Abstract
Background
Recent spread of USA300 methicillin-resistant Staphylococcus aureus (MRSA) to nursing homes has been of particular concern. We sought to predict the ultimate prevalence of USA300 and non-USA300 MRSA and to examine the influence of potential risk factors on MRSA acquisition in community nursing homes.
Methods
The data was collected during a longitudinal MRSA surveillance study that involved 449 residents in six community nursing homes in Wisconsin. The subjects were screened every three months for up to one year. Markov chain models were employed to predict strain-specific prevalence of MRSA at steady state, and to assess the impact of potential risk factors, including recent hospitalizations, invasive medical devices, and antibiotic exposure on MRSA acquisition rates and average duration of colonization.
Results
At steady-state, 20% (95% CI, 15–25%) of residents were predicted to remain colonized with non-USA300 and 4% (95% CI, 2–7%) with USA300 MRSA. Residents who used antibiotics in the previous 3 months were twice more likely to acquire MRSA than those who did not (acquisition rates 0.052 (95% CI, 0.038–0.075) and 0.025 (95% CI, 0.018–0.037), respectively).
Conclusions
Non-USA300 was predicted to remain the dominant MRSA strain in community nursing homes. The higher rate of MRSA acquisition among residents with recent antibiotic exposure suggests that antibiotic stewardship may reduce MRSA colonization in this setting.
Keywords: USA300 MRSA, non-USA300 MRSA, colonization, prevalence, risk factors, acquisition rates
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a common cause of infection in community and healthcare settings.1,2 These infections are responsible for substantial patient morbidity and mortality and significantly increase healthcare utilization relative to infections caused by susceptible strains.1,3–5 Consequently, there has been a great interest in understanding the dynamics and epidemiology of MRSA in healthcare facilities.6–11
Nursing homes are major reservoirs of MRSA with prevalence of colonization among residents approaching 50% in some settings.12–16 Residents in these facilities transition frequently between community and other healthcare settings, and nursing homes likely play a critical role in the regional spread of multidrug-resistant organisms, including MRSA.17–19 While a number of mathematical models have been used to describe the dynamics of MRSA in acute-care settings7,10,11,20 and to predict the role of nursing homes in the regional spread of MRSA,17,20–22 their use in describing the dynamics of MRSA within the nursing home remains limited.
A number of factors could influence the dynamics of MRSA within nursing homes. Prior studies have shown that invasive medical devices, recent antibiotic use, chronic wounds, comorbidity and frailty are risk factors for colonization with MRSA when residents with prevalent MRSA colonization are compared to non-colonized residents in cross-sectional or case-control studies.23 Whether these same conditions and exposures are also risk factors for acquisition of MRSA while residing in nursing homes24,25 and whether different strains of MRSA have an impact on the dynamics of spread in nursing homes remains poorly understood. While healthcare-associated strains (e.g., USA100 CDC pulsotype) have been the dominant MRSA clones identified in most settings,12,14,26 an increasing number of studies suggest that community-associated strains (particularly, the USA300 CDC pulsotype) are becoming more common in these facilities.14,15 This is of particular concern as community-associated MRSA strains may be more virulent and easily spread between individuals as compared to healthcare- associated strains.23,27,28
Given these gaps in knowledge, we sought to describe the transition dynamics of MRSA within nursing homes and assess the influence that MRSA strain-type and potentially modifiable resident characteristics have on this process using a mathematical modeling approach. We hypothesized that resident acquisition of community-acquired MRSA (defined in this study as colonization with any USA300 pulsotype strain) differs from acquisition of healthcare-associated MRSA (defined in this study as colonization with any pulsotype strain other than USA300). We further hypothesized that resident characteristics that influence acquisition of USA300 and non- USA300 MRSA are distinct. To test these hypotheses, we employed Markov chain models to predict the distribution of residents colonized with USA300 and non-USA300 in the long run and to assess how potentially modifiable resident risk factors influence MRSA acquisition stratified by pulsotype strain.
Materials and Methods
Sources of Data
Data employed in this research were derived from a prospective longitudinal study of MRSA colonization performed in six community nursing homes between February 2008 and October 2010.29 The six nursing homes were located in 5 counties in South Central Wisconsin. Subjects participating in this study were screened for MRSA colonization during a baseline examination and every three months thereafter for a period of up to one year or until they were discharged from the study facility, whichever occurred sooner. Surveillance cultures were collected from multiple anatomical locations of each subject, including surface cultures of the nares, skin of the axilla, groin and perirectal region. Additional cultures were obtained from open wounds and the insertion sites of invasive devices, when applicable. A urine specimen was collected from subjects with indwelling urinary devices. Additional data was abstracted from health records during these visits to ascertain subject exposure to risk factors that might potentially influence their colonization status. Subject comorbidity, functional status and cognitive status were assessed at baseline. A Charlson Comorbidity Index30 score ≥3, a Katz Activities of Daily Living (ADL)31 score < 2 and Minimum Data Set (MDS) Cognitive Performance Scale (CPS)32 score ≥5 were used to stratify subjects into severe levels of comorbidity, function and cognition, respectively, and non-severe otherwise. Additional exposure to other potential risk factors, including presence of a chronic wound, invasive medical devices, hospitalization and antibiotic exposure within the preceding 3 months were ascertained at baseline and each subsequent assessment and used as dichotomous variables in our study. This study was reviewed and approved by the Health Sciences Institutional Review Boards of the University of Wisconsin-Madison.
