Abstract
Background
Since aortic diameter is the most significant risk factor for rupture, we sought to identify stress-dependent changes in gene expression to illuminate novel molecular processes in aneurysm rupture.
Materials and Methods
We constructed finite element maps of abdominal computerized tomography scans (CTs) of seven abdominal aortic aneurysm (AAA) patients to map wall stress. Paired biopsies from high- and low-stress areas were collected at surgery using vascular landmarks as coordinates. Differential gene expression was evaluated by Illumina Array analysis, using the whole genome DNA-mediated, annealing, selection, extension, and ligation (DASL) gene chip (n = 3 paired samples).
Results
The sole significant candidate from this analysis, Lamin A/C, was validated at the protein level, using western blotting. Lamin A/C expression in the inferior mesenteric vein (IMV) of AAA patients was compared to a control group and in aortic smooth muscle cells in culture in response to physiological pulsatile stretch. Areas of high wall stress (n = 7) correlate to those regions which have the thinnest walls [778 µm (585–1120 µm)] in comparison to areas of lowest wall stress [1620 µm (962–2919 µm)]. Induced expression of Lamin A/C correlated with areas of high wall stress from AAAs but was not significantly induced in the IMV from AAA patients compared to controls (n = 16). Stress-induced expression of Lamin A/C was mimicked by exposing aortic smooth muscle cells to prolonged pulsatile stretch.
Conclusion
Lamin A/C protein is specifically increased in areas of high wall stress in AAA from patients, but is not increased on other vascular beds of aneurysm patients, suggesting that its elevation may be a compensatory response to the pathobiology leading to aneurysms.
Keywords: Nuclear lamina, Stretch, Smooth muscle cells, Aging
Introduction
Abdominal aortic aneurysm (AAA) is a chronic degenerative vascular disease affecting 5–6% of males and 1–2% of females over 65 years of age [1]. Aneurysmal lesions commonly manifest as focal dilatations of the aorta, increasing in diameter over time, ultimately resulting in aortic rupture, when the strength of the vessel wall fails to withstand blood pressure. What we currently know about the pathological development of AAA is derived from analysis of end-stage human biopsies [2–5], consideration of risk factors [6], genome-wide association studies, and investigating the mechanisms involved in various experimental models of AAA [7].
Initiation is likely to begin with an inflammatory response resulting in activation of matrix-destabilizing metalloproteinases, leading to elastin degradation and vascular remodeling [8–10]. Further expansion is accompanied by subsequent development of luminal thrombus resulting in mural ischemia leading to the production of factors known to induce neovascularization and thus, exacerbation of the inflammatory process [11]. Rarefaction of medial smooth muscle cells (SMC), the result of combined loss from induction of apoptotic cell death and reduction of proliferative expansion, contributes to the production of a cell-poor, matrix-rich vascular lesion that is noncompliant and structurally compromised. Homeostatic mechanisms of repair counter the disease progression, and an increase in the numbers of circulating endothelial progenitor cells has been documented. However, the reparative capacity of such cells is questionable.
Aside from the relationship between maximal aneurismal diameter and risk of rupture, we have few facts that inform our knowledge of the precise mechanism of rupture, currently the 13th most common cause of sudden death in the world [12]. In previously published work, we identified the differential expression and pathological features of aneurysm rupture, by comparing tissue biopsies taken proximal to the rupture site to biopsies collected distal to the rupture. Our data suggested that medial neovascularization preceded rupture and that this local increase in small vessels potentially weakened the vessel wall, leading to rupture [13]. To gain a further understanding of the biological changes occurring in the vessel wall prior to rupture, this study aims to identify stress–dependent modulation in gene expression.
Methods
Patients
The collection of paired AAA biopsies were carried out prospectively and conducted according to the principles of the declaration of Helsinki and approved by the local ethics committee (ref 09/H0803/0011). Patients scheduled for elective open aneurysm repair were recruited and biopsies collected between January 2008 and December 2009. Patients undergoing emergency surgery and those suitable for endovascular repair were excluded. In addition, patients who had had previous surgery or endovascular repair, patients with inflammatory aneurysms and additional connective tissue disorders were also excluded. Inferior mesenteric vein (IMV) samples were collected between December 2008 and January 2010. A segment of IMV was obtained from 16 patients with AAA (> 5 cm in diameter) at the time of open AAA repair. Control samples were obtained from patients undergoing left hemi-colectomy for benign colonic pathology (confirmed by histology). The site of IMV harvesting and specimen size was consistent across both groups. AAA patients were excluded if they had inflammatory AAA, known malignancy, active colitis, or had a previous hemi-colectomy. Control patients were excluded if they had active inflammatory processes, were on current steroid therapy, prior chemotherapy, or an AAA visible on preoperative imaging. Written informed consent was obtained from all patients and consent forms received local ethics committee approval (ref 08/H0803/210). IMV samples were obtained from the colonic mesentery, irrigated with saline, and stripped of accessory connective tissue. Samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until protein analysis.
Biopsy Localization and Sample Harvest
Full thickness biopsies were obtained during elective open repair. Coordinates of regions of highest and lowest wall stress were identified on the Finite Element Analysis (FEA) model by measuring the distance and clock position on the finite element mesh from the proximal starting point (the level of the first CT slice imported into the FEA software). Measurement was performed by using the distance measurement tool in the FEA software interface. A detailed, 3D reconstruction image of the aorta including the major visceral branches was performed (3mensio 3D imaging workstation, 3mensio Medical Imaging B.V., Bilthoven, The Netherlands). The coordinates of regions of interest were localized according to the distance from adjacent visceral branches as determined from the FEA software. Intraoperatively, high and low wall stress regions were marked (according to the above distance) on the aneurysm surface before aortic clamping. Full thickness biopsies were obtained from the marked regions once the sac was opened. The samples were washed in saline to remove any attached intraluminal thrombus (ILT). Mechanical scrapping of ILT was avoided and not necessary as washing was sufficient to remove adjacent ILT as confirmed by microscopic examination. The proximity of areas of high wall stress in relation to maximal aortic diameter (MAD) was assessed visually by inspecting wall stress maps and corresponding computed tomography images.
Wall Thickness Measurements
All samples were measured in saline at room temperature immediately after biopsy. The samples were washed gently to remove excess ILT with no mechanical scraping. Wall thickness was measured by computer assisted micrometry using the AxioVs40 micro telescope system (Carl Zeiss Imaging Solutions, Germany) at 1-mm intervals to allow for aneurysm wall heterogeneity and a mean value for each specimen was obtained.
Finite Element Analysis
FEA is a computational modelling technique used by the engineering industry to simulate wall stress distribution in complex structures. This method has recently been used to predict the risk of rupture in individual AAAs in experimental studies. FEA takes into account detailed aneurysm morphology, hemodynamic forces, and mechanical properties of the AAA to determine the mechanical stress distribution. A detailed 3D model of each AAA is constructed from preoperative computed tomography (CT) imaging to provide a wall stress map that is unique for each patient [14]. Since arterial walls exhibit nonlinear behavior and undergo large strains, a finite strain constitutive equation based on the model derived by Raghavan and Vorp [15] specifically suited for AAA wall was used. Multi-component wall models were built to include the aortic wall and ILT. Appropriate material properties for each component were used. Finite element analyses were performed using ADINA (ADINA R&D Inc., Watertown, Massachusetts, USA) to determine patterns and values of wall stress within the affected aorta.
