel formation (by T7EI assay) for the eight sgRNAs revealed comparable on-target activities with wild-type SpCas9 and SpCas9-HF1 (Extended Data Figs. 3a and 3b, respectively), demonstrating that these GUIDE-seq experiments were working efficiently and comparably with the two different nucleases.
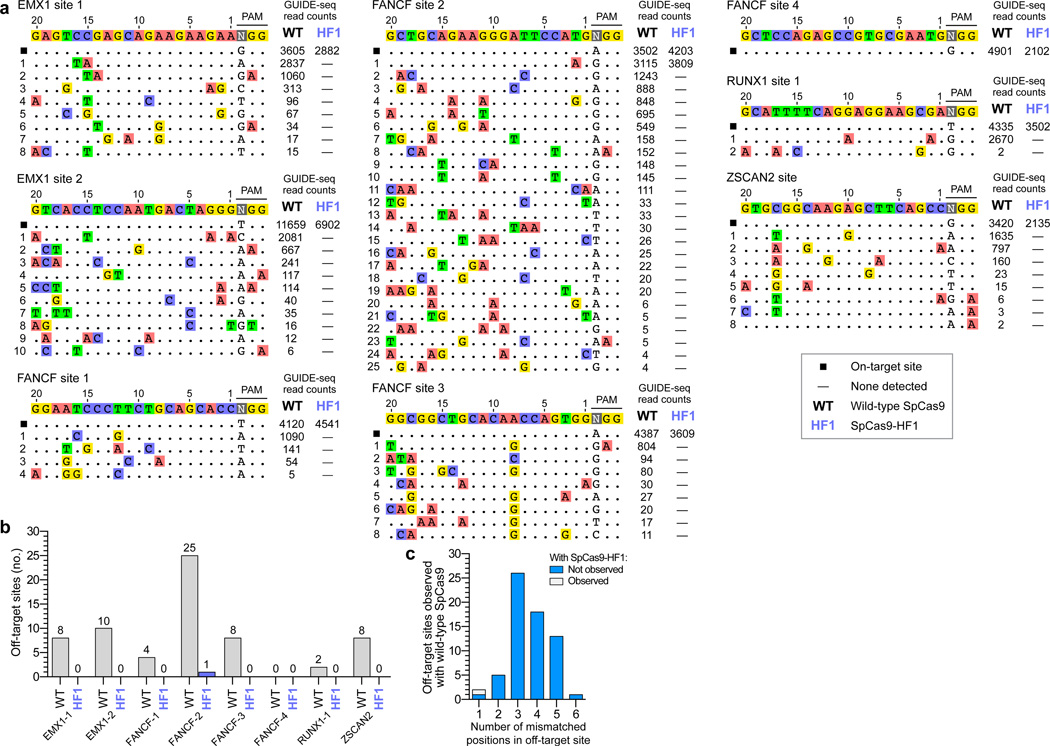
These GUIDE-seq experiments showed that with wild-type SpCas9, seven of the eight sgRNAs induced cleavage at multiple off-target sites (ranging from 2 to 25 per sgRNA), whereas the eighth sgRNA (FANCF site 4) did not yield any detectable off-target sites (Figs. 2a and 2b). The off-target sites identified harbored one to six mismatches distributed throughout various positions in the protospacer and/or PAM sequence (Fig. 2c; Extended Data Fig. 4a). However, with SpCas9-HF1, a complete absence of GUIDE-seq detectable off-target events was observed for six of the seven sgRNAs that induced off-target effects with wild-type SpCas9 (Figs. 2a and 2b). Among these seven sgRNAs, only a single detectable genome-wide off-target was identified, for FANCF site 2, at a site harboring one mismatch within the protospacer seed sequence (Fig. 2a). As with wild-type SpCas9, the eighth sgRNA (FANCF site 4) did not yield any detectable off-target cleavage events when tested with SpCas9-HF1 (Fig. 2a). Notably, with all eight sgRNAs, SpCas9-HF1 did not create any new nuclease-induced off-target sites (i.e., not already observed with wild-type SpCas9) detectable by GUIDE-seq.
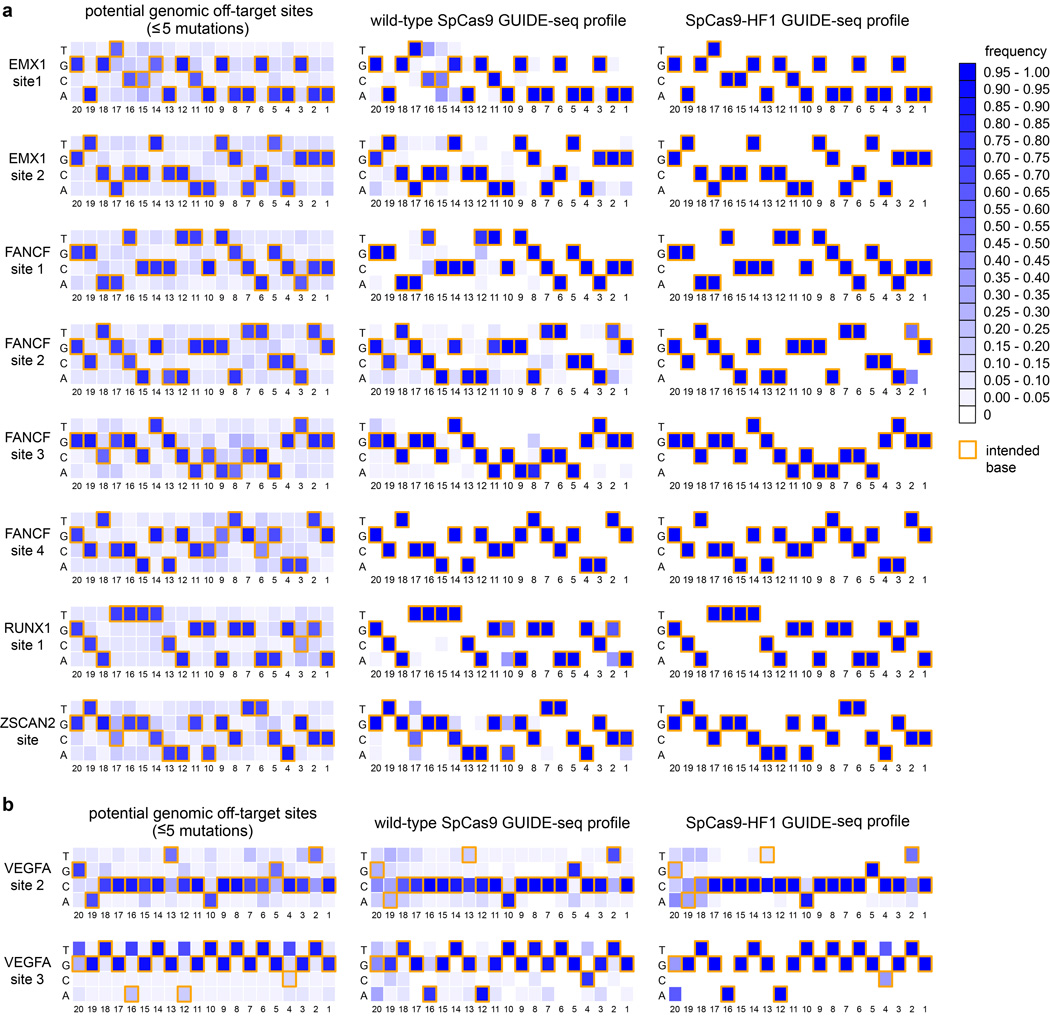
Figure 2. Genome-wide specificities of wild-type SpCas9 and SpCas9-HF1 with sgRNAs targeted to standard, non-repetitive sites.