MRSA Colonization Patterns Observed
Four hundred and forty nine subjects were screened for MRSA colonization at baseline, and 319 of these subjects were screened for MRSA colonization during subsequent follow-up on at least one occasion (Appendix Table 1). A subject’s MRSA status during a particular assessment was considered evaluable only if the results of cultures obtained from the nares, stool or peri-rectal skin, and skin of the axilla or groin was not missing. Of the 1468 subject assessments performed over the course of this study, 12 were considered non-evaluable (0.8% of total sample) due to missing data at one of the three screening body sites (Appendix Table 1). The total of 446 subjects contributed evaluable observations to the study. Ninety-five of the 446 subjects (21.3%) were colonized with MRSA at baseline: 79 subjects (17.7%) were colonized with non-USA300, 13 subjects (2.9%) with USA300 and 3 subjects (0.7%) were co-colonized with both strain-types. An additional 52 subjects (11.7%) who were not identified as colonized at baseline had positive MRSA cultures during one or more of the follow-up assessments. MRSA was never recovered from the remaining 298 subjects (66.8%). In total, 72 MRSA acquisitions were observed over the course of the study: 68 events in subjects who were MRSA colonized following a prior assessment in which they were non-colonized and 4 events in which subjects became colonized with a MRSA strain that was genetically distinct from isolates recovered during prior assessment.
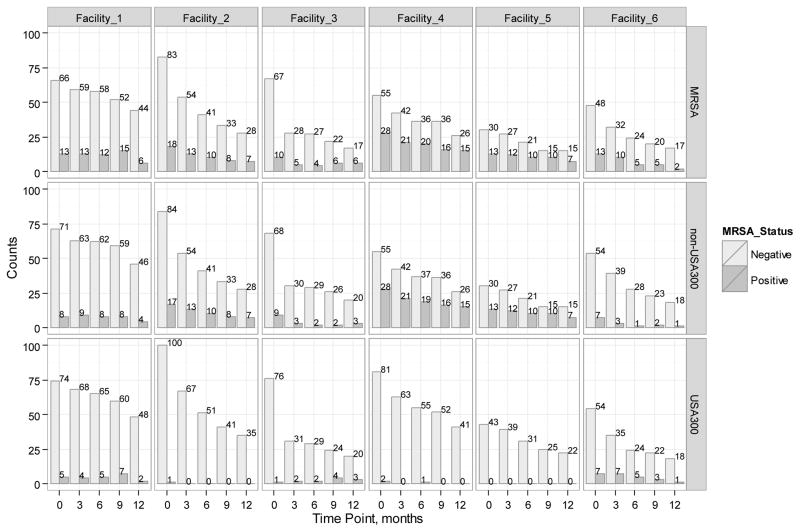
For the purposes of this study, MRSA[+] subjects were classified as colonized with a USA300 if an organism with this pulsotype was recovered from culture, otherwise they were classified as being colonized with a non-USA300 strain (e.g., subjects from whom USA700 was recovered were classified as being colonized with a non-USA300 strain of MRSA). MRSA was recovered from 663 of the cultures. Upon molecular typing, 495 of the isolates were identified as belonging to the USA100 CDC pulsotype group, 114 were assigned to the USA300 CDC pulsotype group, and the remaining 24 isolates were assigned to other CDC pulsotype groups.33 The distribution of the different MRSA strain types, stratified by facility, is included in the appendix (Appendix Figure 1).
Models Overview
We used Markov chain models34 with the assumption of non-state-dependent acquisition rates to describe the dynamics of MRSA in nursing homes. Markov chain models are ideal tools for describing processes in which individuals move through a number of states (e.g., non-colonized, colonized with non-USA300, colonized with USA300) in discrete or continuous time. They allow to model transition rates between the states and predict the population distribution at steady state. The states of our models represented a subject’s possible MRSA colonization status. Colonization data observed in our study was used to fit discrete and continuous time models of differing complexity. Theoretically, a subject in our sample could be free of any MRSA, colonized with USA300 only, colonized with non-USA300 only, or colonized with both strains. Consequently, the model that would describe such a system most closely would have four states. However, due to the low number of transitions observed between some states, models with a fewer number of states (either two or three) were employed in this study (Figure 1A–1B). Over time, a subject could transition to another state or remain in the same state. The probability of a subject’s MRSA colonization status at a particular time point was assumed to only depend on the subject’s MRSA status during the immediate prior assessment and not on the full history of MRSA colonization or the present amount of colonized in the facilities.34 All computations and data analysis were performed in R software environment, version 3.1.0.35
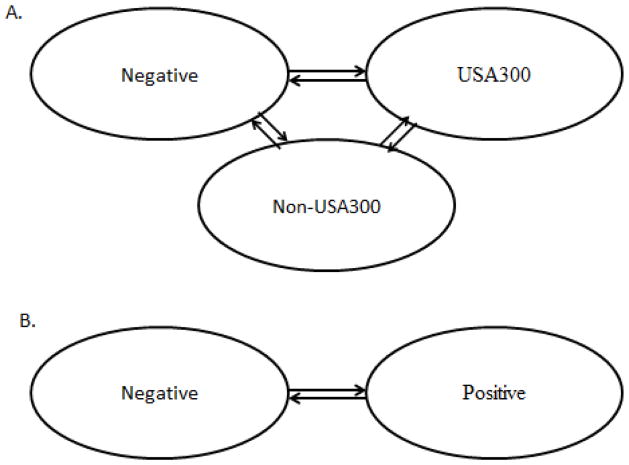
Figure 1.
