Abstract
Autophagy is a lysosomal degradation process that degrades long-lived cellular proteins and damaged organelles as a critical cell survival mechanism in response to stress. We recently reported that acute ethanol induces autophagy, which then reduces ethanol-induced liver injury. However, the mechanisms by which ethanol induces autophagy are not known. In the present study, ethanol treatment significantly increased both mRNA and protein levels of various essential autophagy-related genes in primary cultured mouse hepatocytes and in mouse liver. Both nuclear translocation of FoxO3a and expression of FoxO3a target genes were increased in ethanol-treated primary hepatocytes and mouse liver. Overexpression of a dominant negative form of FoxO3a inhibited ethanol-induced autophagy-related gene expression and enhanced ethanol-induced cell death in primary hepatocytes, which suggests that FoxO3a is a key factor in regulating ethanol-induced autophagy and cell survival. Resveratrol, a well-known SIRT1 agonist, further enhanced ethanol-induced expression of autophagy-related genes, likely via increased deacetylation of FoxO3a. Moreover, acute ethanol–treated Foxo3a−/− mice exhibited decreased autophagy-related gene expression, but enhanced steatosis and liver injury, compared with wild-type mice. FoxO3a thus plays a critical role in ethanol-induced autophagy in mouse liver. Modulating the FoxO3a autophagy pathway may offer novel therapeutic approaches for treating alcoholic liver pathogenesis.
Macroautophagy (hereafter referred to simply as autophagy) is a bulk intracellular degradation system that is mainly responsible for the degradation of long-lived proteins to provide nutrients for survival in response to starvation.1, 2 Accumulating evidence indicates that autophagy can also selectively remove damaged or excess organelles, including mitochondria, endoplasmic reticulum, ribosome, and peroxisome.3, 4, 5 In the liver, autophagy can also help to remove excess lipid droplets to attenuate steatosis (in which case it is termed lipophagy).6
Alcoholic liver disease is a major disease of the liver in Western countries and worldwide. The clinical characteristic of this disease is the accumulation of excess fat in the liver in response to alcohol consumption. In humans, it is known that accumulation of excess fat can progress to more detrimental forms of liver injury, such as inflammation, fibrosis, and cirrhosis. However, it is also well known that only a small portion of alcohol drinkers can develop advanced liver inflammation and fibrosis, suggesting that a cellular protective mechanism or mechanisms could play a critical role in mitigating alcohol-induced liver injury.7, 8 Our research group recently demonstrated that acute ethanol treatment induces autophagy in primary cultured mouse hepatocytes and in mouse liver; pharmacological induction of autophagy attenuated and pharmacological inhibition of autophagy exacerbated ethanol-induced steatosis and liver injury in mice.9 However, the mechanisms by which acute ethanol induces autophagy in hepatocytes are not known.
FoxO3a is a member of the FoxO (forkhead box O) family of transcription factors. FoxO3a regulates expression of genes involved in multiple cellular functions, including oxidative stress, apoptosis, and cell-cycle transition, as well as DNA repair.10, 11 Recent evidence suggests that FoxO3a also regulates expression of autophagy-related (Atg) genes in mouse skeletal muscle12, 13 and cardiomyocytes, which promotes cardiomyocyte survival on induction of oxidative stress.14
FoxO3a is regulated by multiple post-translational modifications, including phosphorylation, acetylation, and ubiquitination.10, 11 FoxO3a is phosphorylated by the serine/threonine protein kinase Akt and becomes sequestered in the cytoplasm, where it is unable to regulate gene expression. In contrast, SIRT1 [sirtuin (silent mating type information regulation 2 homolog) 1 (S. cerevisiae)] deacetylates FoxO3a and modulates its gene transcription and specificity, which increases antioxidative stress genes and suppresses apoptosis genes in response to oxidative stress.10, 15 However, it is not known whether acetylation of FoxO3a affects expression of autophagy-related genes.
In the present study, we demonstrated that acute ethanol treatment activates FoxO3a in mouse liver and in primary cultured mouse hepatocytes. The increased nuclear retention of FoxO3a by ethanol could be mediated by decreased Akt activity. Genetic inhibition of FoxO3a led to the suppression of ethanol-induced expression of autophagy-related genes and exacerbated ethanol-induced liver injury in mice. Moreover, we also found increased autophagy-related gene expression and autophagic flux in ethanol-treated human hepatocytes.
Materials and Methods
Reagents
The following antibodies were used: Atg5 (PM050) from MBL International (Woburn, MA); Beclin 1 (sc11427) and ULK1 (sc33182) from Santa Cruz Biotechnology (Santa Cruz, CA); Bnip3 (Ab10433) and Nix (Ab8399) from Abcam (Cambridge, MA); p62 (H00008878-M01) from Abnova (Walnut, CA); Atg7 (2631), p-FoxO3aS253 (9466), FoxO3a (2497), p-AktS473 (4060), Akt (2966), Sirt1 (2028), anti–acetylated-lysine (9441), and Lamin A/C (2032) from Cell Signaling Technology (Danvers, MA); and β-actin (A5441) and Atg14 (A6358) from Sigma-Aldrich (St. Louis, MO). Horseradish peroxidase–conjugated or Cy3-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Rabbit polyclonal anti-LC3B antibody was generated as described previously.9 Adenovirus GFP-LC3 (Ad-GFP-LC3), Ad-GFP, and adenovirus-dominant negative FoxO3a (Ad-DN-Foxo3a) were generated as described previously.12, 16 Chloroquine (CQ), rapamycin, actinomycin D (ActD), and resveratrol were from Sigma-Aldrich. Ethanol was from Pharmco (Brookfield, CT), and all other chemicals were from Sigma-Aldrich, Life Technologies (Carlsbad, CA), or Calbiochem (EMD Millipore, Billerica, MA).
Animals
Wild-type C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Foxo3a−/− mice were generated as described previously.17 Cryopreserved Foxo3a+/− mouse embryos were purchased from the RIKEN BioResource Center (Ibaraki, Japan) and recovered at the Animal Transgenic Core facility at the University of Kansas Medical Center. The Foxo3a+/− mice were maintained in a B6;129 background. Female Foxo3a−/− mice are infertile; Foxo3a−/− mice were therefore generated by crossing male Foxo3a+/− with female Foxo3a+/− mice. The generated Foxo3a+/+ mice were used as wild-type controls.
All animals received humane care according to the guidelines of the NIH and the University of Kansas Medical Center.
Mouse Ethanol Binge Treatment
Mouse ethanol treatment was modified from the model of Carson and Pruett,18 as we have described previously.9 This model was designed to achieve blood alcohol levels, behavioral effects, and physiological effects comparable to those of human binge drinking. After 6 hours of fasting, male Foxo3a−/− mice and their wild-type littermates were administered 33% (v/v) ethanol at a total cumulative dose of 4.5 g/kg body weight by four equally divided gavages at 15-minute intervals. Control mice received the same volume of double-distilled water. After 6, 12, and 16 hours of treatment, the mice were sacrificed, and blood samples and liver tissues were collected. Liver injury was assessed by determination of serum alanine aminotransferase activity and H&E staining of liver sections, as we have described previously.19 Total liver lysates were prepared using radioimmunoprecipitation assay buffer [1% NP40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl (lauryl) sulfate].