Gene Array Analysis
Tissue biopsies for RNA isolation were transported from the theatre in RNA-stabilizing solution (Ambion, Inc., UK) and processed immediately for total RNA isolation using an RNeasy Mini Fibrous Kit (Qiagen, UK) according to the manufacturer’s recommendations. Briefly, 50 mg of tissue is ground to a fine powder in liquid nitrogen, and the powder then homogenized with lysis buffer using a rotor homogenizer (IKA, Werke GmbH, Germany). The concentration and purity of each sample (ng/µl) was determined by NanoDrop™ ND-3300 (Thermo Scientific, UK) and Agilent BioAnalyser, Agilent Technologies, UK), respectively.
The gene expression of these samples were carried out at The Genome Centre, Queen Mary, University of London using the whole-genome complementary DNA-mediated, annealing, selection, extension, and ligation (DASL) array Human Ref-8 vs. 3.0 Bead Chips (Illumina, San Diego, CA, USA). This chip includes 24,526 probes targeting 18,401 genes. In brief, total RNA was converted into cDNA using the biotinylated random nonamers, oligo-deoxythymidine 18 primers, and Illumina-supplied reagents, according to the manufacturer’s protocol. The biotinylated cDNA was then annealed to assay oligonucleotides and bound to streptavidin-conjugated paramagnetic materials to select RNA at 55°C for 18 hours to the array, followed by washing and staining with streptavidin and conjugation to cyanin 3. Post-hybridization, the chips were scanned using the BeadArray™ Reader system (Illumina, UK). Gene expression data were normalized using quantile normalization and log-transformed. Statistical analysis and hierarchical clustering were performed using the Genespring GX 9 (Agilent Technologies, UK).
Quantitative Real-Time Polymerase Chain Reaction (QRT-PCR)
LMNA expression was validated by QRT-PCR using TaqMan™ Gene expression Assays on Demand (Applied Biosystems, UK) and the Mx4000 Multiplex Quantitative PCR System (Stratagene, UK). The mRNA levels of LMNA was determined by Quantitative Real-Time (QRT)-PCR using TaqMan probes (Assays-on-Demand™ Gene Expression Products, Applied Biosystems, UK), with the carboxyfluorescein fluorescent dye 6-FAM as the 5’-fluorophore and nonfluorescent quencher (NFQ) at the 3’-end of the probe. QRT-PCR reaction mixtures for each sample were 50 µl, containing 25 µl of 2× TaqMan Universal Master Mix (without AmpErase® UNG), 22.5 µl cDNA template, and 2.5 µl of 20× target assay mix. Each sample was run in triplicate and for all reactions, negative controls were run with no template present. In addition, total RNA from patient samples was used for sham reverse transcription reactions with no reverse transcriptase present and then subjected to standard PCR using 18S rRNA primers to verify that no amplification was produced. QRT-PCR was carried out using an Mx4000 Multiplex Quantitative PCR System. The PCR cycle started with an initial 10 min denaturation step at 95°C, followed by 40 cycles of shuttle heating at 95°C for 15 s and 60°C for 1 min. The associated Mx4000 software was used to analyze the data and determine the threshold count (Ct). Ct was determined for the target genes and 18S rRNA. 18S rRNA was chosen as the endogenous control to which we normalized our specimens. Preliminary validation experiments verified that the efficiencies of target gene amplification and the efficiency of the mean value of transferrin receptor and TATA box-binding protein rRNA amplification to be approximately equal; therefore, we validated that the target gene:mean endogenous control rRNA ratio could be calculated using the ΔCt method. For each sample, Cttarget gene and Ctmean endogenous control rRNA were determined, and ΔCt = Cttarget gene – Ctmean endogenous rRNA. The relative level of the target gene normalized to transferrin receptor and TATA box-binding protein was determined by calculating 2-∆Ct.
Western Blotting
Total cell lysates (30 µg protein) were separated on an SDS-PAGE (10–12% running gel, 4% stacking gel. Bio-Rad, Herts, UK). Protein was then transferred to PVDF membrane (Millipore, Watford, UK). Blocking was performed overnight at 4°C in Tris-buffered saline (TBS) plus 5% non-fat dry milk. Blots were incubated with primary antibodies as follows: Lamin A/C (Cell Signaling, clone 4C11 at 1:1000) and Lamin A (Santa Cruz Biotech, clone C20 at 1:500) overnight at 4°C in TBS with 0.05% Tween-20 and 5% non-fat dried milk followed by incubation with HRP-conjugated secondary antibody (1:2000) (New England Biolabs, Herts, UK) for 45 minutes. Immunodetection was accomplished using chemiluminescence (Super Signal-HRP, Pierce Chemical Corp., Chester, UK). Detected bands were scanned on a calibrated densitometer (GS-800, Bio-Rad, Herts, UK) and quantified using Quantity One image analysis software (Bio-Rad). The densitometry of each band was normalized to the abundance of β-actin (ab8227, Abcam, Cambridge, UK) or β-tubulin (ab21057, Abcam, Cambridge, UK) staining using Image J (NIH) software.
Stretch Experiments with Human Aortic Smooth Muscle Cells
Early passages (2–3) human aortic smooth muscle cells (AoSMC) (PromoCell, Heidelberg Germany) were plated in completed growth medium (PromoCell, Heidelberg, Germany) on six-well Bioflex plates coated with collagen Type I (Flexcell International Corporation, NC, USA). Confluent AoSMC were then subjected to no stretch or cyclic sine wave stretch stress using stretch equipment [Flexcell FX-5000T Compression System (Dunn Labortechnik, Ashbach, Germany)] at 37°C and 5% CO2 for different periods of time up to 20 h. Following stretch stress, cells were allowed to relax for 20 h, and soluble proteins were isolated at times (as indicated). Lamin A/C and Prelamin protein expression was measured by western blotting using antibodies of interest. Actin was used as a loading control. Data are the mean ± SD (n = 3 independent experiments at each data point, p* < 0.05).
Statistical Analysis
Data are presented as mean ± SD, unless otherwise stated. Analysis of western blot data was by one-way ANOVA followed by a Bonferronni’s post-hoc test for multiple comparisons. The Benjamini-Hochberg post-hoc test for multiple measures was applied to the differential gene expression data following normalization. Wall thickness was recorded as median and interquartile ranges. Correlation between variables was performed using Spearman’s correlation. Significance was identified as a p value of less than 0.05. Statistical analysis was performed using GraphPad Prism, version 4.0.
Results
Regions of High Stress Correlate with Thinning of the Aortic Wall
Patient-specific wall stress computation was performed on ten patients. It was not possible to obtain paired biopsies from three patients (posterior location in one, near proximal graft anastamosis in two). Seven paired samples were obtained (five males and two females) from regions of highest and lowest wall stress (Figure 1). The seven patients used in this study were all prescribed HMG CoA reductase inhibitor drugs and anti-hypertensive treatments. They were all well nourished, but none were clinically obese. Measurement of seven paired biopsies showed that regions of high wall stress had an approximately two-fold reduction in full wall thickness. The seven patients biopsied had a mean age of 66 (60–82 years) and a median maximal aortic diameter of 60 (51–70 mm).
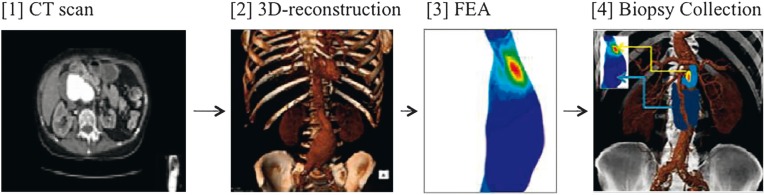
Figure 1.
Schematic diagram illustrating experimental design. CT scans of participating patients (1) were converted into 3D reconstructions (2) and the Diacom™ files analyzed by FEA (3) to provide a three dimensional stress distribution of the AAA. Red color corresponds to highest wall stress whilst blue for lowest wall stress. The FEA were then overlaid onto the 3D CT in the theatre, and the biopsies harvested (4). Of 11 patients recruited, seven paired biopsies were harvested for analysis. In four patients one or other of the biopsy sites were not in an accessible location. AAA = abdominal aortic aneurysm; FEA = finite element analysis.
