Abstract
Purpose
We previously showed that high-density lipoprotein (HDL) radiosensitizes inflammatory breast cancer (IBC) cells in vitro and is associated with better local control after radiotherapy in IBC patients. The microRNA miR-33 family negatively regulates ABCA1. We hypothesized that variation in miR-33a expression in IBC cancer cells vs non-IBC cells correlates with radiation sensitivity following exposure to HDL in vitro.
Materials and Methods
miR-33a expression was analyzed by reverse transcriptase–polymerase chain reaction in four cell lines representing common clinical breast cancer subtypes. Overexpression and knockdown of miR-33a was demonstrated via transfection of miR-33a mimic, or anti-miR-33a constructs in high and low expressing miR-33a cell lines. Clonogenic survival in vitro in these cells was quantified at baseline and following HDL treatment. MiR-33a expression on distant relapse-free survival (DRFS) of 210 cases downloaded from the Oxford breast cancer dataset was determined.
Results
Expression levels of miR-33a was lower in IBC cell lines and IBC tumor samples compared with non-IBC cell lines and normal breast tissue. Cholesterol concentrations in the cell membrane was higher in IBC cells compared with non-IBC cells. Clonogenic survival following 24 hours of HDL treatment was decreased in response to irradiation in the low miR-33a–expressing cell lines, SUM 149 and KPL4, but survival following HDL treatment decreased in the high miR-33a–expressing cell lines, MDA-MB-231 and SUM 159. In the high miR-33a–expressing cell lines, anti-miR-33a transfection decreased radiation resistance in clonogenic assays. Conversely, in the low miR-33a–expressing cell lines, miR-33a mimic reversed the HDL-induced radiosensitization. Breast cancer patients in the top quartile based on miR-33a expression had markedly lower rates of DRFS than the bottom quartile (p= 0.0228, log-rank test). For breast cancer patients treated with radiation, high miR-33a expression predicted for worse overall survival (p=0.06).
Conclusions
Our results reveal miR-33a negatively regulates HDL-induced radiation sensitivity in breast cancer.
Keywords: miR-33, High Density Lipoprotein, Radiation Sensitivity
Introduction
We previously demonstrated inflammatory breast cancer (IBC) patients taking a statin (a drug that lowers serum cholesterol level) had lower local recurrence rates following radiation therapy than patients not taking a statin, and preclinical studies showed that statins radiosensitized both IBC and non-IBC cells [1]. A follow-up study from the same patient cohort demonstrated IBC patients with a higher than normal level of high-density lipoprotein (HDL, which transports cholesterol from peripheral tissues to the liver), had higher local control after radiation than patients with a lower than normal HDL level. On multivariate analysis HDL and vLDL were significant predictors for local control, while statin use was no longer significant once lipoproteins were included in the model. In vitro studies showed that treatment of IBC cell lines with HDL led to increased radiation sensitivity [2]. This work suggested that the influence of statins was through the effect on lipoproteins over intracellular signaling or off-target effects.
Preclinical studies have supported the role of cholesterol regulation in breast cancer progression, mostly in estrogen receptor–positive (ER+) subtypes [3–5]. The mechanism by which cholesterol promotes radiation resistance is at this time not clearly understood. Broadly speaking HDL is expected to mediate reverse cholesterol transport removing both intracellular and membrane cholesterol. The main mediators of cholesterol efflux from intracellular to extracellular HDL as well as extraction of cholesterol from lipid rich membrane regions are the adenosine triphosphate–binding cassette (ABC) transporters ABCA1 and ABCG1 [6].
MicroRNAs (miRNAs) are noncoding small RNAs that repress translation and cleave mRNA by binding to the 3′-untranslated region of their target gene [7]. MiR-33 is known to inhibit the expression of cholesterol transport genes, including ABCA1 and ABCG1 [8]. Given the dependence of HDL on these membrane proteins to extract cholesterol from cells, we hypothesized that expression of miR-33 inhibits HDL-induced breast cancer radiosensitivity by inhibiting ABCA1 and preventing HDL mediated cholesterol extraction. We tested our hypothesis by manipulating the expression of miR-33 in high and low miR-33–expressing breast cancer cell lines and testing radiation sensitivity in these lines in the presence or absence of HDL. Our results indicate that miR-33a decreases HDL-induced radiosensitivity in vitro.
