Abstract
Mass spectrometry (MS) coupled to sample preparation and separation techniques has become a primary tool for proteomics studies. However, due to sample complexity, it is often challenging to achieve fast and efficient sample preparation prior to MS analysis. In recent decades, monolithic materials have been developed not only as chromatographic media, but also as efficient solid supports for immobilizing multiple types of affinity reagents. Herein, the N-acryloxysuccinimide-co-acrylamide-co-N,N'-methylenebisacrylamide (NAS-AAm-Bis) monolith was fabricated within silanized 200 μm i.d. fused-silica capillaries and was used as an immobilized enzyme reactor (IMER). The column was conjugated with trypsin/Lys-C and Lys-N enzymes to allow enzymatic digestions to occur while protein mixture was loaded onto the IMER column followed by MS-based proteomics analysis. Similar MS signal and protein sequence coverage were observed using protein standard bovine serum albumin (BSA) compared to in-solution digestion. Furthermore, mouse serum, yeast, and human cell lysate samples were also subjected to enzymatic digestion by both IMER (in seconds to minutes) and conventional in solution digestion (overnight) for comparison in large-scale proteomics studies. Comparable protein identification results obtained by the two methods highlighted the potential of employing NAS-based IMER column for fast and highly efficient sample preparation for MS analysis in proteomics studies.
Keywords: monolithic column, proteomics, protein identification, ESI-Orbitrap-MS, digestion, IMER, MALDI-MS, database search
1. INTRODUCTION
Proteomics is the large-scale study of proteins which can provide global information of protein identification, characterization, and quantification. Proteomics increasingly plays an important role in major research areas including protein interaction studies, biomarker discovery, cancer remediation, drug treatment, and disease-screening medical diagnostics [1–3]. Mass spectrometry (MS) has become the method of choice for proteomics analysis. Analytes from complex samples can be resolved, fragmented, identified and quantified in various biological samples. Numerous applications have been reported in the field of MS-based peptide and protein analysis [4,5].
Among various approaches in proteomics study, bottom-up proteomics remains the most widely applied method. In the bottom-up approach, proteins are digested into smaller peptide fragments that are often easier to ionize and detect by liquid chromatography-electrospray ionization mass spectrometry (LC-ESI-MS) and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) [6–8]. The classic workflow for MS-based proteomics mainly involves protein solubilization/denaturation, reduction, alkylation, and enzymatic digestion. One of the most time-consuming steps is protein digestion, taking as much as 12h or longer for complex sample digestion. In addition, the introduction of salt requires extra clean up steps in sample preparation, which causes sample loss and higher likelihood of degradation [9,10]. To address this bottleneck in protein identification, characterization and quantification, protein digestion has been improved through the development of novel techniques in order to increase throughput and reproducibility. Over the years, researchers have established various approaches to assist digestion such as microwave irradiation, infrared energy, organic and detergent-assisted methods, power ultrasound with elevated pressure and temperature, etc [11–15]. The improved throughput in protein digestion is also achieved by immobilization of the protease onto a solid support leading to reduced autolysis of enzyme and enhanced digestion efficiency [16,17].
Immobilized enzyme reactors (IMERs) have the ability to increase the throughput and efficiency of sample preparation [18]. Multiple types of solid support have been applied in IMERs fabricated into different formats such as capillary, microchip, pipette tips, etc [19–22]. Monolithic materials that possess a continuous macroporous channel, which features both large surface area and low backpressure, have been demonstrated as an efficient solid support suitable for multiple types of binding ligands [23]. A variety of monoliths have been reported for manufacturing micro-reactors used in proteolytic digestion of proteins into peptides. With active functional groups or leaving groups on the monolithic structure, proteins can be covalently linked to the monolithic surface while retaining their biological activities. Multiple types of monoliths have been employed for protein conjugation including poly(glycidy methacrylate-co-ethylene dimethacrylate), (GMA-EDMA), agarose, cryogel, and silica monoliths [24,25]. The functional groups of monolithic materials make them ideal for multiple immobilization strategies with various ligands. Although monolithic supports feature fast and highly efficient on-column reactions, the covalent-binding reaction cannot be performed under harsh conditions like high temperature in order to retain the bio-activity of enzymes. Typically, excessive amounts of ligands are required for hours or days long reaction to ensure complete binding of enzyme with functional groups [26].
Despite the attractive features of applying fast and highly efficient sample preparation strategies using monolithic support for protein sample preparation, challenges still exist and require technical improvement to enable analysis of low-level analyte containing samples or limited ligand amount. Herein, we constructed an IMER platform incorporated with multi-proteases in an open-tubular type column and employed that for comprehensive identification in complex cell lysates. The inter-connected channel of monolithic materials enables high reaction speed between stationary phase and mobile phase, while the open-tubular structure ensures fast flow rate in the meantime. We immobilized multiple proteases, trypsin/Lys-C mixture and Lys-N onto monolithic column so that proteins can be on-column digested in minutes and delivered to a mass spectrometer for subsequent analysis. Proof-of-concept experiments were performed to demonstrate significant improvement in sample preparation efficiency and in reduction of experimental time with similar protein sequence coverage and protein identification compared to in-solution digestion. Furthermore, IMERs were utilized in large-scale proteomic studies using mouse serum, human and yeast cell lysates. These experiments showed comparable performance to the traditional overnight in-solution digestion method and great potential in micro-scale proteomics analysis.
2. EXPERIMENTAL SECTION
2.1 Reagents and Materials
Sodium chloride, sodium bicarbonate, ammonium hydroxide, acetone, acetonitrile, methanol, ammonium bicarbonate, and urea were purchased from Fisher Scientific (Pittsburgh, PA). Acrylamide (AAm, 99%), N, N’-methylenebisacrylamide (Bis, 99%), ammonium persulfate (APS, 98%), polyethylene glycol (PEG 6000), N,N,N′,N′-Tetramethylethylenediamine (TEMED, 99%), 3-methacryloxypropyltrimethoxysilane (bind-silane), dimethyl sulfoxide (DMSO), trifluoroacetic acid (TFA), iodoacetamide (IAA), α-Cyano-4-hydroxycinnamic acid (CHCA, 99%), mouse serum standard, and bovine serum albumin (BSA) were from Sigma-Aldrich (St. Louis, MO). D/L-dithiothreitol (DTT), trypsin/Lys-C mix, yeast, and human lysate were from Promega (Madison, WI). N-acryloxysuccinimide (NAS) was purchased from Tokyo Chemical Industry (Tokyo, Japan). Lys-N was provided from Thermo Scientific (Rockford, IL). All water used in this study was doubly distilled on a Millipore filtration system (Bedford, MA). Fused-silica capillary with 200 μm i.d. and 360 μm o.d. was purchased from Polymicro Technologies (Phoenix, AZ). Millipore C18 Ziptip column was obtained from Millipore Company and used for sample cleaning.