We found that the median and interquartile range (IQR) for wall thickness from areas of low wall stress were 1620 (962–2912 µm) thick, compared to 778 (585–1120 µm) in regions of high wall stress. No obviously thickened atheromatous plaques were encountered during measurement. No correlation was found between wall thickness (in both high and low stress regions) and maximal aortic diameter (r = 0.2, r = 0.6), respectively.
Gene Array Analysis Identifies Lamin A/C as the Most Significant Stress-Dependent Candidate Gene
Three of the seven paired biopsies provided RNA isolates of sufficient quality (RIN >9) to proceed to gene array analysis. A total of 125 genes (71 upregulated and 54 downregulated) had a greater than 2-fold difference of expression (p < 0.05) in the high wall stress biopsy compared to the low wall-stress biopsy (see Supplementary Table 1). Supplementary Table 2 shows the gene ontology processes associated with the genes identified. However, when the Benjamini- Hochberg multiple testing correction was applied to these data, only Lamin A/C was the sole candidate which was differentially expressed (2.1-fold change, n = 3, p = 0.02). Differential gene expression was validated using QRT-PCR using the transferrin receptor and TATA-box binding protein as reference genes (2.83-fold change, n = 3, p < 0.05).
Supplementary Table 1.
Upregulated genes at regions exposed to high stress (2-fold change threshold).
| Fold change | Symbol | Definition / (Chromosomal Location) | p value |
|---|---|---|---|
| 2.07 | TRUB1 | TruB pseudouridine (psi) synthase homolog 1 (10) | 0.009 |
| 2.11 | MRC2 | Mannose receptor, C Type 2 (17) | 0.038 |
| 2.26 | CA8 | Carbonic anhydrase VIII (8) | 0.016 |
| 2.22 | SORBS3 | Sorbin and SH3 domain-containing 3 (8) | 0.016 |
| 2.24 | AFAP | Actin filament associated protein (4) | 0.034 |
| 2.29 | HSPG2 | Heparan sulfate proteoglycan 2 (perlecan) (1) | 0.029 |
| 2.81 | SOCS5 | Suppressor of cytokine signaling 5 (2) | 0.011 |
| 2.05 | FLJ45337 | FLJ45337 protein (1) | 0.042 |
| 4.61 | TRIM36 | Tripartite motif-containing 36 (5) | 0.017 |
| 2.43 | LUZP2 | Leucine zipper protein 2 (11) | 0.004 |
| 3.54 | GNAO1 | Guanine nucleotide binding protein alpha activating activity O (16) | 0.017 |
| 2.38 | CDKL2 | Cyclin-dependent kinase-like 2 (CDC2-related kinase)(4) | 0.013 |
| 2.06 | HEPH | Hephaestin (X) | 0.015 |
| 2.34 | PODXL2 | Podocalyxin-like 2 (3) | 0.028 |
| 2.26 | LOC654119 | Similar to hypothetical protein FLJ10159 | 0.049 |
| 2.09 | ADD3 | Adducing 3 (gamma) (10) | 0.015 |
| 2.76 | DKK3 | Dickkopf homolog 3 (Xenopus laevis) (11) | 0.016 |
| 2.16 | SBDSP | Shwachman-Bodian-Diamond syndrome pseudogene (7) | 0.006 |
| 2.41 | MXRA8 | Matrix-remodeling associated 8 (1) | 0.035 |
| 2.03 | PPP2R5D | Protein phosphatase 2, regulatory subunit B’, delta isoform (6) | 0.021 |
| 2.27 | ISYNA1 | Myo-inositol 1-phosphatase synthase A1 (19) | 0.038 |
| 2.11 | CDC42EP5 | CDC42 effector protein (Rho GTPase binding) 5 (19) | 0.040 |
| 2.10 | PTPRS | Protein tyrosine phosphatase, receptor type, S (19) | 0.031 |
| 2.01 | PDE5A | Phosphodiesterase 5A, cGMP-specific (4) | 0.032 |
| 2.35 | CKLFSF4 | Chemokine-like factor superfamily 4 (16) | 0.047 |
| 2.86 | VGCNL1 | Voltage gated channel like 1 (13) | 0.015 |
| 2.09 | A2ML1 | Alpha-2-macroglobulin-like 1 (12) | 0.043 |
| 2.06 | CTBP2 | C-terminal binding protein 2 (10) | 0.044 |
| 3.18 | IL17B | Interleukin 17B (5) | 0.018 |
| 3.88 | PCDHB11 | Protocadherin beta 11 (5) | 0.017 |
| 2.78 | ISLR | Immunoglobulin superfamily containing leucine rich repeat (5) | 0.007 |
| 2.11 | MFAP2 | Microfibrillar-associated protein 2 (1) | 0.038 |
| 2.34 | ELN | Elastin (7) | 0.049 |
| 3.36 | RPRM | Reprimo, TP53 dependent G2 arrest mediator candidate (2) | 0.022 |
| 2.34 | PTGER3 | Prostaglandin E receptor 3 (1) | 0.008 |
| 2.06 | NUDT10 | Nudix (nucleoside diphosphate linked moiety X)-type motif 10 (X) | 0.030 |
| 2.26 | DNAJC4 | DnaJ (Hsp40) homolog, subfamily C, member 4 (11) | 0.020 |
| 2.40 | CD151 | CD151 antigen (11) | 0.024 |
| 2.10 | FBLN1 | Fibulin 1 (FBLN1), transcript variant C (22) | 0.028 |
| 2.28 | UNQ1940 | HWKM1940 (UNQ1940) (7) | 0.035 |
| 2.41 | ARHGAP6 | Rho GTPase activating protein 6 (X) | 0.019 |
| 2.54 | KRT7 | Keratin 7 (12) | 0.019 |
| 2.33 | TRIM36 | Tripartite motif-containing 36 (5) | 0048 |
| 2.40 | GLIS2 | GLIS family zinc finger 2 (16) | 0.037 |
| 2.00 | PABPC5 | Poly(A) binding protein, cytoplasmic 5 (X) | 0.028 |
| 2.23 | CNTN4 | Contactin 4 (3) | 0.015 |
| 2.13 | CYP11A1 | Cytochrome P450, family 11, subfamily A, polypeptide 1 (15) | 0.038 |
| 2.21 | KCNMA1 | Potassium large conductance calcium-activated channel, subfamily MA1 (10) | 0.047 |
| 2.15 | RGMB | RGM domain family, member B (5) | 0.037 |
| 2.06 | ZNF704 | Zinc finger protein 704 (8) | 0.043 |
| 2.38 | RND2 | Rho family GTPase 2 (17) | 0.025 |
| 2.42 | ADAMTS2 | ADAM metallopeptidase with thrombospodin Type 1 motif 2 (5) | 0.006 |
| 2.55 | THSD4 | Thrombospondin, Type I, domain containing 4 (15) | 0.027 |
| 2.91 | ANKS1B | Ankyrin repeat and sterile alpha motif domain containing 1B (12) | 0.012 |
| 2.43 | LOC653762 | PREDICTED: similar to B-cell receptor-associated protein 29 (7) | 0.007 |
| 2.01 | INPP5F | Inositol polyphosphate-5-phosphatase F (10) | 0.029 |
| 2.86 | FBXO32 | F-box protein 32 (8) | 0.009 |
| 2.59 | MAP6 | Microtubule-associated protein 6 (11) | 0.033 |
| 2.26 | PGM5P2 | PREDICTED: phosphoglucomutase 5 pseudogene 2 (9) | 0.046 |
| 2.58 | MGC33486 | PREDICTED: hypothetical protein MGC33486 | 0.030 |
| 2.54 | PDE5A | Phosphodiesterase 5A, cGMP-specific (4) | 0.045 |
| 2.85 | MAP1B | Microtubule-associated protein 1B (5) | 0.036 |
| 2.03 | TSC22D3 | TSC22 domain family, member 3 (X) | 0.012 |
| 2.01 | SCARF2 | Scavenger receptor class F, member 2 (22) | 0.048 |
| 3.85 | RTN1 | Reticulon 1 (RTN1), transcript variant 3 (14) | 0.038 |
| 2.41 | CILP | Cartilage intermediate layer protein, nucleotide pyrophosphohydrolase (15) | 0.031 |
| 2.34 | FGF1 | Fibroblast growth factor 1 (acidic) (5) | 0.049 |
| 2.14 | KIAA0773 | KIAA0773 gene product(7) | 0.017 |
| 2.22 | LMO3 | LIM domain only 3 (rhombotin-like 2) (12) | 0.049 |
| 2.85 | LMNA | Lamin A/C (1) | 0.0003 |
Highly significant genes appear in bold.
