Abstract
In this work, we propose DR-TAMAS (Diffeomorphic Registration for Tensor Accurate alignMent of Anatomical Structures), a novel framework for intersubject registration of Diffusion Tensor Imaging (DTI) data sets. This framework is optimized for brain data and its main goal is to achieve an accurate alignment of all brain structures, including white matter (WM), gray matter (GM), and spaces containing cerebrospinal fluid (CSF). Currently most DTI-based spatial normalization algorithms emphasize alignment of anisotropic structures. While some diffusion-derived metrics, such as diffusion anisotropy and tensor eigenvector orientation, are highly informative for proper alignment of WM, other tensor metrics such as the trace or mean diffusivity (MD) are fundamental for a proper alignment of GM and CSF boundaries. Moreover, it is desirable to include information from structural MRI data, e.g., T1-weighted or T2-weighted images, which are usually available together with the diffusion data. The fundamental property of DR-TAMAS is to achieve global anatomical accuracy by incorporating in its cost function the most informative metrics locally. Another important feature of DR-TAMAS is a symmetric time-varying velocity-based transformation model, which enables it to account for potentially large anatomical variability in healthy subjects and patients. The performance of DR-TAMAS is evaluated with several data sets and compared with other widely-used diffeomorphic image registration techniques employing both full tensor information and/or DTI-derived scalar maps. Our results show that the proposed method has excellent overall performance in the entire brain, while being equivalent to the best existing methods in WM.
Keywords: Diffusion tensor imaging, Diffeomorphic image registration, Fiber tractography
1. Introduction
In the past couple of decades, Diffusion Tensor Imaging (DTI) (Basser et al., 1994; Pierpaoli et al., 1996) has been extensively used to investigate the human brain. DTI provides unique microstructural and physiological insight into brain tissue that are informative of processes involved in development, aging, and various diseases. With recent advances in magnet and gradient systems and with the developments of effective echo-planar imaging distortion correction algorithms (Irfanoglu et al., 2015; Andersson et al., 2003), it is now possible to achieve a high degree of anatomical accuracy in DTI data sets. Cross-sectional and longitudinal group analysis of such diffusion MRI (dMRI) data has been gaining popularity, and voxelwise analyses have been performed on numerous developmental and clinical studies (Sadeghi et al., 2015; Maier-Hein et al., 2015; Poudel et al., 2015; Mahoney et al., 2015; Garaci et al., 2015). Accurate spatial normalization is important for a meaningful voxelwise analysis of low resolution imaging data (Bookstein, 2001), but it becomes crucial for the higher-resolution data with fine anatomical details that current DTI scans can provide. Intersubject spatial normalization is also the first step in the creation of population atlases that can be used in normative databases.
There has been an extensive amount of work on spatially normalizing diffusion tensor images (Gee and Alexander, 2006; Mũnoz-Moreno et al., 2009; Wang et al., 2011). Diffusion tensor registration techniques can be broadly categorized into two groups: scalar maps–based or full tensor–based.
Fractional anisotropy (FA) is the metric used by virtually all previously proposed scalar maps–based approaches (Jones et al., 2002; Guimond et al., 2002; Park et al., 2003; Rohde et al., 2004b; Andersson et al., 2007). Scalar-based registration approaches, which are still the most widely used for clinical research applications, perform very well when the goal is to analyze the quantity being used for registration but can be suboptimal to match other DTI metrics such as principal orientations. Several authors have attempted to improve scalar map–based methods by combining several tensor-derived quantities and perform multichannel registration. Yang et al. (2008) proposed a method that integrated geometric features such as prolateness, oblateness, and sphericity of the diffusion tensors and orientation information to perform registration. Wang et al. (2013) used image polynomial expansion strategies on FA and scalar index images to optimize tensor alignment considering shape information. Goh and Vidal (2006) showed an algebraic solution to rigid DTI registration and presented a pointwise feature-based registration algorithm to complement tensor field matching. In their intrarelated works, Wang et al. (2014) and Yap et al. (2009a,b, 2010) employed hierarchical guidance of regional distributions, boundaries, and orientational information derived from diffusion tensors to perform registration.
Full tensor-based approaches appear most promising because they use the entire information available in the data set. These techniques can also be categorized into three classes: algorithms that do not reorient the tensors (e.g. Alexander and Gee (2000); Ruiz-Alzola et al. (2002); Rohde et al. (2004b)), algorithms that disregard the orientation information during optimization but reorient the tensors afterwards (e.g., Irfanoglu et al. (2008, 2009)), and algorithms that also consider tensor reorientation information during optimization. Park et al. (2003) showed that reorienting the tensor field based on the deformation field during optimization or afterwards outperforms “no reorientation” strategy even when tensor reorientation is not explicitly optimized. Curran and Alexander (2003) then showed that considering tensor reorientation during optimization further improved alignment quality using affine transformations. Adluru et al. (2013) showed that tensor reorientation strategies keep anatomically consistent architectures after registration. Zhang et al. (2006) were the first to include tensor orientation information in a diffeomorphic registration framework by modeling the deformation fields with locally affine patches with an explicit rotation component. This method, which was released in the widely used DTITK package, guaranteed diffeomorphism by constraining the neighboring affine patches to be similar, in addition to a subsequent filtering with Gaussian kernels. DTITK’s transformation model with multiaffine patches made tensor reorientation straightforward, but this strategy was not applicable to more general transformation models, such as displacement fields, until Yeo et al. (2008, 2009) proposed an analytical solution to the differentials of the finite-strain tensor reorientation strategy. This method, which is now part of the MEDINRIA package, employed a diffeomorphic Demons-based transformation model using exact finite-strain gradients, which resulted in a very large, sparse linear system that needed to be solved at every optimization iteration. A stationary-velocity parameterization for this method was also proposed (Sweet and Pennec, 2010; Commowick et al., 2012). Using the same differential formulation, Li et al. (2014) proposed another registration strategy that employed local-trust-region techniques that improved the former method’s registration quality in addition to significantly speeding up the processing. Ceritoglu et al. (2009) proposed an extension to the popular large deformation diffeomorphic metric mapping method to support diffusion tensor data, where tensor reorientation was performed by reorienting the eigenvectors of the tensors in their formulation.
DTI-based registration algorithms have traditionally aimed at achieving good alignment of white matter (WM) structures, mostly focusing on diffusion anisotropy and, in some cases, the principal diffusion orientations (Zhang et al., 2006; Jones et al., 2002; Park et al., 2003; Andersson et al., 2007). However, many clinical applications need excellent spatial normalization across all brain structures, WM, as well as gray matter (GM) and cerebrospinal fluid (CSF) filled cavities, such as in stroke, traumatic brain injury (TBI), or in neurodevelopmental and neurodegenerative disorders. Even whole tensor–based approaches currently do not rely on a cost function that would be “locally” optimal. This can cause local misalignment or even global brain morphometry mismatches. Therefore, our primary goal was to develop a DTI registration methodology with locally varying metrics to provide WM pathway alignment capabilities on par with methods that rely on specialized WM metrics but also provide accurate registration of GM regions and CSF spaces. In the following sections, we will first describe the mathematical framework of the proposed method, DR-TAMAS (Diffeomorphic Registration for Tensor Accurate alignMent of Anatomical Structures), which is part of the publicly available TORTOISE diffusion MRI processing package (Pierpaoli et al., 2010). We will then analyze DR-TAMAS’ performance.
2. Materials and Methods
The distinct properties of the DR-TAMAS registration framework are as follows:
Spatially varying multimetrics: DR-TAMAS employs a weighted combination of metrics that favors the tensor deviatoric similarity (Basser and Pierpaoli, 1996) in WM regions, but adds tensor trace (TR) distance and other cross-correlation metrics to quantify the similarities between GM and CSF regions. A smooth spatially varying weighting kernel, which is a function of voxelwise FA values, combines the deformations from these metrics. Fusing different channels of information in this manner guarantees an anatomically faithful alignment of GM and CSF regions in addition to WM fiber-bundle alignment.
Transformation model: DR-TAMAS gives users the possibility for two diffeomorphic transformation models: Symmetric Normalization (SyN) (Avants et al., 2008) used in the ANTS package (Avants et al., 2011) and time-varying velocity-based model (Christensen et al., 1996), both now available in the ITK library. With these transformations, DR-TAMAS is capable of modeling the large deformations necessary to register brains with significant morphological abnormalities.
Analytical finite strain differentials for tensor reorientation without a linear system: We employ the analytical finite strain differentials proposed by Yeo et al. (2008, 2009). However, instead of solving a large, sparse linear system at every optimization iteration, we employ a conjugate-gradient based optimization scheme that considers the effects of changes in the displacement fields on the rotation of neighboring voxels. This formulation significantly speeds up processing and makes it suitable for parallelism.
Combination of other imaging modalities with DTI: DRTAMAS provides the functionality to use other imaging modalities such as T1-weighted or T2-weighted images to further improve the alignment of fine-scale GM regions such as gyri and sulci. The local cross-correlation metric is used to assess the similarity of these additional modalities, and displacement fields are again combined at the metric level with larger voxelwise weights for tensor-based deformation fields for WM regions.
Robust DTI and structural atlas creation: DR-TAMAS gives the functionality to create both DTI and/or anatomical image-based atlases based on a variation of the atlas creation methods proposed by Joshi et al. (2004) and Wu et al. (2011).
In this section, we will describe the mathematical foundations of DR-TAMAS, its atlas creation methodologies, and our validation strategies with several data sets.
2.1. Transformation Model
Every image registration problem consists of a similarity metric and a transformation model. For DR-TAMAS, we opted to provide users the ability to use the time-varying velocity field (TVVF) based transformation and SyN, which we will describe here. SyN (Avants et al., 2008) has been shown several times to be a very powerful and fast transformation model (Klein et al., 2009; Murphy et al., 2011). SyN models the abstract time space of TVVF with just three time points, i.e., the fixed image F at t = 0, the moving image M at t = 1, and a middle image at t = 0.5, where the similarity metric is evaluated. The forward displacement field ϕ1(x, t) maps F onto the middle time point as F′ = F(ϕ1(x, 0.5)), whereas a second displacement field, ϕ2(x, t), maps the moving image M as M′ = M(ϕ2(x, 0.5)). Therefore, when a similarity metric 𝒮 is used, the registration optimization function can be written as follows:
| (1) |
where Ω represents the image domain. Therefore, the backward mapping from the moving image to the fixed image can be described as: , where ◦ represents the deformation combination operator. Optimization of Equation 1 can be performed by using Euler-Lagrange equations, yielding the generic gradient directions:
| (2) |
| (3) |
In these gradient formulations, the second terms ∂𝒮/∂F′ and ∂𝒮/∂M′ depend solely on the employed similarity metric and the last terms ∂x/∂ϕ1 and ∂x/∂ϕ2 are the transformation Jacobians. In traditional image registration, the terms ∂F′/∂x and ∂M′/∂x represent spatial image gradients, but in the case of tensor image registration, these are the terms that might depend on tensor reorientation. The reader is referred to (Avants et al., 2008) for a detailed description of the transformation model and metric gradients for the cross-correlation metric.
The TVVF model we use is a variant of the one provided in the ITK library. The user is still provided the ability to select the number of abstract time points but for each time point t, the registration function gradients are computed using the spatial gradients of both the fixed and moving images for the reasons described in Section 2.3. This formulation can be found in Appendix A.1.
2.2. Mathematical Framework for the Similarity Metrics
DR-TAMAS employs a combination of three types of similarity metrics: the deviatoric tensor similarity metric, tensor trace mean squares similarity, and a cross-correlation metric on all other additional images to be used in registration.
