Abstract
Scope
Exosomes, small vesicles participating in intercellular communication have been extensively studied recently; however, the role of edible plant derived exosomes in interspecies communication has not been investigated. Here, we investigate the biological effects of edible plant derived exosome-like nanoparticles (EPDEN) on mammalian cells.
Methods and results
In this study, exosome-like nanoparticles from four edible plants were isolated and characterized. We show that these EPDENs contain proteins, lipids and microRNA. EPDENs are taken up by intestinal macrophages and stem cells. The results generated from EPDEN transfected macrophages indicate that ginger EPDENs preferentially induce the expression of the anti-oxidation gene, heme oxygenase-1 (HO-1) and the anti-inflammatory cytokine, IL-10; whereas grapefruit, ginger, and carrot EPDENs promote activation of nuclear factor (erythroid-derived 2)-like 2 (Nrf2). Furthermore, analysis of the intestines of canonical Wnt reporter mice, i.e., B6.Cg-Tg(BAT-lacZ)3Picc/J mice, revealed that the numbers of β-galactosidase+ (β-Gal) intestinal crypts are increased, suggesting that EPDEN treatment of mice leads to Wnt mediated activation of the Tcf4 transcription machinery in the crypts.
Conclusion
The data suggest a role for EPDEN mediated interspecies communication by inducing expression of genes for anti-inflammation cytokines, anti-oxidation and activation of Wnt signaling, which are crucial for maintaining intestinal homeostasis.
Keywords: Edible plant exosomes, intestinal macrophages, anti-inflammation, anti-oxidation, Nrf2
Introduction
It is well established that a plant-derived diet has great influence on regulation of mammalian host cell homeostasis, in particular, cells in the digestive system[1–3]. Deregulation of plant-derived diet regulated host cell homeostasis leads to increased susceptibility to infections, chronic inflammatory bowel diseases and cancer[4–10]. However, the cellular and molecular machinery regulating such interspecies mutualism between a plant-derived diet and the mammalian gut is not fully defined.
Exosomes are produced by a variety of mammalian cells including immune, epithelial and tumor cells[11–15]. Exosomes play a role in intercellular communication and can transport mRNA, miRNA, bioactive lipids, and proteins between cells[16–19]. Upon contact, exosomes transfer molecules that can render new properties and/or reprogram their recipient cells. Recently, exosome-like nanoparticles have been identified from grapes and characterized[20]. Whether other plants in our diets also release exosome-like nanoparticles has not been studied. Furthermore, whether those exosome-like nanoparticles play a role in gut homeostasis has not been explored either.
To provide a proof of concept, we selected four edible plants. Ginger or ginger root is a spice from the rhizome of the plant Zingiber officinale, consumed as a delicacy. Carrots are one of the top-ten most economically important vegetables crops in the world. Grapefruit is an excellent source of many nutrients and phytochemicals that contributes to a healthy diet. Grape derived molecules have been shown to possibly inhibit mechanisms of cancer, heart disease, degenerative nerve disease, viral infections and Alzheimer's disease.
In this study, exosome-like nanoparticles were isolated and characterized from all four edible plants. We demonstrate that they possess a similar size and structure to mammalian derived exosomes. Further study showed that these exosome-like nanoparticles are taken up by intestinal macrophages and stem cells, and have biological effects on the recipient cells. Among the four of them, ginger exosome-like nanoparticles strongly induced HO-1 and IL-10 expressed in the macrophages; whereas, fruit-derived exosome-like nanoparticles including grape and grapefruit induced Wnt/TCF4 activation. These results provide a foundation for further understanding how interspecies communication takes place through exosome-like nanoparticles released from different types of edible plants.
Material and Methods
Isolation and purification of EPDENs
Grapes and grapefruit EPDENs were isolated and purified according to the methods as described previously[21]. For isolation and purification of ginger and carrot EPDENs, ginger and carrots were purchased from a local market and washed with water in a plastic bucket 3×, 1–3 min/each time at room temperature (22°C). After the final washing, the ginger or carrots were ground in a mixer to obtain the juice. The juice was sequentially centrifuged at 1000g for 10min, 3000g for 20 min and 10,000g for 40 min to remove large particles. The supernatant was then centrifuged at 150,000g for 90min, the pellet was resuspended in 1mL PBS and transferred to a sucrose step gradient (8%/15%/30%/45%/60%) and centrifuged at 150,000g for 120 min using a previously described protocol[22]. The bands between the 8%/30% layer and 30%/45% layer were harvested separately and noted as EPDENs. The protein concentration of the samples was determined using the Bio-Rad protein quantification assay kit (Bio-Rad, Hercules, CA). The purified specimens were prepared for EM using a conventional procedure[23] and observed using an FEI Tecnai F20 electron microscope operated at 200kV at a magnification of 38,000× and defocus of 2.5 µm. Photomicrographs were taken using a Gatan Ultrascan 4000 CCD camera.
EPDENs particle size and surface charge analysis
The particle size and zeta potential of EPDENs were measured using a previously described method[21]. In brief, the hydrodynamic size and zeta potential of the EPDENs particles were assessed by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). Measurements were made in phosphate buffered solution at pH 7.0 and 25°C after appropriate dilution of the EPDENs preparation. Three independent measurements were performed in each case. Mass distribution of particle size with a polydispersity index was obtained. Sizes are reported as the mean diameter for the hydrodynamic diameter (nm). Zeta potential is the electric potential of a particle in a suspension and was determined by electrophoretic light scattering. For zeta potential analyses, an electric field was applied and surface-charged particles within the dispersion migrated toward the electrode of opposite charge, which is related to zeta potential values. The zeta potential results reported are the mean ± standard deviation. The mean zeta potential values obtained were calculated from at least three different batches.
In vitro digestion of EPDENs
In vitro digestion conditions were based on a previous description [24]. A stomach-like solution was composed of 18.5% w/v HCl (pH 2.0), pepsin solution (80 mg/ml in 0.1 N of HCl, pH 2.0, Sigma), 24mg/ml of bile extract and 4 mg/ml of pancreatin (Sigma) in 0.1 N of NaHCO3. 1 ml of EPDENs in a water solution was incubated with slow rotation at 37°C for 60 min with 1.34 l of stomach-like solution. The pH value of the stomach-like solution was adjusted to 6.5 with 1 N NaHCO3 and was referred to as an intestinal solution. EPDENs were incubated for additional 60 min in the intestinal solution. The stability of EPDENs was evaluated by measuring particle size and surface charge using a previously described method[21].