Materials and Methods
Cell culture and drugs
SUM149 and SUM159 cells were obtained from Asterand (Detroit, MI). Both types of cells were cultured in Ham F12 medium supplemented with 10% fetal bovine serum (FBS). KPL4 IBC cells were maintained in DMEM/F-12 medium supplemented with 10% FBS. For all experiments including lipoproteins, serum was not added to the medium to prevent contamination/confounding of the results by the cholesterol and lipoprotein contained in FBS.
RNA isolation and miRNA PCR
Total RNA was isolated from the cell lines by using Trizol reagent (Invitrogen, Grand Island, NY) according to manufacturer’s protocol. To isolate the miRNAs, RNA isolated from cell lines was reverse-transcribed to cDNA using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) following the manufacturer’s instructions and subjected to polymerase chain reaction (PCR) in a Thermal Cycler (BioRad, Hercules, CA) at 16°C for 30 minutes, 42°C for 30 minutes, and 85°C for 5 minutes. The expression levels of mature miR-33a was measured by quantitative reverse-transcription PCR (qRT-PCR) using TaqMan MicroRNA assays (Applied Biosystems) according to the manufacturer’s instructions. The reaction was performed in a 7300 real-time PCR system (Applied Biosystems) at 95°C for 10 minutes and 40 cycles at 95°C for 15 seconds and 60°C for 60 seconds. U6 was used as a reference.
Immunoblotting
For immunoblotting, cells were subjected to lysis in 1× RIPA lysis buffer containing 1 μM phenylmethylsulfonyl fluoride, and 40 μg of protein was subjected to electrophoresis on sodium dodecyl sulfate–polyacrylamide gels with a concentration gradient of 4% to 20% (Invitrogen). Membranes were incubated with primary antibodies anti-ABCA1 (Novus Biologicals, Littleton, CO). Actin antibody was used as a loading control.
miR-33a, anti-miR-33a, and si-ABCA1 transfection
SUM149 and KPL4 were transfected with 40 nM miRIDIAN miR-33a mimic (miR-33a) and SUM159 and MDA-MB-231 cells with 60 nM miRIDIAN miRNA inhibitor (anti-miR-33a) (Dharmacon, Lafayette, CO), utilizing Oligofectamine (Invitrogen), for 72 hours. All control samples were treated with an equal concentration of a non-targeting control mimic sequence or negative control inhibitor sequence to detect non–sequence-specific effects. miR-33a overexpression and knockdown were verified by using qRT-PCR as described above. siRNA targeting ABCA1 and nontargeting siRNA were purchased from ThermoFisher (Cat# AM16708). SUM149 and KPL4 cells were transfected with the siRNAs using TransIT-TKO Transfection Reagent (Mirus) according to the manufacturer’s protocol. 72 hours after transfection, cells were washed collected for analysis of ABCA1 levels by western blotting and assayed for clonogenic survival.
Clonogenic survival assays
Clonogenic viability of the breast cancer cells was tested in triplicate in standard monolayer conditions. Following 72 hours of miR-33a, anti-miR-33a, or si-ABCA1 transfection, cells were washed with phosphate-buffered saline solution (PBS) and supplemented with serum-free medium with or without 10 μg/mL of HDL for a total of 24 hours. After treatment with HDL, cells were irradiated and then incubated for 14 days. Cells were then subjected to crystal violet staining (10%) to mark colonies with 50 cells or more (≥300-μm diameter). Survival curves were obtained for all groups, and curves were fitted on the basis of the linear-quadratic model. Cells were irradiated by using a 137Cs source (Shepherd Irradiator, J.L. Shepherd and Associates, San Fernando, CA).
Patient outcomes
The Oxford breast cancer dataset [10] was downloaded from the U.S. National Center for Biotechnology Information (NCBI) GEO website (GSE22216). Expression values for miR-33a (Probe set id = ILMN_3167691) and clinical characteristics for 210 cases were extracted. These data were subjected to a Cox regression model for distant recurrence-free survival duration (DRFS) with inclusion of the following covariates: patient age, tumor size and grade, nodal involvement, ER expression, and quartile of miR-33 expression.
Microarray analysis
Fifty-three patients treated between 1997 and 2011 at XXX with available fresh frozen breast tissue were identified from the XXX IBC registry and institutional tissue bank, and used in the microarray study. The miRNA and mRNA microarray analysis were performed with miRNA 3.0 Array and U133 Plus 2.0 Array Human Genome Genechip (Affymetrix) systems according to the manufacturer’s instructions by the Sequencing and Non-coding RNA Program core at XXX. MiRNA microarrays were normalized and quantified using the Robust Multiarray Analysis (RMA) algorithm that borrows strength across arrays. Normalized and quantified intensities were log (base 2) transformed.