2.2.1. Trace Similarity
The trace similarity metric is mathematically the simplest of the two metrics that employ tensor information. Even though individual components of the tensors change after tensor reorientation, the trace value is constant. Therefore, while this metric is used, for processing speed purposes, the tensors are not reoriented, yielding transformed tensors similar to the scalar image case as: F′ = F(ϕ1(x, 0.5)) and similarly for M′. Therefore the registration optimization functions becomes as follows:
| (4) |
| (5) |
where is the ith diagonal tensor component of the fixed tensor image at spatial location x and Tr is the trace operator. The displacement vectors can then be written as follows:
| (6) |
| (7) |
The spatial gradients of the trace are computed by summing the spatial gradients of the individual diagonal components of the tensor.
2.2.2. Deviatoric Tensor Similarity
This metric, which is the default metric in DTITK, has been shown to yield very good alignment in terms of both WM structure and orientation information (Zhang et al., 2006; Wang et al., 2011). The deviatoric tensor of a tensor D is the anisotropic part of D and is defined as with I being the identity matrix (Basser and Pierpaoli, 1996). In compact form, this yields the registration optimization function:
| (8) |
In this metric’s case, however, the transformed and interpolated tensor F′(x) is different from the trace metric as the interpolated tensors also need to be reoriented based on their corresponding deformation fields as follows:
| (9) |
| (10) |
The rotation matrix RF,M is a function of the underlying displacement field ϕ1,2 at voxel locations xℕ, which are in the neighborhood ℕ of x as RF,M(x) = ℛ(ϕ1,2(xℕ)). The finite strain (Alexander et al., 2001) definition of the rotation matrix is as follows: Let JF,M(x) be the 3 × 3 spatial Jacobian matrix of the displacement field ϕ1,2. The differentials are computed with the Jacobian operator 𝒥(ϕ1,2(xℕ)), which in our case is based on the 4th-order centered differences. The local affine matrix A(x) is then defined as A(x) = I + J(x) (Appendix A.2). When the subscripts are omitted, the rotation matrices can be written as:
| (11) |
The formulation for the displacement field vectors can again be computed using Euler-Lagrange equations and chain-rule:
| (12) |
With this formulation, the terms in the second line of Equation 12 are zero because a change in the displacement vector at voxel x does not affect the rotation matrix R at that voxel. However, a change in the displacement vector of a neighboring voxel affects the rotation matrix of the current voxel x. Therefore, the displacement vector update at voxel x results from the cumulative contribution of the “matching term” and “tensor rotation term” that originates from the effect of the change in the current displacement vector to the rotation matrix of a neighboring voxel v ∈ xℕ as follows:
| (13) |
| (14) |
α ∈ [0.1, 1] is a term we introduced to change the relative importance of the tensor rotation term during optimization with respect to the matching term. At the lower levels of the optimization (early iterations of the lower resolutions of the multiresolution image pyramid), α is kept small to favor the correspondence matching, but its value is gradually increased in the later stages of the registration. The detailed derivations of the terms , which are based on the differentials proposed by Yeo et al. (2009), can be found in Appendix A.2.
2.2.3. Anatomical Image Similarity
In addition to the two metrics that consider tensorial information, DR-TAMAS can make use of any number of additional image pairs. These image pairs for the fixed Fs and moving images Ms, are assumed to reside in the exact same anatomical space as their corresponding tensors. Therefore, for such image pairs, the local cross-correlation metric is used on the middle time point images F′s and M′s as described in detail by Avants et al. (2008). Given that a local cross-correlation metric is used to coregister the two images of the image pair, they should be acquired with the same image modality, although acquisition parameters are not required to be identical. F′s and M′s are defined as described in Section 2.1. The CC metric can be defined as follows:
| (15) |
The derivations for the displacement vectors using this metric are described in detail by Avants et al. (2008).
2.2.4. Metric Fusion
DR-TAMAS allows the use of one or both of the tensor metrics in addition to any other optional anatomical image pairs. The information from these different modalities are combined at the metric level, i.e., a displacement field is generated for all the image/tensor/metric pairs and fused at each optimization iteration. This translates into (for ϕ1):
| (16) |
where n is the optimization iteration number, N is the number of optional anatomical images, ◦ is the deformation combination, w is the weight for the corresponding displacement vector, where the sum of all w is one. ϕ2 has its own set of weights to describe the tissue characteristics of the moving image. Theoretically, the default spatially varying weighting that DR-TAMAS provides should represent a good compromise to achieve good registration across all brain regions. However, given that different applications may require a selectively improved performance for specific brain structures, DR-TAMAS includes the possibility of selecting the following customized weighting options:
Default: With the default option, a fractional anisotropy map is generated at every iteration of the optimizer, smoothed with a Gaussian kernel of standard deviation equal to the image spacing and mapped to the range [0, 0.8]. This map is used as a voxelwise weight map for w2, i.e., the deviatoric tensor similarity metric to guarantee that this metric is favored in WM regions and the other metrics/images are favored in GM and CSF regions. The remaining voxelwise weights are distributed evenly among the other employed metrics.
equal: All weights for all images and metrics are set to be equal. With this selection, the metrics are constant throughout the images and are not voxelwise.
WM: w2, the weight for the deviatoric tensor similarity metric, is set to be significantly larger than the other weights (w2 = 0.8) to obtain a better WM alignment. Other metrics are set to be equal.
GM: Similar to the WM case, but this time w2 is set to 0.2, and the remaining weights are distributed evenly among the other metrics in use.
For the default option, the spatially varying weight maps are computed for both the transformed fixed and moving images on the SyN algorithm’s middle space, to capture the current tissue characteristics of both images in each iteration. This approach is significantly different than using a pre-defined spatially varying map on the fixed or moving image’s native space, which is typically the case when an exclusion mask is considered. Therefore, our spatially varying weighting formulation can not be used for such a purpose as the weight maps are computed for both images and evolve over the iterations, therefore are intrinsically dependent on the previous deformation field updates.
2.2.5. Exclusion Masks
Diffeomorphic registration requires that structures, possibly with different shapes, exist in both images. This is not always the case in medical imaging, particularly in the presence of focal abnormalities. Therefore, registration of the patient brain with focal lesions to a control template may be problematic. A typical strategy is to define a mask which excludes the lesion voxels from the metric computation, producing zero displacement for these locations (Avants et al., 2011). However, this is not sufficient when the lesion is in a subregion of the brain that requires large deformations. In such a case, the zero displacement in the lesion voxels and the diffeomorphism constraints would limit the displacements of larger neighboring regions. One other solution is to impose a “rigidity constraint” instead. This can be achieved either by constraining the determinant of the Jacobians within the mask to unity (Sdika, 2008) or by performing additional filtering of the deformation fields to enforce rigidity (Staring et al., 2007). With our unparameterized two-deformation fields model, constraining the Jacobians at subvoxel level is computationally expensive and infeasible. On the other hand, the filtering approach forces the displacement fields to a translation transform rather than a complete rigid transform with a rotation component. These options are not ideal in our case. An elegant solution would be to perform a registration that is designed to be robust to the presence of lesions as proposed by Stefanescu et al. (2004). Instead, for computational speed reasons, we implemented a new algorithm aimed at finding the best rotation and translation that describe the underlying displacements from the deformation fields. The details of this method can be found in Appendix A.3.
2.3. Atlas Creation
DR-TAMAS allows the creation of population atlases for both the diffusion tensor and the additional anatomical images provided by the user. The atlas creation methodology follows the principles proposed by Joshi et al. (2004) as all our tensor-based metrics support the ”averaging property” of Equation 6 in their work, when used individually or in combination (see Appendix A.5 for the proof). Note that this property does not hold for the cross-correlation metric (Avants et al., 2010) used for the registration of additional images, if also a structural atlas is desired by the user. In such a case, we follow the methodologies proposed by Avants et al. (2010) to create a structural atlas.
DTI atlas creation differs from structural image-based atlas creation because typically DT images are premasked to exclude nonbrain tissue regions and sometimes CSF regions. These exclusion masks are likely to differ among subjects, resulting in a fuzzy and poor initial template, which might reduce the quality of the subsequent iterative templates as shown by Wu et al. (2011). Moreover, the log-Euclidean averaging typically performed in tensor registration emphasizes these artifacts produced by holes in the original images. The traditional approach to overcome this issue is to use the voxelwise median instead of the arithmetic mean. However, in our experiments, we observed that the median operator produces very sharp and noisy images, which also tend to affect registration negatively. Therefore, we opted to use a family of smooth functions, which imitate the median function at the earlier stages of the atlas-creation process but which converge to the mean function at the later stages. The details of our atlas creation methodology can be found in Appendix A.4.
2.4. Experimental Setup
The proposed tensor registration methodology was tested using several data sets and compared with other well-known tensor and scalar image registration strategies. In this section we will describe the procedures to assess the performance of DRTAMAS.
2.4.1. Data Sets
Three sets of diffusion MRI data were used for the experimental validation and testing of DR-TAMAS. The first set (Atlas Set) was from 11 healthy volunteers scanned on a 3T MRI scanner (GE Medical Systems) equipped with a 32-channel receive coil. Diffusion weighted images (DWIs) were acquired with a single-shot spin-echo EPI sequence (FOV = 256 × 256 mm, slice thickness = 2 mm, matrix size = 128 × 128, 78 slices, TR/TE=9981/90 ms). An ASSET factor of 2 was used for parallel imaging. Each diffusion data set consisted of 4 image volumes with b = 0 s/mm2, 4 volumes with b = 150 s/mm2, 4 volumes with b = 300 s/mm2, 4 volumes with b = 500 s/mm2 and 62 volumes with b = 1100 s/mm2. High resolution T1-weighted and T2-weighted anatomical images were also acquired with a fast spin-echo sequence.
The second data set (SPG11 Set) consisted of diffusion MRI data collected in an unrelated study from a patient suffering from hereditary spastic paraplegia of type 11. The most salient brain abnormality in this disorder is severe atrophy of the corpus callosum and enlarged ventricles. DWIs were acquired on a Philips 3T Achieva scanner (FOV=296 × 296 mm, slice thickness=2mm, matrix size=160 × 160, 80 slices, TR/TE=11931/95.4 ms). The SENSE factor was 2. Diffusion experiments consisted of 8 volumes with b = 0, 15 volumes with b = 300, and 53 volumes with b = 1100 s/mm2. High resolution T1W and T2W anatomical images were also acquired.
The third data set (Ferret Set) consisted of high quality ex vivo ferret brain data. Two ex vivo ferret brains were scanned on a Bruker 7T vertical bore micro imaging system with the microWB gradient and probe system with a 25mm RF coil. One animal had a brain lesion produced by controlled cortical impact. A T2-weighted structural image was acquired using a multi-slice spin-echo sequence with: TE/TR=30/2000ms, nex=1, and the same spatial parameters described for DTI. For DTI, 94 image volumes were acquired for each specimen using a 3D echo-planar imaging sequence with: TE/TR=36/700ms, nex=1, segments=8 and FOV=40 × 26 × 20mm with matrix size=160 × 104 × 80 for 250 micron resolution. DWIs were acquired with diffusion timings of δ=3ms and Δ = 20ms. The diffusion experimental design consisted of 8 shells with b = 200, 300, 600, 1100, 1800, 3800, 6800, 10000 s/mm2 with N = 18, 18, 18, 18, 32, 32, 56, 105 volumes in each shell. Only the shells up to b = 3800 were used for tensor computations, and all the shells were used for the HARDI model.
2.4.2. DWI Processing
All diffusion data sets were first corrected for motion and eddy-currents distortions using the DIFFPREP tool of the TORTOISE package (Pierpaoli et al., 2010; Rohde et al., 2004a). Every subject’s image in the Atlas Set and also in the Ferret Set was corrected for concomitant field and susceptibility distortions using the “blip-up blip-down” strategy with DR-BUDDI (Irfanoglu et al., 2014, 2015). The images in the SPG11 Set were corrected for these EPI distortions by elastically registering them to a distortion-free structural image (Wu et al., 2008). As is standard processing procedure with the TORTOISE software, all individual imaging data were realigned onto a common standard orientation with the midsagittal plane of the image separating the two hemispheres and the intersection of the anterior and posterior commissures with the sagittal plane lying on the same axial slice (midsagittal and ACPC alignment). This provides a good initialization for tensor registration. The diffusion tensors were computed with DIFFCALC part of TORTOISE using the nonlinear fitting option. DR-TAMAS takes as input the tensor format produced by DIFFCALC.