Primary Hepatocyte Culture
Mouse hepatocytes were isolated by a retrograde, nonrecirculating perfusion of livers with 0.05% collagenase type IV (Sigma-Aldrich), as we have described previously.20 Cells were cultured in Williams’ medium E with 10% fetal bovine serum, but no other supplements, for 2 hours to allow for attachment. Human hepatocytes were isolated and cultured according to methods described previously by Gramignoli et al.20 All cells were maintained in a 37°C incubator with 5% CO2. All human liver specimens were obtained in accordance with the protocol (no. 13513) approved by the University of Kansas Medical Center Human Subjects Committee.
Immunoblot Assay
Cells were washed in PBS and lysed in radioimmunoprecipitation assay buffer. From each sample, 20 μg of protein was separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were immunoblotted with primary antibodies, followed by secondary horseradish peroxidase-conjugated antibodies. The membranes were further developed with Pierce SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific, Rockford, IL). Densitometry analysis was performed using Un-Scan-It software version 6.1 (Silk Scientific, Orem, UT). Densitometry data were further normalized with either β-actin or the respective total protein and expressed as means ± SEM.
qPCR
RNA was isolated from mouse livers using TRIzol reagent (Life Technologies) and was reverse-transcribed into cDNA with Fermentas RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA). Quantitative real-time PCR (qPCR) to quantify gene expression of Atg5, Beclin 1 (Becn1; alias Atg6), Atg7, microtubule-associated protein 1 light chain 3 beta (Map1lc3b; alias LC3b), Ulk1, BCL2/adenovirus E1B interacting protein 3 (Bnip3), and β-actin (Actb) was performed on an ABI Prism 7900HT real-time PCR instrument (Life Technologies) using Fermentas Maxima SYBR Green/ROX qPCR reagents (Thermo Fisher Scientific). The forward and reverse primer sequences were as follows: β-actin, 5′-TGTTACCAACTGGGACGACA-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′; Atg5, 5′-GACCACAAGCAGCTCTGGAT-3′ and 5′-GGTTTCCAGCATTGGCTCTA-3′; Beclin 1 (Atg6), 5′-TGATCCAGGAGCTGGAAGAT-3′ and 5′-CAAGCGACCCAGTCTGAAAT-3′; Atg7, 5′-TCCGTTGAAGTCCTCTGCTT-3′ and 5′-CCACTGAGGTTCACCATCCT-3′; LC3B, 5′-CCGAGAAGACCTTCAAGCAG-3′ and 5′-ACACTTCGGAGATGGGAGTG-3′; ULK1, 5′-GGTGGAGACCTGGCTGACTA-3′ and 5′-TCTTGACTCGGATGTTGCTG-3′; and Bnip3, 5′-AGCTTTGGCGAGAAAAACAG-3′ and 5′-TCCAATGTAGATCCCCAAGC-3′.
Microscopy for Autophagy and Cell Death
To examine autophagy, primary mouse hepatocytes were seeded in a 12-well plate (2 × 105 cells per well) and infected overnight with Ad-GFP-LC3 (100 viral particles per cell). Cells were treated with 80 mmol/L ethanol in the presence or absence of 0.1 μg/mL ActD or with 50 μmol/L resveratrol for 6 hours. After treatment, cells were fixed with 4% paraformaldehyde in PBS for 2 hours at room temperature or were kept at 4°C for microscopy. Fluorescence images were acquired under an Eclipse 200 fluorescence microscope (Nikon, Tokyo, Japan) with MetaMorph software version 6.1 (Downingtown, PA). For the immunostaining assay, fixed primary hepatocytes or cryosections of liver tissues were immunostained with anti-FoxO3a antibody followed by Cy3-conjugated secondary antibody and Hoechst 33342 nuclear staining, as described previously.21 Cell death and morphological changes were analyzed with 1 μg/mL Hoechst 33342 staining and fluorescence imaging. All images were obtained using digital fluorescence microscopy as described above.
Nuclear Fractionation
Mouse liver nuclear proteins were extracted using Pierce NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific) according to the manufacturer’s instructions. In brief, liver tissues were cut into small pieces, homogenized in cytoplasmic extraction reagent I with subsequent addition of cytoplasmic extraction reagent II, and centrifuged. The supernatants resulting from centrifugation were cytoplasmic fractions. The insoluble pellets were suspended in nuclear extraction reagent and centrifuged; the supernatants resulting from this centrifugation contained nuclear proteins.
Hepatic Triglyceride Analysis
Triglyceride level was determined as described previously.9 Frozen liver tissues (50 to 100 mg) were ground using a mortar and pestle. The crushed liver tissues were further incubated with 1 mL of a chloroform/methanol (2:1) mix with vigorous shaking for 1 hour at room temperature. Then, 200 μL of double-distilled water was added into the mix and centrifuged for 5 minutes at 3000 × g. The lower lipid phase was collected and dried at room temperature, and the pellet was redissolved in a tert-butanol and Triton X-114/methanol (2:1) mix. Triglyceride analysis was performed with a colorimetric assay kit according to the manufacturer’s instructions (Sigma-Aldrich).
Statistical Analysis
Experimental data were subjected to a Student’s t-test or one-way analysis of variance analysis with Scheffé’s post hoc test where appropriate. P < 0.05 was considered significant.