Statistical analysis
For in vitro studies, statistical significance of differences between groups was determined by using the Student t-test and calculated by Origin software (OriginLab, Northampton, MA).
Results
miR-33a expression is lower in IBC cells and tissues compared with non-IBC cells and tissues
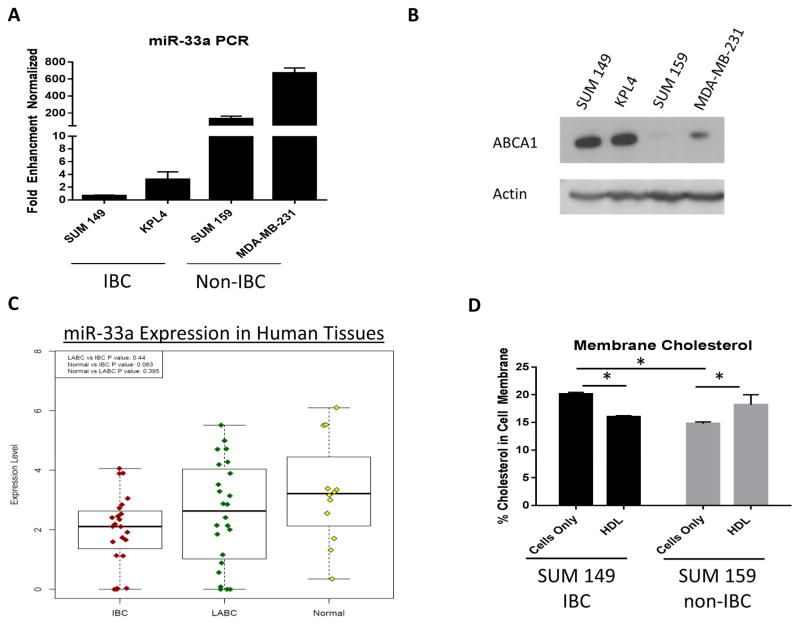
We first set out to determine the expression levels of miR-33a in both IBC and non-IBC breast cancer cell lines. The non-IBC triple negative breast cancer cell lines, SUM 159 and MDA-MB-231, had 132 and 672 fold higher expression of miR-33a by RT-PCR, respectively, compared with the IBC cell lines SUM 149 and KPL4 (Fig 1A). As ABCA1 is a known target of miR-33 [9], we next tested the expression levels of this protein in these cells through western blotting. Expression of ABCA1 protein was inversely correlated with miR-33a RNA expression in both the IBC and non-IBC cells (Fig. 1B). Using a miRNA microarray of miR-33a in human breast tissue samples, miR-33a expression was lower in IBC compared with locally advanced breast cancers (LABC) (p=0.44) and normal breast tissue (p=0.063) (Fig 1C). We next performed lipidomics on the low miR-33a/high ABCA1 SUM 149 cell line and the high miR-33a/low ABCA1 SUM 159 cell line. At baseline, SUM 149 contains more cholesterol in the cellular membrane compared with SUM 159. After 24 hours of treatment with HDL, the percentage of cholesterol is reduced in SUM 149, but is increased in the SUM 159 cells (Fig 1D). These results further show IBC and non-IBC cells have differences in cellular cholesterol regulation.
Figure 1.
Expression of miR-33a and the protein product of its target gene ABCA1 in breast cancer cells. (A) miR-33a expression was higher in non-IBC cell lines (SUM159 and MDA-231) than in IBC cell lines (KPL4 and SUM149). (B) Expression of ABCA1 was higher in KPL4 and SUM149 cells than in SUM159 and MDA-231 cells. (C) miRNA microarray from human tissue showing miR-33a expression in IBC vs LABC vs normal breast. (D) Cholesterol in the cell membrane as a percentage of total lipid content in SUM 149 and SUM 159 cells before and after 24 hours of HDL treatment (10 μg/mL). Significant differences are shown as follows: *, P < 0.05 for unpaired 2-tailed Student’s t-test (n = 3).