Mice
All mice including Lgr5-EGFP-ires-creERT2 mice, TCF/LEF-reporter mice (B6.Cg-Tg (BAT-lacZ)3Picc/J), and C57BL/6j mice at 6–12 weeks of age were obtained from Jackson Laboratories. All animal procedures were approved by the University of Louisville Institutional Animal Care and Use Committee. Mice were administered by gavage EPDENs resuspended in PBS as described previously[25].
Western Blot Analysis and Coomassie Blue staining
Cells were lysed using a method as previously described[25]. Proteins of lysed cells were then separated by SDS-PAGE on 10% polyacrylamide gels. Separated proteins were transferred to nitrocellulose membranes for western blotting using a standard protocol as described previously[26]. A rabbit polyclonal anti-HO-1 antibody was purchased from Santa Cruz Biotechnology. SDS PAGE gels were stained using a Coomassie Blue staining kit (Bio-Rad). Molecular weight markers are based on the mobility of All Blue Protein Standard (Bio-Rad), ranging from 10 kDa to 250 kDa. The gel was imaged with an Odyssey Scanner (LI-COR Bioscience, Lincoln NE).
Cytokine detection
IL-10 in culture supernatants was quantified using ELISA kits from eBioscience.
RNA extraction and real-time PCR
Total RNA was isolated from the RAW 264.7 macrophages cell line (ATCC) with TRIzol according to the manufacturer's specifications (Invitrogen). RNA (1µg) was reverse-transcribed with Superscript III and random primers (Invitrogen). For quantitation of genes of interest, cDNA samples were amplified in a CFX96 Realtime System (Bio-Rad Laboratories, Hercules, CA, USA) and Sso Fasteva green supermixture (Bio-Rad Laboratories) according to the manufacturer's instructions. Fold changes in mRNA expression between treatments and controls were determined by the δCT method as described[27]. Differences between groups were determined using a two-sided Student's t-test and one-way ANOVA. Error bars on plots represent ± SE, unless otherwise noted. The data were normalized to a GAPDH reference. All primers were purchased from Eurofins MWG Operon. The primer pairs for analysis are provided in Supplementary Table 1. All assays were performed in triplicate at least three times.
Electrophoresis of total RNA isolated from EPDENs and plant tissue extracts
Total RNA was extracted from the EPDENs and plant tissues using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The RNA was resuspended in 100 µl of DEPC-treated water. The purified RNA was twice digested with DNase I (Qiagen). The purity and yield of RNA was determined spectrophotometrically by measuring optical density at wavelengths of 260 and 280 nm. Samples were stored at −80°C. To confirm that the nucleic acid isolated from EPDENs was RNA, nucleic acid from EPDENs was treated with 1.0 µg/µl RNase (Sigma) or DEPC treated water as a control for 15 min at 37°C before the samples were loaded on a 12% polyacrylamide gel. A total of 1 µg RNA isolated from EPDENs was resolved on 12% polyacrylamide (acrylamide/bis-acrylamide, 29:1) gels containing 8 M Urea and 1×Tris-Boric Acid-EDTA (TBE, 89 mM tris (pH 7.6), 89 mM boric acid, 2 mM EDTA). A total of 1 µg RNA isolated from plant tissue extracts were resolved on 2% agarose gel. After electrophoresis, the gel was stained with ethidium bromide (0.5 µg/ml) and visualized using a UVP PhotoDoc-It™ Imaging System (UVP, Montpelier, Maryland).
Cell Culture
The murine macrophage cell line RAW 264.7 was obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% FBS and incubated at 37°C in a humidified atmosphere with 5% CO2. The cells were passaged every 2–3 days to maintain them in exponential growth stage. FBS used for culturing RAW 264.7 cell line was pretreated by ultracentrifugation at 100,000g for 16 h to deplete FBS exosomes using a standard protocol as described previously[28].
Nrf2 staining
RAW 264.7 macrophages were cultured in the presence of EPDENs (10 µg/ml) or PBS for 24 hr. Ten µg/ml of EPDENs was chosen and is based on the concentration of EPDENs in the extracted juice, (in addition we assume after passing through the stomach, the concentration of EPDENs in the juice would have been diluted 50-fold in the small intestine). The supernatants from 24 hr cultured cells were harvested for ELISA determination of IL-10 and IL-6. The cells were either lysed for western blot analysis for HO-1 or fixed in cold 4% paraformaldehyde for immunohistological staining of Nrf2. Paraformaldehyde fixed cells were first permeabilized with 1% Triton-X 100 in PBS for 10 min, followed by blocking with 5% BSA in PBS containing 0.1% Triton-X 100 for 1 h, and then stained with a rabbit anti-mouse Nrf2 polyclonal antibody (Santa Cruz Biotechnology) at 22°C for 2hr. After washing, cells were stained with a goat anti-rabbit to fluorescein isothiocyanate–Alexa Fluor 488 (Invitrogen Life Sciences). Slides were mounted with Slow Fade Gold Antifade plus DAPI (4,6-diamidino-2-phenylindole; S36938; Molecular Probes and Invitrogen Life Sciences). Staining of cells was assessed using a Nikon A1R Confocal system.
TLC analysis
EPDEN lipids were extracted with chloroform:methanol (2:1, v/v; 20 ml) using a method as described[20]. TLC was performed according to the method of Masukawa et al.[29]. Briefly, HPTLC-plates (silica gel 60 with concentrating zone, 20 cm × 10 cm; Merck) were used for the separation. Aliquots of concentrated lipid samples extracted from EPDENs were separated on HPTLC-plates, and the plates developed with chloroform/methanol/acetic acid (190:9:1, by vol). After drying, the plates were sprayed with a 10% copper sulfate and 8% phosphoric acid solution and were then charred by heating at 180°C for 5 min. The plate was imaged with an Odyssey Scanner (LI-COR Bioscience, Lincoln NE).
