Abstract
Normalization of feature vector values is a common practice in machine learning. Generally, each feature value is standardized to the unit hypercube or by normalizing to zero mean and unit variance. Classification decisions based on support vector machines (SVMs) or by other methods are sensitive to the specific normalization used on the features. In the context of multivariate pattern analysis using neuroimaging data, standardization effectively up- and down-weights features based on their individual variability. Since the standard approach uses the entire data set to guide the normalization, it utilizes the total variability of these features. This total variation is inevitably dependent on the amount of marginal separation between groups. Thus, such a normalization may attenuate the separability of the data in high dimensional space. In this work we propose an alternate approach that uses an estimate of the control-group standard deviation to normalize features before training. We study our proposed approach in the context of group classification using structural MRI data. We show that control-based normalization leads to better reproducibility of estimated multivariate disease patterns and improves the classifier performance in many cases.
Keywords: Feature Normalization, Multivariate Pattern Analysis, Structural MRI, Support Vector Machine
1. Introduction
Machine learning classification algorithms such as the support vector machine (SVM) [1, 2] are often used to map high-dimensional neuroimaging data to a clinical diagnosis or decision. Structural and functional magnetic resonance imaging (MRI) are promising tools for building biomarkers to diagnose, monitor, and treat neurological and psychological illnesses. Mass-univariate methods such as statistical parametric mapping [3, 4, 5] and voxel- based morphometry [6, 7] test for marginal disease effects at each voxel, ignoring complex spatial correlations and multivariate relationships among voxels. As a result, methods have emerged for performing multivariate pattern analysis (MVPA) that leverage the information contained in the covariance structure of the images to discriminate between the groups being studied [8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29]. Identifying multivariate structural and functional signatures in the brain that discriminate between groups may lead to a better understanding of disease processes and is therefore of great interest in the field of neuroimaging research.
The SVM is a common choice for estimating multivariate patterns in the brain because it is amenable to high-dimensional, low sample size data. Our focus in this work is on patterns in the brain that reflect structural changes due to disease. However, the methods apply more generally to applications of MVPA using BOLD measurements from fMRI data or measures of connectivity across the brain. The SVM takes as input image-label pairs and returns a decision function that is a weighted sum of the imaging features. The estimated weights reflect the joint contribution of the imaging features to the predicted class label.
Machine learning methods in general, and SVMs in particular, are sensitive to differences in feature scales. For example, a SVM will place more importance on a feature that takes values in the range of [1000, 2000] than a feature that takes values in the interval [1, 2]. This is because the former tends to have a stronger influence on the Euclidean distance between feature vector realizations and therefore drives the SVM optimization. To give all voxels or regions of interest equal importance during classifier training, it is common practice to implement feature-wise standardization in some way, either by normalizing each to have mean zero and unit variance or by scaling to a common domain. For example, [30] scale each feature to be in the interval [0, 1], and [31, 32, 33, 34, 35] normalize to mean zero and unit variance. Such a preprocessing step, while common in practice, tends to be applied without weighing the consequent ramifications in a careful manner. Careful consideration must be given to the choice of feature normalization, as it is directly tied to the relative magnitude of the estimated SVM weights and thus the performance and interpretation of the classifier. While the original idea of feature scaling dates back to the universal approximation theorem from the neural network literature, it has not been explored in detail in the context of neuroimaging and MVPA. This is the object of this manuscript.
The rest of this paper is organized as follows: in Section 2, we provide a brief introduction to MVPA using the SVM, review two popular feature normalization methods, and propose an alternative based on the control-group variability. Using simulations, we compare the performance of different feature normalization techniques in Section 3, followed by an investigation of the effects of feature normalization on an analysis of data from healthy controls and patients with Alzheimer’s disease. We include a discussion in Section 4 and concluding remarks in Section 5.
2. Material and Methods
2.1. Multivariate Pattern Analysis using the SVM
Let , i = 1, …, n, denote n independent and identically distributed observations of the random vector (Y, X⊤)⊤, where Y ∈ {−1, 1} denotes the group label, and X ∈ ℝp denotes a vectorized image with p voxels. A popular MVPA tool used in the neuroimaging community is the SVM [1, 2]. SVMs are known to work well for high dimension, low sample size data [36]. Such data are common in the neuroimaging-based diagnostic setting. Henceforth, we focus on MVPA using the SVM.
The hard-margin linear SVM solves the constrained optimization problem
| (1) |
where b ∈ ℝ and v ∈ ℝp are parameters that describe the classification function. For a given set of training data, let the solution to (1) be denoted by . Then, for a new observation Xnew with unknown label Ynew, the classification function returns a predicted group label.
When the data from the two groups are not linearly separable, the soft-margin linear SVM allows some training observations to be either misclassified or fall in the SVM margin through the use of slack variables ξi with associated cost parameter C. In this case, the optimization problem becomes
| (2) |
where C ∈ ℝ is a tuning parameter that penalizes misclassification, and ξ = (ξ1, ξ2, …, ξn)⊤ is the vector of slack variables. For details about solving optimization problems (1) and (2) we refer the reader to [37].
In high-dimensional problems where the number of features is greater than the number of observations, the data are almost always separable by a linear hyperplane [38]. However, when applying MVPA to region of interest (ROI) data such as volumes of subregions in the brain, the data may not be linearly separable. In this case, the choice of C is critical to classifier performance and generalizability. Examples of MVPA using the SVM include classification of multiple sclerosis patients into disease subgroups [39], the study of Alzheimer’s disease [9, 10], and various classification tasks involving patients with depression [40, 41, 42]. This is only a small subset of the relavant literature, which demonstrates the widespread popularity of the approach.
2.2. SVM Feature Normalization for MVPA
The choice of feature normalization affects the estimated weight pattern of a SVM and can lead to vastly different conclusions about the underlying disease process. Two widely implemented approaches are to (i) normalize each feature to have mean zero and unit variance, and (ii) scale each feature to have a common domain such as [0, 1]. Henceforth, we will refer to (i) as standard normalization and (ii) as domain standardization [43].