Results
Acute Ethanol Treatment Increases the Expression of Autophagy Genes in Mouse Liver and in Primary Cultured Mouse Hepatocytes
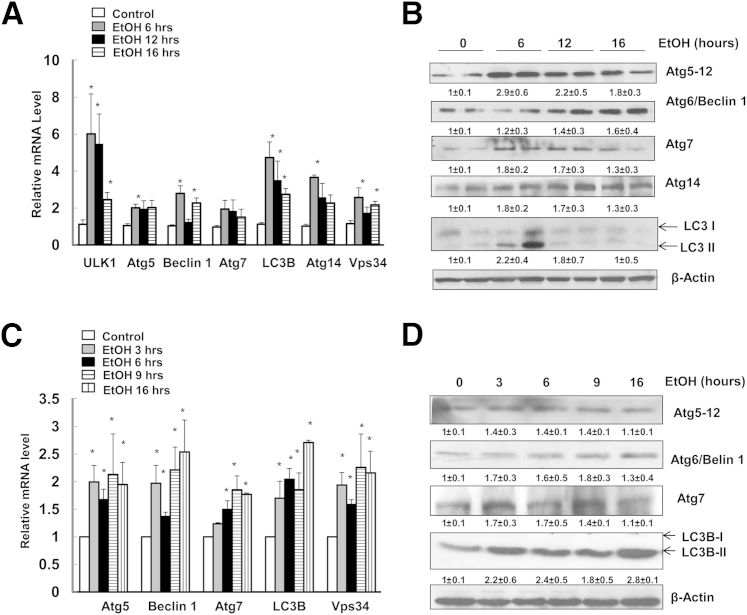
To investigate the expression of autophagy-related genes in mouse liver, male wild-type C57BL/6 mice were administered 4.5 g/kg of water or ethanol for 6, 12, and 16 hours. This animal model, initially established by Carson and Pruett,18 has been widely used to mimic human binge drinking and acute alcohol abuse.22, 23 In this model, an ethanol dose of 4 to 6 g/kg via gavage resulted in a peak blood ethanol concentration of 43.4 to 86.8 mmol/L, which can be commonly achieved in human binge drinkers.22 Using this acute alcohol gavage model, we found that the mRNA levels of various autophagy-related genes, including Ulk1 (yeast Atg1), Atg5, Becn1 (yeast Atg6), Atg7, LC3b (yeast Atg8), Atg14, and Pik3c3 (alias Vps34) were all increased in ethanol-treated mouse liver at all time points. Expression levels of most genes that we assessed (Ulk1, Becn1, LC3b, Atg14, and Vps34) peaked after 6 hours of treatment with ethanol (Figure 1A). Expression levels of these autophagy-related genes started to decrease after 12 and 16 hours of treatment with ethanol, but were still significantly higher than in controls (Figure 1A). Consistent with increased mRNA expression, ethanol treatment also increased Atg5, Beclin 1, Atg7, and LC3B-II protein levels; however, the extent of increase and the respective peak time varied. The expression levels of some proteins (such as Atg5) increased approximately threefold, whereas expression levels of others (such as Atg7 and Beclin 1) increased 60% to 80%, compared with control (Figure 1, B and C). It seems that Atg5, Beclin 1, and Atg14 were maintained at higher levels during the treatment, whereas Atg7 and LC3B-II increased at 6 hours but then slightly declined at 12 and 16 hours of ethanol treatment. We have previously demonstrated an increase in autophagic flux induced by ethanol; in the presence of CQ, which blocks lysosomal degradation, ethanol-induced LC3B-II levels were further enhanced by CQ treatment.9 Therefore, the decreased LC3B-II levels after prolonged ethanol treatment (12 and 16 hours) were likely due to autophagic degradation of LC3B-II. Consistent with in vivo findings, we also found elevated mRNA levels of several autophagy-related genes in ethanol-treated primary mouse hepatocytes as early as 3 hours and sustained to 16 hours (Figure 1C). Moreover, ethanol treatment also increased Atg5, Beclin 1, Atg7, and LC3B-II protein levels in a time-dependent manner (Figure 1D).
Figure 1.
Acute ethanol treatment increases the expression of autophagy-related genes in mouse liver and primary hepatocytes. A: Male C57BL/6 mice were treated with 4.5 g/kg water or ethanol by gavage for 6, 12, or 16 hours. Hepatic mRNA was isolated, and RT-qPCR was performed. B: Mice were treated as for A, and total liver lysates were subjected to Western blot analysis, followed by densitometry analysis. C: Primary cultured mouse hepatocytes were treated with 80 mmol/L ethanol for 3, 6, 9, and 16 hours. mRNA was isolated from cultured hepatocytes, and RT-qPCR was performed. D: Primary cultured mouse hepatocytes were treated as for C, and total cell lysates were subjected to Western blot analysis, followed by densitometry analysis. Data are presented as means ± SEM (fold of control). n = 4–6 (A); n = 4 (B and C); n = 3 (D). ∗P < 0.05, one-way analysis of variance with Scheffé’s post hoc test.
ActD Suppresses Ethanol-Induced Expression of Autophagy-Related Proteins and GFP-LC3 Puncta
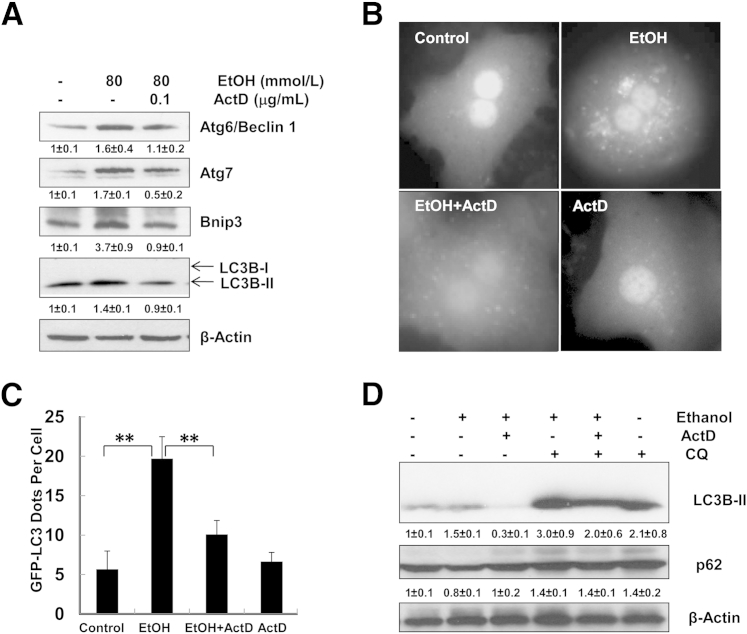
To further investigate the role of gene transcription in ethanol-induced autophagy, we treated primary cultured mouse hepatocytes with ethanol in the presence or absence of ActD, a general transcription inhibitor. Beclin 1, Atg7, Bnip3, and LC3B-II protein levels induced by ethanol decreased in the presence of ActD (Figure 2A). Consistent with our previous reports,9 ethanol treatment increased the number of GFP-LC3 puncta in primary cultured mouse hepatocytes, and these puncta were significantly diminished by ActD (Figure 2, B and C). More importantly, ActD also blocked ethanol-induced autophagic flux, as demonstrated by the decreased LC3B-II levels when cells were treated with ethanol together with ActD and CQ, compared with the cells treated with ethanol and CQ. Furthermore, ethanol treatment also decreased levels of p62, an autophagic substrate protein that is usually degraded by autophagy. Ethanol-induced decrease of p62 was blocked by ActD, indicating that ActD inhibited ethanol-induced autophagic flux in hepatocytes (Figure 2D). These results suggest that ethanol-induced expression of autophagy-related genes could play an important role in ethanol-induced autophagosome formation and autophagic flux.
Figure 2.
ActD inhibits ethanol-induced autophagy in primary mouse hepatocytes. A: Primary cultured hepatocytes were treated with ethanol in the presence or absence of ActD for 6 hours. Total cell lysates were subjected to Western blot analysis, followed by densitometry analysis. B: Ad-GFP-LC3–infected hepatocytes were treated as for A and were examined with fluorescence microscopy. Original magnification, ×60. C: GFP-LC3 puncta were quantified for each experiment, with at least 30 cells counted in each experiment. D: Primary hepatocytes were treated as for A, with or without 20 μmol/L CQ for 6 hours, followed by Western blotting and densitometry analysis. Data are expressed as means ± SEM. n = 3 (A and D); n = 4 (C). ∗∗P < 0.01, one-way analysis of variance with Scheffé’s post hoc test.