2.4.3. Registration Quality Assessment
We compared the performance of DR-TAMAS to those of six well-known tensor and scalar image–based registration methods for diffusion MRI data implemented in publicly available software, using several criteria. These reference registration methods were 1) ANTS-scalar (Avants et al., 2008) using FA and TR images for registration, 2) ANTS-tensor using tensor component images in a multichannel setting, 3) DTITK-dev (Zhang et al., 2006) using the default deviatoric tensor similarity metric, 4) DTITK-full (Zhang et al., 2006) using the full tensor similarity metric, 5) FNIRT (Andersson et al., 2007) from the FSL package using FA images, and 6) DT-REFinD (Yeo et al., 2009), which is the algorithm included in the MEDINRIA package. All these methods along with DR-TAMAS, were used to perform registration for the two sets of human data. Atlas creation was performed with the algorithm provided in the software package, when available, otherwise it was performed by DR-TAMAS but with the metrics produced by the given method. For the registration methods that did not produce reoriented tensor data, the computed deformation fields were applied to the original tensor data, including finite strain reorientation, and the transformed tensors were averaged to generate the final DTI atlas. For each method, default parameters were used for registration and for DR-TAMAS, only tensor data were used for registration without any anatomical images.
For the Atlas Set, registration quality was evaluated by first visual inspection and subsequently, by quantitative assessment by computing the following voxelwise measures in the population:
- Principal Eigenvector Orientation Dispersion (PEOD): With the assumption that a perfect tensor registration algorithm would align the primary orientation, e1, to be identical for all voxels for all subjects, this assessment metric aims to quantify the directional alignment and tensor reorientation capabilities of the methods by examining the voxelwise variance of e1 within the population. Regions of interest (ROIs) for the cingulum bundle (CB), cortical-spinal tracts (CST), genu of corpus callosum (CC) and arcuate fasciculus (AF) were drawn on the final atlases from each method. The FA skeletons were also computed with FSL and used as an additional ROI. As proposed by Basser and Pajevic (2000), representing e1 as a second-order dyadic tensor allows one to quantify the dispersion about the mean within an ROI. Let the mean dyadic tensor at voxel x within the ROI ℝ be as follows:
The eigenvalues β1, β2, β3 of ε̄ can be used to measure the voxelwise dispersion of e1 orientations as:(17)
We report the average PEOD values within each ROI as a measure of orientation quality after registration.(18) - Overlap of the eigenvalue-eigenvector pairs (OVL): OVL (Basser and Pajevic, 2000) measures the similarity of the entire eigenvalue-eigenvector pairs of tensors from two images F and M over a region ℝ. The formulation is:
where Nℝ is the number of voxels in ROI ℝ. For OVL analysis, we used the same four ROIs in addition to the whole brain to quantify each method’s performance. OVL computes the similarity between a pair of images instead of the variance among a set of images. Therefore, we computed the OVL values between each of the 11 images in our population and reported the average of these values as our quantitative measure for each method.(19) FA and Trace Variance: Similarly to PEOD, the variances among the population of voxelwise FA and trace values were also computed. For trace variance, these measures were computed for the whole brain and for FA variance only voxels with FA values greater than 0.2 were used. Maps were also generated to visualize spatial patterns.
- Tensor Covariance (TCov): This measure quantifies the voxelwise variance of the tensors over the population, with smaller variance values indicating better alignment and tensor reorientation. Let the vectorization operator 𝒱 be the operator that converts a tensor into its vector version as . Then the 6 × 6 tensor-covariance matrix, which is equivalent to the underlying 3 × 3 × 3 × 3 higher order covariance tensor (Basser and Pajevic, 2003), can be written as:
where d̄(x) is the average vectorized tensor defined as . We define the TCov metric as the trace of this matrix as TCov(x) = Trace(Cov(x)). For each method, we generated maps of this measure and also reported its average value over the whole brain.(20) - Region Overlaps: Given that one of the goals of our work is to attain good alignment of GM and CSF regions in addition to WM regions, we evaluated the ability of the various methods to achieve good co-registration of these regions in the following way: First, we segmented subcortical GM, cortical GM, cortical WM, CC, and CSF regions using Freesurfer (Fischl et al., 2002) in the native space of the structural MRI of each subject. Subsequently, for each method, the displacement fields mapping each subject onto their corresponding atlases were applied to the segmentation label maps and the Dice overlap measure was computed on the warped label map (Dice, 1945) as:
The Dice coefficient measures how well the segmentation maps of the two images overlap. These coefficients were computed on both individual regions and the whole brain. Similarly to the OVL measure, these coefficients operate on a pair of images hence, they were computed between all the possible pairs in the population, and the means of these pairwise coefficients were reported.(21)
For the SPG11 Set, the diffusion tensor image of the patient was registered to the template generated by DR-TAMAS using the Atlas Set. Our quality assessment strategy involved visual inspection of the Trace, the directionally encoded color (DEC) maps (Pajevic and Pierpaoli, 1999) and the anatomical plausibility of fiber tractography of the cingulum bundle. The reason for choosing the cingulum bundle is as follows: The corpus callosum in the SPG11 patient is atrophic, but the adjacent cingulum bundle is mostly preserved and traceable in its entirety from a single ROI seed on the native patient data. However, when the image of the patient is warped to a healthy control template, it is important to ensure that the large deformation of the corpus callosum would not in turn disrupt the continuity of the cingulum bundle which is in close proximity to the corpus callosum. Therefore, we reasoned that the cingulum bundle’s tractography was a suitable test for the large-deformation modeling and tensor reorientation capabilities of the proposed and reference methods. An ROI of 6 voxels was drawn to the center of the cingulum bundle in the left hemisphere on the coronal plane of the target template image, and this same ROI was used as a seed region for tractography on the registered images from each method. The SPG11 Set was also used to demonstrate the effects of different metric weighting options provided by DR-TAMAS. The same registration was performed with different weight options and the results were visually examined.
In addition to its use with DTI data of the human brain, one can be interested in using DR-TAMAS for the registration of high angular resolution diffusion imaging (HARDI) data, for ex vivo diffusion tensor imaging in animal models, or for registering brains with lesions of relatively large size that may not have corresponding features in the target template. To test how DRTAMAS would perform on these tasks, we used the ferret data set. In this data set, the region affected by the controlled cortical impact did not have a corresponding region in the healthy brain; therefore, an exclusion mask on the impact region was created, and the registration was performed twice, once with and once without this exclusion mask. The resulting TR maps were visually examined. To assess the usability of DR-TAMAS on HARDI data, the MAP-MRI model (Özarslan et al., 2013) that computes features of the diffusion propagator was used for both the healthy and injured ferret brains. The deformation fields obtained from the registration using the mask was applied to the entire DWI set (297 volumes) of the injured brain and the q-vectors were rotated voxelwise based on these deformation fields. These voxelwise q-vectors were used to compute the diffusion tensors and MAP-MRI parameters. The glyphs representing orientation distribution functions (ODFs) derived from the MAP-MRI parameters were visually examined.
3. Results
3.1. Atlas Set Results
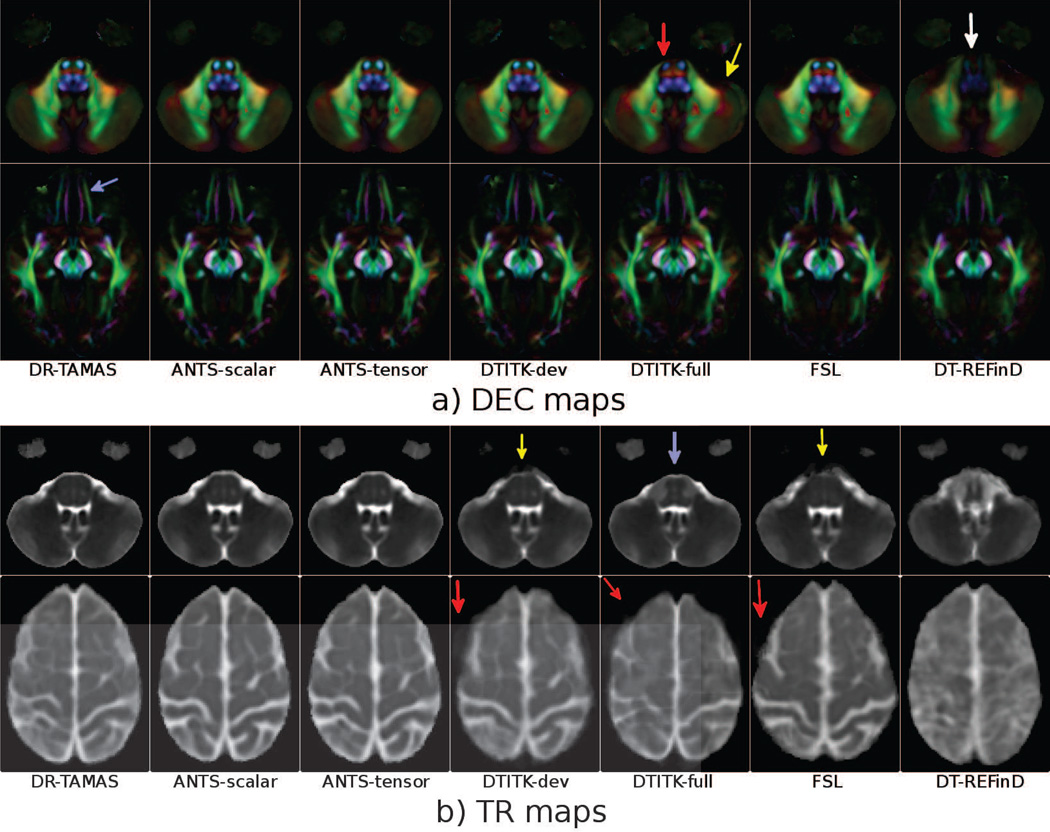
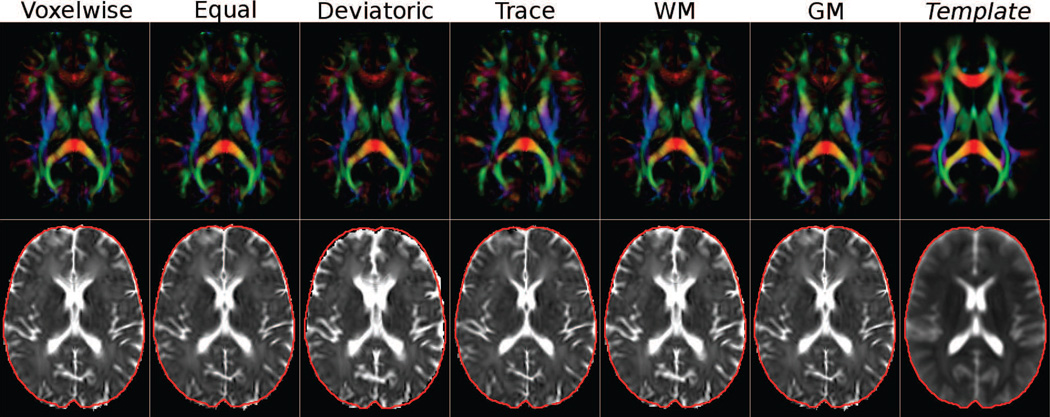
DEC maps for two slices from atlases built by each method can be found in Figure 1 (a). At visual inspection the DEC maps of the various methods appear very similar; however, subtle differences are evident, especially at the brain stem level. DEC maps represent anisotropy and orientation, and all the methods considering some form of anisotropy information (DR-TAMAS, ANTS-scalar, DTITK-dev, FSL) produced similar results; however, two of the methods employing full tensorial information, DTITK-full (red arrow) and DT-REFinD (white arrow) showed some abnormalities. At this level, DT-REFinD failed to align structures resulting in a blurry image. Additionally, DTITK-full was not able to register one of the subjects to the atlas, hence the averaging operation created a ghost-like silhouette outside the brain region (yellow arrow). At the second slice level, the WM of the gyrus rectus and of the medial orbital gyrus are nicely separated with DR-TAMAS (purple arrow), whereas they are slightly blended or merged with the other methods. Additionally, the two methods that only use anisotropy information, DTITK-dev and FSL, produced slightly larger inferior-frontal temporal lobe gray matter regions compared to the methods also using either trace or full tensor information.