a, Off-target cleavage sites of wild-type SpCas9 and SpCas9-HF1 with eight sgRNAs targeted to endogenous human genes, as determined by GUIDE-seq. Read counts represent a measure of cleavage frequency at a given site; mismatched positions within the spacer or PAM are highlighted in color. b, Summary of the total number of genome-wide off-target sites identified by GUIDE-seq for wild-type SpCas9 and SpCas9-HF1 with the sgRNAs used in panel a. c, Off-target sites identified for wild-type SpCas9 and SpCas9-HF1 for the eight sgRNAs, binned according to the total number of mismatches (in the protospacer and PAM) relative to the on-target site.
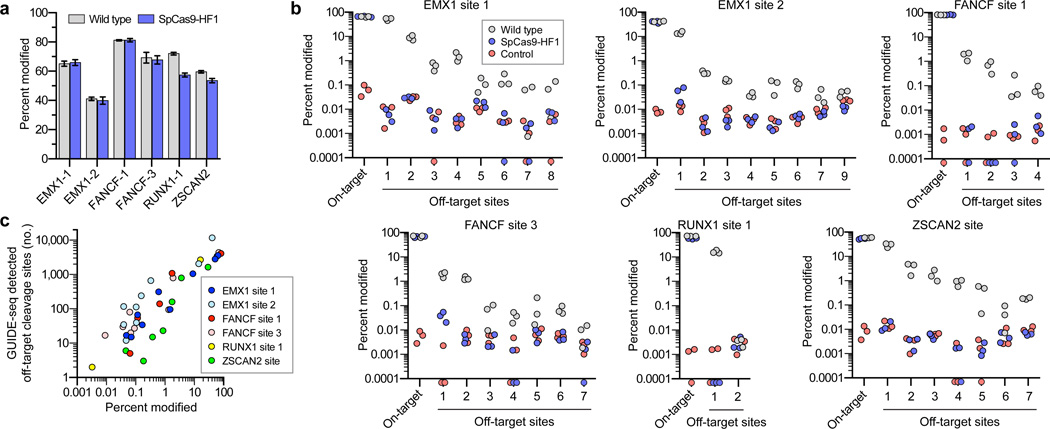
To confirm these GUIDE-seq findings, we used targeted amplicon sequencing to more directly measure the frequencies of indel mutations induced by wild-type SpCas9 and SpCas9-HF1. For these experiments, we transfected human cells only with sgRNA- and Cas9-encoding plasmids (i.e., without the GUIDE-seq tag). We then used next-generation sequencing to examine the on-target sites and 36 of the 40 off-target sites that had been identified for six sgRNAs with wild-type SpCas9 in our GUIDE-seq experiments (four of the 40 sites could not be specifically amplified from genomic DNA). These deep sequencing experiments showed that: (1) wild-type SpCas9 and SpCas9-HF1 induced comparable frequencies of indels at each of the six sgRNA on-target sites, indicating that the nucleases and sgRNAs were functional in all experimental replicates (Figs. 3a and 3b); (2) as expected, wild-type SpCas9 showed statistically significant evidence of indel mutations at 35 of the 36 off-target sites (Fig. 3b) at frequencies that correlated well with GUIDE-seq read counts for these same sites (Fig. 3c); and (3) the frequencies of indels induced by SpCas9-HF1 at 34 of the 36 off-target sites were statistically indistinguishable from the background level of indels observed in samples from control transfections (Fig. 3b). For the two off-target sites that appeared to have statistically significant mutation frequencies with SpCas9-HF1 relative to the negative control, the mean frequencies of indels were 0.049% and 0.037%, levels at which it is difficult to determine whether these are due to sequencing/PCR error or are bona fide nuclease-induced indels. Based on these results, we conclude that SpCas9-HF1 can completely or nearly completely reduce off-target mutations that occur across a range of different frequencies with wild-type SpCas9 to levels generally undetectable by GUIDE-seq and targeted deep sequencing.
Figure 3. Validation of SpCas9-HF1 specificity improvements by deep sequencing of off-target sites identified by GUIDE-seq.
a, Mean on-target percent modification for wild-type SpCas9 and SpCas9-HF1 with six sgRNAs from Fig. 2. Error bars represent s.e.m. for n = 3. b, Percent modification of on-target and GUIDE-seq detected off-target sites with indel mutations. Triplicate experiments are plotted for wild-type SpCas9, SpCas9-HF1, and a negative control; off-target sites are numbered as indicated in Fig. 2a. Filled circles below the x-axis represent replicates for which no insertion or deletion mutations were observed (Supplementary Table 4). Hypothesis testing using a one-sided Fisher exact test with pooled read counts found significant differences (p < 0.05 after adjusting for multiple comparisons using the Benjamini-Hochberg method) for comparisons between SpCas9-HF1 and the control condition only at EMX1-1 off-target 1 and FANCF-3 off-target 1. Significant differences were also found between wild-type SpCas9 and SpCas9-HF1 at all off-target sites, and between wild-type SpCas9 and the control condition at all off-target sites except RUNX1-1 off-target 2. c, Scatter plot of the correlation between GUIDE-seq read counts (from Fig. 2a) and mean percent modification determined by deep sequencing at on- and off-target cleavage sites with wild-type SpCas9.