Ethanol Activates FoxO3a Both in Vivo and in Vitro
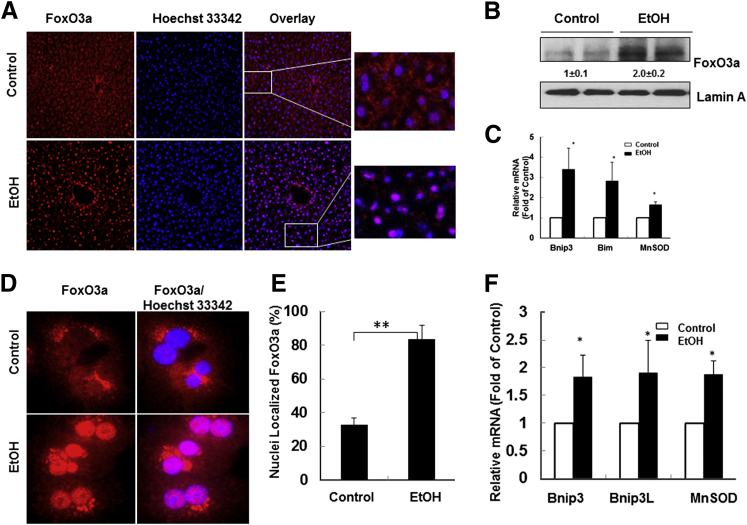
During mouse muscle atrophy, FoxO3a is activated to promote autophagy by up-regulating autophagy-related genes, including LC3b, Atg12, and Becn1.12 In the present study, ethanol treatment increased Beclin 1, Atg7, Bnip3, and LC3B protein levels, and expression of these proteins was suppressed by ActD. We therefore investigated whether ethanol also activates FoxO3a and in turn increases the transcription of Atg genes in hepatocytes. FoxO3a signals were diffuse in both cytosol and nucleus in the liver of control mice, but the FoxO3a nuclear signal dramatically increased after ethanol treatment, indicating that ethanol increased nuclear FoxO3a retention in mouse liver (Figure 3A). Western blot analysis using nuclear fractions showed that ethanol treatment increased nuclear FoxO3a levels approximately twofold (Figure 3B). Moreover, hepatic mRNA levels of three known FoxO3a target genes [Bnip3, Bim (reclassified as Bcl2l11), and manganese superoxide dismutase (Sod2; alias MnSOD)] also increased in ethanol-treated mouse liver (Figure 3C). Similar to the in vivo findings, ethanol treatment also significantly increased nuclear FoxO3a levels in primary cultured mouse hepatocytes (Figure 3, D and E). In addition, ethanol treatment also increased the expression of the known FoxO3a target genes Bnip3, Bnip3L, and MnSOD in vitro (Figure 3F). These findings indicate that ethanol activates FoxO3a in mouse liver and in primary cultured mouse hepatocytes.
Figure 3.
Ethanol activates FoxO3a in mouse liver and primary mouse hepatocytes. A: Male C57BL/6 mice were treated with 4.5 g/kg water or ethanol by gavage for 16 hours. Cryosectioned liver tissue was subjected to immunostaining for FoxO3a using a rabbit anti-FoxO3a antibody, followed by a Cy3-conjugated goat anti-rabbit antibody; nuclei were stained with Hoechst 33342. Representative fluorescence microscopy images are shown. Boxed regions are shown at higher magnification at the right. B: Mice were treated as for A, and nuclear fractions were isolated from control and ethanol-treated mouse liver, followed by Western blotting and densitometry analysis. C: Hepatic mRNA was isolated, and RT-qPCR was performed. D: Primary cultured hepatocytes were treated with 80 mmol/L ethanol for 6 hours and fixed in 4% paraformaldehyde, then immunostained for FoxO3a as for A, and examined with fluorescence microscopy. Representative images are shown. E: Cells with increased nuclear FoxO3a staining were quantified for each experiment, with at least 100 cells counted in each experiment. F: mRNAs were isolated from cultured hepatocytes, and RT-qPCR was performed. Data are presented as means ± SEM. n = 3 (A, B, D, E, and F); n = 4–6 (C). ∗P < 0.05, Student’s t-test; ∗∗P < 0.01, Z-test.
Ethanol Induces Alterations of the Post-Translational Modifications of FoxO3a in Mouse Liver
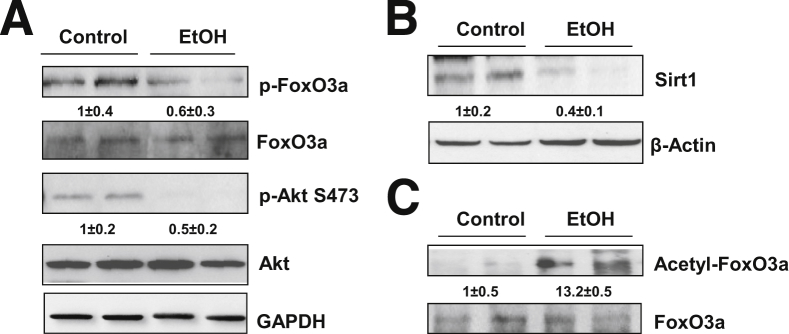
Because FoxO3a is regulated by multiple post-translational modifications, including phosphorylation and acetylation,10 we next investigated whether ethanol treatment affects phosphorylation and acetylation of FoxO3a. Compared with control mice, ethanol treatment decreased the level of p-FoxO3a in mouse liver (Figure 4A). These findings are consistent with our immunostaining data and nuclear Western blot analysis, showing that ethanol increased nuclear FoxO3a (Figure 3A), because p-FoxO3a is liable to be exported to the cytosol from the nucleus. This decreased phosphorylation of FoxO3a could be due to decreased Akt phosphorylation, because Akt phosphorylates FoxOs to promote their nuclear exportation.11 Indeed, the p-Akt levels in ethanol-treated mouse livers were decreased, compared with control (Figure 4A). In addition to phosphorylation, the transcriptional activity of FoxO3a can also be regulated by Sirt1-mediated deacetylation. Ethanol treatment decreased hepatic Sirt1 (Figure 4B), which was correlated with increased acetylated FoxO3a (Figure 4C). These data indicate that acute ethanol treatment decreases the level of phosphorylated FoxO3a, but increases acetylated FoxO3a; the latter is likely associated with a decrease in hepatic Sirt1 induced by ethanol treatment.
Figure 4.
Ethanol induces post-translational modification of FoxO3a in mouse liver. A and B: C57BL/6 mice were treated with 4.5 g/kg water or ethanol by gavage for 16 hours. Total liver lysates were subjected to Western blot analysis for phosphorylated and total FoxO3a and phosphorylated and total Akt (A) and Sirt1 (B), followed by densitometry analysis. C: Mice were treated as for A, and total liver lysates were immunoprecipitated with an anti-FoxO3a antibody, followed by Western blotting using an anti–acetylated lysine antibody and densitometry analysis. Data are expressed as means ± SEM. n = 4.