Figure 1.
DEC maps (a) and Trace maps (b) of the average brains created by each method at two slice levels. The images are rigidly aligned for visualization.
Trace maps for the same data sets are displayed in Figure 1 (b). The trace maps at the brain stem level (top row) reveal that DR-TAMAS, and both ANTS methods performed very well both in the WM region and the surrounding CSF. The two methods employing solely anisotropy, DTITK-dev and FSL, produced very blurry CSF regions outside of the WM (yellow arrows). On the other hand, DTITK-full yielded a sharp WM/CSF interface; however, the WM within the brain stem was artifactual (blue arrow). DTITK-dev, DTITK-full and FSL suffered from mismatches at the superior cortical level (bottom row), which again caused ghost-like artifacts in the trace maps (red arrows).
PEOD measures, which quantify the variances of tensor primary eigenvectors, can be found in Table 1. With this measure, DR-TAMAS and DTITK-dev are the best performing methods with similar performances in all the WM ROIs tested. In WM, the similarity metric used by these two methods is identical and the only differences are the underlying transformation models and the constraints imposed by the surrounding GM and CSF regions. As expected, methods that consider tensor reorientation in their optimization performed well. The ANTS-tensor outperformed the ANTS-scalar method that uses only FA and TR but probably because of the lack of tensor reorientation, trailed behind DTITK-dev and DR-TAMAS.
Table 1.
Principal Eigenvector Orientation Dispersion (PEOD) measures.
| ROIs | DR-TAMAS | ANTS-scalar | ANTS-tensor | DTITK-dev | DTITK-full | FSL | DT-REFinD | Best Method |
|---|---|---|---|---|---|---|---|---|
| CB | 0.086 | 0.135 | 0.130 | 0.088 | 0.167 | 0.155 | 0.163 | DR-TAMAS |
| CST | 0.147 | 0.168 | 0.156 | 0.118 | 0.182 | 0.194 | 0.408 | DTITK-dev |
| CC | 0.059 | 0.147 | 0.136 | 0.060 | 0.078 | 0.151 | 0.075 | DR-TAMAS |
| AF | 0.118 | 0.176 | 0.177 | 0.099 | 0.148 | 0.160 | 0.150 | DTITK-dev |
| skeleton | 0.209 | 0.279 | 0.262 | 0.200 | 0.263 | 0.262 | 0.321 | DTITK-dev |
Lower is better.
Table 2 displays the values for OVL. These results are essentially identical to those found for PEOD, with DR-TAMAS performing the best in CB and CC and DTITK-dev performing the best in CST and AF. ANTS-tensor again performs better than ANTS-scalar with FSL and DT-REFinD trailing behind.
Table 2.
Overlap of eigenvalue-eigenvector pair (OVL) measures.
| ROIs | DR-TAMAS | ANTS-scalar | ANTS-tensor | DTITK-dev | DTITK-full | FSL | DT-REFinD | Best Method |
|---|---|---|---|---|---|---|---|---|
| CB | 0.928 | 0.885 | 0.889 | 0.926 | 0.908 | 0.852 | 0.829 | DR-TAMAS |
| CST | 0.887 | 0.866 | 0.878 | 0.919 | 0.850 | 0.837 | 0.729 | DTITK-dev |
| CC | 0.975 | 0.903 | 0.917 | 0.972 | 0.962 | 0.898 | 0.962 | DR-TAMAS |
| AF | 0.930 | 0.870 | 0.871 | 0.952 | 0.898 | 0.891 | 0.899 | DTITK-dev |
Higher is better.
Table 3 reports the average of voxelwise variance values for FA, TR, and tensors computed across the brain of all subjects for each registration method. Methods that use anisotropy information directly, ANTS-scalar, DTITK-dev, and FSL, produced the lowest FA variances. However, DTITK-dev and FSL achieved this at the cost of very large variance values for the trace. DR-TAMAS showed a very balanced behavior, producing close to optimal results for both metrics. Moreover, DRTAMAS was the best performing method for the tensor variance metric, which measures overall agreement of the diffusion displacement profiles including size, shape, and orientation.
Table 3.
Average FA, TR, and tensor variance measures.
| Var Type | DR-TAMAS | ANTS-scalar | ANTS-tensor | DTITK-dev | DTITK-full | FSL | DT-REFinD | Best Method |
|---|---|---|---|---|---|---|---|---|
| TRvar | 503917 | 481391 | 484210 | 715416 | 559097 | 905980 | 867658 | ANTS-scalar |
| FAvar | 0.0079 | 0.0072 | 0.0090 | 0.0069 | 0.0120 | 0.0077 | 0.0176 | DTITK-dev |
| TCOV | 202146 | 209050 | 206999 | 261177 | 229694 | 347748 | 337949 | DR-TAMAS |
Lower is better.
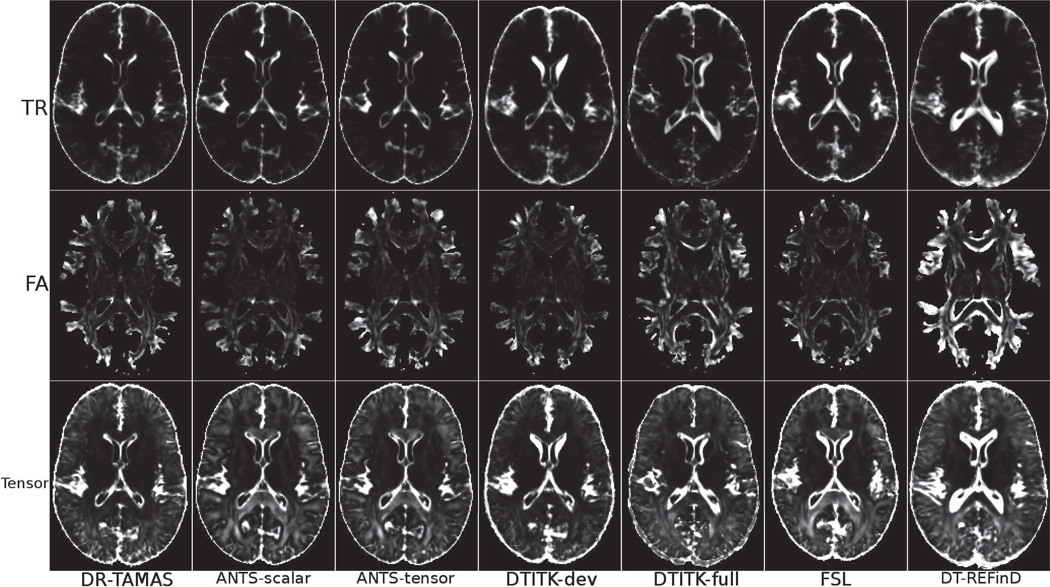
The maps of FA, TR, and tensor variances displayed in Figure 2 provide a more detailed understanding of the origin of the differences reported in Table 3. An examination of the trace variance maps reveals that methods that use only anisotropy information, such as DTITK-dev and FSL show poor performance at the GM/CSF boundaries. This is particularly evident at the level of the head of the caudate nucleus. TCOV maps confirms at the GM/CSF boundaries the suboptimal behavior of methods that rely exclusively on anisotropy information. Moreover, in WM structures such as the splenium of the CC, TCOV is relatively high in spite of low FA variance for the ANTS methods and FSL. Interestingly, these methods are the ones that do not include tensor reorientation in their implementation.
Figure 2.
Variances of FA, TR and tensors computed in the population for each method.
The DICE metrics can be found in Table 4. Results indicate that DR-TAMAS performed very well, being the best method in subcortical and cortical GM, WM, and CSF. ANTS-scalar achieved the best performance in the CC and the whole-brain.
Table 4.
DICE overlap measures.
| ROIs | DR-TAMAS | ANTS-scalar | ANTS-tensor | DTITK-dev | DTITK-full | FSL | DT-REFinD | Best Method |
|---|---|---|---|---|---|---|---|---|
| Subcort GM | 0.821 | 0.797 | 0.794 | 0.794 | 0.777 | 0.789 | 0.738 | DR-TAMAS |
| Cortical GM | 0.615 | 0.598 | 0.566 | 0.557 | 0.525 | 0.549 | 0.499 | DR-TAMAS |
| WM | 0.781 | 0.775 | 0.756 | 0.755 | 0.716 | 0.775 | 0.639 | DR-TAMAS |
| Center CC | 0.705 | 0.736 | 0.687 | 0.691 | 0.674 | 0.713 | 0.652 | ANTS-scalar |
| CSF | 0.839 | 0.808 | 0.809 | 0.800 | 0.812 | 0.800 | 0.731 | DR-TAMAS |
| Whole brain | 0.856 | 0.862 | 0.850 | 0.833 | 0.850 | 0.851 | 0.817 | ANTS-scalar |
Higher is better.
3.2. SPG11 Set Results
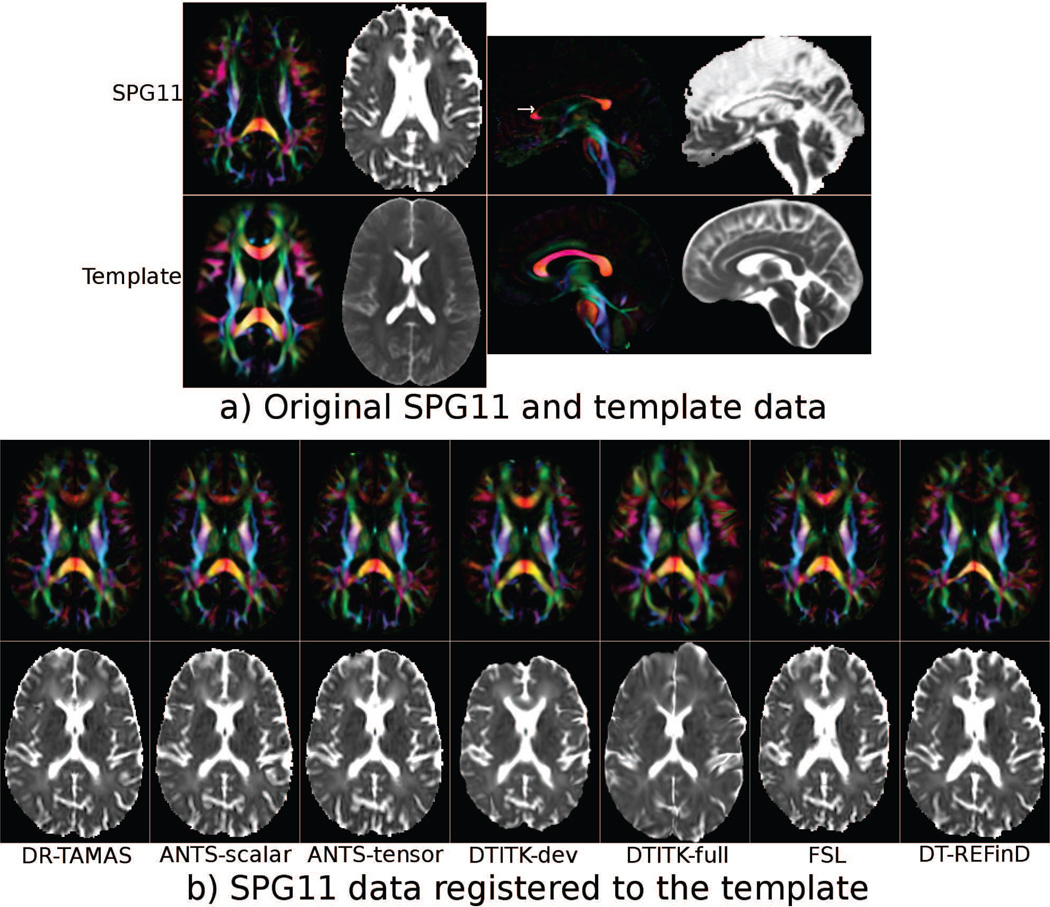
The DEC and TR maps of the brain of the SPG11 subject and the target average brain template are shown in Figure 3 (a). In the patient data, the corpus callosum, especially the body and genu, is severely atrophic, and the ventricle volume size is considerably increased compared with the average brain. The DEC and TR maps from the registration results of each method can be found in Figure 3. It can be noted that the DTITK-dev registration of the genu appears the closest to that of the target template. However, the TR map reveals that the registration of the GM and CSF regions adjacent to CC was unsatisfactory. The anterior horns of the lateral ventricles, for example, were still much larger in the registered patient data than in the template and their shapes did not match. DTITK-full shows smaller ventricles, but the anatomy of the entire frontal lobe appears incorrect. FSL produced reasonable results in the CC, but the ventricles were still too large. The ANTS methods performed similarly with high quality alignment of the ventricles, but poorer performance in the CC. DR-TAMAS was able to achieve a good alignment in both the DEC and TR maps. Even though the genu of the CC in the DEC map was not as completely aligned to the atlas as with DTITK-dev, it was still anatomically plausible in terms of both shape and orientation, and the alignment of the surrounding GM and CSF regions was excellent.