Let μj and σj denote the mean and standard deviation of the jth feature, j = 1, …, p. Denote the corresponding empirical estimates by and . Then, subject i’s standard-normalized jth feature is calculated as
Alternatively, subject i’s domain-scaled jth feature is calculated as
One potential drawback of using domain scaling is the instability of the minimum and maximum order statistics, especially in small sample sizes. This may introduce bias in the estimated weight pattern by up- and down-weighting features in an unstable way. In comparison, the standard normalization may seem relatively stable. However, it implicitly depends on the relative sample size of each group and the separability between groups. To see this, let denote the marginal distribution of Xj, with mean μj and variance . Let denote the conditional distribution of Xj given Y = y with mean μj,y and variance . In addition, let γ = pr(Y = 1). Then, μj = γμj,1 + (1 − γ)μj,−1 and
After simplification, the previous expression can be written as
| (3) |
The right-hand side of expression (3) shows that the variance of feature j depends on a mixture of the conditional variances of both classes and a term that depends on the squared Euclidean distance between their marginal means. Larger marginal separability of feature j will lead to a larger estimate of the pooled standard deviation used for normalization. Thus, normalizing by the pooled standard deviation can in some cases harshly penalize, or down-weight, features that have good separability, leading to a loss in predictive performance. We demonstrate this using simulated data examples in Section 3.1.
The right-hand side of equation (3) also illuminates how normalization is dependent on the relative within-group sample sizes, which may have adverse effects on classifier performance. Suppose data for MVPA are available from a case-control study where the cases have been oversampled. That is, there is one healthy control for each subject with the disease. Suppose further that the true disease prevalence in the population is rare. Then, the estimate of will be an equal mixture of the group variances and , whereas the true in the population depends more heavily on the control-group variance, . Methods for dataset or covariate shift address this issue by weighting individual data points to reflect the distribution of covariates in the population [44, 45]. However, these methods are usually implemented after feature normalization. As a result, the estimated decision rule may be undesirably influenced by the use of a biased estimate of the pooled variance.
As an alternative, we propose normalizing the jth feature as follows:
for all subjects i = 1, …, n, where is the pooled sample mean of feature j, and j is the sample standard deviation of the jth feature calculated using the control-group data only. Note that and are computed using only the control-group, but the normalization is applied to subjects from both groups. We refer to this as control normalization. Note that for features that contribute greatly to the separability of the groups, the control-group standard deviation will be smaller than the pooled-group standard deviation. Scaling by this smaller value will implicitly up-weight the most discriminative features in comparison to the standard-normalization. In some studies or applications, there may not be a control group. In this case, a reference group may be chosen based on expert knowledge or the scientific goals of the study. In Section 3 we demonstrate how the choice of feature normalization technique may lead to a tradeoff of classifier properties such as sensitivity and specificity but results in better overall accuracy in many settings.
Another advantage of the control normalization is the resulting interpretability of the feature values. Fixing all other SVM features at a constant value, the estimated weight corresponding to feature j conveys the magnitude and direction of change in the decision function score for a one unit increase in feature j, where the units are in terms of the control-group standard deviation of that feature. In many studies, it is likely that more knowledge exists about the distribution of values in the normal population, as the disease being studied may be highly heterogeneous, rare, or not yet well-understood. Being able to interpret the estimated disease pattern relative to the healthy control distribution may improve the reproducibility and clinical value of the MVPA results.
Figure 1 displays an example of the influence of feature normalization on the estimated SVM weight pattern. We generated n = 100 independent feature vectors with two signal features and 20 noise features each. The signal features X1 and X2 are generated from a multivariate normal distribution that differs in its parameters between the two diagnostic groups. The signal features jointly have discriminative power, while the remaining noise features are generated independently from the standard normal distribution. The classes are balanced with n0 = 50 control observations and n1 = 50 disease group observations. All features are plotted pre-normalization in the first panel of Figure 1. The correlation between X1 and X2 in the control group is ρ0 = −0.2, and it is ρ1 = −0.6 in the disease-group. The control-normalized, standard-normalized, and domain-scaled versions are plotted in the second, third, and fourth panels. The estimated SVM decision boundary is projected onto the space of these two features by setting the weights of the noise features to zero. The black line in each panel represents this projected decision boundary.
Figure 1. Influence of feature normalization on the SVM decision boundary. From left to right: original feature scales, control-normalized features, standard-normalized features, domain-scaled features.
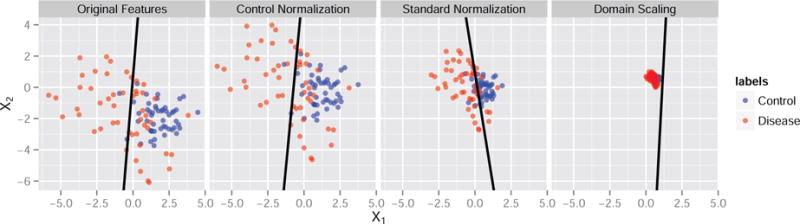
We carefully chose the parameters for the toy example in Figure 1 because they represent a worst-case scenario where the choice of feature normalization changed the sign of the estimated weight associated with feature X1. While changes in magnitude of the estimated SVM weights are expected due to the fact that the SVM is scale-invariant, changes in the direction of the effect of a feature are alarming because this alters the biological interpretation of the feature relative to the overall disease pattern. Using the classifier from the standard normalization, as X2 increases for any fixed X1 it is more likely a subject presenting with the pair (X1, X2) will be classified as healthy. However, using the control normalization the relationship is reversed so that subjects are more likely to be classified as having the disease. Thus, the multivariate pattern in this example is highly dependent on the choice of normalization. While the difference in results and interpretation may not always be so drastic, this example motivates the need for researchers to adopt a single, interpretable technique for feature normalization when performing MVPA using SVMs. We study the effects of feature normalization for a range of parameter settings in Section 3.1.