Resveratrol Further Increases Ethanol-Induced Autophagy Gene Expression by Decreasing FoxO3a Acetylation
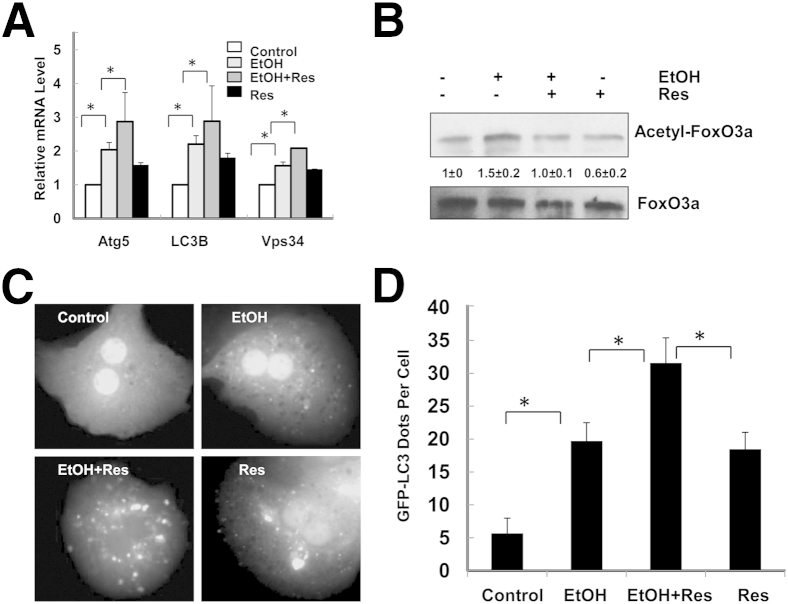
To investigate the influence of acetylation of FoxO3a on ethanol-induced expression of autophagy-related genes, we treated primary mouse hepatocytes with ethanol in the presence or absence of resveratrol, a well-known Sirt1 agonist24 that decreases ethanol-induced liver injury.25 We found that resveratrol alone slightly increased the mRNA levels of Atg5, LC3b, and Vps34, but the combined treatment of ethanol and resveratrol further increased ethanol-induced expression of these autophagy-related genes (Figure 5A). Ethanol-induced acetylation of FoxO3a was suppressed by resveratrol (Figure 5B), suggesting that deacetylated FoxO3a may further promote autophagy-related gene transcription. More importantly, in the presence of resveratrol, the number of ethanol-induced GFP-LC3 puncta was further increased, suggesting that resveratrol enhances ethanol-induced autophagosome formation (Figure 5, C and D). These findings indicate that Sirt1-mediated deacetylation of FoxO3a may promote expression of autophagy-related genes and autophagy induction in ethanol-treated hepatocytes.
Figure 5.
Resveratrol enhances ethanol-induced expression of autophagy-related genes. A: Primary cultured mouse hepatocytes were treated with ethanol in the presence or absence of 50 μmol/L resveratrol (Res) for 6 hours. mRNA was isolated from cultured hepatocytes before RT-qPCR was performed. B: Total cell lysates were immunoprecipitated with an anti-FoxO3a antibody, followed by Western blotting using an anti–acetylated lysine antibody and densitometry analysis. C: Ad-GFP-LC3–infected hepatocytes were treated as for A and then were examined with fluorescence microscopy. Representative GFP-LC3 images are shown. Original magnification, ×60. D: The number of GFP-LC3 puncta per cell was quantified and twenty cells were counted in each experiment. Data are expressed as means ± SEM. n = 3 (A and B); n = 4 (C and D). ∗P < 0.05, one-way analysis of variance with Scheffé’s post hoc test.
Overexpression of a DN-FoxO3a Suppresses Ethanol-Induced Expression of Autophagy-Related Genes and Enhances Ethanol-Induced Apoptosis in Hepatocytes
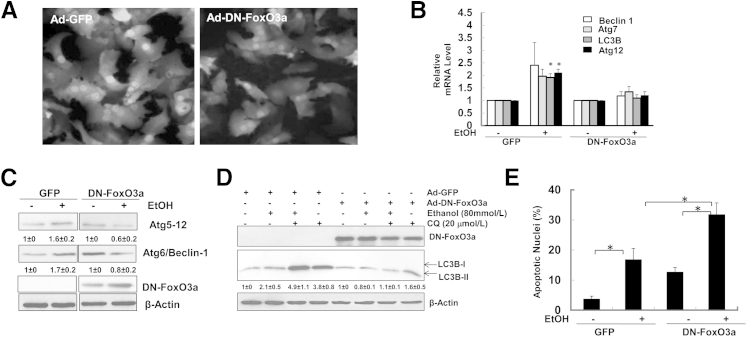
To determine whether overexpression of DN-FoxO3a suppresses ethanol-induced expression of autophagy-related genes, we infected primary mouse hepatocytes with Ad-GFP control and Ad-DN-FoxO3a, which also contains GFP from a separate cytomegalovirus promoter.12 More than 80% of cells showed very high efficiency for the infection after 24 hours of incubation (Figure 6A). After the cells were further treated with ethanol for another 6 hours, ethanol-induced expression of Becn1, Atg7, LC3b, and Atg12 was inhibited by expression of DN-FoxO3a (Figure 6B). Consistent with the changes at the mRNA level, the ethanol-induced increases in Atg5 (Atg12) and Atg6 (Beclin 1) protein levels were also inhibited by DN-FoxO3a (Figure 6C). These results suggest that FoxO3a is responsible for ethanol-induced expression of autophagy-related genes. Furthermore, overexpression of DN-FoxO3a inhibited ethanol-induced autophagic flux in primary hepatocytes (Figure 6D). Interestingly, we also found that CQ treatment alone only mildly increased LC3B-II levels in DN-FoxO3a–infected cells (1.6 versus 1), suggesting that DN-FoxO3a could also inhibit basal autophagy in hepatocytes (Figure 6D). We previously found that inhibition of autophagy led to increased cell death and liver injury in ethanol-treated primary hepatocytes and mice.9 Interestingly, overexpression of DN-FoxO3a also enhanced ethanol-induced apoptosis in primary mouse hepatocytes (Figure 6E). Therefore, the increased cell death caused by expression of DN-FoxO3a could be due to the inhibition of ethanol-induced expression of autophagy-related genes and subsequent autophagy.
Figure 6.
Overexpression of DN-FoxO3a suppresses ethanol-induced expression of autophagy-related genes, but enhances ethanol-induced apoptosis. A: Representative fluorescence images of primary cultured mouse hepatocytes, infected with 10 multiplicity of infection (MOI) Ad-GFP or Ad-DN-FoxO3a, which contain a GFP using separate cytomegalovirus promoters for 24 hours. Original magnification, ×20. B: Primary hepatocytes, infected as in A, were treated with 80 mmol/L ethanol for 6 hours. mRNA was isolated from cultured hepatocytes, and RT-qPCR was performed. C: Total cell lysates were subjected to Western blotting and densitometry analysis. D: Primary hepatocytes were treated as in B, with or without 20 μmol/L CQ, for 6 hours. Total lysates were subjected to Western blotting and densitometry analysis. E: Primary hepatocytes, infected as in A, were treated with 80 mmol/L ethanol for 24 hours. Cells were then stained with Hoechst 33342 for nuclei, and fragmented apoptotic nuclei were quantified. Data are expressed as means ± SEM. n = 3. ∗P < 0.05, one-way analysis of variance with Scheffé’s post hoc test.