Figure 3.
a) DEC and TR maps of the SPG11 data and the target template image. The images are affinely aligned for visualization. The white arrow on the sagittal image indicates the level of the axial slices. b) Registration results of the SPG11 data to the template brain for each method.
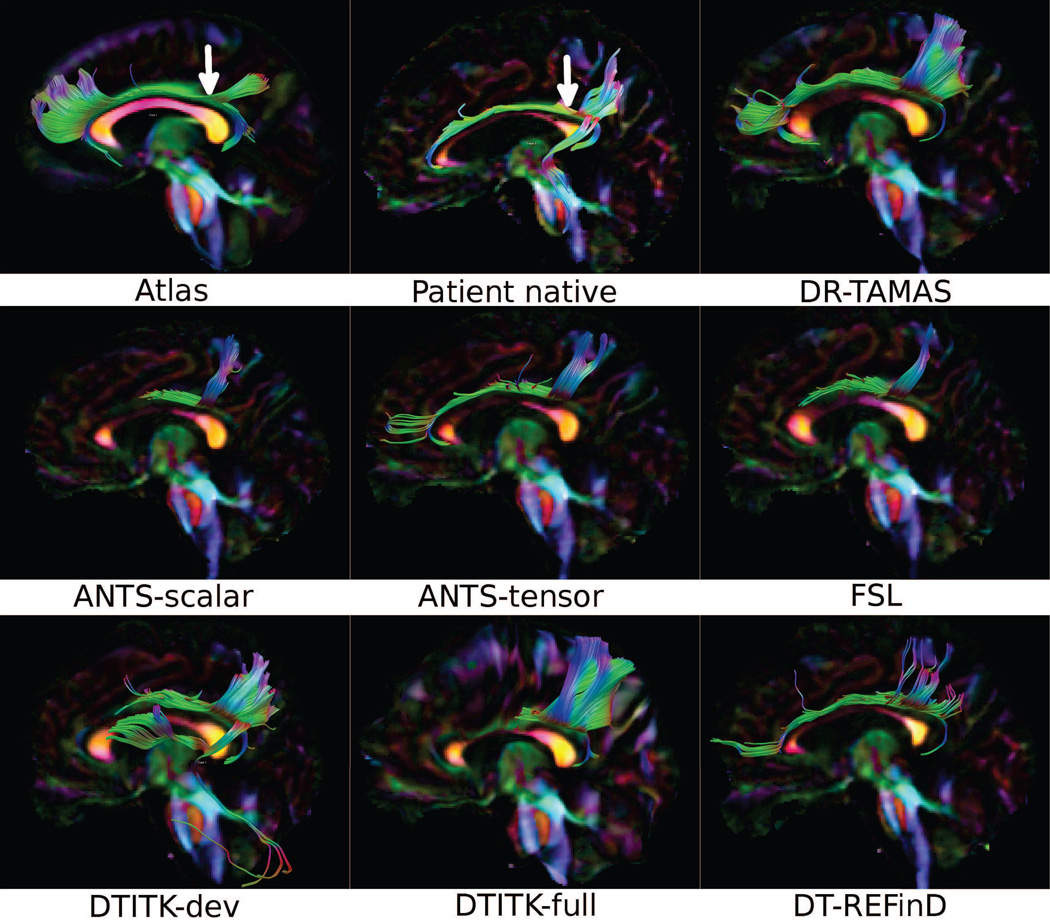
Figure 4 shows tractography results from a single seed ROI in the cingulum bundle on the target population atlas data, the SPG11 patient data in the native space, and the warped patient data for all methods. In the native space patient data, the cingulum was traced for its entire anterior-posterior trajectory. However, in the warped data from most methods, portions of the cingulum bundle remote from the seed could not be traced. This was particularly evident for methods such as FSL and DTITK-dev that produced a good registration of the genu of the corpus callosum. It seems that the deformations required to achieve such high performance in the CC lead to a disruption in the continuity of the cingulum bundle for these methods. DTITK-dev also generated spurious tracks. DR-TAMAS was able to correctly reconstruct this bundle in its entirety, including its most anterior and posterior curving parts. DT-REFinD also produced good tractography results, capturing most of the bundle. Another observation is that ANTS-tensor produced significantly better results than ANTS-scalar.
Figure 4.
Fiber tractography of the cingulum bundle using the target atlas data, the SPG11 patient data at the native space, and the resulting data from registration of SPG11 to the atlas by each method. The seed ROI locations on the coronal plane are indicated by white arrows.
Figure 5 displays the results of performing DR-TAMAS registration with different metric weight configurations. In addition to the four weighting settings described in Section 2.2.4, results from using only the deviatoric metric (third column) and the trace metric (fourth column) are also included. Similar to the case with other techniques, the weight configurations emphasizing white matter favoring metrics (deviatoric and WM) produced larger than normal ventricles in the trace maps. The overall brain size was smaller with the deviatoric metric as well (red contours). On the other hand, the configurations that favor the trace information (Trace and GM) produced incorrect alignment of the genu of the corpus callosum. These two options produced very sharp tissue interfaces on the TR maps as expected. The GM configuration, which also includes the deviatoric similarity metric with a small weight factor of 0.2, was able to produce more white matter tissue in the genu of CC compared to using only the Trace based metric. The more interesting comparisons are among the voxelwise FA, equal and deviatoric metric only configurations. As with DTITK-dev, the genu of the CC was nicely aligned with the target on the DEC maps with the deviatoric metric only but the TR maps showed artifacts. The voxelwise FA option produced a genu that is very similar to the deviatoric case but with no artifacts on the TR maps. The equal weighting between the deviatoric and the TR metrics yielded again a TR map without artifacts, slightly sharper than the voxelwise case, however the shape of the genu was not as nicely aligned compared to the default voxelwise option.
Figure 5.
Results of registering the SPG11 patient data to the template with DR-TAMAS using different metric weight configurations. The target template is displayed in the rightmost column. The red contour was drawn on the TR image of the template data and copied onto the TR images of different registration results.
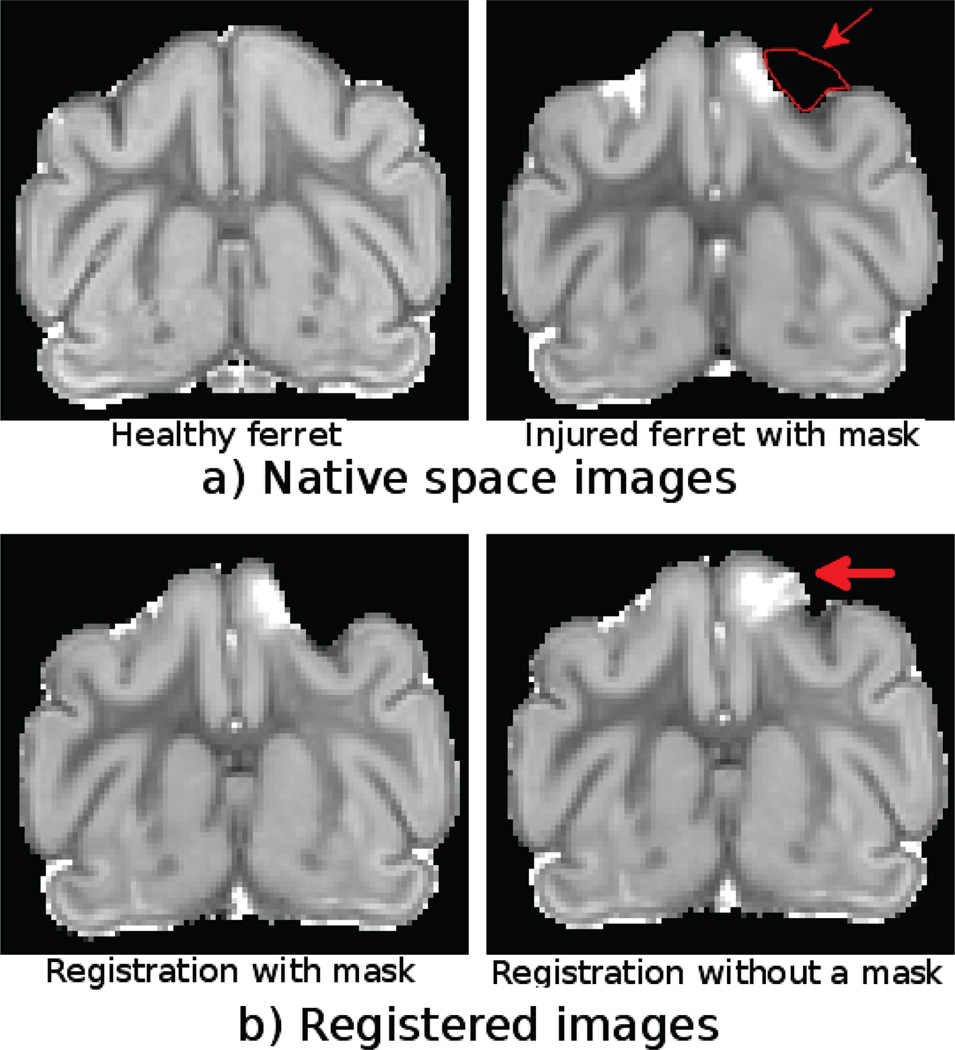
3.3. Ferret Set Results
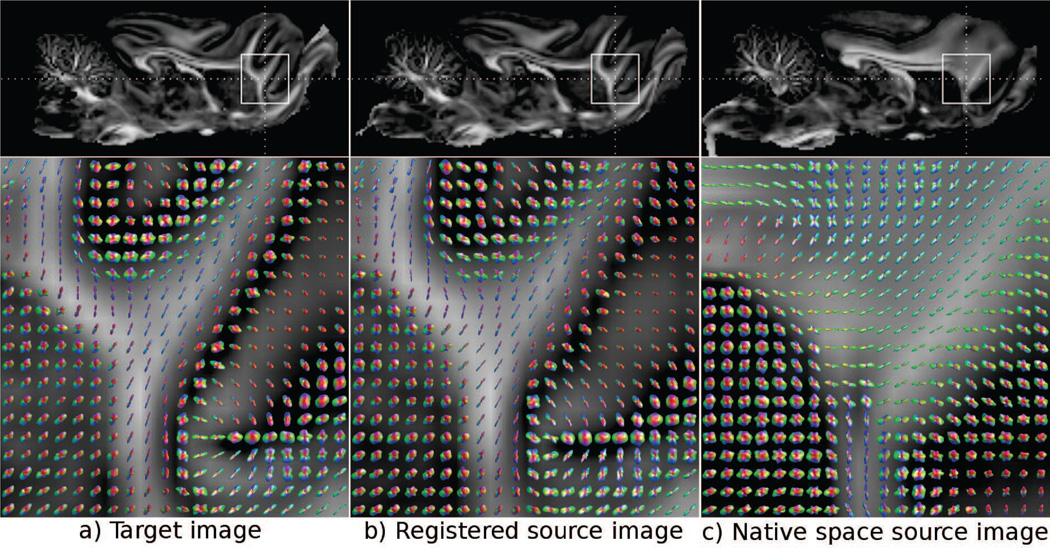
The TR maps of the native space healthy brain and the traumatic brain injury lesion brain from the Ferret Set are displayed in Figure 6 (a), with an area of missing tissue in the core of the lesion delineated by red contours. Figure 6 (b) displays the results of DR-TAMAS registration with and without an exclusion mask. Results of registration using the mask have shown that the mask regions are very similar to the original image with very minor differences due to the in-plane component of the rotation matrix of the enforced rigid transformation. However, in the processed data without a mask, the high trace portion of the lesion is stretched to reach the brain contour of the healthy brain. Figure 7 shows MAP-MRI data from the brain hemisphere contralateral to the lesion to analyze the effects of the registration using the HARDI type of data. The top row shows FA maps, and the bottom row displays the glyphs of the ODFs generated from the MAP-MRI model for the native space target and source data and after application of the deformation fields from DR-TAMAS to the DWIs of the source data. Differences between the two native space images are evident. After registration the regions were aligned, but the glyphs of both WM and GM voxels were also very similar in terms of their shape and orientation.