LacZ staining and immunohistological staining
To assess the treatment effects on expression of lacZ gene regulation by the Tcf4 promoter, B6.Cg-Tg ((BAT-lacZ)3Picc/J) mice were gavage administered EPDENs or PBS as a control vehicle for 3 days. Mice were sacrificed and small intestinal tissue segments were fixed in 0.2% glutaraldehyde, 50 mM EGTA, pH 7.3, 100 mM MgCl2 in PBS. Staining was performed overnight in staining buffer (0.5 mg/ml X-gal, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide in 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Nonidet P40 in PBS; pH 7.3). The next day, intestinal segments were washed in PBS, counterstained with nuclear fast red, and covered in Kaiser’s glycerogelatin. All photomicrographs were taken with an IX71 inverted microscope (Olympus). Swiss-rolled small intestines of (BAT-lacZ)3Picc/J) mice were also fixed and stained with X-gal and examined in a blinded manner by a gastrointestinal pathologist.
To determine whether intestinal macrophages and Lgr5+ intestinal stem cells take up EPDENs, C57B:/6j or Lgr5-EGFP-IRES-CreERT2 mice starved overnight were given 1 mg PKH26 (Sigma) fluorescent dye labeled EPDENs/mouse by gavage in 100 µl of PBS. 6 hr after gavaging mice were sacrificed and intestinal tissues were embedded in OCT compound (Miles Laboratories; Elkhart, IN) and frozen. Tissues were sectioned 5 µm thick with a cryostat and mounted on commercially provided charged slides (Fisher Scientific; Pittsburgh, PA) for immunohistological and DAPI (Molecular Probe) staining. Images were acquired with a Zeiss LSM 510 confocal microscope equipped with a digital image analysis system (Pixera). For macrophage staining, sliced intestinal tissues were stained with rat monoclonal anti-F4/80 (eBioscience). After washing, cells were stained with a goat anti-rat fluorescein isothiocyanate–Alexa Fluor 488 (Invitrogen Life Sciences) for confocal microscope examination.
Grape EPDEN miRNA microarray analysis
Total RNA containing miRNA was extracted from grape EPDENs using the SNC50-kit (Sigma). One hundred nanograms of small RNA enriched samples were submitted to Phalanx Biotech Group, Inc. (Belmont, CA 94002 USA) for miRNA microarray analysis. The miRNA profile [20] of grape exosome-like nanoparticles was used for predicting Grape EPDEN-miRNA homologue sequences to other species using ClustalX2 analysis http://www.ebi.ac.uk/Tools/msa/clustalo/.
Statistical analysis
One-way, two-way analysis of variance (ANOVA) and t-test were used to determine statistical significance (*p<0.05, and **p<0.01).
Results
Characterization of edible plant derived nanoparticles (EPDENs)
Using mammalian exosome purification techniques [30], we isolated edible plant exosome-like nanoparticles from the juice of grapefruit, grapes and the homogenized roots of ginger and carrots. The particles were characterized as exosome-like nanoparticles based on electron microscopic examination (Figure 1a) of a sucrose gradient purified band 2 (Figure 1a, left), size distribution (Figure 1b), charge (Figure 1c), the SDS PAGE gel protein separation profile (Figure 1d, left panels), lipid separation using TLC (Figure 1d, right panels), and the RNAs present (Figure 1e). nanoparticles from bands 1 and 3 (Fig. 1a) were excluded in the following studies because our preliminary data suggest that their role in inducing expression of genes for anti-inflammation cytokines, anti-oxidation and activation of Wnt signaling could not be determined. The migration profile of lipids in grapefruit derived EPDENs is similar to that of the grape-derived EPDEN lipid migration profile (Figure 1d, right panels) but different from the ginger or carrot EPDENs lipid profiles, suggesting that lipids from fruit EPDENs have different polarity in comparison with the lipid profiles of root-derived edible vegetable EPDENs. The presence of nucleic acids was also quantitatively analyzed. RNase treatment lead to the degradation of the nucleic acids purified in EPDEN samples (Figure 1e), suggesting that they are RNAs. Substantial amounts of small sized RNAs were detected by gel electrophoresis. The small sized RNAs enriched in EPDENs were unlikely due the method used for RNA extraction since large sized 25S and 18S were dominantly presented in the total RNAs extracted from plant tissue which is also used for isolating EPDENs. Next, yields of EPDEN RNAs, proteins, and lipids isolated from 100 g of each fruit, ginger or carrot roots were compared. Although the production of total amounts of EPDENs from all four plants were not different, (Figure 1f, left panels), total RNAs extracted from grape or grapefruit EPDENs were much less than amounts of RNAs isolated from ginger or carrot root EPDENs (Figure 1f, middle panels). Mass spectrometry analyses of the grape EPDEN miRNA profile further suggested that grape EPDENs contain miRNAs enriched for the miR169 family [20]. Sequence alignment was performed using ClustalX2 analysis. The sequence of the vvi-miR-169 seed region (AGCCAAG) was used to search for microRNAs that shared a similar seed sequence, which included all known human microRNAs (www.mirbase.org). Two human microRNAs (has-miR-4480 and has-miR-4662a-5p) were found to share a sequence similarity in the seed region with vvi-miR-169 (Figure 1f, right panels). Collectively, these data demonstrated that our preparation contained exosome-like nanoparticles isolated from all four edible plants including both fruits and vegetables.
Figure 1. Identification and characterization of edible plant derived exosomes-like nanoparticles (EPDENs).
(a) Three bands were formed after sucrose grandient ultracentrifugation. EPDENs from the 30%/45% interface were visualized by electromicroscopy. (b) Size distribution and (c) surface Zeta-potential of the particles was determined using the Zetasizer Nano ZS. (d) 50 µg of EPDENs were run on 10% SDS PAGE protein gels and detected with Coomassie Blue staining (left panel). Lipids were detected by TLC (right panel) analysis of the lipid extracts from EPDENs. The lipids extracted from EPDENs were separated on a thin-layer chromatography plate and developed by spraying the plate with a 10% copper sulfate and 8% phosphoric acid solution. A representative image was scanned using an Odyssey Scanner. (e) After electrophoresis on the 12% polyacrylamide gel, EPDEN RNA pretreated with/without RNase was stained with ethidium bromide and visualized with a UVP PhotoDoc-It™ Imaging System. (f) Sucrose purified EPDENs were weighed and expressed as mg of EPDENs /100 g of edible plant (left panel), total RNA from EPDENs was quantified using Nanodrop spectrophotometry to measure absorbance at 260 nm, and expresssed as µg of RNA/100 mg of EPDENs (middel panel). Error bars represent standard deviation (±SD) (** p<0.01, * p<0.05). ClustalX2 analysis of grape EPDEN-miRNA homologue sequencing (right panel). The highlighted nucleotides represent highly conserved regions of the grape EPDEN miRNA domains to human miRNAs. Results (a–e) represent one of four independent experiments.