2.2 Apparatus
An ultrafleXtreme MALDI-TOF/TOF from Bruker Daltonics and MALDI LTQ Orbitrap spectrometer from Thermo Scientific were utilized for IMER column optimization and performance evaluation of standard protein BSA. An UtrafleXtreme MALDI-TOF/TOF (Bruker Daltonics, Bremen, Germany) equipped with a 1000 Hz Smartbeam 2 laser was used for IMER column optimization. The mass spectra were acquired in a positive ion reflectron mode with ion source 1 voltage 25.0 kV, ion source 2 voltage 22.0 kV, reflector 1 voltage 26.6 kV, reflector 2 voltage 13.6 kV and lens voltage 6.0 kV. 500 laser shots were accumulated and all the mass spectra were recorded from m/z 500 to m/z 2500. External calibration was performed by using a standard peptide mixture provided by Bruker Daltonics, including angiotensin II ([M+H]+ 1046.54), angiotensin I ([M+H]+ 1296.68), substance P ([M+H]+ 1347.74), bombesin ([M+H]+ 1619.82), ACTH clip 1–17 ([M+H]+ 2093.09), ACTH clip 18–39 ([M+H]+ 2465.20) and somatostatin ([M+H]+ 3147.47). For evaluating digestion performance of standard protein BSA, the high resolution orbitrap detection was performed with a MALDI-LTQ-Orbitrap XL from Thermo Scientific (Waltham, MA). 10 scans were recorded for each spot with 2 micro-scans each step under the survey CPS (Crystal Positioning System) mode. Mass spectra in positive ion mode from m/z 400 to m/z 3000 were recorded at a resolution of 70000 at m/z 400. ProteoMass MALDI calibration kit from Sigma-Aldrich (St. Louis, MO) was used for instrument to achieve high mass accuracy of ±5 ppm.
A Waters nanoAcquity UPLC system (Milford, MA) coupled to a Thermo Q-Exactive hybrid quadrupole-Orbitrap mass spectrometer was employed for proteomic analysis of complex biological samples. A 75 μm × 100 mm BEH130 C18 column from Waters was used in LC separation using a gradient at 350 nL/min. Mobile phase A was composed of water and 0.1% formic acid. Mobile phase B was composed of ACN and 0.1% formic acid. Separation was performed by ramping solvent B from 3% to 10% over 5 min, then to 40% in the next 60 min, and finally to 95% during the following 10 min. MS spectra were acquired in positive ion mode over m/z 200–2000 at 70,000 resolution (m/z 200) with an AGC target of 1×106 and maximum injection time of 250 ms. Data dependent acquisition selected the top 10 most abundant precursor ions for tandem MS by HCD fragmentation using an isolation width of 2.0 Da, a normalized collision energy of 30, a resolution of 17,500, an AGC target of 2×105, a maximum injection time of 250 ms and a lower mass limit of m/z 100.
2.3 In Solution Protein Digestion
The in solution protein digestion was performed as a comparison to IMER digestion. Briefly, 30 μg of protein mixture was dissolved in 20 μL of 8 M urea (0.96 g urea in 2.0 mL of 25 mM ammonium bicarbonate solution). 1 μL of DTT (1M in 25 mM ammonium bicarbonate) was added to the tube followed by gentle vortex for reduction of disulfide bonds. After reduction at 37 °C for 1 h, 20 μL of IAA (200 mM in 25 mM ammonium bicarbonate) was added to the tube for alkylation for 1 h at room temperature in the dark. To consume residual alkylating reagent, 4 μL of DTT was added followed by 120 μL of 25 mM ammonium bicarbonate solution to dilute urea. In solution digestion was performed overnight at 37 °C after adding 1 μg of trypsin. The next morning, 1 μL of formic acid was added to quench the reaction along with gentle vortexing. The digested 30 μg of BSA was divided into six aliquots and stored at −80 °C. Before usage, one aliquot was dried and reconstituted in 10 μL of 0.1% TFA and desalted with a Ziptip C18 pipette tip to final volume of 17 μL.
2.4 NAS-Based Monolithic Column Fabrication and Ligand Immobilization
A 20 cm fused-silica capillary (200 μm i.d.) was flushed with 1M NaOH for 30 min, followed by water, 0.1M HCl for 30 min, water, and finally acetone. 50% (v/v) of bind-silane in acetone was flushed through the column for 40 min for silanization. The capillary was then rinsed with acetone and water, respectively. IMER column fabrication was derived from a previous report [27]. We modified and improved the polymerization method to reduce ligand consumption. Briefly, a mixture containing 20mg acrylamide, 30mg N, N’-methylenebisacrylamide and 30mg PEG in 1 mL of 0.2 M sodium bicarbonate/0.5 M sodium chloride buffer was heated at 55–60 °C for 15min. Then, 4 μL of 20% (v/v) TEMED was added to the above-mentioned solution and followed by nitrogen de-gassing. 5 μL N-Acryloxysuccinimide (NAS) (140 mg/mL dissolved in DMSO) was then added on top of the solution, and after 1min, 2 μL of 20% (w/v) ammonium persulfate (APS) was added to initiate polymerization. After 30s, 18 μL aliquot of this solution was quickly mixed with 2 μL of ligand solution (1 mg/mL enzyme containing 0.1 M benzamidine) and was pumped through the pre-treated capillary. Therefore, polymerization of IMER column was carried out in parallel with protease conjugation. The entire reaction can be completed within 30 min at room temperature. After reaction, the column was washed with 25 mM ammonium bicarbonate before sample loading.