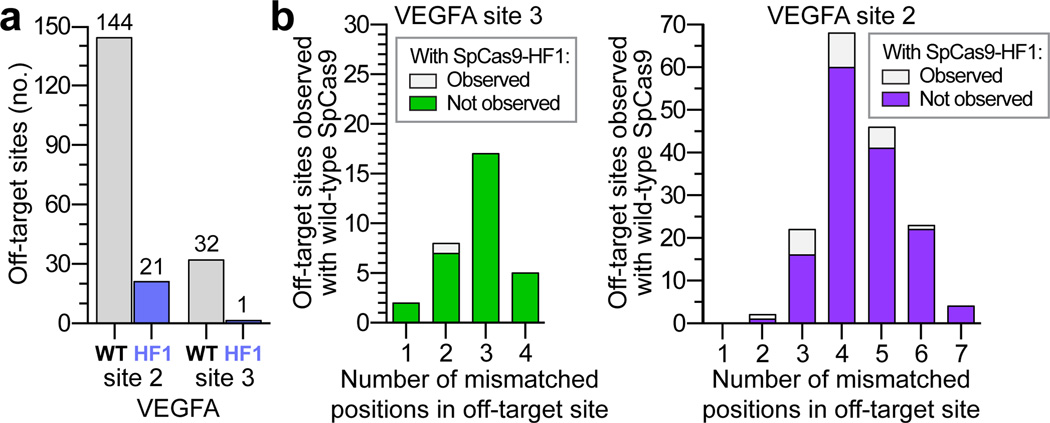
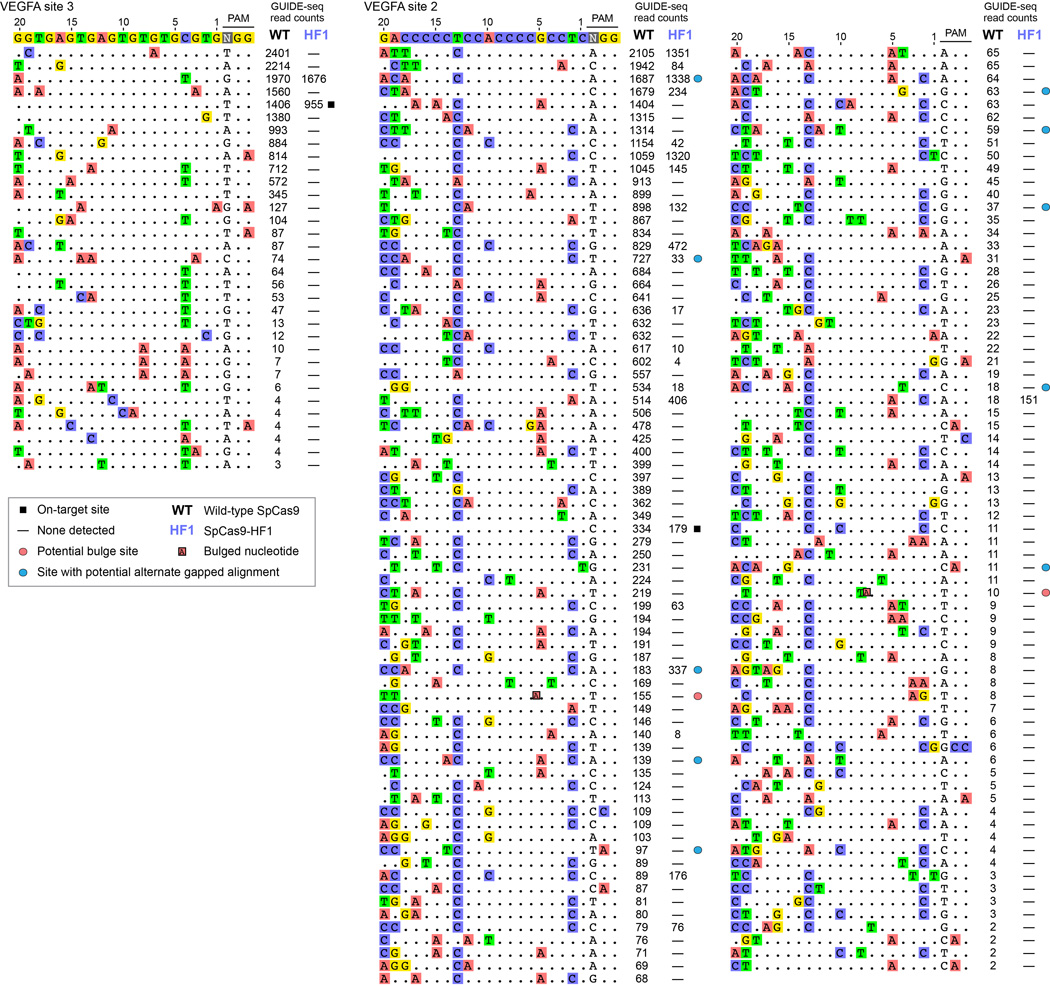
We next assessed the capability of SpCas9-HF1 to reduce genome-wide off-target effects of sgRNAs designed against atypical homopolymeric or repetitive sequences. Although we and other researchers now try to avoid on-target sites with these characteristics due to their relative lack of orthogonality to the genome, we wished to challenge the genome-wide specificity of SpCas9-HF1 with sites that have very large numbers of known off-target sites in human cells. Therefore, we used previously characterized sgRNAs4, 8 that target either a cytosine-rich homopolymeric sequence or a sequence containing multiple TG repeats in the human VEGFA gene (VEGFA site 2 and VEGFA site 3, respectively) (Extended Data Table 1). In control experiments, we again found that each of these sgRNAs induced comparable levels of GUIDE-seq dsODN tag incorporation (Extended Data Fig. 3c) and indel mutations (Extended Data Fig. 3d) with both wild-type SpCas9 and SpCas9-HF1, demonstrating that SpCas9-HF1 is not impaired in on-target activity with either of these sgRNAs. Importantly, these GUIDE-seq experiments revealed that SpCas9-HF1 was highly effective at reducing off-target sites of these sgRNAs, with 123/144 sites for VEGFA site 2 and 31/32 sites for VEGFA site 3 not detected (Fig. 4a and Extended Data Fig. 5). Examination of wild-type SpCas9 off-target sites not detected with SpCas9-HF1 showed that they each possessed a range of total mismatches distributed at various positions within their protospacer and PAM sequences: 2 to 7 mismatches for the VEGFA site 2 sgRNA and 1 to 4 mismatches for the VEGFA site 3 sgRNA (Fig. 4b; Extended Data Fig. 4b); also, nine of these off-targets for VEGFA site 2 may be recognized by an alternate potential base pairing interaction with the sgRNA that might occur with a single bulged base12 at the sgRNA-DNA interface (Extended Data Figs. 5 and 6). Overall, the sites that were still mutated by SpCas9-HF1 possessed a range of 2 to 6 mismatches for the VEGFA site 2 sgRNA and 2 mismatches in the single site for the VEGFA site 3 sgRNA (Fig. 4b), with three of the off-target sites for the VEGFA site 2 sgRNA having an alternative potential single bulge alignment (Extended Data Figs. 5 and 6). Notably, no new nuclease-induced off-target sites were induced by SpCas9-HF1 with either of the two sgRNAs. Collectively, these results demonstrate that SpCas9-HF1 can be highly effective at reducing off-target effects of sgRNAs targeted to simple repeat sequences and can also have substantial impacts on sgRNAs targeted to homopolymeric sequences.