Targeting miR-33a and ABCA1 expression and HDL-induced radiation response
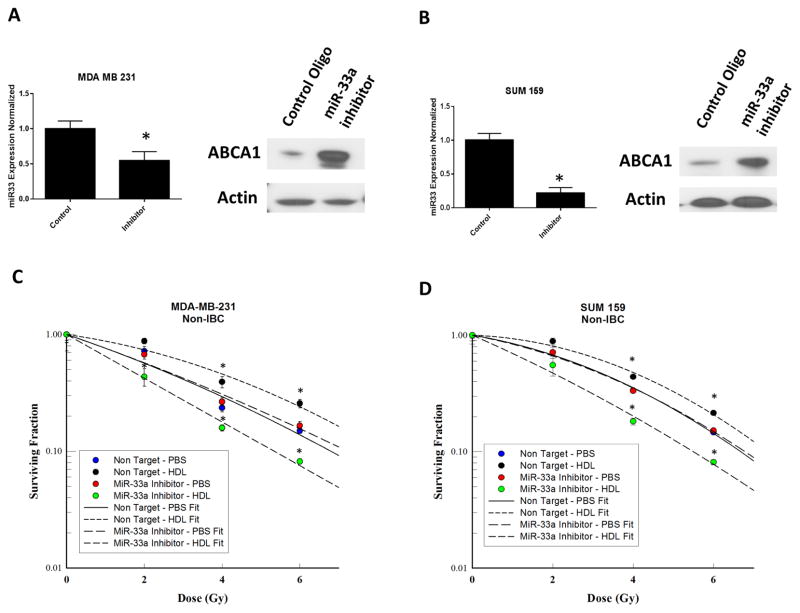
To investigate the role of miR-33a in radiation sensitivity of breast cancer cells in response to HDL, we knocked down miR-33a in SUM159 and MDA-231, two breast cancer cell lines that express miR-33a endogenously (Fig. 1A). Antagonism of endogenous miR-33a decreased miR-33a miRNA levels and increased ABCA1 protein expression in both cell types (Fig. 2A, B). Cells treated with non-target controls showed increased resistance to radiation with treatment of HDL compared with PBS. The transfection of miR-33a inhibitors resulted in radiosensitization of MDA-MB-231 and SUM 159 cells when treated with HDL (Fig. 2C, D).
Figure 2.
Inhibition of miR-33a increases HDL-induced radiosensitization. (A, B) Knockdown of miR-33a in SUM159 and MDA-MB-231 cell lines resulted in greater ABCA1 expression than in cells transfected with control oligonucleotides. (C, D) Clonogenic survival assays of miR-33a knockdown MDA-MB-231 and SUM159 breast cancer cells irradiated at different doses (x axis) with or without HDL pretreatment. HDL pretreatment comprised incubation for 24 hours with HDL 10 μg/mL. After irradiation, colonies were allowed to form for 14 days. The surviving fractions with PBS or HDL at each dose of radiation are shown. Data represented as mean +/− standard deviation. Significant differences are shown as follows: *, P < 0.05 for unpaired 2-tailed Student’s t-test (n = 3).
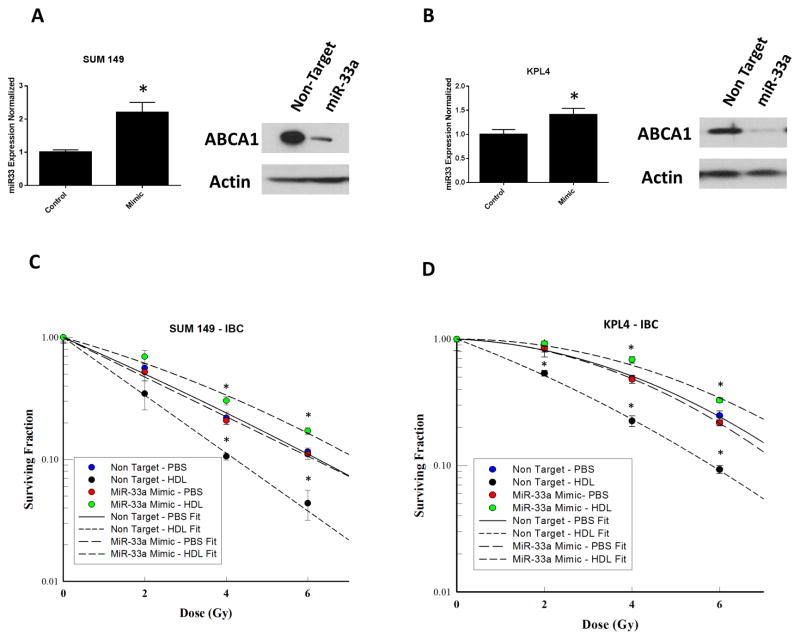
Next, transfection of SUM149 and KPL4 (low miR-33a–expressing cells) with miR-33a mimic increased miR-33a miRNA expression but decreased ABCA1 protein expression (Fig. 3A, B). Cells treated with non-target controls showed decreased resistance to radiation with treatment of HDL compared with PBS. Overexpression of miR-33a resulted in the inhibition of HDL-induced radiosensitization seen previously. Transfection of SUM149 cells with miR-33a mimic increased radiation resistance in the clonogenic survival following HDL treatment (Fig. 3C), and a similar result was observed in KPL4 cells (Fig. 3D). These results demonstrated that miR-33a negatively regulates the radiation response to HDL in the breast cancer cells examined.