Diagrams of the general models used to describe the transition dynamics of MRSA in nursing homes. (A) the model with three states which assumes that a resident may be MRSA free (Negative), colonized with USA300 and, possibly, co-colonized with non-USA300 (USA300) or colonized with non-USA300 (non-USA300); (B) two-state model which considers colonization with any MRSA strain or a specific strain, either non-USA300 or USA300. In the strain-independent model, Positive represents colonized with any MRSA strain and Negative stands for non-colonized with MRSA. In the non-USA300 model, Positive represents colonized with non-USA300 while Negative represents free of non-USA300, irrespective of USA300. Similarly, in the USA300 model, Positive stands for colonized with USA300 and Negative represents free of USA300, irrespective of non-USA300 strain-type.
Predicting Steady-State Distribution of MRSA in Study Nursing Homes
To predict the steady-state distribution of MRSA in nursing homes, a discrete-time Markov chain model with three states was employed (Figure 1A, Appendix Table 2). Due to the small number of transitions observed in each facility, the data from the six study nursing homes were combined, so that the estimated parameters would be representative of the hypothetical “average” nursing home (the characteristics of the individual facilities were published elsewhere29). Models to predict the steady state of MRSA in individual nursing homes, while underpowered, were fit separately. In these models, subjects could be non-colonized (Negative) or colonized either with a USA300 (USA300) or non-USA300 (non-USA300) MRSA strain but not both. The small numbers of co-colonized individuals observed in the study sample were characterized as USA300 in the models. The transition probability matrix34 that was used to calculate the steady-state distribution is shown in Appendix Table 2. Standard errors and 95% confidence intervals around the point estimates were computed by means of bootstrapping with 1000 resampling runs and based on approximate normality.36
Impact of Resident Risk Factors on Predicted MRSA Transition Rates and Average Duration of Colonization in Study Nursing Homes
Continuous-time Markov chain models were used to estimate MRSA transition rates and average duration of colonization (i.e., the average time during which the same MRSA strain remained recoverable from a study subject) in our study of nursing homes. Similar to the approach used for predicting a steady-state distribution, data from the six study nursing homes were combined to increase precision of the estimated model parameters (the characteristics of the participating facilities can be found elsewhere29). A mixture of two- and three-state models was employed (Figure 1A – 1B). A strain-independent two-state model (Figure 1B), in which a subject was either MRSA[+] (Positive) or MRSA[−] (Negative), was used to explore the impact of individual candidate risk factors on MRSA transition rates. Strain-specific models were subsequently fit to identify interaction effects between individual candidate risk factors and strain type (USA300 and non-USA300). Three-state strain-specific models (Figure 1A) failed to converge for most of the candidate risk factors due to the scarcity of data. Consequently, simplified strain-specific two-state models (Figure 1B, Appendix Table 2) were employed. In these simplified two-state models, a subject was designated as non-colonized (Negative) if they did not harbor the MRSA strain being evaluated in the current model and colonized (Positive) otherwise (i.e., a resident colonized with non-USA300 but free of USA300 was considered Negative in the USA300 two-state model). The R package msm (version 1.3) was used for model fitting.35,37
Results
Predicted Steady-State Distribution of MRSA in Study Nursing Homes
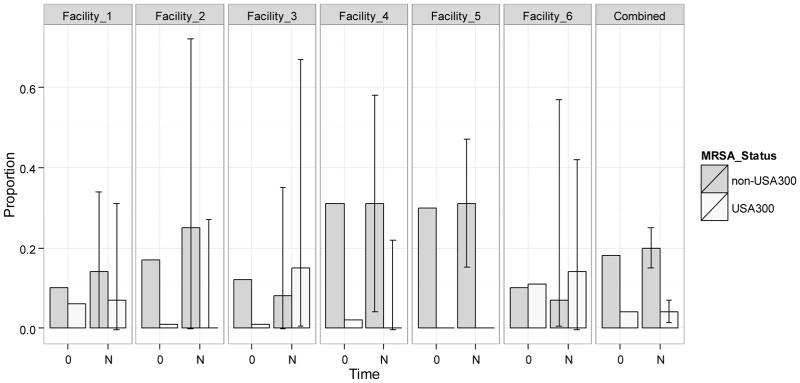
For all facilities combined, that is, for our hypothetical “average” nursing home, the significant differences in the distribution of residents colonized with USA300 and non-USA300 at steady state were observed (Figure 2). In the long run, 4% (95% CI, 2–7%) of nursing home residents were predicted to be colonized with USA300 (and possibly co-colonized with non-USA300), 20% (95% CI, 15–25%) would be colonized with non-USA300, and 75% (95% CI, 70–81%) would remain free of MRSA. Facility-level models predicted that non-USA300 MRSA would dominate over USA300 strains in four of the six study facilities, including Facility 5 which did not have any residents colonized with USA300 over the course of the study. While the predicted prevalence of USA300 exceeded that of non-USA300 in facilities 3 and 6, these differences were not statistically significant.
Figure 2.
Observed distribution of subjects colonized with non-USA300 or USA300 at baseline and predicted distribution with 95% confidence intervals at steady state. USA300 includes those few residents co-colonized with non-USA300. Horizontal axis represents time: 0 stands for baseline, and N stands for steady-state. The 95% confidence intervals at steady-state are depicted with error bars. The columns show the distribution in each facility; the last column labeled Combined presents the distribution in all facilities combined.