Next, we determined whether these EPDENs are stable in stomach-like and intestine-like solutions. We first mimicked in vivo conditions by suspending EPDENs in water or a stomach-like or an intestinal-like solution, and then analyzed the EPDENs for size (Figure 2a) and surface charge (Figure 2b). The results showed (Figure 2a) that the heterogeneity in diameter of grape EPDENs was reduced in both a stomach-like and an intestinal-like solution when compared to the size of grape EPDENs in water. Grape EPDENs became smaller in size in stomach-like and intestine-like solutions. For grapefruit EPDENs, two subsets of EPDENs were separated from the original one in an acidic stomach solution, but not in an intestinal solution. For ginger EPDENs, a subset of the population that enlarged in size was generated in the stomach-like and intestinal-like solution. For carrot EPDENs, the initial population of two subsets was reduced into one population in a stomach-like solution, but not in an intestinal-like solution. Collectively, these data suggest that the size of EPDENs can be altered in a pH dependent manner. We then evaluated the charge of the EPDENs (Figure 2b). Compared to the charge of grape EPDENs in water, when comparing the charge of the all of the EPDENs in an intestinal-like solution there was no change in the charge observed in the EPDENs of grape, grapefruit and carrots; however, the ginger EPDENs had a reduction in negative charge. In contrast, in a stomach-like solution, all four EPDENs had a remarkable reduction in negative charge.
Figure 2. EPDENs are resistant to gastric and intestinal enzymatic digestion.
EPDENs were incubated in water (left column) or a stomach-like solution (middle column) at 37°C for 30 min or first in stomach-like solution at 37°C for 30 min followed by incubation for 30 min in a small intestine-like solution. The change of particle size (a) and surface charge (b) were measured using a Zetasizer. Results (a-b) represent one of five independent experiments.
Orally delivered EPDENs are taken up by intestinal macrophages and stem cells
Macrophages are part of the innate intestinal immune system and have the ability to actively take up synthesized nanoparticles[31–33]. Whether nanoparticles from the diet we daily eat are taken up by intestinal macrophages is not known. Confocal analysis of intestinal tissue sections revealed that the EPDENs were co-localized with F4/80+ macrophages in the lamina propria of both small and large intestine (Figure 3a). Beside intestinal macrophages, we also detected that intestinal stem cells of Lgr5-EGFP-IRES-CreERT2 mice also take up EPDENs (Figure 3b).
Figure 3. Intesinal macrophages and LGR5 stem cells take up orally administrated EPDENs.
Female C57BL/6 mice (a) or Lgr5-EGFP-IRES-CreERT2 mice (b) were gavage administered PKH26 dye labeled EPDENs (1mg per mouse in 200 µl PBS). 6h after the administration, F4/80+/PKH26 cells (a) or Lgr5-EGFP+PKH26+ cells (b) in frozen sections of intestine were examined by confocal microscopy (left) and were quantified (right). Original magnification ×40. A representative image is presented (a,b, left panels, n=5). Error bars represent standard deviation (±SD) (a, b, right panels) (n = 5 mice per group).
EPDENs from different edible plants have distinct biological effects on the induction of expression of anti-inflammatory genes, an anti-oxidation gene and activation of the Wnt/TCF4 signaling pathway
Macrophages are highly activated and increased in numbers in gut related inflammatory disease. Thus determination of the biological effect(s) mediated by EPDENs in terms of induction of anti-inflammatory cytokines may account for the cellular and molecular pathways underlying the beneficial effect of a plant-derived or plant-enhance diet for humans.
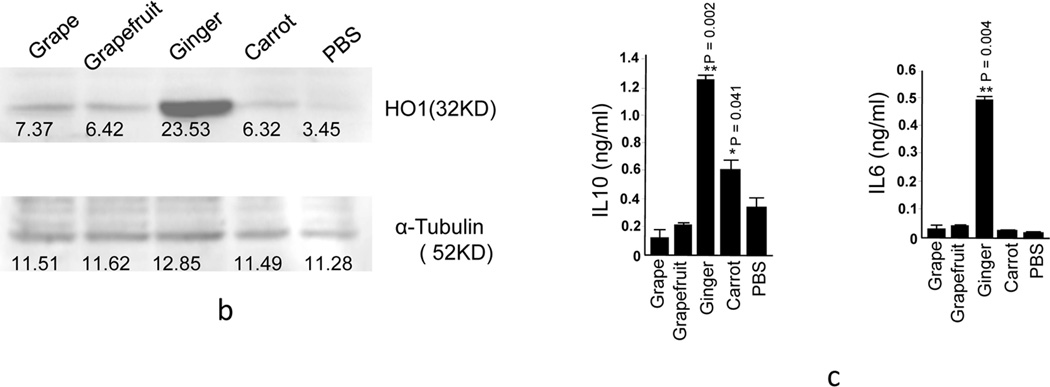
HO-1 [34–36] and IL-10 expressed in macrophages plays a crucial role in preventing colitis due to their potent anti-inflammatory capacity[37, 38][36, 37] [31, 32]. In this study, RAW 264.7 macrophages were cultured in the presence of EPDENs, and a number of cytokines were quantitatively analyzed by real-time PCR. The results showed that the macrophages treated with ginger EPDENs significantly enhanced HO-1 and IL-10 expression (Figure 4a). The augmented expression of HO-1 and IL-10 was further confirmed by western blot (Figure 4b) and ELISA (Figure 4c), respectively. IL-6 has been shown to provide important survival and proliferative signals to many leukocyte populations and orchestrates the development of the immune response. IL-6 plays a vital role in the development of both B cell and T cell responses. Most edible plants have roles in maintaining intestinal immune cell homeostasis. We tested whether EPDENs not only induce immune suppressive molecules like IL-10, but also immune stimulating molecules like IL-6 in order to maintain homeostatic balance. Both real-time PCR (Figure 4a) and ELISA (Figure 4c) results showed that the macrophages treated with ginger EPDENs significantly enhanced IL-6 expression. In contrast with the induction of both IL10 and IL-6 after macrophages were treated with ginger EPDENs, carrot EPDENs tended to induce IL-10 only.