Figure 3.
Ectopic expression of miR-33a inhibits HDL-induced radiosensitization. (A, B) Overexpression of miR-33a via transfection of miR-33 mimic in SUM149 and KPL4 cells resulted in lower ABCA1 expression than non-target transfection. (C, D) Clonogenic survival assays of miR-33a–overexpressing SUM149 and KPL4 breast cancer cells irradiated at different doses (x axis) with or without HDL pretreatment. HDL pretreatment comprised incubation with HDL 10 μg/mL for 24 hours. After irradiation, colonies were allowed to form for 14 days. The surviving fractions at each dose of radiation are shown. Data represented as mean +/− standard deviation. Significant differences are shown as follows: *, P < 0.05 for unpaired 2-tailed Student’s t-test (n = 3).
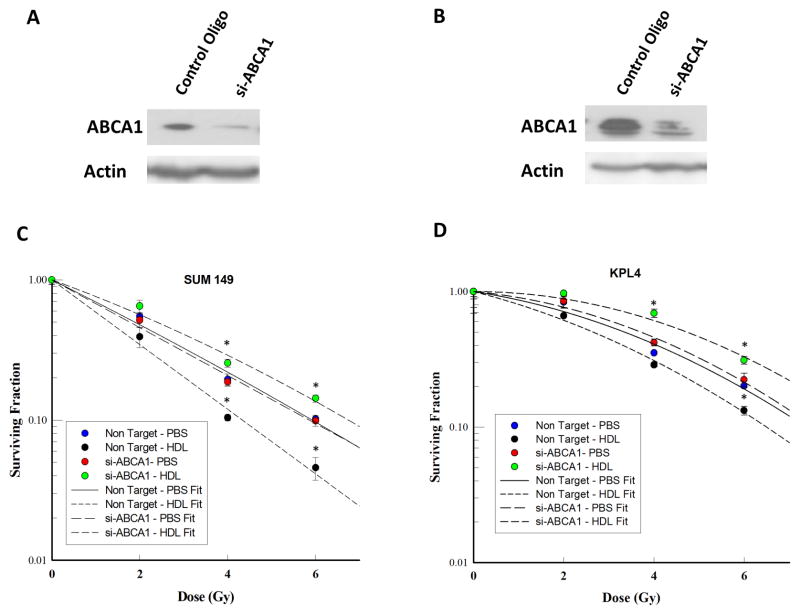
To examine whether the target of miR-33a, ABCA1, is responsible for the effects previously observed, we knocked down ABCA1 in SUM 149 and KPL4 cells. SiRNA targeting of ABCA1 successfully knocked down the protein expression in both SUM 149 and KPL4 (Fig. 4A, B). As observed previously with miR-33 mimics, the knockdown of ABCA1 resulted in increased radiation resistance when treated with HDL in both cell lines. In the non-targeted siRNA transfected cells, treatment with HDL resulted in radiosensitization (Fig 4C, D). These results show that inhibition of ABCA1 is the mechanism through which miR-33a decreases HDL-induced radiosensitization.
Figure 4.