Ethanol-Induced Expression of Autophagy-Related Genes Is Suppressed in Foxo3a−/− Mice, Resulting in Increased Liver Injury and Steatosis
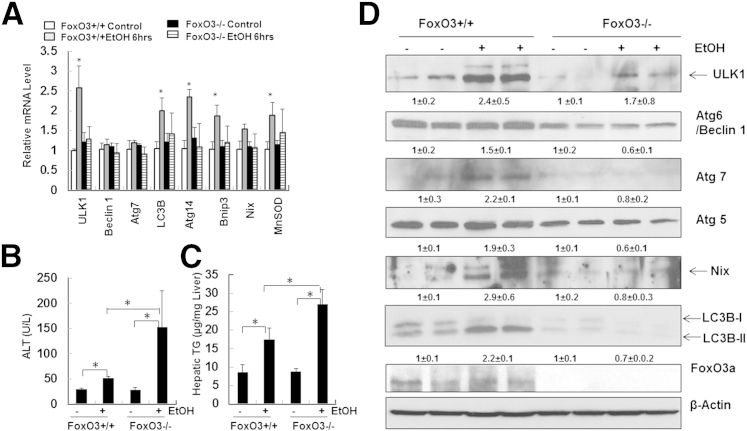
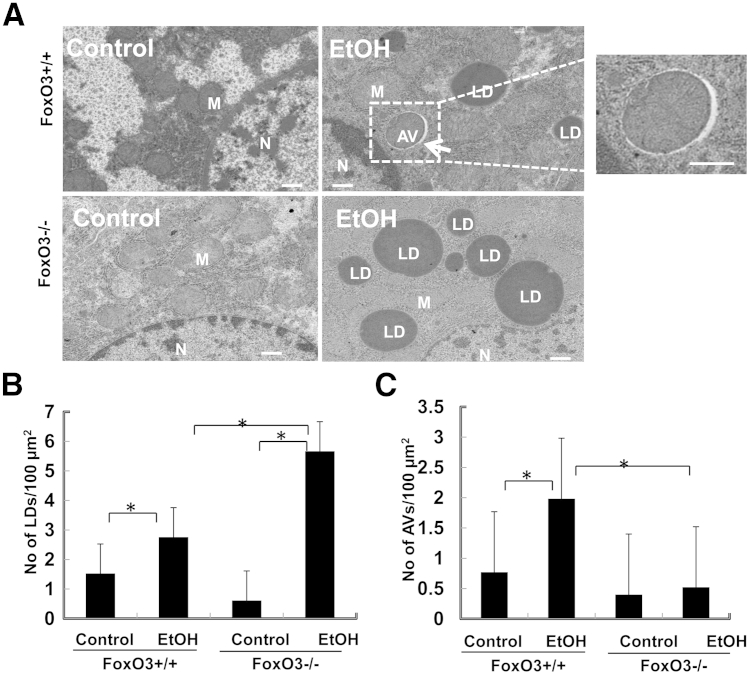
To further investigate the role of FoxO3a in ethanol-induced expression of autophagy-related genes and liver injury in vivo, we treated Foxo3a−/− and age-matched wild-type mice with acute ethanol. Although the basal expression of autophagy genes in Foxo3a−/− mice did not differ significantly from that of wild-type mice, ethanol-induced expression of autophagy-related genes was significantly suppressed in Foxo3a−/− mice (Figure 7A). The expression levels of several autophagy genes in ethanol-treated FoxO3a wild-type mice (B6;129 background), such as Becn1 and Atg7, were not induced as significantly as in ethanol-treated C57BL/6J wild-type mice (Figure 1A). These variations could be due to the difference in genetic background. Similar to findings at the mRNA level, several of the ethanol-induced autophagy proteins were also suppressed in Foxo3a−/− mice (Figure 7D). More importantly, we further found that serum alanine aminotransferase and hepatic triglyceride levels were all significantly increased in ethanol-treated Foxo3a−/− mice, compared with wild-type mice (Figure 7, B and C). Notably, electron microscopy studies revealed that the number of lipid droplets in ethanol-treated Foxo3a−/− mouse liver was significantly higher than that of wild-type mice, whereas the number of autophagosomes was significantly decreased in ethanol-treated Foxo3a−/− mouse liver, compared with wild-type mice (Figure 8). These findings suggest that the decreased expression of autophagy-related genes and autophagy in Foxo3a−/− mice may contribute to the increased steatosis and liver injury after ethanol treatment.
Figure 7.
Foxo3a−/− mice have decreased expression of autophagy-related genes and increased liver injury after ethanol administration. A: Male Foxo3a−/− mice and their wild-type littermates were treated with 4.5 g/kg water or ethanol by gavage for 6 hours. Hepatic mRNA was isolated, and RT-qPCR was performed. B and C: Mice were treated as in A, and serum alanine aminotransferase (ALT) (B) and hepatic triglycerides (TG) (C) were measured. D: Mice were treated as in A, and total liver lysates were subjected to Western blotting and densitometry analysis. Data are expressed as means ± SEM. n = 4–6 (A–C); n = 4 (D). ∗P < 0.05, one-way analysis of variance with Scheffé’s post hoc test.
Figure 8.
Foxo3a−/− mice have decreased number of hepatic lipid droplets and autophagosomes after ethanol treatment. Wild-type and Foxo3a−/− mice were treated with ethanol as reported for Figure 7. A: Representative electron microscopy images are shown. An autophagosome (boxed) is shown at higher magnification to the right. The numbers of lipid droplets (B) and autophagosomes (C) per 100-μm2 cytosolic areas were quantified. Data are expressed as means ± SEM (>20 cell sections). ∗P < 0.05, one-way analysis of variance with Scheffé’s post hoc test. Scale bar = 500 nm. AV, autophagosome; LD, lipid droplet; M, mitochondria; N, nucleus.
Ethanol Induces Expression of Autophagy-Related Genes and Autophagic Flux in Primary Human Hepatocytes
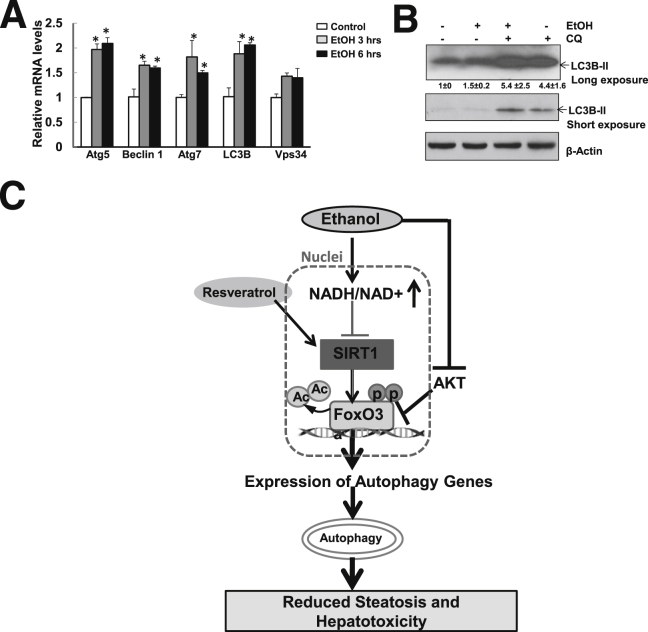
Given that we observed ethanol-induced expression of autophagy-related genes (Figure 1) and autophagic flux in primary mouse hepatocytes,9 we next asked whether ethanol induces similar effects in human hepatocytes. As with mouse hepatocytes, ethanol treatment increased the expression of several autophagy-related genes (Figure 9A), as well as autophagic flux, as demonstrated by further increased levels of LC3B-II in the presence of CQ, compared with either ethanol or CQ treatment alone (Figure 9B).