Figure 4. Induction of anti-inflammation cytokines, anti-oxidation genes expressed in macrophages and actvation of the Wnt pathway in mice treated with EPDENs.
(a–d) Twenty-four hr after RAW 264.7 macrophage treatment with EPDENs (1.0 ug/ml), (a) total RNA was isolated and the mRNA level of HO-1, IL-6, IL-10 and TNFα were determined by real-time PCR. Error bars represent standard deviation (±SD) (** p<0.01, * p<0.05).
(b) EPDEN treated RAW 264.7 macrophages were lysed and the expression of HO-1 was western blot analyzed. A representative image (n=3/treatment) is shown. The density of each western blot band was quantified and is indicated under each band. (c) The supernatants from EPDEN treated RAW 264.7 macrophages were collected for ELISA quantitation of IL-10 and IL-6. Error bars represent standard deviation (±SD) (** p<0.01, * p<0.05).
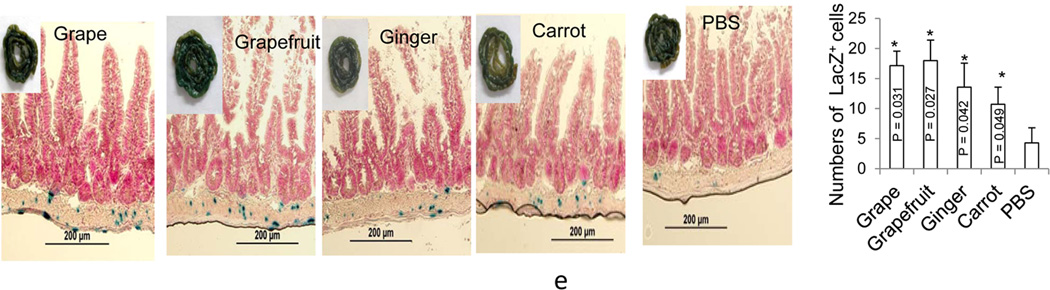
(d) EPDEN treated RAW 264.7 macrophages were stained with anti-Nrf2 and DAPI, and Nrf2+ cells were examined by confocal microscopy. One representative example of at least three independent experiments is shown in paraformaldehyde fixed cells (left) and nuclear positive Nrf2 cells and the total number of cells were quantified by counting 5 arbitrarily selected fields in a double-blinded manner and expressed as the total number of nuclear positive Nrf2 cells/field (right). The results are presented as means ± SD. Significant induction of nuclear positive Nrf2 cells of cells treated with EPDEN was analyzed in compared with the cells treated with PBS control (**P < 0.01, *P < 0.05). (e) Tcf/LEF-reporter mice starved overnight were gavage-administered twice a day for 3 days with 2 mg of EPDENs per mouse in 200 µl PBS or an equal amount of PBS as a control. X-gal staining (blue) of sectioned small intestine of B6.Cg-Tg(BAT-lacZ)3Picc/J mice. Tissues were counterstained with nuclear fast red (red). Original magnification ×40. One representative example of at least three independent experiments is shown (left panel), and number of X-gal+ cells were counted from 10 fields. Data show means ± SEM of three independent experiments with five mice per group. The error bars represent the standard error of the mean. *p < 0.05 compared with the control PBS group.
Nrf2, a key regulator of the HO1 gene, has an important role in anti-inflammation and anti-oxidation[39–44]. Nrf2 nuclear translocation as an indicator for regulating the cellular anti-inflammatory responses is well documented[45–47]. Among the EPDENs tested, ginger EPDENs were the strongest at inducing nuclear translocation of Nrf2 in RAW 264.7 macrophages although grapefruit and carrot EPDENs also increased nuclear translocation of Nrf2 when compared with grape EPDENs or PBS treated RAW 264.7 macrophages (Figure 4d). Collectively, these results suggest that EPDENs from different edible plants have different capacities for promoting Nrf2 nuclear translocation.
Next, we determined whether the EPDENs have an effect on the Wnt/Tcf4 signaling pathway because Wnt/Tcf4 plays an important role in gut homeostasis[48, 49] and immune tolerance[50, 51]. Using the same protocol as used for analysis of the effect of grape EPDENs[20] on gut homeostatis and immune tolerance, B6.Cg-Tg(BAT-lacZ)3Picc/J mice were gavage administered daily for 3 days with EPDENs (2 mg/mouse). Mice were killed 6 h after the last treatment and β-galactosidase activity was determined by staining fresh intestine in X-gal substrate. Characterization of X-gal stained whole intestine (Figure 4e) demonstrated that expression of the β-galactosidase is higher in the crypts of mice treated with the EPDENs than in mice treated with PBS control group.
Discussion
Our findings show that exosome-like nanoparticles are present in edible fruits and vegetables and reveal a previously unrecognized strategy by which plants communicate with mammalian cells via exosome-like nanoparticles in the gut, and in particular intestinal macrophages and stem cells. We found that edible plants contain large amounts of nanoparticles. Like mammalian exosomes, further characterization of the plant nanoparticles led to identifying them as exosome-like nanoparticles based on the nanoparticles being composed of proteins, lipids, and miRNAs. EPDENs from different types of plants have different biological effects on the recipient mammalian cells. This finding opens up a new avenue to further study the molecular mechanisms underlying how the plant kingdom cross-talks with mammalian cells such as intestinal macrophages and stem cells via EPDENs. This information may provide the molecular basis of using multiple plant derived agents for better therapeutic effect than any single plant derived agent.