Figure 4. Genome-wide specificities of wild-type SpCas9 and SpCas9-HF1 with sgRNAs targeted to non-standard, repetitive sites.
a, Summary of the total number of genome-wide off-target cleavage sites identified by GUIDE-seq for wild-type SpCas9 and SpCas9-HF1 with sgRNAs targeted to VEGFA sites 2 and 3. b, Off-target sites identified for wild-type SpCas9 or SpCas9-HF1 with sgRNAs targeted VEGFA sites 2 and 3 binned according to the total number of mismatches (within the protospacer and PAM) relative to the on-target site.
Refining the specificity of SpCas9-HF1
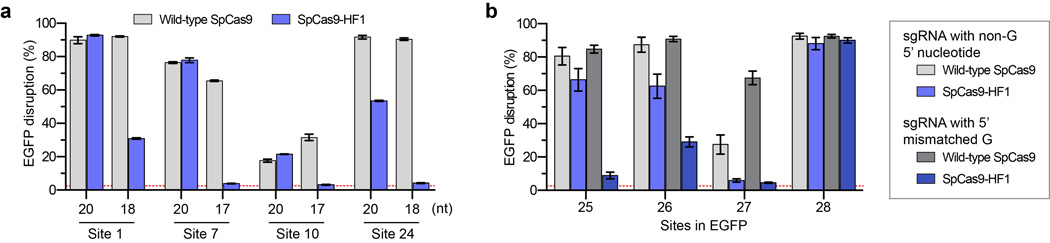
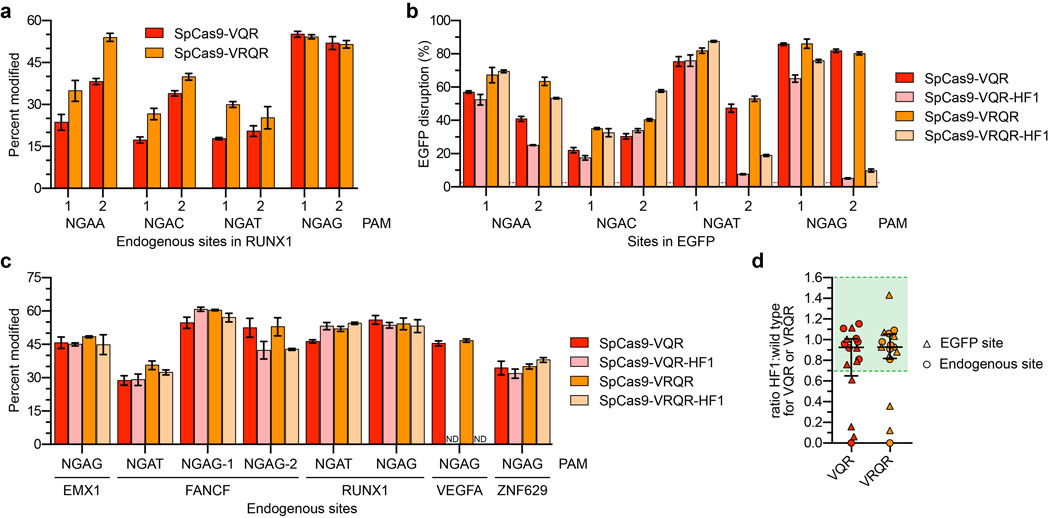
Previously described methods such as truncated sgRNAs14 and the SpCas9-D1135E variant15 can partially reduce SpCas9 off-target effects, and we therefore wondered whether these might be combined with SpCas9-HF1 to further improve its genome-wide specificity. Testing of SpCas9-HF1 with matched full-length and truncated sgRNAs targeted to four sites in the human cell-based EGFP disruption assay revealed that shortening sgRNA complementarity length substantially impaired on-target activities (Extended Data Fig. 7a). By contrast, SpCas9-HF1 with an additional D1135E substitution (a variant we call SpCas9-HF2) retained 70% or more activity of wild-type SpCas9 with six of eight sgRNAs tested using our human cell-based EGFP disruption assay (Figs. 5a and Extended Data Fig. 2b). We also constructed SpCas9-HF3 and SpCas9-HF4 variants harboring additional L169A or Y450A substitutions, respectively, at positions whose side chains are believed to mediate non-specific hydrophobic interactions with the target DNA on its PAM proximal end28, 31 (Fig. 1a). The Y450 residue is notable for participating in a base stacking interaction with the sgRNA31 and undergoing a 120 degree shift upon target binding to create its hydrophobic interaction with the DNA28, 32. SpCas9-HF3 and SpCas9-HF4 retained 70% or more of the activities observed with wild-type SpCas9 with the same six out of eight EGFP-targeted sgRNAs (Figs. 5a and Extended Data Fig. 2b).
Figure 5. Activities of high-fidelity derivatives of SpCas9-HF1 bearing additional substitutions.