Impact of Resident Risk Factors on Predicted MRSA Transition Rates
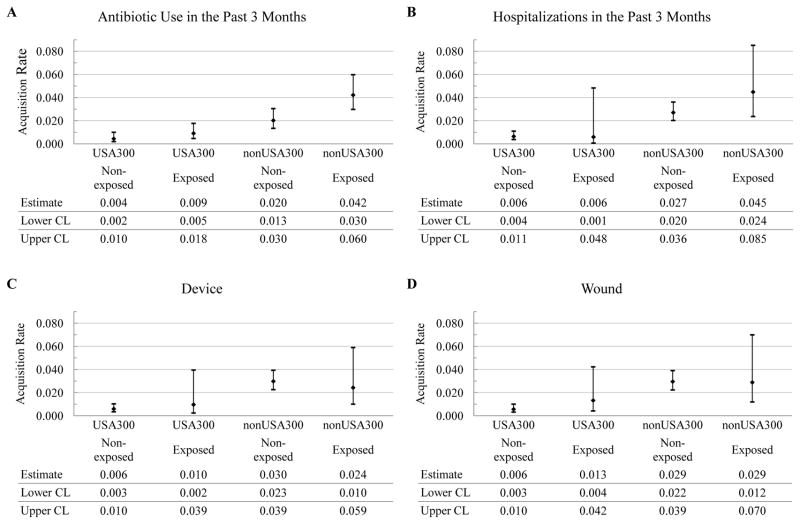
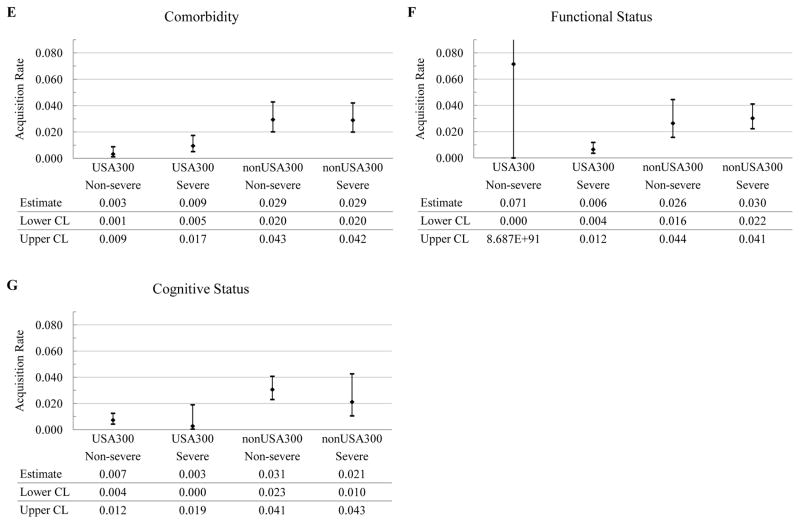
In the strain-independent two-state continuous time Markov model (subjects either MRSA[+] or MRSA[−]), antibiotic use in the past 3 months doubled the acquisition rate of MRSA from 0.025 (95% CI, 0.018 – 0.037) to 0.052 (95% CI, 0.038 – 0.075) (Table 1). None of the other candidate risk factors examined in the study had a statistically significant impact on the transition rates of strain-independent MRSA (Table 1). It is worth noting that the difference between the point estimates of the levels of previous hospitalizations and cognitive status was quite pronounced, even though the statistical significance at 95% confidence level was not achieved in our sample which had low numbers of observations for some subgroups. In the strain-specific two-state models (Figure 1B), recent antibiotic use remained a risk factor for colonization with non-USA300 MRSA, increasing its acquisition rate two-fold, from 0.020 (95% CI, 0.013 – 0.030) to 0.042 (95% CI, 0.030 – 0.060) (Figure 3A). No other potential risk factors influenced the acquisition rates for strain-specific MRSA at 95% confidence level (Figure 3B–3G).
Table 1.
Estimated transition rates (relative frequency per three months) for strain-independent MRSA and their 95% confidence intervals by levels of potential risk factors.
| Risk Factor | Level of Risk Factor | Negative to Positive Estimate (95% CI) |
Positive to Negative Estimate (95% CI) |
|---|---|---|---|
| Antibiotic Use in the past 3 months | Non-exposed | 0.025 (0.018, 0.037)* | 0.122 (0.086, 0.172) |
| Exposed | 0.052 (0.038, 0.075)* | 0.099 (0.069, 0.146) | |
| Hospitalizations in the past 3 months | Non-exposed | 0.034 (0.026, 0.044) | 0.108 (0.083, 0.142) |
| Exposed | 0.056 (0.028, 0.109) | 0.126 (0.059, 0.255) | |
| Invasive Device | Non-exposed | 0.036 (0.028, 0.047) | 0.112 (0.085, 0.146) |
| Exposed | 0.038 (0.017, 0.087) | 0.099 (0.051, 0.187) | |
| Wound | Non-exposed | 0.035 (0.028, 0.045) | 0.121 (0.091, 0.159) |
| Exposed | 0.049 (0.022, 0.105) | 0.069 (0.034, 0.146) | |
| Comorbidity | Non-severe | 0.033 (0.023, 0.048) | 0.121 (0.082, 0.181) |
| Severe | 0.039 (0.027, 0.054) | 0.103 (0.073, 0.143) | |
| Functional Status | Non-severe | 0.032 (0.020, 0.053) | 0.131 (0.074, 0.227) |
| Severe | 0.038 (0.028, 0.050) | 0.105 (0.079, 0.142) | |
| Cognitive Status | Non-severe | 0.038 (0.030, 0.050) | 0.121 (0.092, 0.158) |
| Severe | 0.024 (0.012, 0.047) | 0.048 (0.018, 0.126) |
The difference is statistically significant at the 95% confidence level.
Figure 3.