2.5 On-Column Protein Digestion with IMER-MALDI-MS and IMER-LC-ESI-MS
Before IMER digestion, the protein sample (BSA, mouse serum, yeast lysate, or human lysate dissolved in 25 mM ammonium bicarbonate) was reduced with DTT and alkylated with IAA for 10 min, respectively. The IMER column was conditioned with 25 mM ammonium bicarbonate solution. 10 μL protein sample of 0.3 mg/mL was then loaded and flushed through the column at 1μL/min and collected to a vial from the outlet end of the column. The collected digested sample was then analyzed with either MALDI-MS or LC-ESI-MS without further processing.
3. RESULTS AND DISCUSSION
3.1 Optimization of Monolithic Column
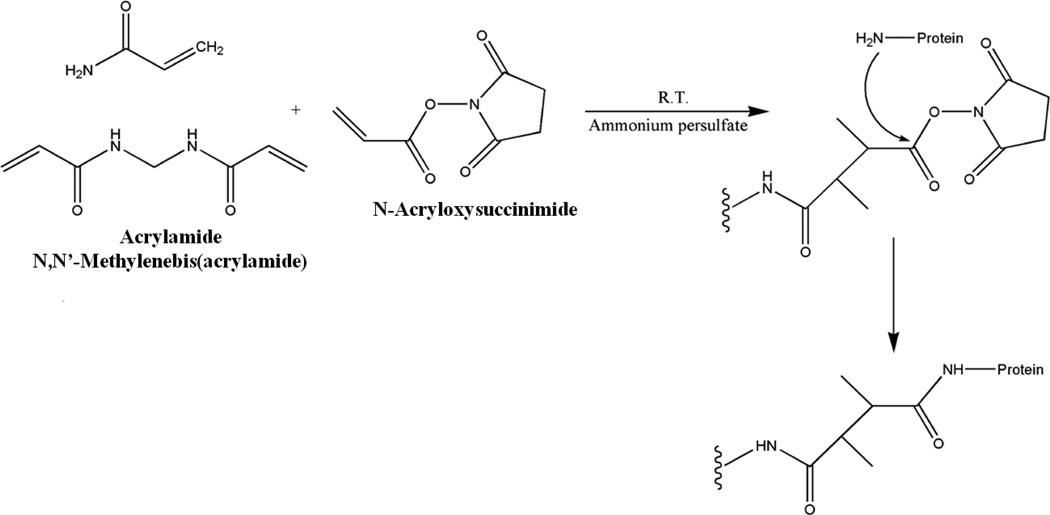
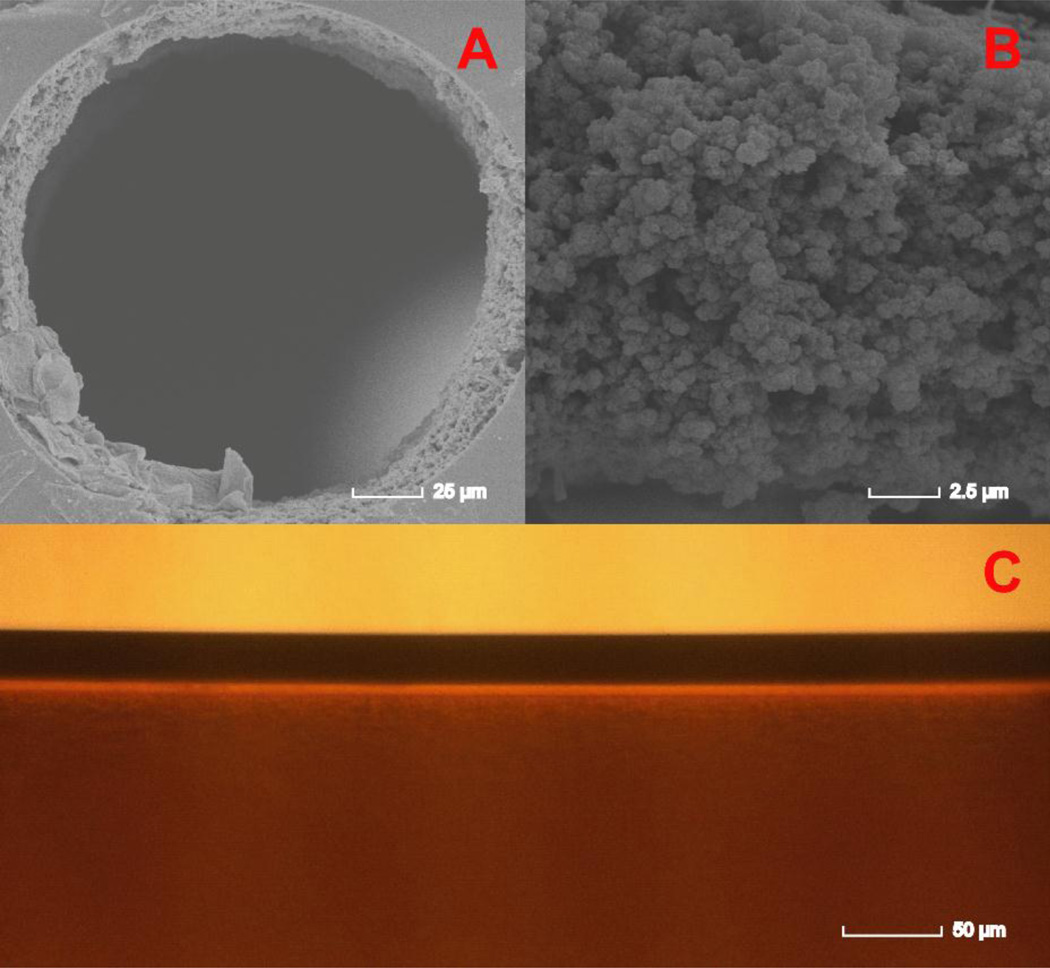
The condition for immobilizing ligands to NAS-AAm-Bis monolith is milder and follows a one-step reaction at room temperature. The electron pair on the N-terminus attacked the carbon of the carbonyl group causing the breaking of the carbon-oxygen bond which led to leaving of the succinimide group. The monomers, porogen, initiator and ligands were mixed in buffer and then filled into silanized capillary. The reactions including both polymerization and immobilization were conducted simultaneously at room temperature for 30 min, and no clean up procedure was needed except conditioning before usage (Figure 1). Compared with other types of monolithic columns, ligand consumption was much lower and the short reaction time without heating greatly preserves the bioactivity for enzymes. The microscopic images of the NAS-based monolithic column are shown in Figure 2. An open tubular column was fabricated with highly uniform NAS-based monolith attached to the silanized capillary inner wall (Figure 2A and 2C), and mass transfer occurred between analytes and the monolith through the pores (Figure 2B). Three different internal dimensions of capillary columns have been tested and each MS profile was summed by three scans. As shown in supplemental Figure S1, enzyme-reactor fabricated in 200 μm i.d. produced higher digestion efficiency, as it provided larger space for monolithic stationary phase and produced the highest available monolithic surface area with intermediate pore size.