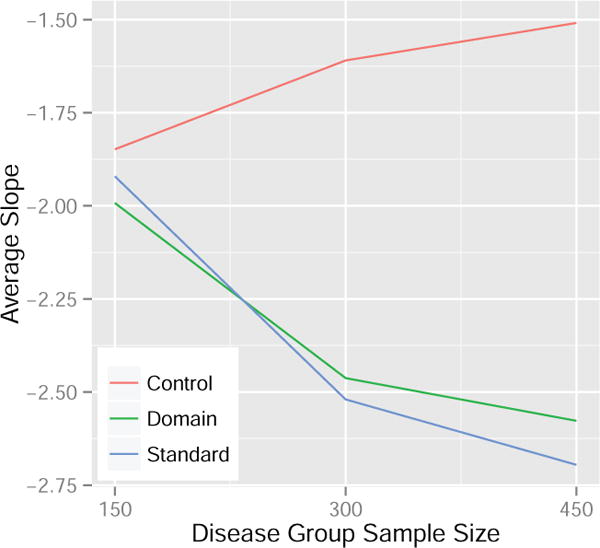
Using the same data generating set-up as the toy example in Figure 1 but restricting the data sets to include only the two signal features, we implemented a small experiment to compare the reproducibility of estimated disease patterns using the control normalization, standard normalization, and domain scaling. Specifically, we studied the effects of disease group sample size and normalization method using a two-way analysis of variance (ANOVA). In particular, we were interested in the effect of increasing samples in the disease group on the average slope of the estimated SVM line for each normalization method. In the ANOVA model, we included terms for the main effects of method and sample size as well as an interaction term, with the interaction effect being of highest interest. A significant interaction term implies that the effect of increasing the sample size of the disease group on the average estimated SVM slope differs among the normalization methods. In our simulation, we estimated the average SVM slope for each level of sample size and normalization method using 100 independent iterations. We then repeated this procedure 20 times to provide replications for the ANOVA model. Results are presented in Figure 2 and the p-value for the interaction term was highly significant (< 0.0001). Based on Figure 2 we are able to conclude that the change in the SVM slope using the control normalization is less than the change observed using the other methods when varying the disease group sample size. Conservatively, the change observed from 150 to 450 disease group samples using the control normalization is approximately 50% and 40% less compared to the standard normalization and domain scaling, respectively. This suggests that results from MVPA may be more reproducible across studies with different sample sizes when the control normalization is used to preprocess the features prior to SVM training.
Figure 2. Average estimated SVM slope for the control normalization, domain scaling, and standard normalization when the disease group sample size is varied. The control group sample size is fixed at 300.
3. Results
3.1. Simulations
In this section, we study a range of data-generating models to compare the performance of the control normalization, standard normalization, and domain scaling when using the linear SVM for MVPA. For all simulations, we generate p features, (X1, X2, …,Xp)⊤, the first two of which have varying levels of joint discriminative power. The remaining p − 2 are independent noise features. The first two features are generated as mixtures of multivariate normal distributions. The following steps describe the procedure used to obtain the results in Figures 3–6. For each of M=1,000 iterations, n0 control subjects are generated as independent draws from the model
| (4) |
Non-control group subjects are generated as n1 independent draws from the model
| (5) |
where , , j = 3, …, p, are all mutually independent. Additionally, we generate t0 = 500 independent control-group samples from model (4) and t1 = 500 independent samples from model (5) for testing. We then train an SVM using the n0 + n1 training samples using the scikit learn library in Python, which internally calls libSVM [46]. When n0 ≠ n1, we train a class-weighted SVM that weights the cost parameter by (n1 + n0)/n0 for the control group and by (n1 + n0)/n1 for the disease group. In Figures 3 and 4 the correlations are fixed at ρ0 = 0, ρ1 = 0, and we vary n0, n1 ∈ {20, 40, …, 200}. In Figures 5 and 6 the sample sizes are fixed at n0 = 50, n1 = 50, and we vary ρ0, ρ1 ∈ {−0.9, −0.8, …, 0.9}.
Figure 3. Average difference in performance measures between the control normalization and standard normalization for a range of sample sizes. Data generated from models 4 and 5. Results reported are percentages.
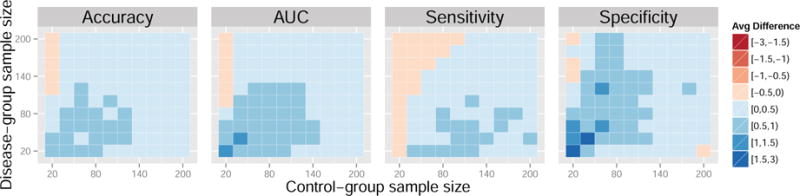
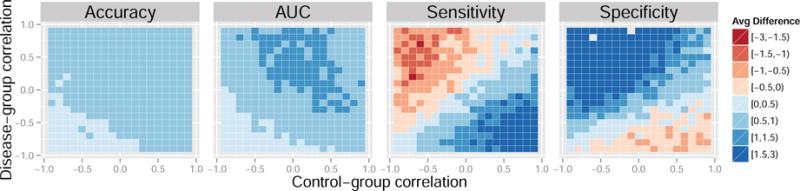
Figure 6. Average difference in performance measures between the control normalization and domain scaling for a range of feature correlations. Data generated from models 4 and 5. Results reported are percentages.
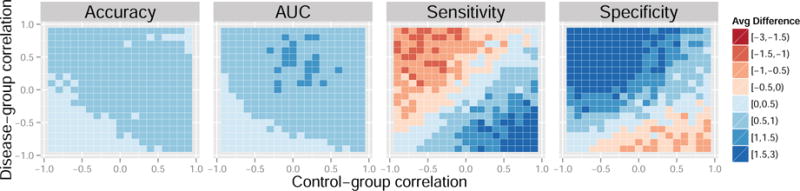
Figure 4. Average difference in performance measures between the control normalization and domain scaling for a range of sample sizes. Data generated from models 4 and 5. Results reported are percentages.
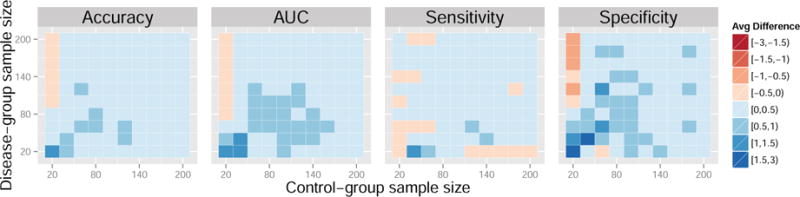
Figure 5. Average difference in performance measures between the control normalization and standard normalization for a range of feature correlations. Data generated from models 4 and 5. Results reported are percentages.
We compare the average difference in accuracy, area under the ROC curve (AUC), sensitivity, and specificity of the SVM on the test set. Given the true test labels, accuracy is defined as the percentage of correct classifications using the SVM decision rule learned from the training data. Sensitivity is the percentage of correct positive predictions, and specificity is the percentage of correct negative predictions. The ROC curve is the proportion of true positives as a function of the false positive rate which ranges in [0, 1] as the SVM intercept b is varied across the real line. Larger values of the criteria are desirable and indicate better classifier performance.