Figure 9.
Ethanol induces autophagy-related gene expression and autophagic flux in primary human hepatocytes. A: Primary human hepatocytes were treated with 80 mmol/L ethanol for 3 or 6 hours. mRNA was isolated, and RT-qPCR was performed. B: Primary human hepatocytes were treated with 80 mmol/L ethanol in the presence or absence of 20 μmol/L CQ for 6 hours. Total cell lysates were subjected to Western blotting and densitometry analysis. C: A proposed model for the role of FoxO3a in ethanol-induced autophagy and liver injury. In ethanol-exposed hepatocytes, ethanol is metabolized by hepatic enzymes, which leads to an increase in the NADH/NAD+ ratio and to subsequent inhibition of Sirt1. Ethanol also leads to AKT inhibition, resulting in FoxO3a dephosphorylation and nuclear retention, which increases the expression of autophagy-related genes and autophagy. Resveratrol activates Sirt1, which leads to FoxO3a deacetylation and enhances expression of autophagy-related genes. FoxO3a-mediated autophagy protects against ethanol-induced liver injury, likely by removal of ethanol-induced accumulation of lipid droplets and damaged mitochondria. Data are expressed as means ± SEM. n = 4 (A); n = 3 (B). ∗P < 0.05, one-way analysis of variance with Scheffé’s post hoc test.
Discussion
The FoxO family of transcription factors regulates multiple cellular processes (including apoptosis, cell cycle, DNA damage repair, oxidative stress, and glucose metabolism) and is highly conserved, from Caenorhabditis elegans to mammals.10, 11 The FoxO family has been shown to regulate autophagy in various systems, including skeletal muscle,12, 13 cardiomyocytes,14 kidney,26 liver,27 and cancer cells.28 Depending on the stress conditions, autophagy can be activated as a cell-survival mechanism, either to provide necessary nutrients on nutrient deprivation or to remove cellular damaged or excess organelles and protein aggregates.1, 2 We have previously demonstrated that acute ethanol treatment induces autophagy as a compensatory mechanism to remove hepatic lipid droplets and damaged mitochondria to attenuate ethanol-induced liver steatosis and injury in mice.9, 29 However, it was not known how acute ethanol modulates hepatic autophagy in mice. Here, we show that acute ethanol treatment promoted nuclear localization of FoxO3a in both primary cultured mouse hepatocytes and in mouse liver. In primary cultured hepatocytes, overexpression of DN-FoxO3a inhibited ethanol-induced up-regulation of autophagy genes and enhanced ethanol-induced cell death. Moreover, acute ethanol treatment led to enhanced steatosis and liver injury in Foxo3a−/− mice, compared with wild-type mice.
It has been suggested that cytosolic FoxO1, particularly the acetylated form, can bind with Atg7 to promote autophagy in certain cancer cells, independent of its transcriptional activity.28 In contrast, it has been demonstrated that the transcriptional activities of FoxOs are required for the induction of autophagy in normal tissues, such as mouse skeletal muscle,12, 13 heart14 and liver.27 In the present study, FoxO3a-mediated transcription of autophagy-related genes played a critical role in alcohol-induced autophagy in mouse liver and primary cultured hepatocytes. This notion is supported by the following evidence: i) ethanol treatment increased the nuclear levels of FoxO3a; ii) suppression of ethanol-mediated expression of autophagy-related genes by ActD inhibited ethanol-induced autophagy; iii) overexpression of DN-FoxO3a inhibited ethanol-induced expression of autophagy-related genes, resulting in reduced autophagy and increased cell death; and iv) Foxo3a−/− mice exhibited decreased expression of autophagy genes in ethanol-treated mouse liver, but increased steatosis and liver injury. Currently, it is not clear why FoxO-mediated transcription of autophagy genes is important for the autophagy process in normal tissues, but cytosolic FoxOs are important for autophagy induction in cancer cells. One possible explanation is that activation of Akt is one of the most frequent alterations observed in human cancer cells, and Akt activation may lead to increased cytosolic FoxO levels because of increased Akt-mediated phosphorylation of FoxOs.
The activity of FoxO3a is largely regulated by its multiple post-translational modifications, including phosphorylation, acetylation, ubiquitination, and methylation.10, 11 FoxOs are phosphorylated by the serine/threonine protein kinase Akt and become sequestered in the cytoplasm, where they are unable to regulate gene expression.11 Acute ethanol treatment decreased the level of phosphorylated Akt and FoxO3a. These findings suggest that acute ethanol-induced nuclear retention of FoxO3a could be mediated by the Akt signaling pathway. In contrast to negative regulation of FoxOs by phosphorylation, the acetylation of FoxOs seems to be more important for determination of specificity of FoxO target genes.15 FoxO acetylation levels are regulated through acetylation by acetyltransferases (eg, p300/CBP) and through deacetylation by deacetylases (eg, SIRT1).10, 30 SIRT1 deacetylates FoxO3a and modulates its gene transcription and specificity, which increases the expression of oxidative stress genes and suppresses the expression of apoptosis genes in response to oxidative stress.10, 15 Ethanol is reported to decrease SIRT1 expression or activity by increasing the NADH/NAD+ ratio in hepatocytes.31 In agreement with these reports, we also found that acute ethanol treatment reduced Sirt1 proteins in mouse liver, but increased acetylation of FoxO3a. However, ethanol increased the expression of autophagy-related genes in primary hepatocytes and mouse liver, suggesting that ethanol-induced nuclear translocation of FoxO3a could be a critical factor, and that this translocation is sufficient to increase FoxO3a-mediated expression of autophagy-related genes. The finding that resveratrol further enhanced ethanol-induced up-regulation of autophagy genes suggests that Sirt1-mediated deacetylation of FoxO3a may favor and promote expression of autophagy-related genes. Further studies are needed to determine whether the deacetylated form of FoxO3a has increased binding affinity for the promoters of autophagy-related genes. Nevertheless, our present findings collectively indicate that FoxO3a is a major target in the regulation of ethanol-induced autophagy-related gene expression and autophagy.