Supplementary Table 2.
Downregulated genes at regions exposed to high stress (2-fold change threshold).
| Fold change | Symbol | Definition / (Chromosomal Location) | p value |
|---|---|---|---|
| 2.11 | SIGLEC5 | Sialic acid binding Ig-like lectin 5 (19) | 0.042 |
| 2.30 | UBASH3A | Ubiquitin associated and SH3 domain-containing, A (21) | 0.049 |
| 4.13 | FCAMR | Fc receptor, IgA, IgM, high affinity (1) | 0.046 |
| 2.18 | LOC349236 | Chromosome 9 open reading frame 164 (9) | 0.004 |
| 2.11 | CYP3A43 | Cytochrome P450, family 3, subfamily A, polypeptide 43 (7) | 0.001 |
| 2.08 | TRIM69 | Tripartite motif-containing 69 (15) | 0.037 |
| 2.03 | CD80 | CD80 antigen (CD28 antigen ligand 1, B7-1 antigen) (3) | 0.009 |
| 4.04 | CHI3L2 | Chitinase 3-like 2 (1) | 0.034 |
| 2.39 | OR5B21 | Olfactory receptor, family 5, subfamily B, member 21 (11) | 0.005 |
| 2.24 | MARCO | Macrophage receptor with collagenous structure (2) | 0.023 |
| 2.76 | LOC158830 | Similar to Ab2-183 (X) | 0.041 |
| 2.25 | HGD | Homogentisate 1,2-dioxygenase (homogentisate oxidase) (3) | 0.021 |
| 2.14 | TTN | Titin (2) | 0.009 |
| 2.42 | DPPA4 | Developmental pluripotency associated 4 (3) | 0.044 |
| 2.11 | TAP2 | Transporter 2, ATP-binding cassette, sub-family B (6) | 0.045 |
| 2.05 | C7orf16 | Chromosome 7 open reading frame 16 (7) | 0.031 |
| 2.40 | TRAF3IP3 | TRAF3 interacting protein 3 (1) | 0.043 |
| 2.24 | ELF3 | E74-like factor 3 (ets domain transcription factor) (1) | 0.034 |
| 2.58 | TLR10 | Toll-like receptor 10 (4) | 0.049 |
| 2.39 | RAB26 | RAB26, member RAS oncogene family (16) | 0.025 |
| 2.43 | SCML4 | Sex comb on midleg-like4 (Drosophila) (6) | 0.034 |
| 2.05 | SLC12A3 | Solute carrier family 12 (sodium/chloride transporters) (16) | 0.028 |
| 2.13 | MCOLN2 | Mucolipin 2 (1) | 0.022 |
| 2.26 | ANGPTL6 | Angiopoietin-like 6 (19) | 0.022 |
| 2.38 | RAB39B | RAB39B, member RAS oncogene family (X) | 0.044 |
| 4.24 | FLJ40919 | Hypothetical protein FLJ40919 (13) | 0.042 |
| 2.28 | CPLX3 | complexin 3 (15) | 0.004 |
| 2.77 | C6orf105 | Chromosome 6 open reading frame 105 (6) | 0.016 |
| 2.31 | RP11-450P7.3 | Kelch-like 34 (Drosophila) (X) | 0.033 |
| 2.11 | TRIM14 | Tripartite motif-containing 14 (9) | 0.047 |
| 2.10 | MGC40069 | Hypothetical protein MGC40069 | 0.033 |
| 2.78 | CCL22 | Chemokine (C-C motif) ligand 22 (16) | 0.017 |
| 2.01 | TM7SF2 | Transmembrane 7 superfamily member 2 (11) | 0.035 |
| 2.33 | CCL20 | Chemokine (C-C motif) ligand 20 (2) | 0.040 |
| 2.47 | CCR6 | Chemokine (C-C motif) ligand 6 (6) | 0.034 |
| 2.05 | EMR2 | EGF-like module containing, mucin-like hormone rec (19) | 0.019 |
| 2.83 | KIAA1189 | KIAA1189 (2) | 0.047 |
| 2.48 | KIR3DL2 | Killer cell immunoglobulin-like receptor (19) | 0.012 |
| 2.05 | C9orf128 | Chromosome 9 open reading frame 128 (9) | 0.048 |
| 2.11 | ATP2A3 | ATPase, Ca + + transporting, ubiquitous (17) | 0.048 |
| 2.26 | ANKRD55 | Ankyrin repeat domain 55 (5) | 0.046 |
| 2.97 | TXK | TXK tyrosine kinase (4) | 0.041 |
| 2.75 | TMEM16J | Transmembrane protein 16J (11) | 0.011 |
| 2.64 | FCRL3 | Fc receptor-like 3 (1) | 0.009 |
| 2.76 | KEL | Kell blood group, metallo-endopeptidase (7) | 0.040 |
| 2.07 | C20orf174 | Chromosome 20 open reading frame 174 (20) | 0.042 |
| 2.63 | CXADR | Coxsackie virus and adenovirus receptor (21) | 0.044 |
| 2.06 | FLJ43752 | FLJ43752 protein (6) | 0.001 |
| 2.91 | ZAP70 | Zeta-chain (TCR) associated protein kinase 70 kDa (2) | 0.028 |
| 3.03 | IBSP | Integrin-binding sialoprotein (4) | 0.022 |
| 2.62 | TDO2 | Tryptophan 2,3-dioxygenase (4) | 0.043 |
| 2.78 | FCRL3 | Fc receptor-like 3 (1) | 0.047 |
| 2.37 | FLJ11795 | FLJ11795 protein (5) | 0.049 |
| 2.33 | COL4A3 | Collagen, Type IV, alpha 3 (2) | 0.033 |
Highly significant genes appear in bold.