Figure 1. Synthesis of NAS-AAm-Bis monolithic column with immobilized ligand.
NAS, AAm and Bis were mixed with PEG and initiator. The ligand immobilization was conducted simultaneously with polymerization at room temperature for 30 min.
Figure 2. Microscopic images of NAS-AAm-Bis monolith fabricated within 200 μm i.d. capillary.
A: A scanning electron microscopic (SEM) image showing the NAS-AAm-Bis monolith fabricated within a 200 μm silanized capillary. An open tubular column was formed with a uniform layer of monolith attached to the capillary inner wall. B: A zoom-in of the SEM image to show the detailed structure of NAS-based monolith. C: An optical microscopic image taken from outside of the column showing the capillary wall (upper) and monolith (lower).
3.2 Evaluation of IMER Digestion with BSA
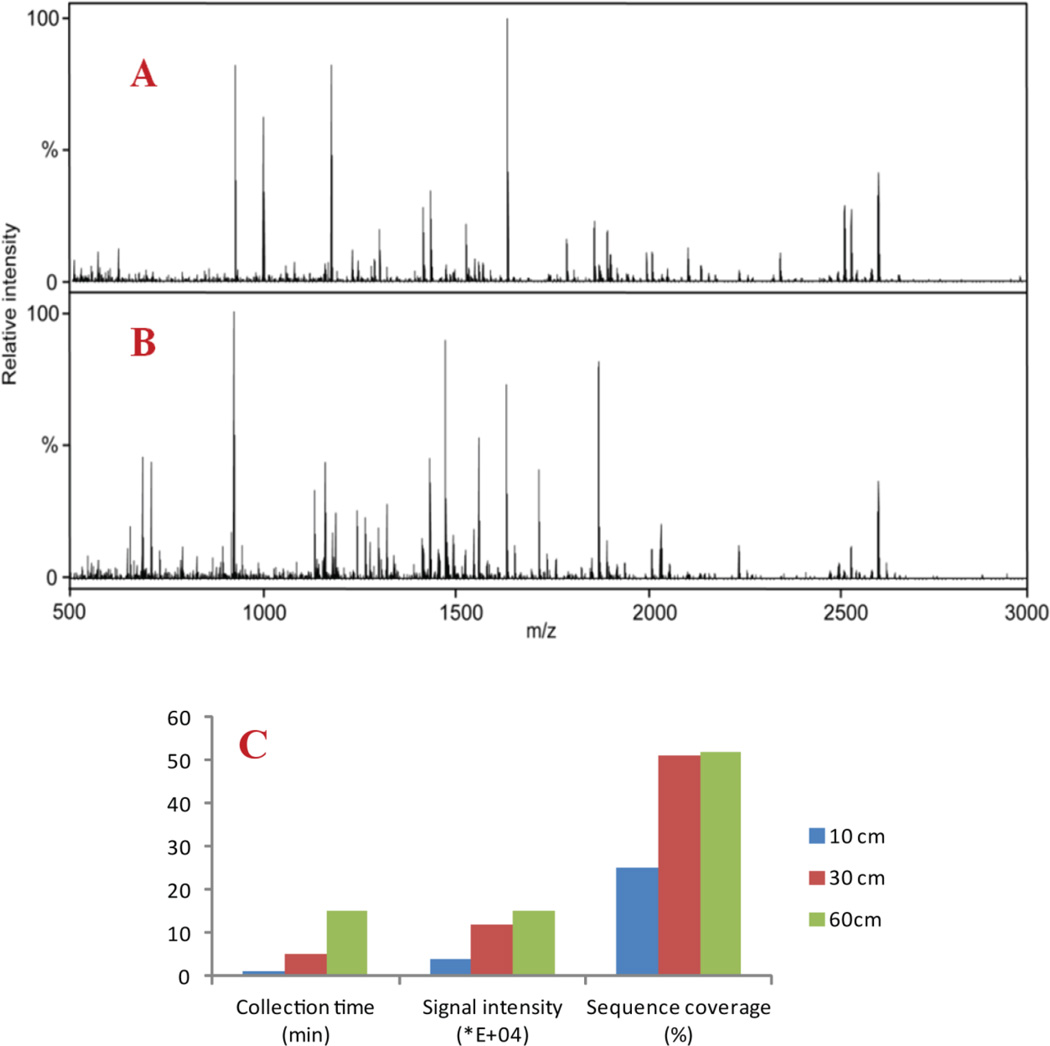
We immobilized trypsin onto the monolith as an IMER and compared its performance with in solution digestion via MALDI-MS detection. The same concentration (5 μM) of BSA was digested by in-solution method and IMER columns of different lengths, including 10 cm, 30 cm, and 60cm. A 60 cm column had higher backpressure and longer distance for flowing through which gave rise to increased collection time of digested peptides from IMER. Therefore, we balanced digestion time with peptides’ intensities and sequence coverage which demonstrated 30 cm to be most efficient. As outlined in Figure 3C, the 30 cm IMER column yielded signal intensity above 1×105 and a sequence coverage at 50% while keeping digestion time within 5 min to produce sufficient amount of peptides for MALDI MS detection. Figure 3A and 3B show the mass spectral comparison between in solution and IMER digestion with MALDI-LTQ-Orbitrap detection. MALDI MS spectra were averaged by 20 scans and full MS were subjected to Mascot fingerprint search. Carbamidomethyl (C) and oxidation (M) were set as variable modifications, peptide mass tolerance for precursor was set as 25 ppm, and one missed cleavage was allowed in database search. Similar protein coverage at 48–52% was observed for both methods while slightly higher MS signal intensity was obtained with IMER digestion. Compared to other IMERs, this one-step polymerized IMER features minimized preparation time of enzyme reactor with reduced proteases consumption, in the meantime retains good sequence coverage. Considering the significantly reduced digestion time (within 5 min for IMER vs. overnight for in solution digestion) and the capability for online coupling to other preparation/separation dimensions, IMER digestion provides a new avenue for fast and efficient proteomics sample preparation.