Knockdown of ABCA1 inhibits HDL-induced radiosensitization. (A, B) SUM 149 and KPL4 transfected with si-ABCA1 resulted in knockdown of ABCA1 protein expression measured by western blotting. (C, D) Clonogenic survival assays of ABCA1 knockdown SUM149 and KPL4 breast cancer cells irradiated at different doses (x axis) with PBS or HDL pretreatment (10 μg/mL). After irradiation, colonies were allowed to form for 14 days. The surviving fractions at each dose of radiation are shown. Data represented as mean +/− standard deviation. Significant differences are shown as follows: *, P < 0.05 for unpaired 2-tailed Student’s t-test (n = 3).
miR-33a expression in breast cancer patients predicts for survival outcomes
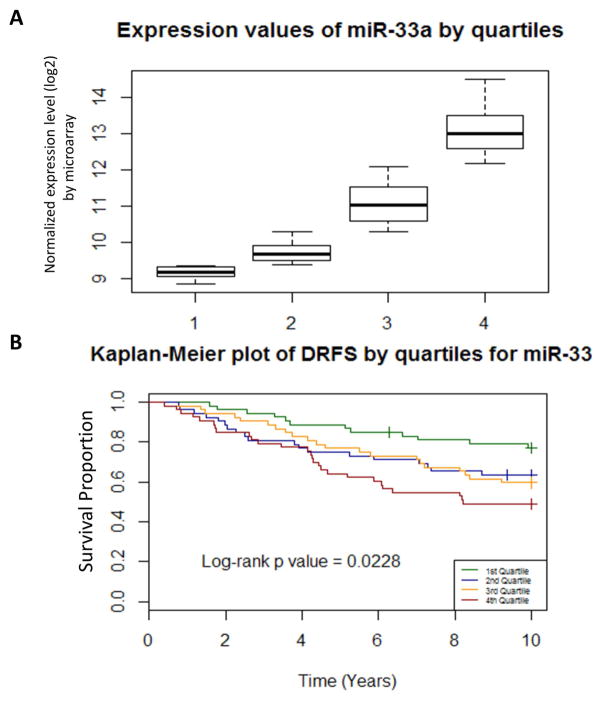
To explore the clinical significance of miR-33 expression, we examined a previously published dataset from the NCBI GEO [10]. Radiotherapy was delivered in 84% of cases. The 210 cases analyzed were separated into four quartiles based on miR-33a expression (Fig. 5A). We tested for differences (log-rank test) in distant recurrence free survival (DRFS) by miR-33a expression quartile and found a significant difference in DRFS between the highest and lowest quartiles (p=0.0228) (Fig. 5B). Cox regression model was performed for DRFS with inclusion of following covariates: age, size, nodes, ER status, grade, and quartile of miR-33. Multivariate Cox regression analysis for DRFS revealed that patients in quartile 4 (greatest miR-33a expression) had a DRFS hazard ratio of 2.0 relative to quartile 1. This trend approached significance (p=0.076).
Figure 5.
High serum miR-33a level is associated with worse outcome in breast cancer patients. (A) Expression of miR-33a in tumors from 210 patients whose data were downloaded from the Oxford breast cancer dataset was separated into four quartiles. (B) Kaplan-Meier curves representing distant recurrence–free survival (DRFS) for each miR-33a quartile.
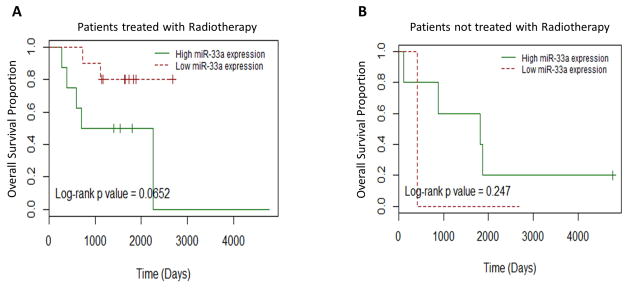
Furthermore, total RNA isolated from fresh frozen tissue of the primary tumors of 47 breast cancer tissue and 12 normal breast tissue samples from 53 patients was subjected to miRNA microarray analysis. Clinical outcome data and radiation treatment information was available for the breast cancer patients. MiR-33a expression in the breast cancer samples was compared with normal breast tissue. High miR-33a expression in patients predicted for shorter overall survival in breast cancer patients treated with radiation therapy (Fig. 6A) (p=0.065). There was no difference in overall survival in patients not treated with radiation between the two groups (Fig. 6B) (p=0.247).
Figure 6.
High miR-33a expression levels are associated with poor overall survival in breast cancer patients treated with radiation therapy. (A) Kaplan-Meier curves showing high miR-33a levels (green curves) were associated with shorter overall survival in breast cancers treated with radiation. (B) Kaplan-Meier curves for overall survival in the breast cancer patients not treated with radiation comparing high vs low miR-33a expression.