Lamin A/C Protein is Significantly Increased in Regions of High Walls Stress
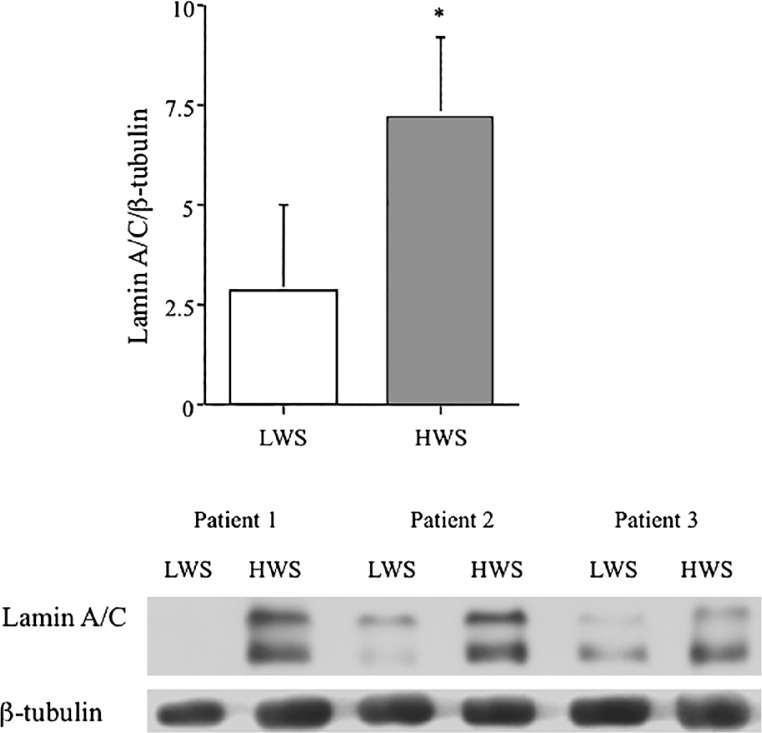
To establish if the differential expression of Lamin A/C was reiterated at the protein level, we analyzed relative protein abundance by western blotting. We found a two-fold induction of Lamin A/C protein (0.99 ± 0.28 vs. 1.88 ± 0.72, n=3, p < 0.05) in aortic biopsies from regions of high wall stress compared to low wall stress (Figure 2). Western blotting data of Lamin A/C for each of the three samples is included in the panel below the histogram.
Figure 2.
Lamin A/C protein abundance is differentially induced in human aneurysmal biopsies from areas of high wall-stress. Histogram shows relative abundance of Lamin A/C from protein lysates of aneurysmal biopsies from areas of low wall stress (LWS) and high wall stress (HWS) relative to β-tubulin. Western blotting gel data for each of three patients is shown in the panel below. Data represent mean ± SD (n = 3, p < 0.05).
Lamin A/C Protein Concentration is Not Elevated In Non-Aneurysmal Vascular Beds of AAA Patients
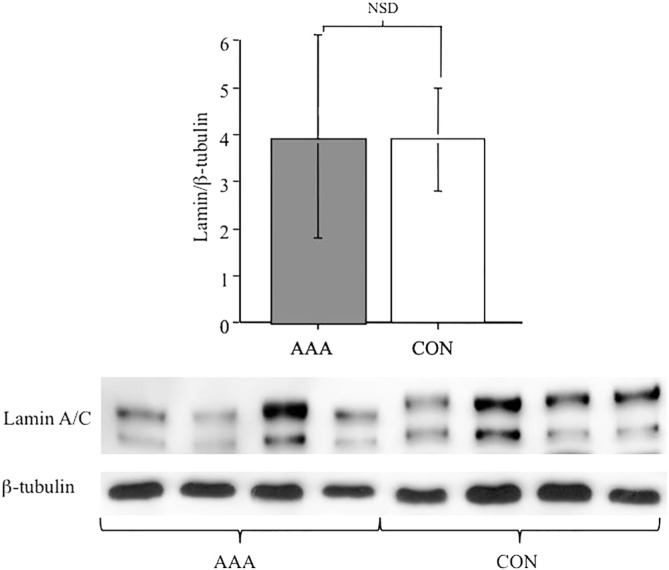
Accruing evidence supports the idea that early aneurismal changes can also be manifest in vascular beds without signs of dilatation. Analysis of non-aneurysmal vessels from AAA patients allow us to explore the question as to whether elevation of Lamin A/C expression represents an early systemic event. We measured the protein abundance in the IMV of AAA patients removed following surgery for other conditions and compared the level of expression to IMV from control patients without AAA. Figure 3 shows no significant difference between the abundance of Lamin A/C from IMV of patients with AAA compared to controls (3.32 ± 2.17 vs. 3.94 ± 1.12, n = 16, p = NSD).
Figure 3.
Lamin A/C is not induced in other vascular beds of aneurysm patients. Protein abundance of Lamin A/C relative to β-tubulin in the inferior mesenteric vein of control (no aneurysm) and aneurysmal patients shows no significant difference. Histogram represents data as mean ± SD (n = 16), with a representative example of four samples from each group as western blotting gel data in panel below; NSD = not significantly different.
Physiological Pulsatile Stretch of SMC in Culture Induces a Time-Dependent Elevation of Lamin A/C and Prelamin A Expression
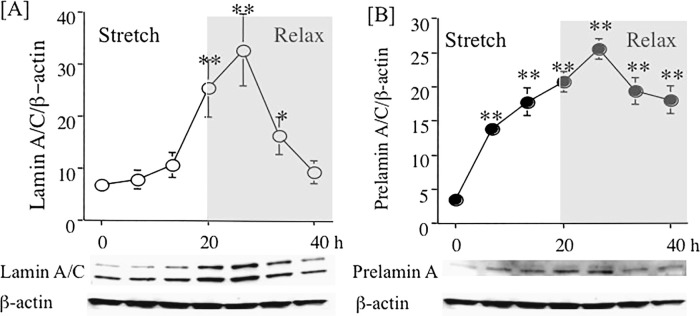
Since SMC of the normal human vasculature are subjected to constant pulsatile stretch, we explored whether expression of Lamin A/C could be influenced by pulsatile stretch in vitro. We seeded SMC onto Flexcell™ plates, and once the cells had reached confluence, the complete growth medium was changed and the cultures subjected to pulsatile stretch at 15% elongation, 1 Hz for 20 hours, followed by 20-hours relaxation. Figure 4 shows western blotting data from protein lysates prepared at indicated times, confirming that the expression of [A] Lamin A/C and [B] Prelamin A steadily increased, reaching a peak at 24 hours after the start of the experiment (approximately five-fold higher than basal levels), 4 hours following the beginning of relaxation. Following a 20-hour period of relaxation, Lamin A/C expression was down to normal resting state and the Prelamin A remained significantly elevated (approximately 3-fold increased) at this time (n = 3, *p < 0.05, **p < 0.01).
Figure 4.
Stretch-dependent effects on Lamin A/C and Prelamin A in human aortic smooth muscle cells. Human aortic smooth muscle cells (SMC) were grown to confluence and following a change of complete medium were then exposed to 20 hours stretch (15% elongation, 1 Hz) followed by 20-hours relaxation. Protein lysates were prepared at 6, 12, 20, 24, 32, and 40 hours. Panel A. Time-course of changes in Lamin A/C expression. Panel B. Time-course of changes in Prelamin A expression relative to β-actin in SMCs. Data are representative of three experiments (*p < 0.05, **p < 0.01).
Discussion
A number of human aneurysm gene expression profile arrays have previously been reported [16–19] and analysis of paired biopsies, using the presence or absence of thrombus over the aneurysmal wall as a discriminator, identified MMP−1, −7, −9, and −12 as being differentially expressed [20]. However, this is the first time that wall stress has been used to discriminate paired biopsies for gene expression profiling. Identification of stress-dependent expression of Lamin A/C in AAA represents the first time that attention has been drawn to the nuclear lamina in this degenerative vascular pathology.
Differential gene array analyses notoriously reveal many divergent candidate genes. Since following rigorous statistical adjustment for multiple measures, Lamin A/C was revealed as our sole candidate, it is most likely that either our selection criteria were overly stringent, or we have too small a sample. If the Benjamini-Hochberg post-hoc test for multiple comparison is not applied to the array data, reducing the level of statistical stringency, a broad register of differential genes is generated (see Supplementary Table 3). Using a two-fold change as a cut-off and a p value less than 0.005 we identify the expression of seventy one genes which are induced and fifty four reduced. This gene array represents molecules that play a role in the immune response, regulation of cell growth, gene expression, angiogenesis, cell matrix synthesis, degradation, and inflammation. Two other candidates have been shown to have a particular relevance to aneurysm biology: ADAMTS2, an extracellular matrix protease known to be involved in Ehlers-Danlos syndrome [21], and also identified as having a role in arterial tortuosity syndrome [22]; and prostaglandin E2 receptor, previously shown to have a role in aneurysm biology [23] and recently reintroduced as a potential therapeutic candidate for use in cardiovascular inflammation.