Figure 6.
Trace maps of healthy and injured ferret brains in the native space with the region to be excluded from the registration outlined in red.
Figure 7.
Effect of application of tensor registration based deformation fields from DR-TAMAS to HARDI models. Top row images display the FA maps with the ROIs indicated by white rectangles. Images in the bottom row display the ODF glyphs computed in these regions. The native space source image in (c) is registered to the target image in (a) with the registration output displayed in (b). The glyphs representing the complex diffusion process in the gray matter regions are directionally and shape-wise very similar between the registered source image and the target image.
4. Discussion
Our main objective in developing a novel image registration method for diffusion tensor images (DTI) was to attain high quality alignment of white matter WM structures without compromising the alignment of gray matter and cerebro-spinal fluid filled regions. Unlike previously proposed methods, DRTAMAS uses a spatially varying similarity metric, which is designed to favor orientation and anisotropy information in WM and to favor trace and structural MRI information in regions with isotropic diffusion properties.
We compared the performance of DR-TAMAS to that of other available diffusion MRI-based registration methods using different data sets. The accuracy of alignment of various brain structures was determined by using a combination of several metrics that included voxelwise variance of anisotropy, trace, and orientation maps, as well as degree of overlap of segmented WM, GM and CSF compartments obtained from structural MRIs. Not surprisingly, all registration methods showed good performances when alignment was tested on metrics that where used in their registration cost function. For example, FSL, a method that uses only FA in its similarity metric, performed well when alignment was assessed using FA maps; however, it showed poor performance when alignment was assessed using metrics of orientational coherence of eigenvectors or variance of trace. Also, the best correspondence based on the variance of-trace metric was achieved by the two methods that explicitly used trace information in their cost function: DR-TAMAS and ANTS-scalar. Thus one would have expected methods that use full tensor information, eigenvector information, and tensor reorientation during the registration to perform well when tested with metrics such as principal eigenvector dispersion and tensor covariance measures. This was generally true; however, large differences in performance could be found among methods.
If one considers all brain regions and assessment metrics, DR-TAMAS showed a very good overall performance; therefore it should be considered as the method of choice if one is interested in overall registration accuracy across the brain. For this reason, DR-TAMAS would also be an ideal method to create population brain atlases that faithfully represent the average anatomy for the population in all brain regions. The question arises as to what would be the method of choice if one were interested in registering only WM structures, for example, when the goal is to perform tractography. For instance, although DTITK-dev did not perform well in registering GM and CSF regions, it showed excellent performance in WM (see Tables 1 and 2). In this regard interesting observations can be made from the data of the hereditary spastic paraplegia type SPG11 patient. With the SPG11 data, DTITK-dev was able to warp the thin corpus callosum of the patient to match the corpus callosum of the target brain atlas of healthy subjects with very high morphological quality and high FA values, therefore assuring good traceability. However the excellent alignment of the CC was obtained at the expenses of a poor alignment of adjacent structures that had low anisotropy, such as the CSF spaces and the cingulate cortex, which in turn affected the continuity of the cingulum bundle, as can be seen in Figure 4. This leads us to conclude that although the registration of WM structures is in theory optimal with methods that give preference to anisotropy, and this is true for some WM regions, other WM structures are negatively affected by the poor registration of adjacent GM or CSF structures. For this reason, we would suggest using balanced methods, such as DR-TAMAS, even if the goal is the alignment of WM for tractography purposes.
Another important issue that the registration in the genu of the corpus callosum in the SPG11 data helps us analyze is defining the goal that a tensor-based registration algorithm should achieve. As Figure 3 indicates, DTITK-dev and FSL generated a very good visual alignment in this region with both morphological similarity to the target template region and high anisotropy values. However, in the original native space patient data, this region was severely atrophic and had relatively low anisotropy at the level displayed in the axial slice shown in Figure 3 (a). Therefore, the high anisotropy in the registration results of these two methods originated from pulling up voxels with high anisotropy, which were located several slices more caudally in the native data. This high anisotropy region is clearly visible on the sagittal DEC map displayed in Figure 3 (a). From a mathematical standpoint, the one-to-one mapping required by a diffeomorphism between the target template and the native patient data existed, and the registration algorithm pulled up these locations that were not spatially in correspondence to minimize its cost function. Such a mapping of noncorresponding regions could have negative implications on voxel-based morphometry analysis if used with tensor-derived quantities such as FA. With this type of analysis, mapping the genu of CC to a high anisotropy region would lead to a reduced statistical difference between the control and the patient data, which is obviously undesirable. In the extreme case of completely absent regions in one of the images, such as the data presented in the Ferret Set in this study, a one-to-one mapping does not exist, hence the solution requires exclusion of noncorresponding regions with a mask. However, this solution is not suitable for the SPG11 data because the CC does exist, and excluding the CC from registration would also affect the surrounding regions. Therefore, we believe “the ideal” tensor-based registration method should be capable of aligning images with a good morphological correspondence while preserving the diffusion characteristics of the tissue in its native images. In this respect, in the genu of the CC on the SPG11 data set, DR-TAMAS appeared to have reached good morphological similarity to the template while preserving the relatively low anisotropy of the original data.
An additional aspect that is worth discussing is the smoothing of the deformation fields. DR-TAMAS’ strategy for field smoothing is borrowed from the ITK library and is identical to that used by the ANTS algorithm. DR-TAMAS smooths the displacement vector update fields and the combined field (Equation 16) with two separate isotropic Gaussian kernels, where the kernel sizes are user-defined parameters. The choice of these smoothing parameters can significantly affect the quality of the final registration, where with data such as the SPG11 Set, small smoothing factors can lead to negative Jacobian determinants. Negative Jacobian determinants are inconsistent with diffeomorphism and would indicate that a structure has been torn apart by the registration, which is clearly undesirable. However, with well-behaved images, excessive smoothing will blur tissue interfaces, which is also undesirable. Currently, the user needs to experiment with these parameters based on the nature of the data. A strategy to help with this issue is to inherently apply an anisotropic smoothing kernel based on partial differential equations, where the structure tensor required by these methods is computed from the diffusion tensor images themselves and applied to the deformation fields. We are planning to investigate this approach in future research.
In addition to the deformation field smoothing Gaussian kernel sizes, another user-defined parameter that might affect the registration quality is the α term from Equation 14. In our early experiments, we observed with some data that the norm of the rotation term differential was significantly larger than the norm of the matching term differential. We hypothesized that this could be problematic for registrations that require large deformation mapping, where finite strain rotation and its differential are known to be problematic. The parameter alpha was introduced to be essentially a tweak to make the registration more robust if such large deformation mapping is required between the fixed and moving images. The use of this term results in a gradient direction that is the convex combination of the gradient direction obtained when tensor reorientation is considered in the optimization and the gradient direction when it is not. Therefore, the gradient direction obtained with this term does not originate from any theoretical formulation, however, in our experiments (data not shown), we achieved empirically robust results in terms of both tensor reorientation and large deformation mapping when this parameter was employed.
In this study, we showed an example of the use of DR-TAMAS with the HARDI type of data in the form of MAP-MRI. Application of a tensor-based registration to HARDI data was expected to be accurate in the WM, but we observed improvements even in GM regions. The ODFs in the GM regions surrounding the fiber bundles have similar directionality and shape in the registered source image and the target image. The improvements in these regions can be attributed to the observed anisotropy in ex-vivo tissue and the diffeomorphic movements of the surrounding regions, where DR-TAMAS has enough information to operate with. To further improve the alignment of such regions, one can start with an initial tensor-based registration with DR-TAMAS, and subsequently, further fine-tune the registration with similarity metrics specialized for MAP-MRI or the HARDI model in consideration.
One weakness of this study is that all the tested reference methods and DR-TAMAS were used with their default settings to perform registration on all data sets. It is likely that for each method, better performances may be achieved by fine-tuning the registration parameters. Therefore, the results presented in this study should not be perceived as “the best achievable performance” of a particular method but rather as representative of what can be achieved by a typical user. It would also be interesting to analyze the best possible performances in a future study or a conference workshop challenge, with the help and contribution from the authors of each method to clarify their strengths and weaknesses.
Highlights.
DR-TAMAS is a novel diffusion tensor image registration and atlas creation method.
Deviatoric tensor and trace differences are considered for tensor-driven metrics.
Optional anatomical images can complement tensor-based registration.
All considered metrics are combined at a voxel level based on tissue type.
DR-TAMAS performs robustly in all brain regions including WM, GM, and CSF.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development (NICHD), National Institutes of Health (NIH). Support included funding from the Department of Defense through the Henry Jackson Foundation (HJF Award#: W81XWH-13-2-0019) with the U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick, Maryland, 21702-5014, being the awarding office. The contents of this work do not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred. Special thanks go to Dr. Alex Martin, Section on Cognitive Neuropsychology, NIMH, for providing the MRI data used for the Atlas Set, Dr. Filippo Arrigoni of the Eugenio Medea Institute in Bosisio Parini, Italy, for providing the diffusion tensor data acquired on the HSP SPG11 subject, and Drs. Susan Schwerin and Sharon Juliano who are partner investigators in the Ferret study from which a sample dataset was used in this work. We also thank Liz Salak for editing this manuscript.
A. Appendix
A.1. Time-Varying Velocity Field Gradients
Let v1(x, t) be the tangential velocity field of ϕ1 that forward maps the fixed image to the moving image such that computed using the spatial gradients of the fixed image as in Equation 2 and v2(x, t) be the velocity field of the forward field ϕ2(x) generated using the movingimage gradients. In typical TVVF formulations, as in the ITK implementation, only v1(x, t) is computed using the fixed-image gradients. In the tensor registration case, due to the undesired holes in the images, in which there is no spatial gradient information, it is more robust to employ both sets of spatial gradients as follows:
| (22) |
where the velocity field at time point t is computed using images warped to that point as follows:
| (23) |
where ‖·‖ signifies the determinant of the spatial Jacobian of the displacement field up to time point t.