Each colored square in the heatmaps represents a self-contained simulation with 1,000 iterations. The color indicates the average difference between a given performance measure between the control-normalized SVM and either the standard-normalized or domain-scaled SVM. Dark blue indicates superior performance of the control normalization. Across the simulations summarized in Figures 3 and 4, average accuracies varied approximately between 60% and 80%, average AUCs varied approximately between 70% and 80%, average sensitivities varied approximately between 55% and 80%, and average specificities varied approximately between 55% and 80%. In Figures 3 and 4, the control normalization performs better on average than the standard normalization and domain scaling for most combinations of within-group sample size. Notable exceptions are when the sample size of the control-group is much smaller than that of the disease-group. The standard normalization appears to improve sensitivity when the disease-group sample size is large but seemingly at the cost of reduced specificity. Overall, the results appear similar when comparing the control normalization to domain scaling.
Next, we present a case where the control normalization demonstrates significant improvement over the alternative feature standardizations. The following procedure was used to obtain the results in Figures 7–8. For each of M=1,000 iterations, n0 control subjects are generated as independent draws from the model
| (6) |
Non-control group subjects are generated as n1 independent draws from the model
| (7) |
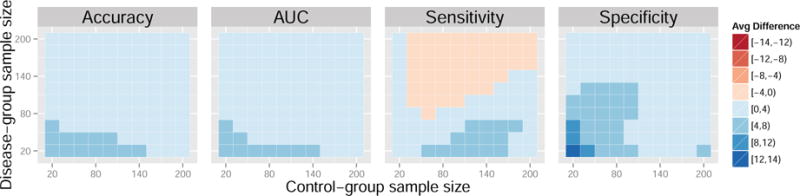
Additionally, we generate t0 = 500 independent control-group samples from model (6) and t1 = 500 independent samples from model (7) for testing. We vary the group sample sizes, n0, n1 ∈ {20, 40, …, 200}. For this set of model parameters, the control-normalization demonstrates improvement in accuracy, AUC, sensitivity, and specificity for a majority of sample sizes when compared to the standard normalization and domain scaling. In some cases, the gains surpass four percent. While there is a tradeoff between sensitivity and specificity with larger disease-group sample sizes, the overall accuracy and AUC favor the control normalization.
Figure 7. Average difference in performance measures between the control normalization and standard normalization for a range of sample sizes. Data generated from models 6 and 7.
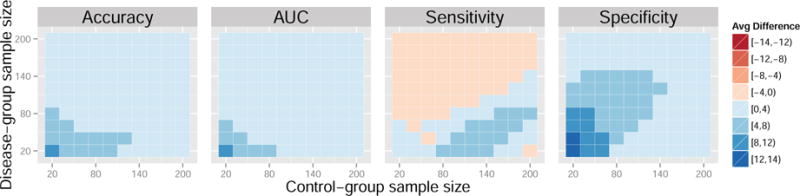
Figure 8. Average difference in performance measures between the control normalization and domain scaling for a range of sample sizes. Data generated from models 6 and 7.
3.2. Case Study
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) (http://www.adni.loni.usc.edu) is a multimillion dollar study funded by a number of public and private resources from the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), Food and Drug Administration (FDA), the pharmaceutical industry, and non-profit organizations. Aims of the study include developing sensitive and specific image-based biomarkers for early diagnosis of Alzheimer’s disease (AD), as well as monitoring the progression of mild cognitive impairment (MCI) and AD. Understanding and predicting disease trajectories is imperative for the discovery of effective treatments that intervene in the early stages of the disease to prevent irreversible damage to the brain.
The ADNI data are publicly available and as a result have been thoroughly analyzed in the neuroimaging literature [47]. A detailed comparison of SVM classification results using different categories of imaging features is given in [9]. In this section, we compare the performance of different SVM feature normalization techniques using volumes obtained from a multi-atlas segmentation pipeline applied to structural MRIs from the ADNI database [48].
The final dataset used for this analysis consists of labels indicating the presence or absence of AD and the volumes of 137 regions of interest (ROIs) in the brain for each subject. Each region is divided by the subject’s total intracranial volume to adjust for differences in individual brain size. The data consist of 230 healthy controls (CN) and 200 patients diagnosed with AD with ages ranging between 55 and 90. Table 2 displays the number (N) and average age of subjects in each diagnosis by sex group. The overall p-value for the mean difference in age between diagnosis groups was not significant (p = 0.59), nor were the p-values significant when calculated separately by sex (p = 0.24, Female; p = 0.68, Male). AD is associated with atrophy in the brain, and thus the AD group has smaller volumes on average in particular ROIs compared to the CN group.
Table 2.
Demographic summary of the ADNI data. The number (N) and average age of subjects in each group is given.
| Diagnosis | Sex | N | Average Age |
|---|---|---|---|
| CN | Female | 112 | 76.15 |
| AD | Female | 97 | 75.05 |
| CN | Male | 118 | 75.83 |
| AD | Male | 103 | 76.20 |
To give intuition about the differences between the control and standard normalization procedures in the ADNI data, we plot the densities of six features, stratified by group, in Figure 9. Whereas the pooled and control-group estimated variability is nearly identical for features such as the white matter parietal lobe and occipital pole, the control-group variability is less than the pooled variability for more marginally separable features such as the amygdala, hippocampus, inferior lateral ventricle, and parahippocampal gyrus. Thus, we expect a SVM trained after control normalization to place relatively heavier weights on these marginally discriminative features than a SVM trained after standard normalization. Figure 10 displays SVM weight patterns from the three methods. Based on Figure 10, it appears all methods obtain similar estimated disease patterns with a few subtle differences. Table 1 lists the top 10 features in order of the magnitude of their weights. As anticipated, the control normalization places more emphasis on the two amygdala regions because their marginal separability ensures a smaller denominator is used in the control-group normalization step, up-weighting these features compared to the standard normalization.
Figure 9. Density plots of ROI volumes by group.
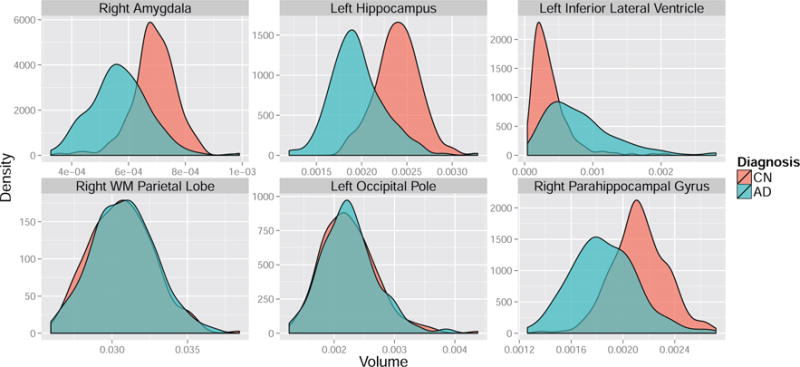
Figure 10. SVM weight patterns for discriminating between AD and CN subjects by feature standardization method.