Predicted acquisition rates for non-USA300 and USA300 (relative frequency per three months) and their 95% confidence intervals by potential risk factors. The candidate risk factors are antibiotic use in the past 3 months (Antibiotic), previous hospitalizations in the past 3 months (Hospitalizations), presence of invasive device (Device) or wound (Wound), comorbidity (Comorbidity), functional status (Functional), and cognitive status (Cognitive). The levels of the candidate risk factors are indicated parenthetically and represent: (1) absence (No) or presence (Yes) of the risk factor for antibiotic use, previous hospitalizations, invasive device, and wound and; (2) severity (Non-severe or Severe) for comorbidity, functional status and cognitive status. CL, confidence limit.
Average Duration of Colonization
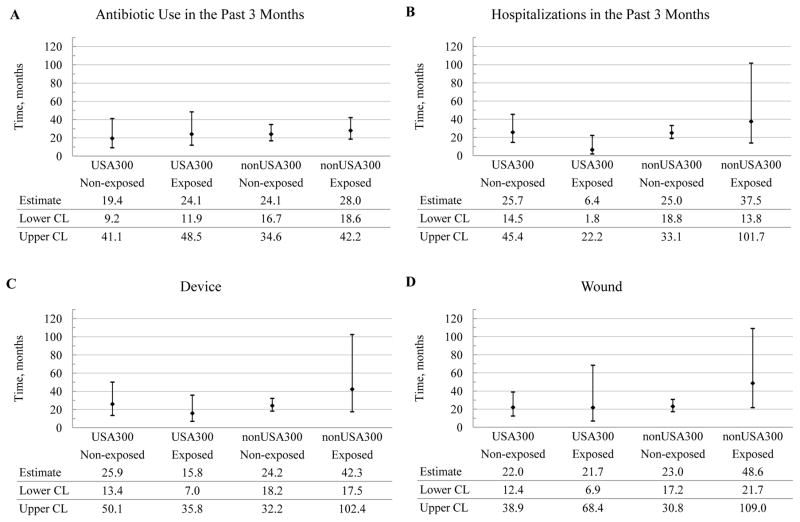
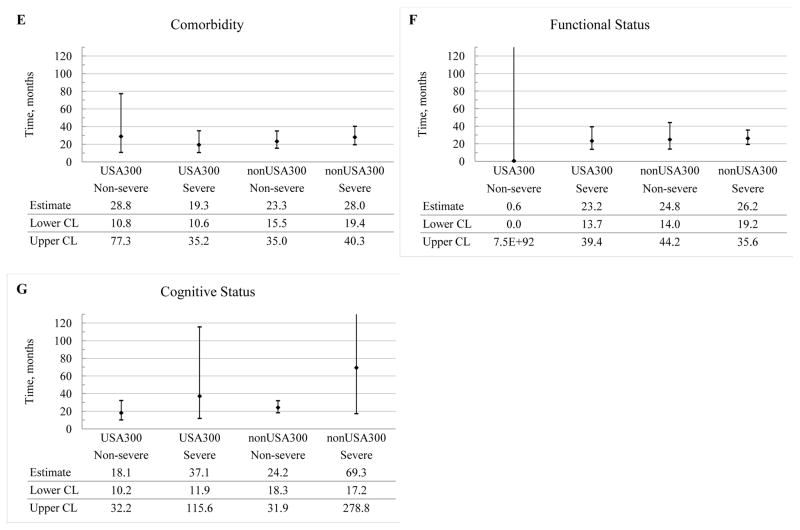
The average duration of colonization with either stain type was longer than a year for most of the candidate risk factors considered (Figure 4). Furthermore, no significant differences were observed in the average duration of colonization between non-USA300 and USA300, regardless of the risk factors being examined (Figure 4). For example, for the subjects who used antibiotics in the past 3 months, the average duration of colonization with non-USA300 was not significantly different from the one with USA-300. Furthermore, the use of antibiotics did not significantly impact the duration of colonization for residents colonized with either strain.
Figure 4.
Predicted average duration of colonization with non-USA300 and USA300 and their 95% confidence intervals, expressed in months, by potential risk factors. The average duration of colonization is defined as the average period of time during which the same MRSA strain remained recoverable from a study subject. The candidate risk factors are antibiotic use in the past 3 months (Antibiotic), previous hospitalizations in the past 3 months (Hospitalizations), presence of invasive device (Device) or wound (Wound), comorbidity (Comorbidity), functional status (Functional), and cognitive status (Cognitive). The levels of the candidate risk factors are indicated parenthetically and represent: (1) absence (No) or presence (Yes) of the risk factor for antibiotic use, previous hospitalizations, invasive device, and wound and; (2) severity (Non-severe or Severe) for comorbidity, functional status and cognitive status. CL, confidence limit.
Discussion
The emergence of community-associated MRSA and subsequent introduction into healthcare settings, including nursing homes,14,15,26 has become a growing concern in recent years.1,2,8,9 To our knowledge, the current study represents the first attempt to model strain-specific long-term prevalence of MRSA, its acquisition rates and the duration of colonization in community nursing homes while accounting for factors that potentially impact these dynamics.
Our study showed that, on average, healthcare-associated strains of MRSA would continue to dominate in nursing homes in our region although we could not exclude the possibility that USA300 could achieve dominance in individual facilities. These findings are consistent with other studies demonstrating a preponderance of healthcare-associated MRSA strains in nursing homes within the same region.16 In these studies, USA300 was identified as the dominant circulating strain only rarely, even in regions with high levels of community-acquired strains circulation in the surrounding population.
Antibiotic exposure within the previous 3 months was the only factor that, when entered into our model, significantly increased the acquisition rate of strain-independent MRSA. It also increased the acquisitions rates of strain-specific MRSA, although statistical significance at a 95% confidence level was only achieved for non-USA300. This finding is consistent with other studies which have found that recent antibiotic use increased nursing home resident risk of MRSA acquisition.13,25 Antibiotics are among the most frequently prescribed medications in nursing homes38 and much of this use is inappropriate.38,39 Our findings in this study suggest that antibiotic stewardship40 may represent a powerful tool to reduce the spread of MRSA in nursing homes.