Discussion
In this study, we report for the first time that miR-33a inhibits cholesterol transport protein ACBA1 in breast cancer cell lines as has been reported in normal cells such as macrophages. Further, we demonstrate that expression of miR-33a was inversely correlated with HDL-induced radiation sensitivity in breast cancer cells. Our results support the importance of HDL mediated reverse cholesterol transport in breast cancer cell response to radiation therapy. Additional studies of ABCA1 as a biomarker for response to HDL directed radiosensitization strategies are warranted. ABCA1 deficiency in cancer cells was shown to increase cellular cholesterol, leading to increased cell survival [11]. In prostate cancer, hypermethylation of the ABCA1 promoter, which led to elevated cholesterol levels, was detected in high-grade prostate cancers but not in normal prostate tissues [12]. MiR-33 silencing in vivo increased hepatic expression of ABCA1 and plasma HDL levels [13]. Additional studies showed that miR-33 can promote glioma-initiating cells [14] and was upregulated in patients with chemoresistant osteosarcoma [9, 15].
The regulation of intracellular cholesterol is complex and is influenced by genetic factors and by posttranscriptional mechanisms [16]. MiR-33 is expressed in various cell types and tissues, and is a key regulator of cholesterol homeostasis by regulating the cholesterol transporters that remove cholesterol stores from cells to HDL lipoproteins [9]. Transcription of miR-33 is stimulated under low sterol conditions and represses genes, including ABCA1 and ABCG1, which are involved in cholesterol export [13]. Indeed, miR-33 control of both liver HDL biosynthesis and cellular efflux of cholesterol to nascent HDL particles has been shown by in vivo manipulation of miR-33 levels leading to changes in both circulating HDL levels and cellular cholesterol concentrations [13]. ABCA1 functions as the primary gatekeeper in regulating removal of excess free intracellular cholesterol from cells such as macrophages by effluxing cellular cholesterol to lipid-free apoA-1, resulting in the formation of HDL particles [17]. Some studies suggest these ABCA1 and a similar cholesterol efflux transporter ABCG1 are preferentially expressed in lipid rich membrane regions and also facilitate removal of lipid from the membrane surrounding these proteins in addition to channeling cholesterol form the intracellular space [17]. ABCA1 is known to be associated with HDL from the observation that patients with ABCA1 mutations (i.e., Tangier disease) have a deficiency in plasma HDL [18].
Another important observation was that HDL induced radioprotection in high miR-33a expressing cell lines, but radiosensitization in low miR-33a cell lines. This was also confirmed in the low miR-33a expressing cells that were transfected with miR-33a mimics or siRNA targeting ABCA1. The possible reasoning is complex. First, the miRNA microarray on breast cancer tissues and PCR for miR-33a on cells lines showed IBC express less miR-33a than both normal tissue and other advanced breast tumors. Secondly, we sent IBC and non IBC cells, treated and untreated with HDL, to a lipidomics core that specializes in measuring lipid content of cell membranes. The IBC cell line had at baseline higher percentage of cholesterol in the plasma membrane compared with the non-IBC cells. The IBC cells, low miR-33a expressing, decreased the percentage content of cholesterol in the cellular membrane following HDL treatment. In contrast, the non IBC cell line, high miR-33a, increased the percentage of cholesterol in the cellular membrane following the HDL treatment (Fig 1D). Third, we know from other studies that IBC cells have a unique regulation of cellular cholesterol compared to non IBC cells. IBC cells have the ability to maintain intracellular cholesterol in lipid depleted environments [19]. Fourth, our group and others have shown that increased membrane cholesterol concentrations enhance downstream signaling pathways that increase DNA repair and increase radiation resistance [2]. We believe that cells that have high miR-33a expression downregulate the ABCA1 transporter and therefore in the presence of HDL the cholesterol is inserted into the membrane instead of effluxed out of the cell to be picked up by HDL. This can lead to increased radiation resistance. This is further supported by our clonogenic assays of miR-33a mimics and ABCA1 knockdown IBC cells that had the highest radiation resistance with HDL treatment.
In conclusion, our results reveal that miR-33 decreases HDL-induced radiosensitivity in vitro, and cholesterol transport might be a novel way to target the disease of IBC.