Our results demonstrated that the EPDENs induce nuclear translocation of macrophage Nrf2 and intestinal Wnt/TCF4 activation of mice orally administrated edible plant EPDENs. Both nuclear translocation of Nrf2 [52, 53] and Wnt/Tcf4 activation[54] play a critical role in anti-inflammatory responses. Consistent with those effects, IL-10 and HO-1 are preferentially induced in macrophages stimulated with ginger EPDENs whereas no induction of IL-10 and HO-1 when macrophages were stimulated with grapefruit or carrot EPDENs. Nuclear translocation of Nrf2 is involved in HO-1 activation and in the activation of IL10[55]. The discrepancy of results generated from different EPDENs in terms of induction of IL-10 and HO-1 is likely due to unidentified molecules in grapefruit or carrot EPDENs that activate different pathways. The activated pathways may have a role in preventing the expression of IL-10 and HO-1.
Interestingly, the anti-inflammatory cytokine IL-10 and the proinflammatory cytokine IL-6 are also slightly induced in ginger EPDEN-treated macrophages. IL-6 plays an instrumental role in innate immunity and antibody production[56–59]. After cells receive an inflammatory stimulus, the inflammation process is followed by an anti-inflammatory response that prevents excessive damage to the host. IL-6 and IL-10 are key players in these processes. The necessity of these counteractive processes for balancing an inflammatory event is evident in the IL-10 knock-out mouse which spontaneously acquires IBD[60–62], as well in the development of RA in humans due to impaired IL-10 production[63, 64]. It is conceivable that EPDENs carrying a variety of different molecules that could stimulate both pro- and anti-inflammatory cytokines that could be capable of inducing the release of pro- as well as anti-inflammatory cytokines from targeted cells. Therefore, ingesting ginger EPDENs may have a role in maintaining intestinal homeostasis in terms of production of pro- and anti-inflammatory cytokines. Further study of ginger EPDEN-mediated induction of pro- and anti-inflammatory cytokines may lead to identification of EPDEN molecules that induce both IL-6 and IL-10.
From a therapeutic standpoint, crude extracts of ginger are commonly used to treat various types of “stomach problems,” pain relief from arthritis or muscle soreness, menstrual pain, upper respiratory tract infections, cough, and bronchitis. Ginger EPDENs may have a better therapeutic effect than crude ginger extract since ginger EPDENs are preferentially taken up by intestinal macrophages or monocytes. Moreover, ginger EPDENs could also be considered a better delivery vehicle for therapeutic agents such as anti-inflammatory drugs. Preferential up take of ginger EPDENs by macrophages might provide a means to combine or attach other molecules to the ginger EPDENs so as to deliver targeted molecules to EPDEN recipient cells and transform ginger EPDENs into a therapeutic agent carrier. This approach would be advantageous over the conventional clinical strategies of attempting to neutralize pro-inflammatory cytokines or by administration of anti-inflammatory cytokines. Therapeutic application of the aforementioned strategies is problematic due to the inherent limitation of specificity directed against a single target in the cells, which typically leads to toxicity.
Our data show that fruit-derived EPDENs have dominate effects on modulation of Wnt/TCF4 activity and ginger EPDEN have an effect on Nrf2 activation or induction of anti-inflammatory molecules. It has been known for decades that people eating a variety of edible plants daily are the recipients of many beneficial health effects when compared to subjects that ingest fewer types of edible plants. Ingesting EPDENs from a variety of fruits and vegetables daily would be expected to provide greater beneficial effects for maintaining gut homeostasis than ingesting EPDENs from single edible plant.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH) (UH2TR000875, RO1AT004294); the Louisville Veterans Administration Medical Center (VAMC) Merit Review Grants (H.-G.Z.). We thank Dr. Jerald Ainsworth for editorial assistance.
References
- 1.Patrick C, Wang GS, Lefebvre DE, Crookshank JA, et al. Promotion of autoimmune diabetes by cereal diet in the presence or absence of microbes associated with gut immune activation, regulatory imbalance, and altered cathelicidin antimicrobial Peptide. Diabetes. 2013;62:2036–2047. doi: 10.2337/db12-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozuka C, Yabiku K, Sunagawa S, Ueda R, et al. Brown rice and its component, gamma-oryzanol, attenuate the preference for high-fat diet by decreasing hypothalamic endoplasmic reticulum stress in mice. Diabetes. 2012;61:3084–3093. doi: 10.2337/db11-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orr JS, Puglisi MJ, Ellacott KL, Lumeng CN, et al. Toll-like receptor 4 deficiency promotes the alternative activation of adipose tissue macrophages. Diabetes. 2012;61:2718–2727. doi: 10.2337/db11-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pope JL, Bhat AA, Sharma A, Ahmad R, et al. Claudin-1 regulates intestinal epithelial homeostasis through the modulation of Notch-signalling. Gut. 2013 doi: 10.1136/gutjnl-2012-304241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moossavi S, Zhang H, Sun J, Rezaei N. Host-microbiota interaction and intestinal stem cells in chronic inflammation and colorectal cancer. Expert Rev Clin Immunol. 2013;9:409–422. doi: 10.1586/eci.13.27. [DOI] [PubMed] [Google Scholar]
- 6.Chen K, Liu M, Liu Y, Yoshimura T, et al. Formylpeptide receptor-2 contributes to colonic epithelial homeostasis, inflammation, and tumorigenesis. J Clin Invest. 2013;123:1694–1704. doi: 10.1172/JCI65569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker C, Watson AJ, Neurath MF. Complex roles of caspases in the pathogenesis of inflammatory bowel disease. Gastroenterology. 2013;144:283–293. doi: 10.1053/j.gastro.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 8.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 9.Kant P, Sainsbury A, Reed KR, Pollard SG, et al. Rectal epithelial cell mitosis and expression of macrophage migration inhibitory factor are increased 3 years after Roux-en-Y gastric bypass (RYGB) for morbid obesity: implications for long-term neoplastic risk following RYGB. Gut. 2011;60:893–901. doi: 10.1136/gut.2010.230755. [DOI] [PubMed] [Google Scholar]