Supplementary Table 3.
Gene Ontology for candidate genes identified as differentially expressed in response to high wall stress.
| Count1 | % | |
|---|---|---|
| Genes with increased expression | ||
| Extracellular region | 16 | 25.0 |
| Cell adhesion | 9 | 14.1 |
| Extracellular space | 8 | 12.5 |
| Extracellular region & proteinaceous extracellular matrix | 7 | 10.9 |
| Nervous system development | 6 | 9.4 |
| Cell adhesion & protein binding | 5 | 7.8 |
| Cell adhesion & integral to plasma membrane | 4 | 6.3 |
| Cell adhesion, membrane & protein binding | 4 | 6.3 |
| Metal ion binding & response to drug | 3 | 4.7 |
| Cell adhesion, membrane, protein binding & integral to plasma membrane | 3 | 4.7 |
| Plasma membrane | 17 | 34.0 |
| Receptor activity | 10 | 20.0 |
| Plasma membrane & receptor activity | 9 | 18.0 |
| Extracellular space & extracellular response | 6 | 12.0 |
| Immune response | 5 | 10.0 |
| Signal transduction, immune response | 4 | 8.0 |
| Protein binding, interspecies interaction between organisms & plasma membrane | 3 | 6.0 |
| Nucleotide binding, plasma membrane, protein transport | 3 | 6.0 |
| Inflammatory response, signal transduction & immune response | 3 | 6.0 |
| Signal transduction, immune response & chemotaxis | 3 | 6.0 |
Number of differentially expressed genes annotated under term.
2 Percent of differentially expressed genes annotated under term.
The identification of Lamin A/C as a stress-induced candidate gene is consistent with several aspects of the pathobiology of this disease. Lamins form a thin (20 nm) intermediate filament meshwork which lines the nucleoplasmic face of the nuclear envelope (NE). The nuclear lamina is associated with the inner NE and the underlying chromatin and also provides an anchoring point for the nuclear pore complex. In mammalian somatic cells, Lamins A and C (Lamin A/C) are generated by differential splicing of a single gene (LMNA). These products differ by virtue of a unique C-terminal extension, 99 amino acids for Lamin A and 6 for Lamin C [24]. Lamin A goes through multiple steps of post-translational processing at its C-terminal CaaX motif, involving 3–4 enzymes. First, a farnesyl moiety is added to cysteine by farnesyl transferase 1. The cleavage of the last three amino acids (aaXing) by either Rce1 or ZmpSte2 occurs. Then, the C-terminal cysteine is carboxymethylated by isoprenylcysteine carboxymethyltransferase, resulting in carboxymethylated Prelamin A. Within 90 minutes of synthesis, ZmpSte24 cleaves Prelamin A (74 kDa), 14 amino acids upstream of the C-terminus, resulting in mature Lamin A (72 kDa) [25–30].
Mechanotransduction, the phenomenon by which cells respond to applied force, is critical to several physiological and pathological processes [31]. The exact mechanisms which govern how forces can influence biological processes have not been determined, but putative direct force effects on the genome must require transduction through the nuclear lamina. Lamins not only provide a nuclear scaffold of intermediate filaments important in mechanotransduction but are also found in the nucleoplasm, being part of the intranuclear reticulum, thought to play an important role in the maintenance of nuclear structural integrity, in DNA replication, transcriptional regulation, and apoptosis [32–34]. Expression and accumulation of a mutant form of Lamin A associated with Hutchinson-Gilford Progeric Syndrome (HGPS) (Lamin A D50) causes changes in the nuclear lamina network organization [35] and an increase in the accumulation of Prelamin and nuclear stiffness [36] with implications for mechanisms of normal ageing and atherosclerotic vascular changes. Recent studies have demonstrated that accumulation of Prelamin accelerates normal smooth muscle cell aging [37].
Mutation in the LMNA gene have been responsible for at least nine classes of disorders, collectively called laminopathies, including muscular dystrophies Emery-Dreifuss and limb-girdle muscular dystrophy, Type 1B [38], in accelerated aging, HGPS [39, 40] or atypical Werner’s Syndrome [41], and dilated cardiomyopathy [42], Charcot-Marie-Tooth disease Type 1 [43], Dunnigan-type familial partial lipodystrophy [44], mandibuloacral dysplasia [45], and restrictive dermopathy [46]. A number of the clinical features of these diseased phenotypes are suggestive of accelerated aging, and studies in vitro have shown that overexpression of mutant and wild-type Lamin A accelerates telomere shortening and replicative senescence [47]. We and others have shown that there is an elevated number of circulating progenitor cells and that the telomeres of such cells are shorter in comparison to age matched controls, suggesting that AAAs represent focal lesions of accelerated aging [48, 49]. This being so, the demonstration of focal induction of Lamin A/C expression warrants investigative consideration as a potential player in generating these chronic degenerative vascular lesions.
Apoptosis, programmed cell death, plays a fundamental role in aneurysm development [50]. In a recent study where aneurysm formation was perturbed by elevation of plasma high-density lipoprotein concentration, we show that site-specific apoptosis was inhibited [51]. In cells programmed to die, the caspase-dependent degradation of Lamins has been recognized as a prelude to nuclear destruction [52]. Recent work suggests that not only is Lamin degradation a feature of apoptosis but that failure to correctly assemble nuclear lamina is a trigger for apoptosis [53]. The overexpression of Lamin A/C that we observe in the high wall-stress regions of AAA may represent a mechanism to counter apoptotic induction, but further work is needed to support this idea.
The predominant weakness of these experiments is the small number of paired biopsies available for analysis. With an increase in application of endovascular repair, the numbers of elective open repair are reduced. To address this issue, we are archiving additional material over a longer period of time for further studies.
We have identified an increase in Lamin A/C protein expression as a wall stress-dependent change observed in AAAs. Since these changes are not observed in non-aneurysmal vascular beds of AAA patients, it is most likely to be a compensatory response to the accelerated ageing process that is characteristic of aneurysm pathobiology. The reason that induction of expression of Lamin A/C may be desirable and represent a homeostatic mechanism to counter the degenerative progression of the vessel may relate to the ability of Lamin A/C to protect chromatin structure and prevent apoptosis. Although it will be challenging to develop Lamin A/C as a molecule of any clinical use in the management/or treatment of AAA, our findings support the validity of this experimental design as a means of further discovery. Larger numbers of patients are required to allow us to gain a significant number of good quality paired samples for similar analysis. However, since aneurysms share common features of biology with advanced aging, and treatment with farnesyltransferase inhibitors has the potential to perturb this process in vitro and in vivo [54, 55], these drugs may provide an interesting new therapeutic strategy for management of AAA growth. Further investigation of target genes, such as Lamin A/C, that impact on mechanotransduction may have importance in thoracic aortic aneurysms, since although the etiology of these pathologies differs from abdominal aortic aneurysms the role of mechanodysfunction on molecular changes in cell biology may be similar [56–58]. Such ideas warrant further investigation. The ability to mimic induction of Lamin A/C in vitro will facilitate more molecular analyses in the future.