Figure 3. Mass spectral comparison between in solution digestion and IMER digestion with MALDI-MS detection.
A: Mass spectra generated from MALDI-LTQ-Orbitrap with BSA tryptic peptides via in solution digestion. B: Mass spectra generated from MALDI-LTQ-Orbitrap with BSA tryptic peptides via IMER digestion. C: Histogram showed optimized IMER condition of BSA digestion. The blue column represented that a 10 cm IMER yielded 25% sequence coverage of BSA and signal intensity of 4.00×104 [a.u.] over 1 min sample collection time. The red column represented that a 30 cm IMER yielded 51% sequence coverage of BSA and signal intensity of 1.20×105 [a.u.] over 5 min sample collection time. The green column represented that a 60 cm IMER yielded 52% sequence coverage of BSA and signal intensity of 1.50×105 [a.u.] over 15 min sample collection time.
3.3 IMER Digestion of Mouse Serum, Yeast Cell Lysate and Human Cell Lysate Samples
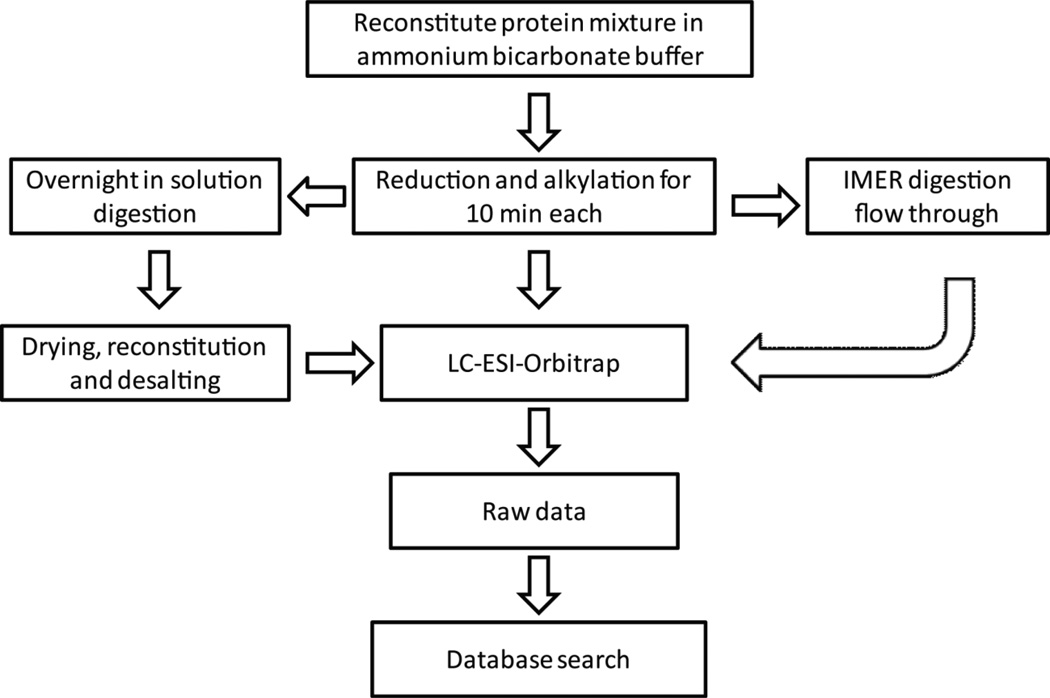
Compared to other previously published work on IMER columns, the NAS-AAm-Bis column features mild fabrication condition and an open tubular inner structure with higher protein sequence coverage as observed with BSA analysis [28,29]. We further applied IMER digestion to more complex samples followed by LC-ESI-Orbitrap detection to test if IMER digestion can be applied to large-scale proteomics studies. As outlined in Figure 4, the mouse serum, yeast and human cell lysate samples with final concentration of 0.3μg/μL were dissolved in ammonium bicarbonate buffer for reduction and alkylation for 10 min each, and then loaded onto a 30-cm IMER column for on-column digestion. The flow-through was collected without any further cleaning step. Alternatively, the protein mixtures were also digested by conventional in solution method overnight followed by drying, reconstitution, and desalting. The digested samples using both methods were loaded onto the LC column and analyzed with high resolution Q-Exactive Orbitrap mass analyzer, respectively.
Figure 4. Scheme of offline coupling IMER digestion and conventional in solution digestion to LC-MS for large-scale proteomics studies.