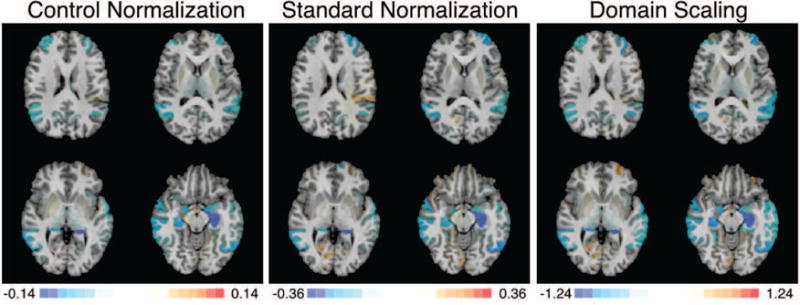
Table 1.
Top 10 ranked features by the SVM weights in decreasing absolute value.
| Rank | Control Normalization | Standard Normalization | Domain Scaling |
|---|---|---|---|
| 1 | Left Hippocampus | Left Hippocampus | Left Hippocampus |
| 2 | Right Hippocampus | Left Inferior Temporal Gyrus | Right Hippocampus |
| 3 | Left Inferior Lateral Ventricle | Left Inferior Lateral Ventricle | Left Inferior Lateral Ventricle |
| 4 | Left Inferior Temporal Gyrus | Left Middle Frontal Gyrus | Left Inferior Temporal Gyrus |
| 5 | Left Amygdala | Right Hippocampus | Left Middle Frontal Gyrus |
| 6 | Right Amygdala | Left Superior Frontal Gyrus | Right Middle Temporal Gyrus |
| 7 | Right Middle Temporal Gyrus | Right Middle Temporal Gyrus | Left Superior Temporal Gyrus |
| 8 | Left Middle Frontal Gyrus | Left Amygdala | Right Amygdala |
| 9 | Right Angular Gyrus | Left Superior Temporal Gyrus | Left Amygdala |
| 10 | Right Inferior Lateral Ventricle | Right Calcarine Cortex | Left Middle Temporal Gyrus |
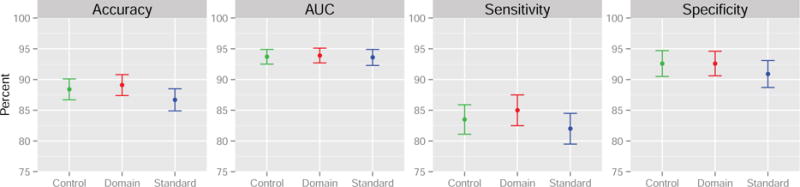
We compare the control normalization proposed in Section 2.2 to the standard normalization and domain scaling using 5-fold cross-validated estimates of classifier accuracy, area under the curve (AUC), sensitivity, and specificity. Results are shown in Figure 11. The control normalization and domain scaling outperform the standard normalization across all performance measures, increasing the cross-validated accuracy, sensitivity, and specificity by more than one percent. By a small margin, domain-scaling performs best in terms of prediction for this dataset but at the cost of fitted model interpretability. To quantify the uncertainty in the estimates in Figure 11, we repeated the 5-fold cross-validation proceedure 1000 times using random subsamples of 140 patients and 140 controls. The point estimates are shown with a single standard error on each side. For this particular data set, the performance differences are not statistically significant across the three methods.
Figure 11. 5-fold cross-validation results with measures of uncertainty estimated by sub-sampling the original data.
Finally, to compare the generalizabilitiy of the control normalization to other methods and study robustness of the results of this section to possible confounding by gender, we performed a gender-stratified analysis. Table 2 gives the counts of gender by diagnosis and the average age in these categories. An overall t-test of the mean age across diagnosis groups was not significant, and similarly t-tests of mean age between diagnosis groups calculated separately by gender were not significant. However, it is still possible that disease patterns vary across gender. In addition, the data are sparse for subjects less than 70 years of age. Thus, to study generalizability and robustness of our results to possible age and gender confounding, we split the data by gender and restricted our analysis to subjects 70 and older. Using this subset of the ADNI data, we trained the SVMs for each normalization method on 100 randomly sampled females and applied the model to classify male subjects as AD or CN. We repeated this process 1000 times and report average results in Table 3. We also report average results from the same procedure where we used random subsets of male subjects to train the models and diagnose the female group as AD or CN. On average, we observe slight gains in accuracy and area under the ROC curve when using the control normalization. Gains of one to two percentage points are along the lines of the improvements observed in our simulation study in Section 3.1. While not statistically significant, it is notable that the control normalization outperformed the other methods across all performance measures with the exception of displayed equivalence in sensitivity to the standard normalization.
Table 3.
Gender-stratified analysis of ADNI data.
| Training | Method | Accuracy | AUC | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Control Normalization | 87% | 89% | 85% | 88% | |
| Females | Domain Scaling | 86% | 88% | 86% | 86% |
| Standard Normalization | 86% | 88% | 85% | 87% | |
|
| |||||
| Control Normalization | 86% | 89% | 78% | 92% | |
| Males | Domain Scaling | 85% | 87% | 76% | 91% |
| Standard Normalization | 85% | 88% | 78% | 90% | |
4. Discussion
Interpretability of estimated disease patterns is a desirable quality for most applications of MVPA to neuroimaging data. Standardizing features by the control-group variability leads to estimated support vector weights whose individual interpretation relies solely on a sample of normal subjects. In some cases, this may increase the reliability of image-based biomarkers for disease classification. Indeed, the use of normative samples to develop cognitive tests is commonplace in the neuropsychological literature, and we refer the reader to [49] and references therein for many examples. Along these lines, we advocate for control-group feature standardization when estimating disease patterns in the brain and developing image-based biomarkers. Additionally, as demonstrated in Section 2.2, the control normalization improves the interpretability of the SVM weight pattern due to the relative stability of the weights across different disease group sample sizes. It is difficult to interpret estimated disease patterns that are affected by sample size, since the underlying “true” SVM weights should not depend on the number of samples in each group.
We showed in Section 2.2 that the standard normalization method depends on the relative sample size of the two groups as well as the marginal separability; in contrast, the control normalization is unaffected by these qualities of the data and hence provides better generalizability across samples. We believe that including a control normalization step in the MVPA preprocessing pipeline is a simple alternative to current practice that promises increased interpretability, generalizability, and performance of the results.