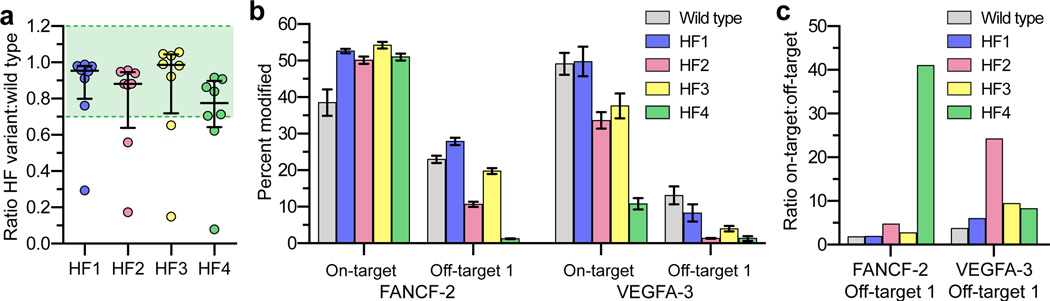
a, Summary of the on-target EGFP disruption activities of various SpCas9-HF variants compared to wild-type SpCas9 (from the data in Extended Data Fig. 2b). SpCas9-HF1 contains N497A, R661A, Q695, and Q926A substitutions; HF2 = HF1 + D1135E; HF3 = HF1 + L169A; HF4 = HF1 + Y450A. The median and interquartile range are shown; the interval showing >70% of wild-type activity is highlighted in green. b, Mean percent modification by SpCas9 and HF variants at the FANCF site 2 and VEGFA site 3 on-target sites, as well as off-target sites from Figs. 2a and Extended Data Fig. 5 resistant to the effects of SpCas9-HF1. Percent modification determined by T7E1 assay; background indel percentages were subtracted for all experiments; error bars represent s.e.m. for n = 3. c, Specificity ratios of wild-type SpCas9 and HF variants with the FANCF site 2 or VEGFA site 3 sgRNAs, plotted as the ratio of on-target to off-target activity (from panel b).
We next sought to determine whether SpCas9-HF2, -HF3, or -HF4 could reduce indel frequencies at two off-target sites that remained susceptible to modification by SpCas9-HF1, one with the FANCF site 2 sgRNA and another with the VEGFA site 3 sgRNA. For the FANCF site 2 off-target, which bears a single mismatch in the seed sequence of the protospacer, we found that SpCas9-HF4 (containing the additional Y450A substitution) reduced indel mutation frequencies to near background level as judged by T7EI assay while also beneficially increasing on-target activity (Fig. 5b), resulting in the greatest increase in specificity among the three variants (Fig. 5c). For the VEGFA site 3 off-target site, which bears two protospacer mismatches (one in the seed sequence and one at the nucleotide most distal from the PAM sequence), SpCas9-HF2 (containing the additional D1135E substitution) showed near background levels of indel formation as determined by T7E1 assay while showing modest effects on on-target mutation efficiency (Fig. 5b), leading to the greatest increase in specificity for this off-target site from among the three variants tested (Fig. 5c).
Discussion
The SpCas9-HF1 variant characterized in this report reduces all or nearly all genome-wide off-target effects to undetectable levels as judged by GUIDE-seq and targeted next-generation sequencing, with the most robust and consistent effects observed with sgRNAs designed against standard, non-repetitive target sequences. Our observations suggest that off-target mutations might be minimized by using SpCas9-HF1 to target non-repetitive sequences that do not have closely matched sites (e.g., bearing 1 or 2 mismatches) elsewhere in the genome; such sites can be easily identified using existing publicly available software programs33. An interesting question will be to determine whether SpCas9-HF1 induces off-target mutations at frequencies below the detection limit of existing unbiased genome-wide methods (Supplementary Discussion). We also discuss other practical considerations for targeting sites of interest with SpCas9-HF1, including the use of sgRNAs with non-G or mismatched 5’ nucleotides (Extended Data Fig. 7b) and altering the PAM recognition specificity of SpCas9-HF1 (Extended Data Fig. 8), in the Supplementary Discussion.
Further biochemical experiments and structural characterization will be required to define the mechanism by which SpCas9-HF1 achieves its high genome-wide specificity. We do not believe that the four substitutions we introduced alter the stability or steady-state expression level of SpCas9 in human cells, because titration experiments with decreasing concentrations of expression plasmids suggest that wild-type SpCas9 and SpCas9-HF1 behave comparably as their amounts are lowered (Extended Data Fig. 9). Although our initial rationale for making the substitutions in SpCas9-HF1 was to decrease the energetics of interaction between the Cas9-sgRNA and the target DNA (as has been previously proposed to explain the increased specificities of transcription activator-like effector nucleases bearing substitutions at positively charged residues34), recent work has provided greater mechanistic insights into SpCas9 recognition and cleavage. These studies suggest alternative and more detailed models (e.g., formation of an active cleavage complex through conformational changes or kinetics of off-target site recognition35, 36 that might be affected by the substitutions in our SpCas9-HF1 variant (Supplementary Discussion).
More broadly, our results validate a general strategy for the engineering of additional high-fidelity variants of CRISPR-associated nucleases. We found that introducing substitutions at additional non-specific DNA contacting residues can further reduce some of the very small number of residual off-target sites that persist for certain sgRNAs with SpCas9-HF1. Thus, we envision that variants such as SpCas9-HF2, SpCas9-HF4, and others might be used in a customized fashion to eliminate any potential off-target sites that might be resistant to the specificity improvements of SpCas9-HF1. In addition, our variants might be combined with substitutions in residues that contact the non-target DNA strand, alterations that have been shown to reduce SpCas9 off-target effects while our manuscript was under review37. Overall, our results demonstrate that the approach of mutating non-specific DNA contacts is highly effective at increasing SpCas9 specificity and suggest it might be extended to other naturally occurring and engineered Cas9 orthologues38–42 as well as other CRISPR-associated nucleases43, 44.
METHODS
Plasmids and oligonucleotides
DNA sequences of plasmids used in this study can be found in Supplementary Information. sgRNAs target sites are available in Supplementary Table 1, and oligonucleotides used in this study can be found in Supplementary Table 2. SpCas9 expression plasmids containing amino acid substitutions were generated by standard PCR and molecular cloning into JDS2464. sgRNA expression plasmids were constructed by ligating oligonucleotide duplexes into BsmBI cut BPK152015. Unless otherwise indicated, all sgRNAs were designed to target sites containing a 5’-guanine nucleotide.