Previous studies showed the mediator of cholesterol transport, high-density lipoprotein (HDL), increased radiation sensitivity in vitro of breast cancer cells and high HDL levels predicted for protective outcomes in breast cancer patients. This study investigates a mediator in cholesterol export, miR-33a, and its influence on breast cancer response to high-density lipoprotein (HDL)-induced radiosensitization. We showed that miR-33a regulates HDL-induced radiation sensitivity in breast cancer and high miR-33a expression predicted for worse distant relapse-free survival.
Acknowledgments
Supported by the National Institutes of Health Grants RO1CA180061 and RO1CA138239, a grant from the State of Texas Rare and Aggressive Breast Cancer Research Program, and the National Center for Clinical and Translational Science Grant TL1-TR000369.
Footnotes
Ethical Standards
The experiments comply with the current laws of the U.S.A.
Conflict of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.XXX
- 2.XXX
- 3.Llaverias G, Danilo C, Mercier I, Daumer K, Capozza F, Williams TM, Sotgia F, Lisanti MP, Frank PG. Role of cholesterol in the development and progression of breast cancer. Am J Pathol. 2011;178(1):402–12. doi: 10.1016/j.ajpath.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson ER, Wardell SE, Jasper JS, Park S, Suchindran S, Howe MK, Carver NJ, Pillai RV, Sullivan PM, Sondhi V, Umetani M, Geradts J, McDonnell DP. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science. 2013;342(6162):1094–8. doi: 10.1126/science.1241908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonnell DP, Park S, Goulet MT, Jasper J, Wardell SE, Chang CY, Norris JD, Guyton JR, Nelson ER. Obesity, cholesterol metabolism, and breast cancer pathogenesis. Cancer Res. 2014;74(18):4976–82. doi: 10.1158/0008-5472.CAN-14-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yvan-Charvet L, Wang N, Tall AR. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol. 2010;30(2):139–43. doi: 10.1161/ATVBAHA.108.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: origin, function and application. Journal of Experimental & Clinical Cancer Research : CR. 2012;31(1):38–38. doi: 10.1186/1756-9966-31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Hernando C, Suárez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Current opinion in lipidology. 2011;22(2):86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore KJ, Rayner KJ, Suárez Y, Fernández-Hernando C. microRNAs and cholesterol metabolism. Trends in endocrinology and metabolism: TEM. 2010;21(12):699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71(17):5635–45. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 11.Smith B, Land H. Anti-cancer activity of the cholesterol exporter ABCA1 gene. Cell reports. 2012;2(3):580–590. doi: 10.1016/j.celrep.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee BH, Taylor MG, Robinet P, Smith JD, Schweitzer J, Sehayek E, Falzarano SM, Magi-Galluzzi C, Klein EA, Ting AH. Dysregulation of cholesterol homeostasis in human prostate cancer through loss of ABCA1. Cancer Res. 2013;73(3):1211–8. doi: 10.1158/0008-5472.CAN-12-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–3. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Sun T, Hu J, Zhang R, Rao Y, Wang S, Chen R, McLendon RE, Friedman AH, Keir ST, Bigner DD, Li QJ, Wang H, Wang XF. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J Clin Invest. 2014;124(10):4489–502. doi: 10.1172/JCI75284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Huang Z, Wu S, Zang X, Liu M, Shi J. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J Exp Clin Cancer Res. 2014;33:12. doi: 10.1186/1756-9966-33-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wellington CL, Walker EK, Suarez A, Kwok A, Bissada N, Singaraja R, Yang YZ, Zhang LH, James E, Wilson JE, Francone O, McManus BM, Hayden MR. ABCA1 mRNA and protein distribution patterns predict multiple different roles and levels of regulation. Lab Invest. 2002;82(3):273–83. doi: 10.1038/labinvest.3780421. [DOI] [PubMed] [Google Scholar]
- 17.Oram JF, Lawn RM. ABCA1. The gatekeeper for eliminating excess tissue cholesterol. J Lipid Res. 2001;42(8):1173–9. [PubMed] [Google Scholar]
- 18.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, Kaminski WE, Hahmann HW, Oette K, Rothe G, Aslanidis C, Lackner KJ, Schmitz G. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22(4):347–51. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 19.Martin BJ, van Golen KL. A comparison of cholesterol uptake and storage in inflammatory and noninflammatory breast cancer cells. Int J Breast Cancer. 2012;2012:412581. doi: 10.1155/2012/412581. [DOI] [PMC free article] [PubMed] [Google Scholar]