We have focused attention on classification of subjects into groups based on medical diagnosis. In principle, the idea of control-based feature normalization for MVPA naturally extends to classification of events within subjects. For example, MVPA is increasingly applied to fMRI data in order to decode which patterns of activity correspond to a specific cognitive task or mental state [11, 50, 51]. In this setting, one could normalize the fMRI data using the subset of time points where the subject is at rest or performing the control task. Application of these techniques to within-subject timeseries data would have the potential for increasing task decoding accuracy in the face of performance variability that exists both between [52] and within individuals [53, 54], and is known to have a substantial relationship to patterns of activation [55, 56]. Performance variability is a particularly important confound in studies of neuropsychiatric conditions [57, 58] as well as lifespan studies of [59] and aging [60]. Such an application would represent a natural extension of the current work, and the overarching idea remains the same: feature normalization as a data preprocessing step in MVPA should be applied in an intentional way with an understanding of the way in which it might affect the results of the analysis.
5. Conclusion
The roots of feature scaling for preprocessing lie in the neural network literature of the 1990s. The universal approximation theorem, which broadly states that simple neural networks can approximate a rich set of functions, was initially proven for functions defined on a unit hypercube domain using a multilayer perceptron constructed with sigmoidal neurons [61]. It became natural to assume that centering and scaling of data would lead to a faster convergence even though neural networks are theoretically affine invariant [62]. For the most part, the optimization turned out to be more graceful with these scaled inputs, since it slowed down network saturation and avoided the vanishing gradients problem to a certain extent.
However, applying scaling to kernel methods such as SVMs or distance-based methods such as k-means tends to yield completely different results depending on the scaling method used. This is because these methods are not transformation-invariant. In such a case, scaling essentially imposes a form of soft feature selection since it implicitly changes the metric used for computing the kernel matrix. This fact is important in the context of image-based diagnosis using SVMs with region of interest (ROI) data. Scaling implicitly enforces the fact that variation in the amygdala, which is a relatively small structure in terms of volume, is as important as that in the prefrontal lobe, which is much larger in volume. Thus, appropriate scaling of features is an important but under-emphasized issue that we have attempted to call attention to in this manuscript.
It is critical for researchers wishing to interpret the results of MVPA from SVMs to understand how the choice of feature normalization influences the results, as well as how to determine the best method for their scientific question. We have proposed a control-based normalization and demonstrated several advantages of the approach for classifying subjects into groups, for example, by medical diagnosis. Most notably, we have highlighted the possibility of improved classifier performance according to criteria such as accuracy and AUC for a comprehensive set of data generating distributions. The control normalization improves classifier performance by giving higher weight, relative to other standardization techniques, to features with greater marginal separability between groups. Depending on the underlying data generating distribution and relative sample size between groups, different classifiers will experience tradeoffs between sensitivity and specificity. The optimal choice of feature normalization may depend on the unknown data generating distribution as well as certain clinical considerations. As a result, the interpretability of the control normalization is an attractive property that makes it amenable to a vast majority of clinical applications. Considered along with overall increases in accuracy and AUC demonstrated by the control normalization in the simulations and data examples, we advocate for its adoption as standard practice in MVPA using the SVM.
Highlights.
A control-based feature normalization is proposed for support vector classification.
Classifier performance is improved over two common feature normalization methods.
Estimated multivariate patterns are more interpretable using the proposed method.
Acknowledgments
The authors would like to acknowledge funding by NIH grant R01 NS085211 and a seed grant from the Center for Biomedical Image Computing and Analytics at the University of Pennsylvania. This work represents the opinions of the researchers and not necessarily that of the granting institutions.
Data used in the preparation of this article were obtained from the Alzheimers Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimers disease (AD). For up-to-date information, see www.adni-info.org.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cortes C, Vapnik V. Support-vector networks. Machine learning. 1995;20(3):273–297. [Google Scholar]
- 2.Vapnik V. The nature of statistical learning theory. Springer Science & Business Media; 2013. [Google Scholar]
- 3.Frackowiak R, Friston K, Frith C, Dolan R, Mazziotta J, editors. Human Brain Function. Academic Press; USA: 1997. URL http://www.fil.ion.ucl.ac.uk/spm/doc/books/hbf1/ [Google Scholar]
- 4.Friston KJ, Frith C, Liddle P, Frackowiak R. Comparing functional (PET) images: the assessment of significant change. Journal of Cerebral Blood Flow & Metabolism. 1991;11(4):690–699. doi: 10.1038/jcbfm.1991.122. [DOI] [PubMed] [Google Scholar]
- 5.Friston KJ, Holmes AP, Worsley KJ, Poline JP, Frith CD, Frackowiak RS. Statistical parametric maps in functional imaging: a general linear approach. Human brain mapping. 1994;2(4):189–210. [Google Scholar]
- 6.Ashburner J, Friston KJ. Voxel-based morphometry – the methods. Neuroimage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 7.Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the RAVENS maps: Methods and validation using simulated longitudinal atrophy. NeuroImage. 2001;14(6):1361–1369. doi: 10.1006/nimg.2001.0937. doi: http://dx.doi.org/10.1006/nimg.2001.0937. URL http://www.sciencedirect.com/science/article/pii/S1053811901909371. [DOI] [PubMed] [Google Scholar]