A.2. Deviatoric Metric Differentials
In their work, Yeo et al. (2009) showed that under finite strain formulation, the differential of a rotation matrix R with respect to a component of the Jacobian matrix J can be described as:
| (24) |
where Tr is the trace operator, I is the identity matrix, A is the affine matrix as A = J + I, (.)ii is the iith column of a matrix, and K is a matrix for which the i jth coefficient is one and the rest is zero. The operator ⊡ takes in a vector m = [m1,m2,m3]T and converts it to a skew-symmetric matrix as follows:
Let x be the voxel of interest, where the displacement vectors need to be computed. Let v be a voxel in the neighbourhood of x as: v ∈ xℕ : {{x−−, x−, x+, x++}, {y−−, y−, y+, y++}, {z−−, z−, z+, z++}} and the weights for the spatial gradient kernel: w = [1/12, −8/12, 8/12, −1/12]. Given a deformation field ϕ with 3 components, ϕx, ϕy, ϕz, the Jacobian matrix J(x) computed with the Jacobian operator 𝒥(ϕ (v)) can be written as follows (with l ∈ {−−, −, +, ++}):
| (25) |
As Equation 14 states, the term is needed to compute displacement differentials. By chain-rule this term can be written as follows:
| (26) |
The first term in this equation is defined in Equation 24. Please note that this term is computed in the neighboring voxel location. The second term can be computed from Equation 25. However, in Equation 25, the Jacobian is computed at voxel x using neighboring voxels v, whereas in Equation 26, the required term is computed at voxel v using the voxel x. This can be accomplished with a simple sign change as if the voxel v is at the ++ location relative to x; then x is at the −− location relative to v. The third term is simply the component of the Jacobian matrix computed at x. For example, for the x component of the deformation field, and the neighboring voxel v being x++, i.e., , this equation becomes:
| (27) |
and for :
| (28) |
A.3. Rigidity for Exclusion Masks
The simple heuristic idea behind our masking method is determining the best rotation matrix and translation vector describing the underlying displacements. Let ℳ be an exclusion mask defined on the moving-image space and ℳD is the morphologically dilated ℳ with a square structural element of size d, a parameter to DR-TAMAS. Let ℳE be the set difference as ℳE = ℳD\ℳ. Let the weight function be w(x) = 0.2 if x ∈ ℳ and x ∉ ℳD and w(x) = 1 if x ∈ ℳE. The weights were chosen this way to have the regions surrounding the exclusion mask have more of an effect on the rigid transformation than the voxels inside. The best rotation matrix R and translation vector t then can be computed by minimizing the error function as follows:
| (29) |
with the rotation matrix R constructed using Euler angles α, β, γ this equation can be simply solved with a gradient descent algorithm with gradients as follows:
| (30) |
| (31) |
The gradients for β and γ can be similarly computed, and the differential for is straightforward. After the optimum translation vector tf and rotation matrix Rf are computed, the new displacement vectors can be computed in a straightforward way, and the new final deformation field can be computed by applying Gaussian smoothing along the interface of the exclusion mask to guarantee diffeomorphism.
This process guarantees that the exclusion mask defined on the moving image will move rigidly to the middle time point image. However, the other part towards the fixed image needs to be constrained as well. Let ℳ′ be the transformed exclusion mask to the middle image with ℳ′(x) = ℳ(ϕ2(x)). Then the procedure described above needs to be repeated for and ℳ′, and the final ϕ1 can be computed by determining the inverse of the deformation field .
A.4. Iterative Template Averaging Functions
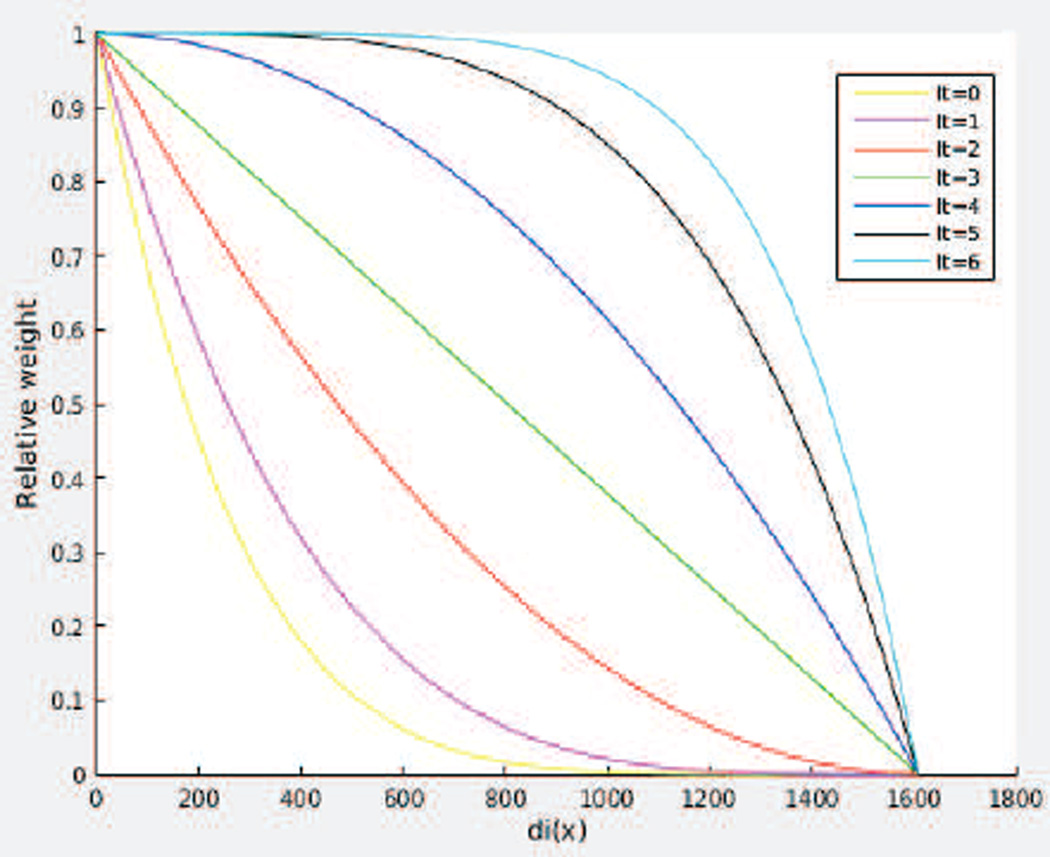
DR-TAMAS uses 6 iterations of atlas creation, Let it denote the current iteration, and let 𝒜0 be the initial template before registration, 𝒜6 be the final template, and 𝒜it the current template. Let Tri(x) be the trace of the subject i in the population at voxel x. The median trace for that voxel is denoted med(x) = median(Tri(x)) for all i. Let the deviation of the trace for each subject be di(x) = |Tri(x) − med(x)|. We use a function of these di(x) and it to determine the weighting contribution wi(x) of each subject to the template as follows:
This family of functions is plotted in Figure 8.
Figure 8.
Weighting functions of the subjects with respect to the current iteration for template averaging (It). di(x) is the voxelwise deviation of the trace of an image from the median trace at that voxel.
As the plots indicate for the early iterations (yellow, blue curves), only the subjects close to the median in terms of their trace values, i.e., low di(x) have larger weights and as the template creation process progresses, the weighting function becomes almost independent of di(x) and weighs all subjects almost equally, converging to an arithmetic mean operator.
A.5. Proof of Tensor Averaging for Atlas Creation
In their work, Joshi et al. (2004) showed that one can compute the diffeomorphic atlas by iterative registration followed by averaging. The averaging property (Equation 6 in their work) held true only if the similarity metric used for the registration produced such an optimal equation, which was the case for their L2-norm difference metric. Because we use the same averaging operator in our atlas creation routines, we have to show that the metrics we proposed in this work also follow the same ”averaging equation” for completeness. Let us denote the ith transformed tensor image at iteration n as for simplicity and as the jkth component of this tensor image. Then the average tensor image Ī can be found using the combined tensor-derived metrics as:
In this representation, the first summation term originates from the Trace similarity, the second summation term originates from the off-diagonal component similarity when using the deviatoric distance metric, and the third and last term (third and fourth lines) originates from the diagonal components when again using the deviatoric metric. When we take the derivative w.r.t. Ī12 to find this average off-diagonal component, the first and third summation terms do not contribute as they consist of only the diagonal terms. Therefore, equating the derivative to zero yields the formulation:
The other off-diagonal components can similarly be shown to be the average of the corresponding components in the tensor image set. Therefore, off-diagonal components follow the description of Joshi et al. (2004).
Deriving the formulations for the diagonal components is a little bit more involved because the first and the third terms contribute to the derivatives. Performing the differentiation, equating the system to zero and rearranging the terms with straightforward algebraic manipulations lead to the following linear system of equations:
When this system of equations is solved either for the extreme cases where either only the trace or the deviatoric metric is used (i.e. w1 = 0 or w2 = 0), or for its general case, it can be shown that is satisfied. Therefore, for both the diagonal and off-diagonal components, the averaging property holds when both our proposed metrics used alone or in combination.