Human cell culture and transfection
U2OS cells (a gift from Toni Cathomen, Freiburg) and U2OS.EGFP cells (containing a single integrated copy of an EGFP-PEST reporter gene)30 were cultured in Advanced DMEM supplemented with 10% HI FBS, 2 mM GlutaMax, and penicillin/streptomycin at 37°C with 5% CO2. The growth media for U2OS.EGFP cells was additionally supplemented with 400 µg ml−1 Geneticin. All cell culture reagents were obtained from Life Technologies. Cell line identity was validated by STR profiling (ATCC) and deep-sequencing, and cells were tested bi-weekly for mycoplasma contamination. Unless otherwise noted, cells were co-transfected with 750 ng of Cas9 plasmid and 250 ng of sgRNA plasmid. For negative control experiments, Cas9 plasmids were co-transfected with a U6-null plasmid. Nucleofections were performed using the DN-100 program on a Lonza 4-D Nucleofector with the SE Cell Line Kit according to the manufacturer’s protocol (Lonza). For T7E1 assays, GUIDE-seq experiments, and targeted deep sequencing, genomic DNA was extracted ~72 hours post-transfection using the Agencourt DNAdvance Genomic DNA Isolation Kit (Beckman Coulter Genomics).
Human cell EGFP disruption assay
EGFP disruption experiments, in which cleavage and induction of indels by non-homologous end-joining (NHEJ)-mediated repair within a single integrated EGFP reporter gene leads to loss of cell fluorescence, were performed as previously described4, 30. Briefly, transfected cells were analyzed ~52 hours post-transfection for loss of EGFP expression using a Fortessa flow cytometer (BD Biosciences). Background EGFP loss was determined using negative control transfections gated at ~2.5% for all experiments (represented as a red dashed line in figures). P values for comparisons between SpCas9 variants were calculated using a one-sided t-test with equal variances and adjusted for multiple comparisons using the method of Benjamini and Hochberg (Supplementary Table 3).
T7E1 assays
To quantify mutagenesis frequencies at desired genomic loci, T7E1 assays were performed as previously described30. Briefly, on- or off-target sites were amplified from ~100 ng of genomic DNA using Phusion Hot-Start Flex DNA Polymerase (New England Biolabs) using the primers listed in Supplementary Table 2. An Agencourt Ampure XP cleanup (Beckman Coulter Genomics) was performed prior to the denaturation and annealing of ~200 ng of the PCR product, followed by digestion with T7E1 (New England Biolabs). Purified digestion products were quantified using a QIAxcel capillary electrophoresis instrument (Qiagen) to approximate the mutagenesis frequencies induced by Cas9-sgRNA complexes. P values for comparisons between SpCas9 variants were calculated using a one-sided t-test with equal variances and adjusted for multiple comparisons using the method of Benjamini and Hochberg (Supplementary Table 3).
GUIDE-seq
GUIDE-seq relies on the integration of a short dsODN tag into DNA breaks to enable amplification and sequencing of adjacent genomic sequence, with the number of tag integrations at any given site providing a quantitative measure of cleavage efficiency8. GUIDE-seq experiments were performed and analyzed essentially as previously described8. Briefly, U2OS cells were transfected with 750 ng of Cas9 and 250 ng sgRNA plasmids as described above, along with 100 pmol of a GUIDE-seq end-protected dsODN that contains an NdeI restriction site8. Restriction fragment length polymorphism (RFLP) assays were used to estimate GUIDE-seq tag integration frequencies at the intended on-target sites as previously described15, using the primers listed in Supplementary Table 2. The overall on-target mutagenesis frequencies of GUIDE-seq tag-treated samples was determined by T7E1 assay as described above. Tag-specific amplification and library preparation8 were performed prior to high-throughput sequencing on an Illumina MiSeq instrument. GUIDE-seq data was analyzed as previously described8 using open-source GUIDE-seq analysis software (http://www.jounglab.org/guideseq) and the summarized results can be found in Supplementary Table 4. Genomic sites were excluded from analysis on the basis of overlap with background genomic breakpoint regions detected in any of four oligo-only control samples, overlap with previously identified Cas9-sgRNA independent breakpoints in human U2OS cells8, or as neighboring genomic window consolidation artifacts likely due to extensive end-resection around breakpoints (Supplementary Table 4). Potential RNA- or DNA-bulge sites12 (Extended Data Fig. 6) were identified by sequence alignment with Geneious version 8.1.6 (http://www.geneious.com)45. Sequencing data was corrected for U2OS cell-type specific SNPs with the site encoding the smallest edit distance to the intended sgRNA site used as the most likely off-target (Supplementary Table 4). Differences in number of GUIDE-seq identified off-target sites between this work and previous studies8, 15 are likely due to different experimental conditions (e.g., different promoters, quantity of plasmids used for transfection) and/or to sampling effects at the limit of detection of these particular experiments (Supplementary Table 4), and most likely not due to depth of sequencing which was similar between experiments.
Positional profiles generated from GUIDE-seq data (Extended Data Fig. 4) were made by weighting each nucleotide at each on/off-target site by the number of GUIDE-seq read counts. Sites containing gapped alignments relative to the human genome were not considered. Positional profiles for potential genomic off-target sites were restricted to sequences containing five or fewer mutations relative to the on-target site and to sequences containing NGG PAMs. Heat maps were generated with R 3.2.2 and the image function, with colors determined using the function colorRampPalette(c("white","blue"))(2500).