The present results support a novel mechanism for FoxO-mediated hepatoprotection against acute ethanol administration in mice through activation of expression of autophagy-related genes. This notion is supported by the observation that overexpression of DN-FoxO3a enhanced ethanol-induced apoptosis and that Foxo3a−/− mice exhibited exacerbated liver injury and steatosis after acute ethanol administration. Our results are also consistent with a recent report that overexpression of Atg14, a target gene of FoxOs, decreases hepatic triglyceride contents.27 Another significant finding from our present study was that ethanol also increased autophagy-related gene expression and autophagic flux in primary human hepatocytes. However, in addition to regulating autophagy-related genes, FoxO3a also regulates expression of genes that have multiple functions, such as antioxidant and DNA repair genes, which could also attenuate ethanol-induced liver injury.30
Controversial evidence suggests that autophagy status could differ between chronic and acute ethanol treatment in mice.32, 33 Although it has been suggested that autophagy could be decreased in the chronic ethanol feeding model, the evidence to support this argument is still not clear, because autophagic flux data in these models are lacking (probably because the dynamic nature of the autophagic process and of the constitutive synthesis and turnover of LC3 makes it difficult to assess autophagic flux in this model). In a recent study by Lin et al,34 autophagic flux analysis was performed after the mice were fed a Lieber–DeCarli liquid diet; the results indicated increased autophagic flux after chronic ethanol feeding, at least at the 4-week time point. These results were generally consistent with the acute alcohol binge model. Although more careful autophagic flux assays should be performed (and with multiple time points) to further dissect the autophagy process in the chronic ethanol feeding model, the Lin et al34 study also demonstrated that pharmacological promotion of autophagy by rapamycin and carbamazepine attenuated ethanol-induced liver injury in this 4-week feeding model. Regardless of the acute or chronic ethanol treatment, therefore, our findings, taken together with those of others, suggest that autophagy modulation could be one of the ideal approaches to treat alcoholic liver diseases. Although modulation of Akt or Sirt1 may indirectly enhance FoxO3a activity, the specific pharmacological activators for FoxOs remain to be identified. Developing specific FoxO activators will be a challenge, but could lead to new approaches for treatment of alcoholic diseases.
In summary, our data suggest that FoxO3a-mediated autophagy-related gene expression plays a critical role in acute ethanol-induced autophagy. Genetic loss of FoxO3a enhanced acute ethanol-induced liver injury and steatosis in mice. The cellular and molecular events regulating ethanol-induced FoxO3a-mediated autophagy are summarized in Figure 9C.
Acknowledgments
We thank Drs. Jinghui Zhao and Alfred L. Goldberg (Harvard Medical School) for the reagents, Jessica Williams for critical reading of this manuscript, Dr. Melissa Larson [University of Kansas Medical Center (KUMC) Transgenic Core Facility] for the recovery of FoxO3a knockout mice from cryopreserved embryos, and Ken Dorko for providing the human hepatocytes. The hepatocytes used in this study were derived from samples collected and provided by the KUMC Department of Pharmacology, Toxicology and Therapeutics Cell Isolation Core lab and the University of Kansas Liver Center.
Footnotes
Supported in part by NIH grants R01-AA020518-01 and P20-RR021940 (W.X.D), R01-AA015951 and R01-AA013623 (M.Y.), and P20-GM103549 (COBRE grant to the University of Kansas Medical Center Cell Isolation Core laboratory).
References
- 1.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shintani T., Klionsky D.J. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding W.X., Yin X.M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cebollero E., Reggiori F., Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol. 2012;2012:182834. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao B., Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141:1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding W.X., Manley S., Ni H.M. The emerging role of autophagy in alcoholic liver disease. Exp Biol Med (Maywood) 2011;236:546–556. doi: 10.1258/ebm.2011.010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding W.X., Li M., Chen X., Ni H.M., Lin C.W., Gao W., Lu B., Stolz D.B., Clemens D.L., Yin X.M. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H., Tindall D.J. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 11.Tzivion G., Dobson M., Ramakrishnan G. FoxO transcription factors; regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011;1813:1938–1945. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Sengupta A., Molkentin J.D., Paik J.H., DePinho R.A., Yutzey K.E. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. J Biol Chem. 2011;286:7468–7478. doi: 10.1074/jbc.M110.179242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Hu L.S., Cheng H.L., Jedrychowski M.P., Gygi S.P., Sinclair D.A., Alt F.W., Greenberg M.E. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 16.Ding W.X., Ni H.M., Gao W., Yoshimori T., Stolz D.B., Ron D., Yin X.M. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyamoto K., Araki K.Y., Naka K., Arai F., Takubo K., Yamazaki S., Matsuoka S., Miyamoto T., Ito K., Ohmura M., Chen C., Hosokawa K., Nakauchi H., Nakayama K., Nakayama K.I., Harada M., Motoyama N., Suda T., Hirao A. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Carson E.J., Pruett S.B. Development and characterization of a binge drinking model in mice for evaluation of the immunological effects of ethanol. Alcohol Clin Exp Res. 1996;20:132–138. doi: 10.1111/j.1530-0277.1996.tb01055.x. [DOI] [PubMed] [Google Scholar]
- 19.Ni H.M., Bockus A., Boggess N., Jaeschke H., Ding W.X. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55:222–232. doi: 10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gramignoli R., Green M.L., Tahan V., Dorko K., Skvorak K.J., Marongiu F., Zao W., Venkataramanan R., Ellis E.C., Geller D., Breite A.G., Dwulet F.E., McCarthy R.C., Strom S.C. Development and application of purified tissue dissociation enzyme mixtures for human hepatocyte isolation. Cell Transplant. 2012;21:1245–1260. doi: 10.3727/096368911X600939. [DOI] [PubMed] [Google Scholar]
- 21.Ding W.X., Ni H.M., DiFrancesca D., Stolz D.B., Yin X.M. Bid-dependent generation of oxygen radicals promotes death receptor activation-induced apoptosis in murine hepatocytes. Hepatology. 2004;40:403–413. doi: 10.1002/hep.20310. [DOI] [PubMed] [Google Scholar]
- 22.Shukla S.D., Pruett S.B., Szabo G., Arteel G.E. Binge ethanol and liver: new molecular developments. Alcohol Clin Exp Res. 2013;37:550–557. doi: 10.1111/acer.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Ajmo J.M., Liang X., Rogers C.Q., Pennock B., You M. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295:G833–G842. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kume S., Uzu T., Horiike K., Chin-Kanasaki M., Isshiki K., Araki S., Sugimoto T., Haneda M., Kashiwagi A., Koya D. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong X., Tao R., Depinho R.A., Dong X.C. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y., Yang J., Liao W., Liu X., Zhang H., Wang S., Wang D., Feng J., Yu L., Zhu W.G. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 29.Ding W.X., Li M., Yin X.M. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy. 2011;7:248–249. doi: 10.4161/auto.7.2.14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daitoku H., Sakamaki J., Fukamizu A. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim Biophys Acta. 2011;1813:1954–1960. doi: 10.1016/j.bbamcr.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 31.You M., Cao Q., Liang X., Ajmo J.M., Ness G.C. Mammalian sirtuin 1 is involved in the protective action of dietary saturated fat against alcoholic fatty liver in mice. J Nutr. 2008;138:497–501. doi: 10.1093/jn/138.3.497. [DOI] [PubMed] [Google Scholar]
- 32.Dolganiuc A., Thomes P.G., Ding W.X., Lemasters J.J., Donohue T.M., Jr. Autophagy in alcohol-induced liver diseases. Alcohol Clin Exp Res. 2012;36:1301–1308. doi: 10.1111/j.1530-0277.2012.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donohue T.M., Jr. Autophagy and ethanol-induced liver injury. World J Gastroenterol. 2009;15:1178–1185. doi: 10.3748/wjg.15.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin C.W., Zhang H., Li M., Xiong X., Chen X., Dong X.C., Yin X.M. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]