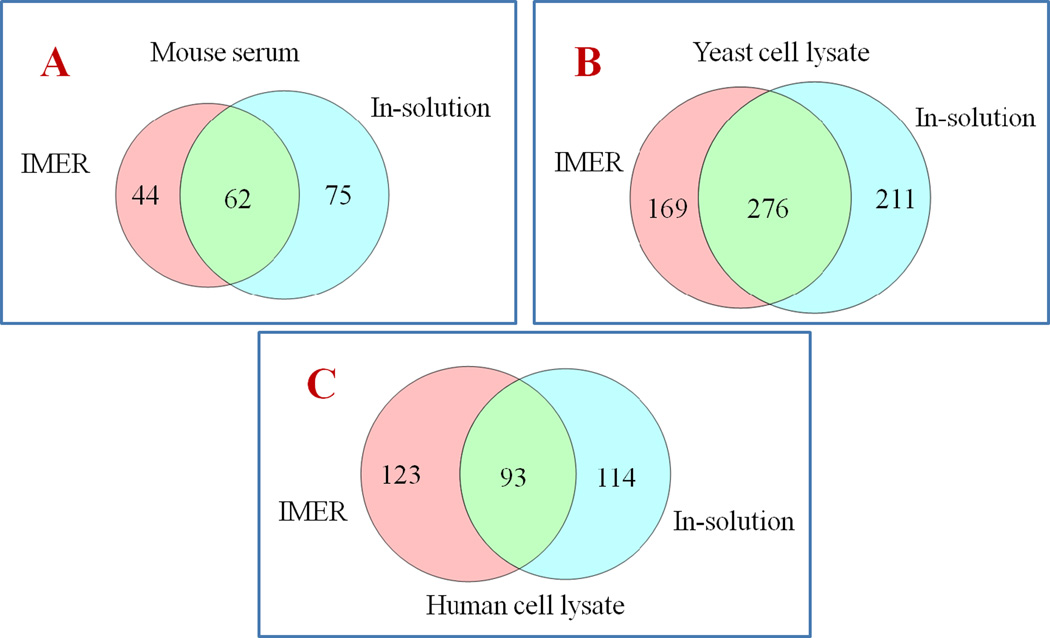
The comparison of LC chromatograms from IMER and conventional in-solution digestion is shown in Figure 5. Immobilized enzymes exhibited similar digestion efficiency as free enzymes. Peptides digested by IMER showed a clean chromatogram, indicating no extra clean-up step was needed and complex protein mixtures were completely digested compared to in solution digestion. Raw data files were processed through Mascot MS/MS ion search and Proteome Discoverer 1.4 (PD) using Sequest HT algorithm. By using Mascot MS/MS ion search for mouse serum, 106 proteins with significant scores (p<0.05) have been matched with IMER digestion, while 137 matches were observed from the digestion in solution plus sample cleaning. Data analysis for yeast and human cell lysates using PD also showed a comparable performance for IMER and in-solution digestion in terms of total identified protein numbers. With a precursor mass tolerance of 25 ppm and a fragment mass tolerance of 0.02 Da, identified peptides were additionally filtered with a false discovery rate (FDR) better than 1% as high confident peptides. As summarized in Table 1, 445 protein groups (with 5072 merged proteins and 12922 peptides) were identified in yeast lysate sample by IMER and 487 protein groups (with 5697 merged proteins and 10021 peptides) for in-solution method, while 216 protein groups (with 33846 merged proteins and 18052 peptides) were observed from human cell lysate samples by IMER and 207 protein groups (with 39010 merged proteins and 20696 peptides) by conventional method. As expected, the majority of the proteins identified are commonly shared while each digestion method also obtained unique protein IDs as shown in the Venn diagram in Figure 6, indicating its ability in improving complementary protein identifications as well as being an attractive alternative for global analysis of peptides and proteins from complicated biological matrix.
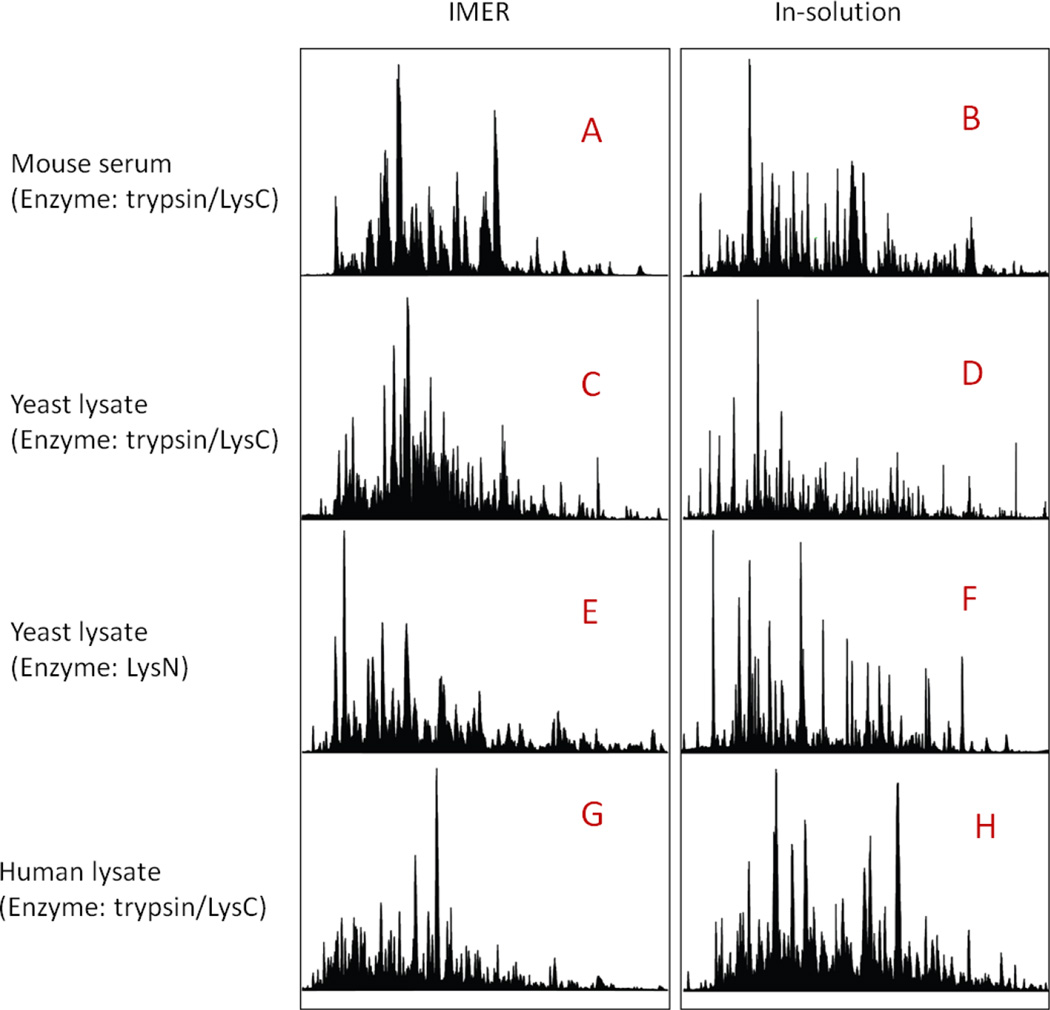
Figure 5. LC chromatograms of IMER and in-solution digestion prior to large-scale proteomics studies.