In contrast to other studies,12,13 a number of candidate risk factors we examined in this study did not achieve statistical significance at a 95% confidence level. For example, the increase in MRSA colonization predicted by our model in the presence of recent hospitalizations (acquisition rate: 0.034 to 0.056) or a wound (acquisition rate: 0.035 to 0.049) was substantial in absolute terms but did not achieve statistical significance. Likewise, impaired cognition appeared to be associated with a reduced risk of MRSA acquisition (transition rate: 0.038 to 0.024) but, again, this did not achieve statistical significance. It is possible that a number of these risk factors would have achieved statistical significance using models based on a larger dataset or those that incorporated a multivariate approach rather than the bivariate approach employed in this study.
The limitations of our study were mainly driven by modeling assumptions and scarcity of data. For example, while MRSA acquisition rates were facility-specific, the data from the six facilities were combined due to its paucity in separate facilities. Hence, our estimates would be representative of a hypothetical “average” nursing home. Likewise, we examined one candidate risk factor at a time. Such model simplifications allowed comparisons within each risk factor but restricted comparisons across the risk factors. Furthermore, due to the scarcity of data, all of the potential risk factors were considered as dichotomous variables in our models. This was likely to reduce the discriminatory power of the models to detect the true level of association between the candidate risk factors and acquisition of MRSA. Thus, the true level of association between recent antibiotic exposure and MRSA acquisition was likely to be attenuated by dichotomizing its variable (yes or no), since residents who received a single dose of antimicrobials and those who received a long course of antibiotics over the previous 3 months were assigned to the same group. Similarly, residents were classified into non-severe and severe levels of comorbidity, functional status and cognitive status based on the arbitrarily chosen cutoffs for the associated raw scores. It may have resulted in assigning residents with similar amounts of exposure to different levels of the respective candidate risk factors. Moreover, our study did not account for changes in the admission prevalence of strain-specific MRSA that may occur over time, for example, due to changes in the prevalence of MRSA in referring hospitals or the surrounding community. Such changes may affect the epidemiology of MRSA in the nursing homes over time.
Additional work is needed to better understand the transmission dynamics of MRSA in community nursing homes. It includes determining the condition under which MRSA could be reduced or eliminated from the facilities and evaluating the risk of MRSA outbreak in this setting. Understanding associations between potentially modifiable resident characteristics and acquisition and persistence of MRSA strains may contribute to adopting better informed infection control strategies aimed at decreasing the burden of MRSA in nursing homes.
Conclusions
Our study suggests that under current conditions non-USA300 MRSA will continue to dominate USA300 in community nursing homes. Antibiotic exposure in the previous 3 months was the only factor associated with a significant increase in MRSA acquisition rates in the study facilities. Consequently, antibiotic stewardship may reduce the burden of MRSA in nursing homes.
Highlights.
Non-USA300 was predicted to remain the dominant MRSA strain in community nursing homes. At steady-state, 20% (95% CI, 15–25%) of residents were predicted to remain colonized with non-USA300 MRSA and 4% (95% CI, 2–7%) with USA300 MRSA.
Residents who used antibiotics in the previous 3 months were twice more likely to acquire MRSA than those who did not (acquisition rates 0.052 (95% CI, 0.038–0.075) and 0.025 (95% CI, 0.018–0.037), respectively).
Antibiotic stewardship may reduce MRSA colonization in community nursing homes.
Acknowledgments
Financial Support. Christopher J. Crnich activity on this project was supported by the Clinical and Translational Science Award (CTSA) program, through the NIH National Center for Advancing Translational Sciences (NCATS) (Grant UL1TR000427; the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH); the University of Wisconsin School of Medicine and Public Health, through the Wisconsin Partnership Program; and a research scholarship from the ASP/IDSA T. Franklin Williams Program.. David F. Anderson was supported through NSF grants DMS-1009275 and DMS-1318832 and Army Research Office grant W911NF-14-1-0401.
Thank you notes. We would like to express our great appreciation to Drs. Vicki Bier, Molly Carnes and David Zimmerman for their constructive suggestions, invaluable feedback, and support during different stages of the study. We are very grateful to Dr. Bret Hanlon for his methodological advice. We would also like to thank Ms. Helena Tsotsis for her help with data retrieval.
Appendix
Appendix Table 1.
Study attrition over time.
| Time Point | No. of Subjects Screened for MRSA at Assessment | No. of Subjects Evaluable* at Assessment |
|---|---|---|
| Baseline | 449 | 444† |
| 3 months | 319 | 316 |
| 6 months | 271 | 268 |
| 9 months | 238 | 238 |
| 12 months | 191 | 190 |
MRSA status of a subject at a particular assessment was considered evaluable for the purpose of the study if all three culture results obtained from nares, stool or peri-rectal skin and skin of the axilla or groin were valid (e.g., if a subject’s cultures obtained from nares and skin of the axilla or groin were both negative for MRSA, while the culture obtained from stool or peri-rectal skin was missing, the assessment was considered not evaluable).
Two subjects with non-evaluable observations at baseline had at least one evaluable observation during subsequent assessments (one subject had an evaluable observation at 3 months, and the other subject had evaluable observations at 3, 6 and 9 months). That is, the total of 446 subjects had evaluable observation during the study period.
Appendix Table 2.