Targeted deep-sequencing
Off-target sites identified by GUIDE-seq were amplified using Phusion High-Fidelity DNA polymerase (New England Biolabs) using the primers listed in Supplementary Table 2 for the genomic amplicons listed in Supplementary Table 5. PCR products were generated for each on- and off-target site from ~100 ng of genomic DNA extracted from U2OS cells. Products were generated from triplicate transfections for each of three experimental conditions: 1) control (wild-type SpCas9 + pSL695, a control sgRNA expression plasmid that does not encode a functional sgRNA), 2) wild-type SpCas9 + sgRNA, and 3) SpCas9-HF1 + sgRNA. PCR products were purified with Ampure XP magnetic beads (Agencourt), normalized in concentration, and pooled into nine samples (individual triplicate experiments for each of the three conditions listed above). Illumina Tru-seq compatible deep-sequencing libraries were prepared using ~500ng of each pooled sample using a ‘with-bead’ HTP library preparation kit (KAPA BioSystems), and sequenced via 150-bp paired-end sequencing on an Illumina MiSeq instrument. High-throughput sequencing data was analyzed essentially as previously described18. Breifly, paired reads were mapped to the human genome (reference sequence GRChr37) using the bwa mem algorithm with default parameters. High-quality reads (average quality score ≥ 30) were analyzed for the presence of two or more bp indels that overlapped to the on- or off-target sites (Supplementary Table 5). One bp indel mutations were only included if they occurred directly adjacent to the predicted cleavage site. P-values for comparisons between control, wild-type SpCas9 + sgRNA, and SpCas9-HF1 + sgRNA (Supplementary Table 5) were obtained on pooled triplicate data using a one-sided Fisher exact test in the R 3.2.2 software package. P-values for each set of comparisons were adjusted for multiple comparisons using the method of Benjamini and Hochberg (function p.adjust(method = “BH”) in R).
Code Availability
Scripts for GUIDE-seq analysis (v0.9) can be found at http://jounglab.org/guideseq. The scripts used for indel calling on deep sequencing data and GUIDE-seq profiles are available upon request.
Extended Data
Extended Data Figure 1. SpCas9 interaction with the sgRNA and target DNA.
a, Schematic illustrating the SpCas9:sgRNA complex, with base pairing between the sgRNA and target DNA. b, Structural representation of the SpCas9:sgRNA complex bound to the target DNA, from PDB: 4UN3 (ref. 29). The four residues that form hydrogen bond contacts to the target-strand DNA backbone are highlighted in blue; the HNH domain is hidden for visualization purposes.
Extended Data Figure 2. On-target activities of high-fidelity SpCas9 variants.
a and b, EGFP disruption activities of wild-type SpCas9 and SpCas9-HF1 (panel a) and SpCas9-HF1-derivative variants (panel b) in human cells. SpCas9-HF1 contains N497A, R661A, Q695, and Q926A substitutions; HF2 = HF1 + D1135E; HF3 = HF1 + L169A; HF4 = HF1 + Y450A. Error bars represent s.e.m. for n = 3; mean level of background EGFP loss represented by the red dashed line.
Extended Data Figure 3. On-target activity comparisons of wild-type and SpCas9-HF1 with various sgRNAs used for GUIDE-seq experiments.
a and c, Mean GUIDE-seq tag integration at the intended on-target site for GUIDE-seq experiments shown in Figs. 2a and Extended Data Fig. 5 (panels a and c, respectively), quantified by restriction fragment length polymorphism assay. Error bars represent s.e.m. for n = 3. b and d, Mean percent modification at the intended on-target site for GUIDE-seq experiments shown in Figs. 2a and Extended Data Fig. 5 (panels b and d, respectively), detected by T7E1 assay. Error bars represent s.e.m. for n = 3.
Extended Data Figure 4. Positional summary of off-target sites identified by GUIDE-seq.
Heat maps derived from GUIDE-seq data with sgRNAs targeting a, non-repetitive, or b, repetitive or homopolymeric sites in the genome are shown. Base frequencies in the set of all potential genomic off-target sites (weighted equally) with NGG PAMs and five or fewer mutations for each sgRNA are shown on the left. Summaries of off-target sites identified by GUIDE-seq for wild-type SpCas9 and SpCas9-HF1 (both weighted by read count) are shown on the right. Yellow box outlines denote on-target bases at each position. Positions (20-1) are shown below the heat maps, with 1 being the most PAM-proximal position. Note the presence of mismatches that would be expected to create potential wobble interactions (G→A or T→C) at certain positions among the off-target sites induced by wild-type SpCas9 and that SpCas9-HF1 appears to improve off-target sites without any obvious positional bias.
Extended Data Figure 5. Genome-wide cleavage specificity of wild-type SpCas9 and SpCas9-HF1 with sgRNAs targeted to non-standard, repetitive sites.
a, GUIDE-seq profiles of wild-type SpCas9 and SpCas9-HF1 using two sgRNAs known to cleave large numbers of off-target sites4, 8. GUIDE-seq read counts represent a measure of cleavage efficiency at a given site; mismatched positions within the spacer or PAM are highlighted in color; red circles indicate sites likely to have the indicated bulge12 at the sgRNA-DNA interface; blue circles indicate sites that may have an alternative gapped alignment relative to the one shown (see Extended Data Fig. 6). Off-target sites marked with red circles are not included in the counts of Fig. 4b; sites marked with blue circles are counted with the number of mismatches in the non-gapped alignment for Fig. 4b.
Extended Data Figure 6. Potential alternate alignments for VEGFA site 2 off-target sites.
Ten VEGFA site 2 off-target sites identified by GUIDE-seq (left) that may potentially be recognized as off-target sites with single nucleotide gaps12 (right), aligned using Geneious45 version 8.1.6 (http://www.geneious.com).
Extended Data Figure 7. Activities of wild-type SpCas9 and SpCas9-HF1 with truncated and 5’ mismatched sgRNAs14.
a, EGFP disruption activities of wild-type SpCas9 and SpCas9-HF1 using full-length or truncated sgRNAs. b, EGFP disruption activities of wild-type SpCas9 and SpCas9-HF1 using sgRNAs that encode a matched 5’ non-G nucleotide or an intentionally mismatched 5’ G nucleotide. For both panels, error bars represent s.e.m. for n = 3, and the mean level of background EGFP loss observed in control experiments is represented by the red dashed line.
Extended Data Figure 8. Altering the PAM recognition specificity of SpCas9-HF1.
a, Comparison of the mean percent modification of on-target endogenous human sites by the SpCas9-VQR variant (ref. 15) and an improved SpCas9-VRQR variant using 8 sgRNAs, quantified by T7E1 assay. Both variants are engineered to recognize an NGAN PAM. Error bars represent s.e.m. for n = 3. b, On-target EGFP disruption activities of SpCas9-VQR and SpCas9-VRQR compared to their -HF1 counterparts using eight sgRNAs. Error bars represent s.e.m. for n = 3; mean level of background EGFP loss in negative controls represented by the red dashed line. c, Comparison of the mean on-target percent modification by SpCas9-VQR and SpCas9-VRQR compared to their -HF1 variants at eight endogenous human gene sites, quantified by T7E1 assay. Error bars represent s.e.m. for n = 3; ND, not detectable. d, Summary of the fold-change in on-target activity when using SpCas9-VQR or SpCas9-VRQR compared to their corresponding -HF1 variants (from panels b and c). The median and interquartile range are shown; the interval showing greater than 70% of wild-type activity is highlighted in green.