A, C, E and G: LC chromatograms for IMER digestion of mouse serum standard, yeast cell lysate (immobilized with trypsin/LysC and LysN) and human cell lysate, respectively. B, D, F, H: LC chromatograms of mouse serum standard, yeast and human cell lysate generated by traditional in-solution method.
Table 1.
Summary of the performance of IMER and in-solution digestion using complex protein mixture.a
| Protein mixture | Number of proteins groups identified with in-solution digestion |
Number of proteins groups identified with IMER |
Enzyme immobilized |
Database search engine |
|---|---|---|---|---|
| Mouse serum std | 137 | 106 | Trypsin/LysC | Mascot |
| Yeast lysate | 487 | 445 | Trypsin/LysC | PD |
| Yeast lysate | 473 | 368 | LysN | Mascot |
| Human lysate | 207 | 216 | Trypsin/LysC | PD |
IMER digestion time was 5 min at room temperature. LC-MS/MS analysis for in-solution and IMER digestion was identical and parameters were described in apparatus session with same protein loading amount of 300 ng.
Figure 6. Comparisons between proteins identified by IMER and in-solution digestion for different biological samples.
A: Comparison of the number of proteins detected by IMER and in-solution digestion of mouse serum standard. B: Comparison of the number of proteins detected by IMER and in-solution digestion of yeast cell lysate. C: Comparison of the number of proteins detected by IMER and in-solution digestion of human cell lysate.
3.4 Comprehensive Protein Identification Using Multi-IMERs
Unlike trypsin which cleaves at the carboxyl side of lysine and arginine, LysN protease cleaves at the amino-terminus of lysine residues. As a result, the peptides generated by LysN are longer than those generated by trypsin and have more prevalent charged amino terminal peptide fragments. Thus, the combination of using two types of enzymes greatly improved the peptide coverage and protein identification. A complementary identification of yeast lysate was performed using two types of IMER immobilized with trypsin/Lys-C mixture and Lys-N, respectively. The digested peptides were further injected onto LC-ESI-Orbitrap and the resulting data were processed through Mascot MS/MS ion search. As a result of complementary digestion, 559 proteins were matched in total, whereas 220 proteins were overlapped by both IMERs. In comparison to previous IMER studies, we demonstrated highly efficient one-step preparation of open-tubular IMER which accomplished polymerization and enzyme conjugation in 30 min. On-column digestion time was reduced to 5 min and acquired comparable sequence coverage without excessive amount of enzyme or immobilization time. Additionally, the combination of multiple proteases on IMER improves the analysis of different sub-groups of peptides, thus enhancing identifications for “shotgun” proteomics.
4. CONCLUSIONS
In this work, we report on the development of an NAS-AAm-Bis monolithic enzyme reactor and its offline coupling to MALDI-TOF/TOF and MALDI-LTQ-Orbitrap platforms for feasibility evaluation using protein standard, and ESI-Orbitrap for proteomics study. The results showed that monolithic support enables highly specific and effective enzymatic reactions between analytes and immobilized enzymes with improved reproducibility and throughput. In comparison with conventional protein digestion in solution, IMER exhibits similar protein and peptide coverage but with significantly reduced reaction time and sample loading amount. In addition, no extra steps for cleaning are needed, highlighting the great potential for online coupling to MS analysis. The present work demonstrated the advantages of applying monolithic affinity support to complex peptidomics and proteomics studies.
Supplementary Material
Highlights.
Highly efficient fabrication of NAS-based monolithic IMER column is developed
Complete protein digestion by IMER column in minutes with high sequence coverage
Fast IMER digestion offers complementary identification in large-scale proteomics
Acknowledgments
This work is supported by National Institutes of Health grants (1R01DK071801 and S10RR029531). The authors thank Thermo Pierce for providing LysN enzyme. We also thank the Biological and Biomaterials Preparation, Imaging and Characterization Facility at UW-Madison for taking the scanning electron microscope image.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cheng G, Chen P, Wang Z-G, Sui X-J, Zhang J-L, Ni J-Z. Immobilization of trypsin onto multifunctional meso-/macroporous core-shell microspheres: A new platform for rapid enzymatic digestion. Anal. Chim. Acta. 2014;812:65–73. doi: 10.1016/j.aca.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi H, Miyazaki M. Enzyme-immobilized reactors for rapid and efficient sample preparation in MS-based proteomic studies. Proteomics. 2013;13:457–466. doi: 10.1002/pmic.201200272. [DOI] [PubMed] [Google Scholar]
- 3.Switzar L, Giera M, Niessen WMA. Protein digestion: An overview of the available techniques and recent developments. J Proteome Res. 2013;12:1067–1077. doi: 10.1021/pr301201x. [DOI] [PubMed] [Google Scholar]
- 4.Feng S, Ye M, Jiang X, Jin RW, Zou H. Coupling the immobilized trypsin microreactor of monolithic capillary with muRPLC-MS/MS for shotgun proteome analysis. J. Proteome Res. 2006;5:422–428. doi: 10.1021/pr0502727. [DOI] [PubMed] [Google Scholar]
- 5.Boonen K, Landuyt B, Baggerman G, Husson SJ, Huybrechts J, Schoofs L. Peptidomics: The integrated approach of MS, hyphenated techniques and bioinformatics for neuropeptide analysis. J. Sep. Sci. 2008;31:427–445. doi: 10.1002/jssc.200700450. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi H, Miyazaki M, Kawazumi H, Maeda H. Multidigestion in continuous flow tandem protease-immobilized microreactors for proteomic analysis. Anal. Biochem. 2010;407:12–18. doi: 10.1016/j.ab.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, Wang X, Liu Y, Liu B, Wu H, Yang P. Trypsin entrapped in poly(diallyldimethylammonium chloride) silica sol-gel microreactor coupled to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2008;22:1257–1264. doi: 10.1002/rcm.3478. [DOI] [PubMed] [Google Scholar]