Acknowledgements
The authors would like to thank Dr. Ken Laing for his helpful advice on the gene array experiment and for the bioinformatics support necessary for analysis. Thanks to Jan Polonieki for statistical advice and our thanks to all the patients who consented to collection and donation of their tissue for this study. The authors are grateful to Zhuo Cheng and Anenta Ratneswaren who assisted in the finite element analysis presented in this study. This work was supported by an EU FP7-HEALTH integrated project grant (Fighting Aneurysmal Disease), agreement number 200467. AM was the recipient of a Royal College of Surgeons Fellowship.
Conflict of Interest
The authors have no conflicts of interest relevant to this publication.
References
- 1.Collaborative Aneurysm Screening Group A comparative study of the prevalence of abdominal aortic aneurysms in the United Kingdom, Denmark and Australia. J Med Screen. 2001;8:46–84. DOI: 10.1136/jms.8.1.46 [DOI] [PubMed] [Google Scholar]
- 2.Allaire E, Hasenstab D, Kenagy RD, Starcher B, Clowes MM, Clowes AW.. Prevention of aneurysm development and rupture by local overexpression of plasminogen activator inhibitor-1. Circulation. 1998;98:249–255. DOI: 10.1161/01.CIR.98.3.249 [DOI] [PubMed] [Google Scholar]
- 3.Curci JA, Liao S, Huffman MD, Shapiro SD, Thompson RW.. Expression and localization of macrophage elastase (matrix metalloproteinase-12) in abdominal aortic aneurysms. J Clin Invest. 1998;102:1900–1910. DOI: 10.1172/JCI2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freestone T, Turner RJ, Coady A, Higman DJ, Greenhalgh RM, Powell J.. Inflammation and matrix metalloproteinase’s in the enlarging abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. 1995;15:1145–1151. DOI: 10.1161/01.ATV.15.8.1145 [DOI] [PubMed] [Google Scholar]
- 5.Wilson WR, Anderton M, Schwalbe EC, Jones JL, Furness PN, Bell PR, et al. Matrix metalloproteinase’s-8 and -9 are increased at the site of abdominal aortic aneurysm rupture. Circulation. 2006;113: 438–445. DOI: 10.1161/CIRCULATIONAHA.105.551572 [DOI] [PubMed] [Google Scholar]
- 6.Lederle FA, Johnson GR, Wilson SE, Chute EP, Hye RJ, Makaroun MS, et al. The aneurysm detection and management study screening program: Validation cohort and final results. Arch Intern Med. 2000;160:1425–1430. DOI: 10.1001/archinte.160.10.1425 [DOI] [PubMed] [Google Scholar]
- 7.Tsui JC. Experimental models of abdominal aortic aneurysms. Open Cardiovasc Med J. 2010;4:221–230. DOI: 10.2174/1874192401004010221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordon IM, Hinchliffe RJ, Loftus IM, Thompson MM.. Pathophysiology and epidemiology of abdominal aortic aneurysms. Nat Rev Cardiol. 2011;8:92–102. DOI: 10.1038/nrcardio.2010.180 [DOI] [PubMed] [Google Scholar]
- 9.Shimizu K, Mitchell RN, Libby P.. Inflammation and cellular immune responses in abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol. 2006;26:987–994. DOI: 10.1161/01.ATV.0000214999.12921.4f [DOI] [PubMed] [Google Scholar]
- 10.Shin IS, Kim JM, Kim KL, Jang SY, Jeon ES, Choi SH, et al. Early growth response factor-1 is associated with intraluminal thrombus formation in human abdominal aortic aneurysm. J Am Coll Cardiol. 2009;53:792–799. DOI: 10.1016/j.jacc.2008.10.055 [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Pinna R, Madrigal-Matute J, Tarin C, Burillo E, Esteban-Salan M, Pastor–Vargas C, et al. Proteomic analysis of intraluminal thrombus highlights complement activation in human abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol. 2013;33:2013–2020. DOI: 10.1161/ATVBAHA.112.301191 [DOI] [PubMed] [Google Scholar]
- 12.Filardo G, Powell JT, Martinez MA, Ballard DJ.. Surgery for small asymptomatic abdominal aortic aneurysms. Cochrane Database Syst Rev. 2012;3:CD001835 DOI: 10.1002/14651858.cd001835.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choke E, Thompson MM, Dawson J, Wilson WR, Sayed S Loftus IM, et al. Abdominal aortic aneurysm rupture is associated with increased medial neovascularization and overexpression of proangiogenic cytokines. Arterioscler Thromb Vasc Biol. 2006;26:2077–2082. DOI: 10.1161/01.ATV.0000234944.22509.f9 [DOI] [PubMed] [Google Scholar]
- 14.Xu XY, Borghi A, Nchimi A, Leung J, Gomez P, Cheng Z, et al. High levels of 18-FDG uptake in aortic aneurysm wall are associated with high wall stress. Eur J Vas Endovasc Surg. 2010;39:295–301. DOI: 10.1016/j.ejvs.2009.10.016 [DOI] [PubMed] [Google Scholar]
- 15.Raghavan ML, Vorp DA.. Toward a biomechanical tool to evaluate rupture potential of abdominal aortic aneurysm: Identification of a finite strain constitutive model and evaluation of its applicability. J Biomech. 2000;33:475–482. DOI: 10.1016/S0021-9290(99)00201-8 [DOI] [PubMed] [Google Scholar]
- 16.Lenk GM, Tromp G, Weinsheimer S, Gatalica Z,Berguer R, Kuivaniemi H.. Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms. BMC Genomics. 2007;8:237–242. DOI: 10.1186/1471-2164-8-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choke E, Thompson MM, Jones A, Torsney E, Dawson J, Laing K, et al. Gene expression profile of abdominal aortic aneurysm rupture. Ann NY Acad Sci. 2006;1085:311–314. DOI: 10.1196/annals.1383.007; [DOI] [PubMed] [Google Scholar]
- 18.Tung WS, Lee JK, Thompson RW.. Simultaneous analysis of 1176 gene products in normal human aorta and abdominal aortic aneurysms using a membrane-based complementary DNA expression array. J Vasc Surg. 2001;34:143–150. DOI: 10.1067/mva.2001.113310 [DOI] [PubMed] [Google Scholar]
- 19.Tilson MD, Fu C, Xia SX, Syn D, Yoon Y, McCaffrey T.. Expression of molecular messages for angiogenesis by fibroblasts from aneurysmal abdominal aorta versus dermal fibroblasts. Int J Surg Invest. 2000;1:453–457. PMID: [PubMed] [Google Scholar]
- 20.Kazi M, Zhu C, Roy J, Paulsson-Berne G, Hamsten A, Swedenborg J, et al. Difference in matrix-degrading protease expression and activity between thrombus-free and thrombus-covered wall of abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. 2005;25:1341–1346. DOI: 10.1161/01.ATV.0000166601.49954.21 [DOI] [PubMed] [Google Scholar]
- 21.Colige A, Nuytinck L, Hausser I, van Essen AJ, Thiry M, Herens C, et al. Novel types of mutation responsible for the dermatosparactic type of Ehlers-Danlos syndrome (Type VIIC) and common polymorphisms in the ADAMTS2 gene. J Invest Dermatol. 2004;123:656–663. DOI: 10.1111/j.0022-202X.2004.23406.x [DOI] [PubMed] [Google Scholar]
- 22.Gardella R, Zoppi N, Assanelli D, Muiesan ML, Barlati S, Colombi M.. Exclusion of candidate genes in a family with arterial tortuosity syndrome. Am J Med Genet A. 2004;126A:221–228. DOI: 10.1002/ajmg.a.20589 [DOI] [PubMed] [Google Scholar]
- 23.Bayston T, Ramessur S, Reise J, Jones KG, Powell JT.. Prostaglandin E2 receptors in abdominal aortic aneurysm and human aortic smooth muscle cells. J Vasc Surg. 2003;38:354–359. DOI: 10.1016/S0741-5214(03)00339-2 [DOI] [PubMed] [Google Scholar]
- 24.Lin F, Warman HJ.. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem.1993;268:16321–16326. PMID: [PubMed] [Google Scholar]