- 10.Asquith M, Powrie F. An innately dangerous balancing act: intestinal homeostasis, inflammation, and colitis-associated cancer. J Exp Med. 2010;207:1573–1577. doi: 10.1084/jem.20101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Souza-Schorey C, Clancy JW. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012;26:1287–1299. doi: 10.1101/gad.192351.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez-Vazquez C, Villarroya-Beltri C, Mittelbrunn M, Sanchez-Madrid F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev. 2013;251:125–142. doi: 10.1111/imr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rak J. Microparticles in cancer. Seminars in thrombosis and hemostasis. 2010;36:888–906. doi: 10.1055/s-0030-1267043. [DOI] [PubMed] [Google Scholar]
- 15.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends in cell biology. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nature reviews. Immunology. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buzas EI, Gyorgy B, Nagy G, Falus A, Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nature reviews. Rheumatology. 2014 doi: 10.1038/nrrheum.2014.19. [DOI] [PubMed] [Google Scholar]
- 18.Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. The American journal of pathology. 2014;184:28–41. doi: 10.1016/j.ajpath.2013.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayner KJ, Hennessy EJ. Extracellular communication via microRNA: lipid particles have a new message. Journal of lipid research. 2013;54:1174–1181. doi: 10.1194/jlr.R034991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju S, Mu J, Dokland T, Zhuang X, et al. Grape Exosome-like Nanoparticles Induce Intestinal Stem Cells and Protect Mice From DSS-Induced Colitis. Mol Ther. 2013;21:1345–1357. doi: 10.1038/mt.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, Zhuang X, Deng ZB, Jiang H, et al. Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Mol Ther. 2013 doi: 10.1038/mt.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Yu S, Zinn K, Wang J, et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. Journal of immunology. 2006;176:1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 23.Spilman MS, Welbon C, Nelson E, Dokland T. Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J Gen Virol. 2009;90:527–535. doi: 10.1099/vir.0.007674-0. [DOI] [PubMed] [Google Scholar]
- 24.Hermida LG, S-X M, Barnadas-Rodríguez R. Combined strategies for liposome characterization during in vitro digestion. Journal of Liposome Research. 2009;19:207–219. doi: 10.1080/08982100902740847. [DOI] [PubMed] [Google Scholar]
- 25.Sun D, Zhuang X, Xiang X, Liu Y, et al. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18:1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Shah SV, Xiang X, Wang J, et al. COP9-associated CSN5 regulates exosomal protein deubiquitination and sorting. Am J Pathol. 2009;174:1415–1425. doi: 10.2353/ajpath.2009.080861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang X, Zhuang X, Ju S, Zhang S, et al. miR-155 promotes macroscopic tumor formation yet inhibits tumor dissemination from mammary fat pads to the lung by preventing EMT. Oncogene. 2011;30:3440–3453. doi: 10.1038/onc.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Shah SV, Xiang X, Wang J, et al. COP9-associated CSN5 regulates exosomal protein deubiquitination and sorting. The American journal of pathology. 2009;174:1415–1425. doi: 10.2353/ajpath.2009.080861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masukawa Y, Narita H, Sato H, Naoe A, et al. Comprehensive quantification of ceramide species in human stratum corneum. J Lipid Res. 2009;50:1708–1719. doi: 10.1194/jlr.D800055-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang X, Liu Y, Zhuang X, Zhang S, et al. TLR2-mediated expansion of MDSCs is dependent on the source of tumor exosomes. Am J Pathol. 2010;177:1606–1610. doi: 10.2353/ajpath.2010.100245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He C, Yin L, Tang C, Yin C. Multifunctional polymeric nanoparticles for oral delivery of TNF-alpha siRNA to macrophages. Biomaterials. 2013;34:2843–2854. doi: 10.1016/j.biomaterials.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 32.Coco R, Plapied L, Pourcelle V, Jerome C, et al. Drug delivery to inflamed colon by nanoparticles: comparison of different strategies. International journal of pharmaceutics. 2013;440:3–12. doi: 10.1016/j.ijpharm.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Collnot EM, Ali H, Lehr CM. Nano- and microparticulate drug carriers for targeting of the inflamed intestinal mucosa. J Control Release. 2012;161:235–246. doi: 10.1016/j.jconrel.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 34.Onyiah JC, Sheikh SZ, Maharshak N, Steinbach EC, et al. Carbon monoxide and heme oxygenase-1 prevent intestinal inflammation in mice by promoting bacterial clearance. Gastroenterology. 2013;144:789–798. doi: 10.1053/j.gastro.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheikh SZ, Hegazi RA, Kobayashi T, Onyiah JC, et al. An anti-inflammatory role for carbon monoxide and heme oxygenase-1 in chronic Th2-mediated murine colitis. J Immunol. 2011;186:5506–5513. doi: 10.4049/jimmunol.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegazi RA, Rao KN, Mayle A, Sepulveda AR, et al. Carbon monoxide ameliorates chronic murine colitis through a heme oxygenase 1-dependent pathway. J Exp Med. 2005;202:1703–1713. doi: 10.1084/jem.20051047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee T-S, Chau L-Y. Heme oxygenase-1 mediates the anti-inflammatory effect of interleukin-10 in mice. Nat Med. 2002;8:240–246. doi: 10.1038/nm0302-240. [DOI] [PubMed] [Google Scholar]