- 8.Duan J, Liang Z, Yang C, Zhang J, Zhang L, Zhang W, Zhang Y. Rapid protein identification using monolithic enzymatic microreactor and LC-ESI-MS/MS. Proteomics. 2006;6:412–419. doi: 10.1002/pmic.200500234. [DOI] [PubMed] [Google Scholar]
- 9.López-Ferrer D, Cañas B, Vázquez J, Lodeiro C, Rial-Otero R, Moura I, Capelo JL. Sample treatment for protein identification by mass spectrometry-based techniques, TrAC. Trends Anal. Chem. 2006;25:996–1005. [Google Scholar]
- 10.Capelo JL, Carreira R, Diniz M, Fernandes L, Galesio M, Lodeiro C, Santos HM, Vale G. Overview on modern approaches to speed up protein identification workflows relying on enzymatic cleavage and mass spectrometry-based techniques. Anal. Chim. Acta. 2009;650:151–159. doi: 10.1016/j.aca.2009.07.034. [DOI] [PubMed] [Google Scholar]
- 11.Shin S, Yang H-J, Kim J, Kim J. Effects of temperature on ultrasound-assisted tryptic protein digestion. Anal. Biochem. 2011;414:125–130. doi: 10.1016/j.ab.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 12.Vale G, Santos HM, Carreira RJ, Fonseca L, Miró M, Cerdà V, Reboiro-Jato M, Capelo JL. An assessment of the ultrasonic probe-based enhancement of protein cleavage with immobilized trypsin. Proteomics. 2011;11:3866–3876. doi: 10.1002/pmic.201100200. [DOI] [PubMed] [Google Scholar]
- 13.Pramanik BN, Mirza UA, Ing YH, Liu Y-H, Bartner PL, Weber PC, Bose AK. Microwave-enhanced enzyme reaction for protein mapping by mass spectrometry: a new approach to protein digestion in minutes. Protein Sci. 2009;11:2676–2687. doi: 10.1110/ps.0213702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Zhang L, Yang P, Chen G. Infrared-assisted tryptic proteolysis for peptide mapping. Proteomics. 2008;8:2579–2582. doi: 10.1002/pmic.200800086. [DOI] [PubMed] [Google Scholar]
- 15.Li F, Schmerberg CM, Ji QC. Accelerated tryptic digestion of proteins in plasma for absolute quantitation using a protein internal standard by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2009;23:729–732. doi: 10.1002/rcm.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girelli AM, Mattei E. Application of immobilized enzyme reactor in on-line high performance liquid chromatography: A review. J. Chromatogr. B. 2005;819:3–16. doi: 10.1016/j.jchromb.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 17.Massolini G, Calleri E. Immobilized trypsin systems coupled on-line to separation methods: Recent developments and analytical applications. J. Sep. Sci. 2005;28:7–21. doi: 10.1002/jssc.200401941. [DOI] [PubMed] [Google Scholar]
- 18.Long Y, Wood TD. Immobilized pepsin microreactor for rapid peptide mapping with nanoelectrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2014;26:194–197. doi: 10.1007/s13361-014-1015-8. [DOI] [PubMed] [Google Scholar]
- 19.Svec F. Organic polymer monoliths as stationary phases for capillary HPLC. J. Sep. Sci. 2004;27:1419–1430. doi: 10.1002/jssc.200401825. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, He B, Yu A, Zhou W. Synthesis and antibacterial activity of substituted oxotriazolylphenyl oxazolidinones. Chem. Bio. Drug. Des. 2007;70:65–69. doi: 10.1111/j.1747-0285.2007.00531.x. [DOI] [PubMed] [Google Scholar]
- 21.Altun Z, Hjelmström A, Abdel-Rehim M, Blomberg LG. Surface modified polypropylene pipette tips packed with a monolithic plug of adsorbent for high-throughput sample preparation. J. Sep. Sci. 2007;30:1964–1972. doi: 10.1002/jssc.200600523. [DOI] [PubMed] [Google Scholar]
- 22.Barut M, Podgornik A, Brne P, Štrancar A. Convective interaction media short monolithic columns: enabling chromatographic supports for the separation and purification of large biomolecules. J. Sep. Sci. 2005;28:1876–1892. doi: 10.1002/jssc.200500246. [DOI] [PubMed] [Google Scholar]
- 23.Safdar M, Spross J, Jänis J. Microscale immobilized enzyme reactors in proteomics: lastest developments. J. Chromatogr. A. 2014;1324:1–10. doi: 10.1016/j.chroma.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 24.Connolly D, Currivan S, Paull B. Polymric monolithic materials modified with nanoparticles for separation and detection of biomolecules: a review. Proteomics. 2012;12:2904–2917. doi: 10.1002/pmic.201200142. [DOI] [PubMed] [Google Scholar]
- 25.Walsh Z, Paull B, Macka M. Inorganic monoliths in separation science: a review. Anal. Chim. Acta. 2012;750:28–47. doi: 10.1016/j.aca.2012.04.029. [DOI] [PubMed] [Google Scholar]
- 26.Krenkova J, Svec F. Less common applications of monoliths: IV. Recent developments in immobilized enzyme reactors for proteomics and biotechnology. J. Sep. Sci. 2009;32:706–718. doi: 10.1002/jssc.200800641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palm AK, Novotny MV. Analytical characterization of a facile porous polymer monolithic trypsin microreactor enabling peptide mass mapping using mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:1374–1382. doi: 10.1002/rcm.1500. [DOI] [PubMed] [Google Scholar]
- 28.Liang Y, Tao DY, Ma JF, Sun LL, Liang Z, Zhang LH, Zhang YK. Hydrophilic monolith based immobilized enzyme reactors in capillary and on microchip for high-throughput proteomic analysis. J. Chromatogr. A. 2011;1218:2898–2905. doi: 10.1016/j.chroma.2011.02.073. [DOI] [PubMed] [Google Scholar]
- 29.Celebi B, Bayraktar A, Tuncel A. Synthesis of a monolithic, micro-immobilised enzyme reactor via click-chemistry. Anal. Bioanal. Chem. 2012;403:2655–2663. doi: 10.1007/s00216-012-6075-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.