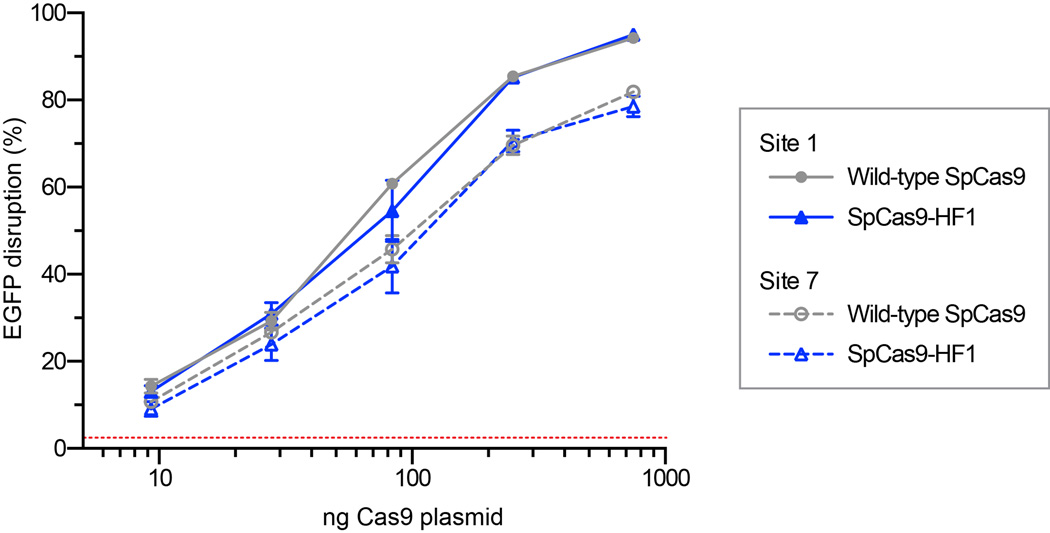
Extended Data Figure 9. Titrations of wild-type SpCas9 and SpCas9-HF1 expression plasmid amounts.
Human cell EGFP disruption activities from transfections with varying amounts of wild-type and SpCas9-HF1 expression plasmids. For all transfections, the amount of sgRNA-containing plasmid was fixed at 250 ng. Two sgRNAs targeting different sites were used; Error bars represent s.e.m. for n = 3; mean level of background EGFP loss in negative controls is represented by the red dashed line.
Extended Data Table 1.
Summary of potential mismatched sites in the reference human genome for the ten sgRNAs examined by GUIDE-seq
| mismatches to on-target site* | ||||||||
|---|---|---|---|---|---|---|---|---|
| site | spacer with PAM | 1 | 2 | 3 | 4 | 5 | 6 | total |
| EMX1-1 | GAGTCCGAGCAGAAGAAGAAGGG | 0 | 1 | 18 | 273 | 2318 | 15831 | 18441 |
| EMX1-2 | GTCACCTCCAATGACTAGGGTGG | 0 | 0 | 3 | 68 | 780 | 6102 | 6953 |
| FANCF-1 | GGAATCCCTTCTGCAGCACCTGG | 0 | 1 | 18 | 288 | 1475 | 9611 | 11393 |
| FANCF-2 | GCTGCAGAAGGGATTCCATGAGG | 1 | 1 | 29 | 235 | 2000 | 13047 | 15313 |
| FANCF-3 | GGCGGCTGCACAACCAGTGGAGG | 0 | 0 | 11 | 79 | 874 | 6651 | 7615 |
| FANCF-4 | GCTCCAGAGCCGTGCGAATGGGG | 0 | 0 | 6 | 59 | 639 | 5078 | 5782 |
| RUNX1-1 | GCATTTTCAGGAGGAAGCGATGG | 0 | 2 | 6 | 189 | 1644 | 11546 | 13387 |
| ZSCAN2 | GTGCGGCAAGAGCTTCAGCCGGG | 0 | 3 | 12 | 127 | 1146 | 10687 | 11975 |
| VEGFA-2 | GACCCCCTCCACCCCGCCTCCGG | 0 | 2 | 35 | 456 | 3905 | 17576 | 21974 |
| VEGFA-3 | GGTGAGTGAGTGTGTGCGTGTGG | 1 | 17 | 383 | 6089 | 13536 | 35901 | 55927 |
determined using Cas-OFFinder (http://www.rgenome.net/cas-offinder/)
Supplementary Material
Acknowledgments
B.P.K. is supported by a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship. V.P. was supported by the Massachusetts General Hospital (MGH) Department of Pathology. S.Q.T. is supported by an MGH Tosteson and Fund for Medical Discovery Fellowship. J.K.J. is supported by a US National Institutes of Health (NIH) Director’s Pioneer Award, NIH R01 GM107427, NIH R01 GM088040, and the Jim and Ann Orr MGH Research Scholar Award.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/XXXXXX.
Author Contributions
B.P.K., V.P., and J.K.J. conceived of and designed experiments. B.P.K., V.P., and M.S.P. performed all experiments. N.T.N. contributed to GUIDE-seq library preparation. B.P.K., V.P., M.S.P., S.Q.T., and Z.Z. analyzed the data. B.P.K., V.P., and J.K.J. wrote the manuscript with input from all the authors.
Plasmids encoding the high-fidelity SpCas9, VQR, and VRQR variants described in this manuscript have been deposited with the non-profit plasmid distribution service Addgene (http://www.addgene.org/crispr-cas). All sequencing data from this study is available through the NCBI Sequence Read Archive (SRA) under accession number SRP066862.
Competing financial interests
J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine, Hera Testing Laboratories, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. A patent application has been filed for high-fidelity Cas9 variants.
References
- 1.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 4.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 11.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. 231 p following 243. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum Gene Ther. 2015;26:425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimasu H, et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell. 2014 doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 32.Jinek M, et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science. 2014 doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guilinger JP, et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat Methods. 2014;11:429–435. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight SC, et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 37.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2015 doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou Z, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleinstiver BP, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015 doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zetsche B, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shmakov S, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Molecular Cell. 60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.