Counts of the observed transitions and transition probabilities (parenthetically) for all facilities combined for the three-state model and strain-specific two-state models.
| From State | To State | ||
|---|---|---|---|
| Negative | USA300 | non-USA300 | |
| Three-State Model* | |||
| Negative | 711 (0.913) | 13 (0.017) | 55 (0.071) |
| USA300 | 10 (0.222) | 30 (0.667) | 5 (0.111) |
| non-USA300 | 52 (0.278) | 2 (0.011) | 132 (0.711) |
| Two-State Strain-Specific Models | |||
| USA 300 Model† | |||
| Negative | 950 (0.984) | 15 (0.016) | NA |
| USA300 | 15 (0.333) | 30 (0.667) | NA |
| Non-USA300 Model‡ | |||
| Negative | 762 (0.929) | NA | 58 (0.071) |
| non-USA300 | 54 (0.284) | NA | 136 (0.716) |
In the three-state model, subjects can be MRSA-free (Negative), colonized with USA300 and possibly co-colonized with non-USA300 (USA300) or colonized with non-USA300 only (non-USA300).
In this model, a subject can be either colonized with USA300 (USA300) or non-colonized with USA (Negative). Subjects classified as Negative in this model can be colonized with non-USA300 strains.
In this model, a subject can be either colonized with non-USA300 (non-USA300) or free from non-USA300 (Negative). Subjects classified as Negative in this model can be colonized with USA300.
Appendix Figure 1.
Counts of residents colonized with strain-independent and strain-specific MRSA in each facility over time. Rows represent MRSA strains: MRSA (any strain of MRSA), non-USA300 and USA300. Columns represent facilities. Bars show the number of subjects colonized with the corresponding MRSA strain (Positive) or non-colonized with this MRSA strain (Negative).
Footnotes
Disclosure of Potential Conflicts of Interest. All authors report no conflicts of interest associated with this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA : the journal of the American Medical Association. 2007 Oct 17;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Dantes R, Mu Y, Belflower R, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA internal medicine. 2013 Nov 25;173(21):1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capitano B, Leshem OA, Nightingale CH, Nicolau DP. Cost effect of managing methicillin-resistant Staphylococcus aureus in a long-term care facility. Journal of the American Geriatrics Society Jan. 2003;51(1):10–16. doi: 10.1034/j.1601-5215.2002.51003.x. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America Feb. 2005;26(2):166–174. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- 5.Anderson DJ, Kaye KS, Chen LF, et al. Clinical and financial outcomes due to methicillin resistant Staphylococcus aureus surgical site infection: a multi-center matched outcomes study. PloS one. 2009;4(12):e8305. doi: 10.1371/journal.pone.0008305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naimi TS, LeDell KH, Como-Sabetti K, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA : the journal of the American Medical Association. 2003 Dec 10;290(22):2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 7.Cooper BS, Medley GF, Stone SP, et al. Methicillin-resistant Staphylococcus aureus in hospitals and the community: stealth dynamics and control catastrophes. Proceedings of the National Academy of Sciences of the United States of America. 2004 Jul 6;101(27):10223–10228. doi: 10.1073/pnas.0401324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Annals of internal medicine. 2006 Mar 7;144(5):309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 9.Klein EY, Sun L, Smith DL, Laxminarayan R. The changing epidemiology of methicillin-resistant Staphylococcus aureus in the United States: a national observational study. American journal of epidemiology. 2013 Apr 1;177(7):666–674. doi: 10.1093/aje/kws273. [DOI] [PubMed] [Google Scholar]
- 10.Bootsma MC, Diekmann O, Bonten MJ. Controlling methicillin-resistant Staphylococcus aureus: quantifying the effects of interventions and rapid diagnostic testing. Proceedings of the National Academy of Sciences of the United States of America. 2006 Apr 4;103(14):5620–5625. doi: 10.1073/pnas.0510077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temime L, Opatowski L, Pannet Y, Brun-Buisson C, Boelle PY, Guillemot D. Peripatetic health-care workers as potential superspreaders. Proceedings of the National Academy of Sciences of the United States of America. 2009 Oct 27;106(43):18420–18425. doi: 10.1073/pnas.0900974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mody L, Kauffman CA, Donabedian S, Zervos M, Bradley SF. Epidemiology of Staphylococcus aureus colonization in nursing home residents. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2008 May 1;46(9):1368–1373. doi: 10.1086/586751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garazi M, Edwards B, Caccavale D, Auerbach C, Wolf-Klein G. Nursing homes as reservoirs of MRSA: myth or reality? Journal of the American Medical Directors Association Jul. 2009;10(6):414–418. doi: 10.1016/j.jamda.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Murphy CR, Hudson LO, Spratt BG, et al. Predicting high prevalence of community methicillin-resistant Staphylococcus aureus strains in nursing homes. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America Mar. 2013;34(3):325–326. doi: 10.1086/669519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tattevin P, Diep BA, Jula M, Perdreau-Remington F. Methicillin-resistant Staphylococcus aureus USA300 clone in long-term care facility. Emerging infectious diseases Jun. 2009;15(6):953–955. doi: 10.3201/eid1506.080195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds C, Quan V, Kim D, et al. Methicillin-resistant Staphylococcus aureus (MRSA) carriage in 10 nursing homes in Orange County, California. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America Jan. 2011;32(1):91–93. doi: 10.1086/657637. [DOI] [PubMed] [Google Scholar]