When additional anatomical images are also used for registration, the term is added to the above metric formulation. Because this term does not include any tensor components, it does not affect the above equations for tensor atlas creation. To create an anatomical atlas, however, the formulations defined in (Avants et al., 2010) need to be used due to the cross-correlation metric.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adluru N, Zhang H, Tromp DPM, Alexander AL. Effects of DTI spatial normalization on white matter tract reconstructions. Proceedings of SPIE. 2013;8669 doi: 10.1117/12.2007130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DC, Gee JC. Elastic matching of diffusion tensor images. Computer Vision and Image Understanding. 2000;72(2):233–250. [Google Scholar]
- Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial transformations of diffusion tensor magnetic resonance images. IEEE Transactions on Medical Imaging. 2001;20(11):1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: Application to diffusion tensor imaging. NeuroImage. 2003;20(2):870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- Andersson JLR, Jenkinson M, Smith S. Tech. rep. FMRIB Oxford University; 2007. Non-linear registration, aka spatial normalisation. [Google Scholar]
- Avants B, Epstein C, Grossman M, Gee J. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC. The optimal template effect in hippocampus studies of diseased populations. NeuroImage. 2010;49(3):2457–2466. doi: 10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, Le Bihan D. Estimation of the effective selfdiffusion tensor from the NMR spin echo. Journal of Magnetic Resonance. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S. Statistical artifacts in diffusion tensor MRI (DTMRI) caused by background noise. Magnetic Resonance in Medicine. 2000;44:41–50. doi: 10.1002/1522-2594(200007)44:1<41::aid-mrm8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S. A normal distribution for tensor-valued random variables: applications to diffusion tensor MRI. IEEE Transactions on Medical Imaging. 2003;202:785–794. doi: 10.1109/TMI.2003.815059. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. Journal of Magnetic Resonance. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Bookstein FL. Voxel-based morphometry should not be used with imperfectly registered images. NeuroImage. 2001;14(6):1454–1462. doi: 10.1006/nimg.2001.0770. [DOI] [PubMed] [Google Scholar]
- Ceritoglu C, Oishi K, Li X, Chou M-C, Younes L, Albert M, Lyketsos C, van Zijl PC, Miller MI, Mori S. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. NeuroImage. 2009;47(2):618–627. doi: 10.1016/j.neuroimage.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen G, Rabbitt R, Miller M. Deformable templates using large deformation kinematics. Image Processing, IEEE Transactions on. 1996;5(10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- Commowick O, Wiest-Daessle N, Prima S. Automated diffeomorphic registration of anatomical structures with rigid parts: application to dynamic cervical MRI. 2012;15:163–170. doi: 10.1007/978-3-642-33418-4_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran KM, Alexander DC. Diffusion tensor orientation matching for image registration. SPIE Medical Imaging. 2003;5032:149–156. [Google Scholar]
- Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Garaci F, Toschi N, Lanzafame S, Marfia GA, Marziali S, Meschini A, Di Giuliano F, Simonetti G, Guerrisi M, Massa R, Floris R. Brain MR diffusion tensor imaging in Kennedy’s disease. The Neuroradiology Journal. 2015;28(2):126–132. doi: 10.1177/1971400915581740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee JC, Alexander DC. Diffusion-tensor image registration. In: Weickert J, Hagen H, editors. Visualization and Processing of Tensor Fields. Mathematics and Visualization. Springer: Berlin Heidelberg; 2006. pp. 327–342. [Google Scholar]
- Goh A, Vidal R. Algebraic methods for direct and feature based registration of diffusion tensor images. In: Leonardis A, Bischof H, Pinz A, editors. Computer Vision ECCV 2006. Vol. 3953 of Lecture Notes in Computer Science. Springer: Berlin Heidelberg; 2006. pp. 514–525. [Google Scholar]
- Guimond A, Guttmann CRG, Warfield SK, Westin CF. Deformable registration of DT-MRI data based on transformation invariant tensor characteristics. IEEE International Symposium on Biomedical Imaging: Macro to Nano. 2002:761–764. [Google Scholar]
- Irfanoglu MO, Koay CG, Pajevic S, Machiraju R, Basser PJ. Diffusion tensor field registration in the presence of uncertainty. Proceedings of MICCAI. 2009;1:181–189. doi: 10.1007/978-3-642-04268-3_23. [DOI] [PubMed] [Google Scholar]
- Irfanoglu MO, Machiraju R, Sammet S, Pierpaoli C, Knopp MV. Automatic deformable diffusion tensor registration for fiber population analysis. Proceedings of MICCAI. 2008;2:1014–1022. doi: 10.1007/978-3-540-85990-1_122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu MO, Modi P, Nayak A, Hutchinson EB, Sarlls J, Pierpaoli C. DR-BUDDI: (Diffeomorphic registration for blip-up blip-down diffusion imaging) method for correcting echo planar imaging distortions. Neuroimage. 2015;106:284–289. doi: 10.1016/j.neuroimage.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu MO, Modi P, Nayak A, Knutsen A, Sarlls J, Pierpaoli C. DR-BUDDI: Diffeomorphic registration for blip up-down diffusion imaging. Proceedings of MICCAI. 2014;17:218–226. doi: 10.1007/978-3-319-10404-1_28. [DOI] [PubMed] [Google Scholar]
- Jones DK, Griffin LD, Alexander DC, Catani M, Horsfield MA, Howard R, Williams SCR. Spatial normalization and averaging of diffusion tensor MRI data sets. NeuroImage. 2002;17:592–617. [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004;23(Supplement 1):151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang M-C, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shi Y, Tran G, Dinov I, Wang D, Toga A. Fast local trust region technique for diffusion tensor registration using exact reorientation and regularization. IEEE Transactions on Medical Imaging. 2014 May;33(5):1005–1022. doi: 10.1109/TMI.2013.2274051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney CJ, Simpson IJA, Nicholas JM, Fletcher PD, Downey LE, Golden HL, Clark CN, Schmitz N, Rohrer JD, Schott JM, Zhang H, Ourselin S, Warren JD, Fox NC. Longitudinal diffusion tensor imaging in frontotemporal dementia. Annals of Neurology. 2015;77(1):33–46. doi: 10.1002/ana.24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein KH, Westin C-F, Shenton ME, Weiner MW, Raj A, Thomann P, Kikinis R, Stieltjes B, Pasternak O. Widespread white matter degeneration preceding the onset of dementia. Alzheimer’s & Dementia. 2015;11(5):485–493. doi: 10.1016/j.jalz.2014.04.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Moreno E, Cárdenes-Almeida R, Martin-Fernandez M. Review of techniques for registration of diffusion tensor imaging. In: Aja-Fernndez S, de Luis Garca R, Tao D, Li X, editors. Tensors in Image Processing and Computer Vision. Advances in Pattern Recognition. London: Springer; 2009. pp. 273–297. [Google Scholar]
- Murphy K, van Ginneken B, Reinhardt J, Kabus S, Ding K, Deng X, Cao K, Du K, Christensen G, Garcia V, Vercauteren T, Ayache N, Commowick O, Malandain G, Glocker B, Paragios N, Navab N, Gorbunova V, Sporring J, de Bruijne M, Han X, Heinrich M, Schnabel J, Jenkinson M, Lorenz C, Modat M, McClelland J, Ourselin S, Muenzing S, Viergever M, De Nigris D, Collins D, Arbel T, Peroni M, Li R, Sharp G, Schmidt-Richberg A, Ehrhardt J, Werner R, Smeets D, Loeckx D, Song G, Tustison N, Avants B, Gee J, Staring M, Klein S, Stoel B, Urschler M, Werlberger M, Vandemeulebroucke J, Rit S, Sarrut D, Pluim J. Evaluation of registration methods on thoracic CT: The EMPIRE10 challenge. Medical Imaging, IEEE Transactions on. 2011 Nov;30(11):1901–1920. doi: 10.1109/TMI.2011.2158349. [DOI] [PubMed] [Google Scholar]
- Özarslan E, Koay CG, Shepherd TM, Komlosh ME, Irfanoglu MO, Pierpaoli C, Basser PJ. Mean apparent propagator (MAP) MRI: A novel diffusion imaging method for mapping tissue microstructure. NeuroImage. 2013;78:16–32. doi: 10.1016/j.neuroimage.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: Application to white matter fiber tract mapping in the human brain. Magnetic Resonance in Medicine. 1999;42:526–540. [PubMed] [Google Scholar]
- Park H, Kubicki M, Shenton ME, Guimond A, McCarley RW, Maier SE, Kikinis R, Jolesz FA, Westin CF. Spatial normalization of diffusion tensor MRI using multiple channels. NeuroImage. 2003;20:1995–2009. doi: 10.1016/j.neuroimage.2003.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Chiro GD. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201:637–648. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Walker L, Irfanoglu MO, Barnett AS, Chang LC, Koay CG, Pajevic S, Rohde GK, Sarlls J, Wu M. Tortoise: An integrated software package for processing of diffusion MRI data. Proceedings of International Society of Magnetic Resonance in Medicine. 2010:1597. [Google Scholar]
- Poudel GR, Stout JCDJFD, Churchyard A, Chua P, Egan GF, Georgiou-Karistianis N. Longitudinal change in white matter microstructure in Huntington’s disease: The IMAGE-HD study. Neurobiology of Disease. 2015;74(0):406–412. doi: 10.1016/j.nbd.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Rohde GK, Barnett AS, Basser PJ, Marenco S, Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion weighted MRI. Magnetic Resonance in Medicine. 2004a;51(1):103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- Rohde GK, Pajevic S, Pierpaoli C. Nonlinear registration of diffusion tensor images using directional information. Proceedings of International Society for Magnetic Resonance in Medicine. 2004b;11:1213. [Google Scholar]
- Ruiz-Alzola J, Westin CF, Warfield SK, Alberola C, Maier S, Kikinis R. Nonrigid registration of 3D tensor medical data. Medical Image Analysis. 2002;6:143–161. doi: 10.1016/s1361-8415(02)00055-5. [DOI] [PubMed] [Google Scholar]
- Sadeghi N, Nayak A, Walker L, Irfanoglu MO, Albert P, Pierpaoli C, Group BDC. Analysis of the contribution of experimental bias, experimental noise, and inter-subject biological variability on the assessment of developmental trajectories in diffusion MRI studies of the brain. NeuroImage. 2015;109:480–492. doi: 10.1016/j.neuroimage.2014.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdika M. A fast nonrigid image registration with constraints on the jacobian using large scale constrained optimization. Medical Imaging, IEEE Transactions on. 2008;27(2):271–281. doi: 10.1109/TMI.2007.905820. [DOI] [PubMed] [Google Scholar]
- Staring M, Klein S, Pluim JPW. Nonrigid registration with tissue-dependent filtering of the deformation field. Physics in Medicine and Biology. 2007;52(23):6879. doi: 10.1088/0031-9155/52/23/007. [DOI] [PubMed] [Google Scholar]
- Stefanescu R, Commowick O, Malandain G, Bondiau P-Y, Ayache N, Pennec X. Non-rigid atlas to subject registration with pathologies for conformal brain radiotherapy; Proceedings of MICCAI. Vol. 3216 of Lecture Notes in Computer Science; 2004. pp. 704–711. [Google Scholar]
- Sweet A, Pennec X. Log-domain diffeomorphic registration of diffusion tensor images; WBIR: Workshop on Biomedical Image Registration; 2010. pp. 198–209. [Google Scholar]
- Wang Q, Yap P-T, Wu G, Shen D. Diffusion tensor image registration using hybrid connectivity and tensor features. Human Brain Mapping. 2014;35(7):3529–3546. doi: 10.1002/hbm.22419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen Z, Nie S, Westin C-F. Diffusion tensor image registration using polynomial expansion. Physics in Medicine and Biology. 2013;58(17):6029. doi: 10.1088/0031-9155/58/17/6029. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Liu Z, Zhang H, Escolar ML, Gilmore JH, Gouttard S, Fillard P, Maltbie E, Gerig G, Styner M. {DTI} registration in atlas based fiber analysis of infantile Krabbe disease. NeuroImage. 2011;55(4):1577–1586. doi: 10.1016/j.neuroimage.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Jia H, Wang Q, Shen D. Sharpmean: Groupwise registration guided by sharp mean image and tree-based registration. NeuroImage. 2011;56(4):1968–1981. doi: 10.1016/j.neuroimage.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Chang LC, Walker L, Lemaitre H, Barnett AS, Marenco S, Pierpaoli C. Comparison of EPI distortion correction methods in diffusion tensor MRI using a novel framework. Proceedings of MICCAI. 2008;11:321–329. doi: 10.1007/978-3-540-85990-1_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Shen D, Davatzikos C, Verma R. Diffusion tensor image registration using tensor geometry and orientation features. Proceedings of MICCAI. 2008;5242:905–913. doi: 10.1007/978-3-540-85990-1_109. [DOI] [PubMed] [Google Scholar]
- Yap P, Zhu H, Lin W, Shen D. Hierarchical diffusion tensor image registration based on tensor regional distributions. Proceedings of International Society of Magnetic Resonance in Medicine. 2009a;17:1426. [Google Scholar]
- Yap P-T, Wu G, Zhu H, Lin W, Shen D. Timer: Tensor image morphing for elastic registration. NeuroImage. 2009b;47(2):549–563. doi: 10.1016/j.neuroimage.2009.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap P-T, Wu G, Zhu H, Lin W, Shen D. F-timer: Fast tensor image morphing for elastic registration. IEEE Transactions on Medical Imaging. 2010 May;29(5):1192–1203. doi: 10.1109/TMI.2010.2043680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo B, Vercauteren T, Fillard P, Pennec X, Gotland P, Ayache N, Clatz O. DTI registration with exact finite-strain differential. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI. 2008;14:700–703. [Google Scholar]
- Yeo B, Vercauteren T, Fillard P, Peyrat J, Pennec X, Golland P, Ayache N, Clatz O. DT-REFinD: Diffusion tensor registration with exact finite-strain differential. IEEE Transactions on Medical Imaging. 2009;28(12):1914–1928. doi: 10.1109/TMI.2009.2025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yushkevich PA, Alexander DC, Gee JC. Deformable registration of diffusion tensor MR images with explicit orientation optimization. Medical Image Analysis. 2006;10(5):764–785. doi: 10.1016/j.media.2006.06.004. [DOI] [PubMed] [Google Scholar]