- 25.Gerace L, Comeau C, Benson M.. Organization and modulation of nuclear lamina structure. J Cell Sci Suppl. 1984;1:137–160. DOI: 10.1242/jcs.1984.Supplement_1.10 [DOI] [PubMed] [Google Scholar]
- 26.Varela I, Cadiñanos J, Pendás AM, Gutiérrez-Fernández A, Folgueras AR, Sánchez LM, et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. DOI: 10.1038/nature04019 [DOI] [PubMed] [Google Scholar]
- 27.Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc Natl Acad Sci USA. 2002;99:13049–13054. DOI: 10.1073/pnas.192460799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corrigan DP, Kuszczak D, Rusinol AE, Thewke DP, Hrycyna CA, Michaelis S, et al. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem J. 2005;387: 129–318. DOI: 10.1042/BJ20041359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber K, Plessmann U, Traub P.. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 1989;257:411–414. DOI: 10.1016/0014-5793(89)81584-4 [DOI] [PubMed] [Google Scholar]
- 30.Beck LA, Hosick TJ, Sinensky M.. Isoprenylation is required for the processing of the lamin A precursor. J Cell Biol. 1990;110:1489–1499. DOI: 10.1083/jcb.110.5.1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shyu KG. Cellular and molecular effects of mechanical stretch on vascular cells and cardiac myocytes. Clin Sci (Lond). 2009;116:377–389. DOI: 10.1042/CS20080163 [DOI] [PubMed] [Google Scholar]
- 32.Fuchs E, Weber K.. Intermediate filaments: Structure, dynamics, function and disease. Annu Rev Biochem. 1994;63:345–382. DOI: 10.1146/annurev.bi.63.070194.002021 [DOI] [PubMed] [Google Scholar]
- 33.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. DOI: 10.1172/JCI200419670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson KL, Zastrow MS, Lee KK.. Lamins and disease: Insights into nuclear infrastructure. Cell. 2001;104:647–650. DOI: 10.1016/S0092-8674(02)01452-6; [DOI] [PubMed] [Google Scholar]
- 35.Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101:8963–8968. DOI: 10.1073/pnas.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, Misteli T.. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2006;103: 10271–10276. DOI: 10.1073/pnas.0601058103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ragnauth CD, Warren DT, Liu Y, McNair R, Tajsic T, Figg N, et al. Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging. Circulation. 2010;121:2200–2210. DOI: 10.1161/CIRCULATIONAHA.109.902056 [DOI] [PubMed] [Google Scholar]
- 38.Muchir A, Bonne G, van der Kool AJ, van Meegan M, Baas F, Bolhuis PA, et al. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD18). Hum Mol Genet. 2000;9:1453–1459. DOI: 10.1093/hmg/9.9.1453 [DOI] [PubMed] [Google Scholar]
- 39.De Sandre-Giovannoli A, Chaouch M, Kozlov S, Vallat JM, Tazir M, Kassouri N, et al. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am J Hum Genet. 2002;70:726–736. DOI: 10.1086/339274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. DOI: 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Lee L, Kudlow BA, Dos Santos HG, Sletvold O, Shafeghati Y, et al. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:440–445. DOI: 10.1016/S0140-6736(03)14069-X [DOI] [PubMed] [Google Scholar]
- 42.Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–1724. DOI: 10.1056/NEJM199912023412302 [DOI] [PubMed] [Google Scholar]
- 43.De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055 DOI: 10.1126/science.1084125 [DOI] [PubMed] [Google Scholar]
- 44.Shackelton S, Lloyd D, Jackson SN, Evans R, Niermeijer MF, Singh BM, et al. LMNA, encoding Lamin A/C is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–156. DOI: 10.1038/72807 [DOI] [PubMed] [Google Scholar]
- 45.Novelli G, Muchir A, Sangiuolo F, Helbling-Leclerc A, D’Apice MR, Massart C, et al. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. Am J Hum Genet. 2002;71:426–431. DOI: 10.1086/341908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navarro CL, De Sandre-Giovannoli A, Bernard R, Boccaccio I, Boyer A, Geneviève D, et al. Lamin A and ZMPSTE24 (FACE-1) defects cause nuclear disorganization and identify restrictive dermopathy as a lethal neonatal laminopathy. Hum Mol Genet. 2004;13:2493–2503. DOI: 10.1093/hmg/ddh265 [DOI] [PubMed] [Google Scholar]
- 47.Huang S, Risques RA, Martin GM, Rabinovitch PS, Oshima J.. Accelerated telomere shortening and replicative senescence in human fibroblasts overexpressing mutant and wild-type lamin A. Exp Cell Res. 2008;314:82–91. DOI: 10.1016/j.yexcr.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dawson J, Tooze J, Cockerill G, Choke E, Loftus I, Thompson MM.. Endothelial progenitor cells and abdominal aortic aneurysms. Ann NY Acad Sci. 2006;1085:327–330. DOI: 10.1196/annals.1383.011 [DOI] [PubMed] [Google Scholar]
- 49.Cafueri G, Parodi F, Pistorio A, Bertolotto M, Ventura F, Gambini C, et al. Endothelial and smooth muscle cells from abdominal aortic aneurysm have increased oxidative stress and telomere attrition. PLoS One. 2012;7:e35312 DOI: 10.1371/journal.pone.0035312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson RW, Liao S, Curci JA.. Vascular smooth muscle cell apoptosis in abdominal aortic aneurysms. Coron Artery Dis. 1997;8:623–631. DOI: 10.1097/00019501-199710000-00005 [DOI] [PubMed] [Google Scholar]
- 51.Torsney E, Pirianov G, Charolidi N, Shoreim A, Gaze D, Petrova S, et al. Elevation of plasma high-density lipoproteins inhibits development of experimental abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. 2012;32:2678–2686. DOI: 10.1161/ATVBAHA.112.00009 [DOI] [PubMed] [Google Scholar]
- 52.Lazebnik Y, Takahashi A, Moir R, Goldman R, Poiriser G, Kaufmann S, et al. Studies of lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc Natl Acad Sci USA. 1995;92:9042–9046. DOI: 10.1073/pnas.92.20.9042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steen RL, Collas P.. Mistargeting of B-type lamins at the end of mitosis: implications on cell survival and regulation of lamins A/C expression. J Cell Biol. 2001;153:621–626. DOI: 10.1083/jcb.153.3.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capell BC, Olive M, Erdos MR, Cao K, Faddah DA, Tavarez UL, et al. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc Natl Acad Sci USA. 2008;105:15902–15907. DOI: 10.1073/pnas.0807840105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang SH, Bergo MO, Toth JI, Qiao X, Hu Y, et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc Natl Acad Sci USA. 2005;102:10291–10296. DOI: 10.1073/pnas.0504641102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeremy RW, Robertson E, Lu Y, Hambly BD.. Perturbations of mechanotransduction and aneurysm formation in heritable aortopathies. Int J Cardiol. 2013;169:7–16. DOI: 10.1016/j.ijcard.2013.08.056 [DOI] [PubMed] [Google Scholar]
- 57.Bäck M, Gasser TC, Michel JB, Caligiuri G.. Biomechanical factors in the biology of aortic wall and aortic valve diseases. Cardiovasc Res. 2013;99:232–241. DOI: 10.1093/cvr/cvt040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humphrey JD, Milewicz DM, Tellides G, Schwartz MA.. Cell biology. Dysfunctional mechanosensing in aneurysms. Science. 2014;344:477–479. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]