- 38.Sheikh SZ, Hegazi RA, Kobayashi T, Onyiah JC, et al. An Anti-Inflammatory Role for Carbon Monoxide and Heme Oxygenase-1 in Chronic Th2-Mediated Murine Colitis. The Journal of Immunology. 2011;186:5506–5513. doi: 10.4049/jimmunol.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo C, Wang L, Zhang J, Numazawa S, et al. The Crosstalk between Nrf2 and AMPK Pathways Is Important for the Anti-inflammatory Effect of Berberine in LPS-Stimulated Macrophages and Endotoxin-Shocked Mice. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2012.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu W, Qiu Q, Wang H, Whitman SA, et al. Nrf2 is crucial to graft survival in a rodent model of heart transplantation. Oxid Med Cell Longev. 2013;2013:919313. doi: 10.1155/2013/919313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao XD, Zhou YT, Lu XJ. Sulforaphane enhances the activity of the Nrf2-ARE pathway and attenuates inflammation in OxyHb-induced rat vascular smooth muscle cells. Inflamm Res. 2013;62:857–863. doi: 10.1007/s00011-013-0641-0. [DOI] [PubMed] [Google Scholar]
- 42.Davidson BA, Vethanayagam RR, Grimm MJ, Mullan BA, et al. NADPH oxidase and Nrf2 regulate gastric aspiration-induced inflammation and acute lung injury. J Immunol. 2013;190:1714–1724. doi: 10.4049/jimmunol.1202410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng AS, Cheng YH, Chiou CH, Chang TL. Resveratrol upregulates Nrf2 expression to attenuate methylglyoxal-induced insulin resistance in Hep G2 cells. Journal of agricultural and food chemistry. 2012;60:9180–9187. doi: 10.1021/jf302831d. [DOI] [PubMed] [Google Scholar]
- 44.Juurlink BH. Dietary Nrf2 activators inhibit atherogenic processes. Atherosclerosis. 2012;225:29–33. doi: 10.1016/j.atherosclerosis.2012.08.032. [DOI] [PubMed] [Google Scholar]
- 45.Lee JA, Lee MY, Shin IS, Seo CS, et al. Anti-inflammatory effects of Amomum compactum on RAW 264.7 cells via induction of heme oxygenase-1. Archives of pharmacal research. 2012;35:739–746. doi: 10.1007/s12272-012-0419-x. [DOI] [PubMed] [Google Scholar]
- 46.Lee IS, Lim J, Gal J, Kang JC, et al. Anti-inflammatory activity of xanthohumol involves heme oxygenase-1 induction via NRF2-ARE signaling in microglial BV2 cells. Neurochemistry international. 2011;58:153–160. doi: 10.1016/j.neuint.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Theiss AL, Vijay-Kumar M, Obertone TS, Jones DP, et al. Prohibitin is a novel regulator of antioxidant response that attenuates colonic inflammation in mice. Gastroenterology. 2009;137:199–208. doi: 10.1053/j.gastro.2009.03.033. 208 e191-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim BM, Mao J, Taketo MM, Shivdasani RA. Phases of canonical Wnt signaling during the development of mouse intestinal epithelium. Gastroenterology. 2007;133:529–538. doi: 10.1053/j.gastro.2007.04.072. [DOI] [PubMed] [Google Scholar]
- 49.Sancho R, Nateri AS, de Vinuesa AG, Aguilera C, et al. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. Embo J. 2009;28:1843–1854. doi: 10.1038/emboj.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oderup C, LaJevic M, Butcher EC. Canonical and noncanonical Wnt proteins program dendritic cell responses for tolerance. J Immunol. 2013;190:6126–6134. doi: 10.4049/jimmunol.1203002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng ZB, Zhuang X, Ju S, Xiang X, et al. Intestinal mucus-derived nanoparticle-mediated activation of Wnt/beta-catenin signaling plays a role in induction of liver natural killer T cell anergy in mice. Hepatology. 2013;57:1250–1261. doi: 10.1002/hep.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim SW, Lee HK, Shin JH, Lee JK. Up-down Regulation of HO-1 and iNOS Gene Expressions by Ethyl Pyruvate via Recruiting p300 to Nrf2 and Depriving It from p65. Free radical biology & medicine. 2013;65:468–476. doi: 10.1016/j.freeradbiomed.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 53.Luo C, Urgard E, Vooder T, Metspalu A. The role of COX-2 and Nrf2/ARE in anti-inflammation and antioxidative stress: Aging and anti-aging. Medical hypotheses. 2011;77:174–178. doi: 10.1016/j.mehy.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 54.Gersemann M, Stange EF, Wehkamp J. From intestinal stem cells to inflammatory bowel diseases. World journal of gastroenterology : WJG. 2011;17:3198–3203. doi: 10.3748/wjg.v17.i27.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piantadosi CA, Withers CM, Bartz RR, MacGarvey NC, et al. Heme oxygenase-1 couples activation of mitochondrial biogenesis to anti-inflammatory cytokine expression. J Biol Chem. 2011;286:16374–16385. doi: 10.1074/jbc.M110.207738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mora JR, Iwata M, Eksteen B, Song SY, et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314:1157–1160. doi: 10.1126/science.1132742. [DOI] [PubMed] [Google Scholar]
- 57.Jin JO, Han X, Yu Q. Interleukin-6 induces the generation of IL-10-producing Tr1 cells and suppresses autoimmune tissue inflammation. J Autoimmun. 2013;40:28–44. doi: 10.1016/j.jaut.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujihashi K, Kono Y, Kiyono H. Effects of IL6 on B cells in mucosal immune response and inflammation. Res Immunol. 1992;143:744–749. doi: 10.1016/0923-2494(92)80015-d. [DOI] [PubMed] [Google Scholar]
- 59.McGhee JR, Fujihashi K, Lue C, Beagley KW, et al. Role of IL-6 in human antigen-specific and polyclonal IgA responses. Adv Exp Med Biol. 1991;310:113–121. doi: 10.1007/978-1-4615-3838-7_13. [DOI] [PubMed] [Google Scholar]
- 60.Rennick D, Davidson N, Berg D. Interleukin-10 gene knock-out mice: a model of chronic inflammation. Clin Immunol Immunopathol. 1995;76:S174–S178. doi: 10.1016/s0090-1229(95)90144-2. [DOI] [PubMed] [Google Scholar]
- 61.Coombes JL, Robinson NJ, Maloy KJ, Uhlig HH, Powrie F. Regulatory T cells and intestinal homeostasis. Immunol Rev. 2005;204:184–194. doi: 10.1111/j.0105-2896.2005.00250.x. [DOI] [PubMed] [Google Scholar]
- 62.Barbara G, Xing Z, Hogaboam CM, Gauldie J, Collins SM. Interleukin 10 gene transfer prevents experimental colitis in rats. Gut. 2000;46:344–349. doi: 10.1136/gut.46.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neidhart M, Jungel A, Ospelt C, Michel BA, et al. Deficient expression of interleukin-10 receptor alpha chain in rheumatoid arthritis synovium: limitation of animal models of inflammation. Arthritis Rheum. 2005;52:3315–3318. doi: 10.1002/art.21274. [DOI] [PubMed] [Google Scholar]
- 64.Cairns AP, Crockard AD, Bell AL. Interleukin-10 receptor expression in systemic lupus erythematosus and rheumatoid arthritis. Clin Exp Rheumatol. 2003;21:83–86. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.