- 8.Craddock RC, Holtzheimer PE, Hu XP, Mayberg HS. Disease state prediction from resting state functional connectivity. Magnetic Resonance in Medicine. 2009;62(6):1619–1628. doi: 10.1002/mrm.22159. URL http://dx.doi.org/10.1002/mrm.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuingnet R, Rosso C, Chupin M, Lehricy S, Dormont D, Benali H, Samson Y, Colliot O. Spatial regularization of SVM for the detection of diffusion alterations associated with stroke outcome. Medical Image Analysis. 2011;15(5):729–737. doi: 10.1016/j.media.2011.05.007. special Issue on the 2010 Conference on Medical Image Computing and Computer-Assisted Intervention. doi: http://dx.doi.org/10.1016/j.media.2011.05.007. URL http://www.sciencedirect.com/science/article/pii/S1361841511000594. [DOI] [PubMed] [Google Scholar]
- 10.Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiology of Aging. 2011;32(12):2322.e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. doi: http://dx.doi.org/10.1016/j.neurobiolaging.2010.05.023. URL http://www.sciencedirect.com/science/article/pii/S019745801000237X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davatzikos C, Ruparel K, Fan Y, Shen D, Acharyya M, Loughead J, Gur R, Langleben DD. Classifying spatial patterns of brain activity with machine learning methods: application to lie detection. Neuroimage. 2005;28(3):663–668. doi: 10.1016/j.neuroimage.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Davatzikos C, Resnick S, Wu X, Parmpi P, Clark C. Individual patient diagnosis of AD and FTD via high-dimensional pattern classification of MRI. NeuroImage. 2008;41(4):1220–1227. doi: 10.1016/j.neuroimage.2008.03.050. doi: http://dx.doi.org/10.1016/j.neuroimage.2008.03.050. URL http://www.sciencedirect.com/science/article/pii/S1053811908002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davatzikos C, Xu F, An Y, Fan Y, Resnick SM. Longitudinal progression of Alzheimer’s-like patterns of atrophy in normal older adults: the SPARE-AD index. Brain. 2009;132(8):2026–2035. doi: 10.1093/brain/awp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Martino F, Valente G, Staeren N, Ashburner J, Goebel R, Formisano E. Combining multivariate voxel selection and support vector machines for mapping and classification of fMRI spatial patterns. Neuroimage. 2008;43(1):44–58. doi: 10.1016/j.neuroimage.2008.06.037. [DOI] [PubMed] [Google Scholar]
- 15.Fan Y, Shen D, Gur RC, Gur RE, Davatzikos C. Compare: classification of morphological patterns using adaptive regional elements, Medical Imaging. IEEE Transactions on. 2007;26(1):93–105. doi: 10.1109/TMI.2006.886812. [DOI] [PubMed] [Google Scholar]
- 16.Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RSJ. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131(3):681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koutsouleris N, Meisenzahl EM, Davatzikos C, Bottlender R, Frodl T, Scheuerecker J, Schmitt G, Zetzsche T, Decker P, Reiser M, et al. Use of neuroanatomical pattern classification to identify subjects in at-risk mental states of psychosis and predict disease transition. Archives of general psychiatry. 2009;66(7):700–712. doi: 10.1001/archgenpsychiatry.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langs G, Menze BH, Lashkari D, Golland P. Detecting stable distributed patterns of brain activation using gini contrast. NeuroImage. 2011;56(2):497–507. doi: 10.1016/j.neuroimage.2010.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mingoia G, Wagner G, Langbein K, Maitra R, Smesny S, Dietzek M, Burmeister HP, Reichenbach JR, Schlösser RG, Gaser C, et al. Default mode network activity in schizophrenia studied at resting state using probabilistic ICA. Schizophrenia research. 2012;138(2):143–149. doi: 10.1016/j.schres.2012.01.036. [DOI] [PubMed] [Google Scholar]
- 20.Mourão-Miranda J, Bokde AL, Born C, Hampel H, Stetter M. Classifying brain states and determining the discriminating activation patterns: support vector machine on functional MRI data. NeuroImage. 2005;28(4):980–995. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- 21.Pereira F. Beyond brain blobs: machine learning classifiers as instruments for analyzing functional magnetic resonance imaging data. ProQuest; 2007. [Google Scholar]
- 22.Richiardi J, Eryilmaz H, Schwartz S, Vuilleumier P, Van De Ville D. Decoding brain states from fmri connectivity graphs. Neuroimage. 2011;56(2):616–626. doi: 10.1016/j.neuroimage.2010.05.081. [DOI] [PubMed] [Google Scholar]
- 23.Sabuncu MR, Van Leemput K. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2011. Springer; 2011. The Relevance Voxel Machine (RVoxM): a Bayesian method for image-based prediction; pp. 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr Alzheimer’s disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage. 2008;39(3):1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkataraman A, Rathi Y, Kubicki M, Westin CF, Golland P. Joint modeling of anatomical and functional connectivity for population studies, Medical Imaging. IEEE Transactions on. 2012;31(2):164–182. doi: 10.1109/TMI.2011.2166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Childress AR, Wang J, Detre JA. Support vector machine learning-based fMRI data group analysis. NeuroImage. 2007;36(4):1139–1151. doi: 10.1016/j.neuroimage.2007.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L, Groth KM, Pearlson G, Schretlen DJ, Calhoun VD. Source-based morphometry: The use of independent component analysis to identify gray matter differences with application to schizophrenia. Human brain mapping. 2009;30(3):711–724. doi: 10.1002/hbm.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiss PT, Ogden RT. Functional generalized linear models with images as predictors. Biometrics. 2010;66(1):61–69. doi: 10.1111/j.1541-0420.2009.01233.x. [DOI] [PubMed] [Google Scholar]
- 29.Gaonkar B, Davatzikos C. Analytic estimation of statistical significance maps for support vector machine based multi-variate image analysis and classification. NeuroImage. 2013;78(0):270–283. doi: 10.1016/j.neuroimage.2013.03.066. doi: http://dx.doi.org/10.1016/j.neuroimage.2013.03.066. URL http://www.sciencedirect.com/science/article/pii/S1053811913003169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng X, Lin P, Zhang T, Wang J. Extreme learning machine-based classification of ADHD using brain structural MRI data. doi: 10.1371/journal.pone.0079476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanke M, Halchenko YO, Sederberg PB, Olivetti E, Fründ I, Rieger JW, Herrmann CS, Haxby JV, Hanson SJ, Pollmann S. PyMVPA: a unifying approach to the analysis of neuroscientific data. Frontiers in neuroinformatics. :3. doi: 10.3389/neuro.11.003.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zacharaki EI, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem ER, Davatzikos C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magnetic Resonance in Medicine. 2009;62(6):1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etzel JA, Valchev N, Keysers C. The impact of certain methodological choices on multivariate analysis of fMRI data with support vector machines. Neuroimage. 2011;54(2):1159–1167. doi: 10.1016/j.neuroimage.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Shen H, Tang F, Zang Y, Hu D. Combined structural and resting-state functional MRI analysis of sexual dimorphism in the young adult human brain: an MVPA approach. Neuroimage. 2012;61(4):931–940. doi: 10.1016/j.neuroimage.2012.03.080. [DOI] [PubMed] [Google Scholar]