- 17.Lee BY, Bartsch SM, Wong KF, et al. The importance of nursing homes in the spread of methicillin-resistant Staphylococcus aureus (MRSA) among hospitals. Medical care Mar. 2013;51(3):205–215. doi: 10.1097/MLR.0b013e3182836dc2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiener J, Quinn JP, Bradford PA, et al. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA : the journal of the American Medical Association. 1999 Feb 10;281(6):517–523. doi: 10.1001/jama.281.6.517. [DOI] [PubMed] [Google Scholar]
- 19.Won SY, Munoz-Price LS, Lolans K, Hota B, Weinstein RA, Hayden MK. Emergence and rapid regional spread of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America Sep. 2011;53(6):532–540. doi: 10.1093/cid/cir482. [DOI] [PubMed] [Google Scholar]
- 20.Barnes SL, Harris AD, Golden BL, Wasil EA, Furuno JP. Contribution of interfacility patient movement to overall methicillin-resistant Staphylococcus aureus prevalence levels. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America Nov. 2011;32(11):1073–1078. doi: 10.1086/662375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesosky M, McGeer A, Simor A, Green K, Low DE, Raboud J. Effect of patterns of transferring patients among healthcare institutions on rates of nosocomial methicillin-resistant Staphylococcus aureus transmission: a Monte Carlo simulation. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America Feb. 2011;32(2):136–147. doi: 10.1086/657945. [DOI] [PubMed] [Google Scholar]
- 22.Macal CM, North MJ, Collier N, et al. Modeling the transmission of community-associated methicillin-resistant Staphylococcus aureus: a dynamic agent-based simulation. Journal of translational medicine. 2014;12:124. doi: 10.1186/1479-5876-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crnich CJ. Impact and Management of MRSA in the Long-Term Care Setting. Curr Transl Geriatr and Exp GErontol Rep. 2013;2:125–135. [Google Scholar]
- 24.Furuno JP, Shurland SM, Zhan M, et al. Comparison of the methicillin-resistant Staphylococcus aureus acquisition among rehabilitation and nursing home residents. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America Mar. 2011;32(3):244–249. doi: 10.1086/658667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone ND, Lewis DR, Johnson TM, 2nd, et al. Methicillin-resistant Staphylococcus aureus (MRSA) nasal carriage in residents of Veterans Affairs long-term care facilities: role of antimicrobial exposure and MRSA acquisition. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America Jun. 2012;33(6):551–557. doi: 10.1086/665711. [DOI] [PubMed] [Google Scholar]
- 26.Shurland SM, Stine OC, Venezia RA, et al. USA300 methicillin-resistant S. aureus (USA300 MRSA) colonization and the risk of MRSA infection in residents of extended-care facilities. Epidemiology and infection. 2012 Mar;140(3):390–399. doi: 10.1017/S0950268811001324. [DOI] [PubMed] [Google Scholar]
- 27.Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. The Journal of antimicrobial chemotherapy Sep. 2009;64(3):441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- 28.Desai R, Pannaraj PS, Agopian J, Sugar CA, Liu GY, Miller LG. Survival and transmission of community-associated methicillin-resistant Staphylococcus aureus from fomites. American journal of infection control Apr. 2011;39(3):219–225. doi: 10.1016/j.ajic.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Crnich CJ, Duster M, Hess T, Zimmerman DR, Drinka P. Antibiotic resistance in non-major metropolitan skilled nursing facilities: prevalence and interfacility variation. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America Nov. 2012;33(11):1172–1174. doi: 10.1086/668018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. Journal of chronic diseases. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 31.Katz S, Ford AB, Moskowitz RW, Jackson BA, Jaffe MW. Studies of Illness in the Aged. The Index of Adl: A Standardized Measure of Biological and Psychosocial Function. JAMA : the journal of the American Medical Association. 1963 Sep 21;185:914–919. doi: 10.1001/jama.1963.03060120024016. [DOI] [PubMed] [Google Scholar]
- 32.Morris JN, Fries BE, Mehr DR, et al. MDS Cognitive Performance Scale. Journal of gerontology Jul. 1994;49(4):M174–182. doi: 10.1093/geronj/49.4.m174. [DOI] [PubMed] [Google Scholar]
- 33.Crnich CJ, Duster M, Warrack S, Maki D, Safdar N. Comparison of Pulsed-Gel Electrophoresis and a Commercial Repetitive-Element PCR Method for Assessment of Methicillin-Resistant Staphylococcus aureus Clustering in Different Health Care Facilities. Journal of clinical microbiology Jun. 2014;52(6):2027–2032. doi: 10.1128/JCM.03466-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawler GF. Introduction to stochastic processes. 2. Boca Raton: Chapman & Hall/CRC; 2006. [Google Scholar]
- 35.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. http://www.R-project.org/ [Google Scholar]
- 36.Wasserman L. All of statistics : a concise course in statistical inference. New York: Springer; 2004. [Google Scholar]
- 37.Jackson CH. Multi-State Models for Panel Data: The msm Package for R. Journal of Statistical Software. 2011;38(8):1–29. http://www.jstatsoft.org/v38/i08/ [Google Scholar]
- 38.Nicolle LE, Bentley DW, Garibaldi R, Neuhaus EG, Smith PW. Antimicrobial use in long-term-care facilities. SHEA Long-Term-Care Committee. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America. 2000 Aug;21(8):537–545. doi: 10.1086/501798. [DOI] [PubMed] [Google Scholar]
- 39.Zimmer JG, Bentley DW, Valenti WM, Watson NM. Systemic antibiotic use in nursing homes. A quality assessment. Journal of the American Geriatrics Society. 1986 Oct;34(10):703–710. doi: 10.1111/j.1532-5415.1986.tb04301.x. [DOI] [PubMed] [Google Scholar]
- 40.Dellit TH, Owens RC, McGowan JE, Jr, et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007 Jan 15;44(2):159–177. doi: 10.1086/510393. [DOI] [PubMed] [Google Scholar]