- 35.Sato JR, Kozasa EH, Russell TA, Radvany J, Mello LE, Lacerda SS, Amaro E., Jr Brain imaging analysis can identify participants under regular mental training. PloS one. 2012;7(7):e39832–e39832. doi: 10.1371/journal.pone.0039832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schölkopf B, Tsuda K, Vert JP. Kernel methods in computational biology. MIT press; 2004. [Google Scholar]
- 37.Hastie T, Tibshirani R, Friedman J. Elements of Statistical Learning. Vol. 1. Springer; Berlin: 2001. (Springer series in statistics). [Google Scholar]
- 38.Orrù G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neuroscience & Biobehavioral Reviews. 2012;36(4):1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Bendfeldt K, Klöppel S, Nichols TE, Smieskova R, Kuster P, Traud S, Mueller-Lenke N, Naegelin Y, Kappos L, Radue EW, et al. Multivariate pattern classification of gray matter pathology in multiple sclerosis. Neuroimage. 2012;60(1):400–408. doi: 10.1016/j.neuroimage.2011.12.070. [DOI] [PubMed] [Google Scholar]
- 40.Costafreda SG, Chu C, Ashburner J, Fu CH. Prognostic and diagnostic potential of the structural neuroanatomy of depression. PLoS One. 2009;4(7):e6353. doi: 10.1371/journal.pone.0006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong Q, Wu Q, Scarpazza C, Lui S, Jia Z, Marquand A, Huang X, McGuire P, Mechelli A. Prognostic prediction of therapeutic response in depression using high-field MR imaging. Neuroimage. 2011;55(4):1497–1503. doi: 10.1016/j.neuroimage.2010.11.079. [DOI] [PubMed] [Google Scholar]
- 42.Liu F, Guo W, Yu D, Gao Q, Gao K, Xue Z, Du H, Zhang J, Tan C, Liu Z, et al. Classification of different therapeutic responses of major depressive disorder with multivariate pattern analysis method based on structural MR scans. PLoS One. 2012;7(7):e40968. doi: 10.1371/journal.pone.0040968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 44.Quionero-Candela J, Sugiyama M, Schwaighofer A, Lawrence ND. Dataset shift in machine learning. The MIT Press; 2009. [Google Scholar]
- 45.Moreno-Torres JG, Raeder T, Alaiz-RodríGuez R, Chawla NV, Herrera F. A unifying view on dataset shift in classification. Pattern Recognition. 2012;45(1):521–530. [Google Scholar]
- 46.Chang CC, Lin CJ. LIBSVM: A library for support vector machines, ACM Transactions on Intelligent. Systems and Technology. 2011;2:27:1–27:27. [Google Scholar]
- 47.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer’s & Dementia. 2013;9(5):e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doshi J, Erus G, Ou Y, Davatzikos C. MICCAI Challenge Workshop on Segmentation: Algorithms, Theory and Applications. Nagoya, Japan: 2013. Ensemble-based medical image labeling via sampling morphological appearance manifolds. [Google Scholar]
- 49.Lezak MD. Neuropsychological assessment. Oxford university press; 2004. [Google Scholar]
- 50.Haynes JD, Rees G. Decoding mental states from brain activity in humans. Nature Reviews Neuroscience. 2006;7(7):523–534. doi: 10.1038/nrn1931. [DOI] [PubMed] [Google Scholar]
- 51.Norman KA, Polyn SM, Detre GJ, Haxby JV. Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends in cognitive sciences. 2006;10(9):424–430. doi: 10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Gur RC, Richard J, Calkins ME, Chiavacci R, Hansen JA, Bilker WB, Loughead J, Connolly JJ, Qiu H, Mentch FD, et al. Age group and sex differences in performance on a computerized neurocognitive battery in children age 8–21. Neuropsychology. 2012;26(2):251. doi: 10.1037/a0026712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacDonald SW, Li SC, Bäckman L. Neural underpinnings of within-person variability in cognitive functioning. Psychology and aging. 2009;24(4):792. doi: 10.1037/a0017798. [DOI] [PubMed] [Google Scholar]
- 54.Roalf DR, Gur RE, Ruparel K, Calkins ME, Satterthwaite TD, Bilker WB, Hakonarson H, Harris LJ, Gur RC. Within-individual variability in neurocognitive performance: Age-and sexrelated differences in children and youths from ages 8 to 21. Neuropsychology. 2014;28(4):506. doi: 10.1037/neu0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roalf DR, Ruparel K, Gur RE, Bilker W, Gerraty R, Elliott MA, Gallagher RS, Almasy L, Pogue-Geile MF, Prasad K, et al. Neuroimaging predictors of cognitive performance across a standardized neurocognitive battery. Neuropsychology. 2014;28(2):161. doi: 10.1037/neu0000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fox MD, Snyder AZ, Vincent JL, Raichle ME. Intrinsic fluctuations within cortical systems account for intertrial variability in human behavior. Neuron. 2007;56(1):171–184. doi: 10.1016/j.neuron.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 57.Callicott JH, Bertolino A, Mattay VS, Langheim FJ, Duyn J, Coppola R, Goldberg TE, Weinberger DR. Physiological dysfunction of the dorsolateral prefrontal cortex in schizophrenia revisited. Cerebral Cortex. 2000;10(11):1078–1092. doi: 10.1093/cercor/10.11.1078. [DOI] [PubMed] [Google Scholar]
- 58.Wolf DH, Satterthwaite TD, Calkins ME, Ruparel K, Elliott MA, Hopson RD, Jackson CT, Prabhakaran K, Bilker WB, Hakonarson H, et al. Functional neuroimaging abnormalities in youth with psychosis spectrum symptoms. JAMA psychiatry. 2015;72(5):456–465. doi: 10.1001/jamapsychiatry.2014.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satterthwaite TD, Wolf DH, Erus G, Ruparel K, Elliott MA, Gennatas ED, Hopson R, Jackson C, Prabhakaran K, Bilker WB, et al. Functional maturation of the executive system during adolescence. The Journal of Neuroscience. 2013;33(41):16249–16261. doi: 10.1523/JNEUROSCI.2345-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gazzaley A, Cooney JW, Rissman J, D’Esposito M. Top-down suppression deficit underlies working memory impairment in normal aging. Nature neuroscience. 2005;8(10):1298–1300. doi: 10.1038/nn1543. [DOI] [PubMed] [Google Scholar]
- 61.Cybenko G. Approximation by superpositions of a sigmoidal function, Mathematics of control. signals and systems. 1989;2(4):303–314. [Google Scholar]
- 62.Perantonis SJ, Lisboa PJ. Translation, rotation, and scale invariant pattern recognition by high-order neural networks and moment classifiers, Neural Networks. IEEE Transactions on. 1992;3(2):241–251. doi: 10.1109/72.125865. [DOI] [PubMed] [Google Scholar]


