Abstract
Sponges are considered poor invaders, and no genetic studies on introduced sponges have been performed up to now. Paraleucilla magna is the first calcareous sponge introduced to the Mediterranean and Northeastern Atlantic. The study aimed at investigating the genetic makeup and connectivity of the introduced populations of P. magna and at exploring signs of local phenotypic adaptation, to gain insight on the species invasive potential. Ten populations along the species introduction range (Brazil, Açores, Madeira, and continental Europe) were genetically characterized by using nine microsatellite markers. Most populations were genetically structured as suggested by significant Dst and Fst values, significant differences among populations (AMOVA) and the presence of private alleles. The analyzed populations belonged to three genetically homogeneous groups (K) according to the Bayesian algorithm (structure software) and the UPGMA dendrogram. Genetic diversity within populations was higher than expected. Recurrent introductions of non-randomly selected individuals from the native sources may have contributed to the heterozygote deficit found in all populations by forming pedigree structures with mating among relatives. Moreover, the species biological cycle was monitored in a population established on native Mediterranean assemblages (41°40′27″N, 2°47′25″E) and compared with the species cycle in other habitats. Contrasting life spans, growth habits, and reproduction cycles, depending on the habitat conditions, were recorded. To summarize, high genetic diversity, phenotypic local adaptation, and high reproduction rates altogether allow predicting the fast proliferation of P. magna in newly colonized regions and point to its strong invasive potential.
Electronic supplementary material
The online version of this article (doi:10.1007/s00227-016-2862-6) contains supplementary material, which is available to authorized users.
Introduction
Marine habitats support an ever-growing suite of allochthonous species mainly introduced by marine traffic and aquiculture activities (Naylor et al. 2001; Ruesink et al. 2005; Coutts and Dodgshun 2007; Haupt et al. 2009), which may harm ecosystems, economies, and/or public health (e.g. Ruiz et al. 1997; Grosholz 2002; Orensanz et al. 2002; Ruiz and Carlton 2003; Galil 2007; Carlton 2009; Rilov and Croocks 2009; Zenetos et al. 2010; Pitta et al. 2011; National Invasive Species Council (NISC), http://www.invasivespecies.gov/global/ISAC/ISAC_whitepapers.html).
Although most introduced marine species remain confined to particularly favorable habitats, such as harbors, marinas or sea farms (Zibrowius 1991; Robinson et al. 2005; Glasby et al. 2007; Tyrrell and Byers 2007; Dafforn et al. 2009; Bulleri and Chapman 2010) and only a small percentage (ca. 0.1 %) modifies the structure and function of native communities (Mack et al. 2000; Colautti and MacIsaac 2004; Blackburn et al. 2011; Thomsen et al. 2011), they may cause important economic losses by fouling ships’ hulls, clogging intake pipes, and competing for resources with cultured species and thus lowering culture yields (Ruiz et al. 1997; Pimentel et al. 2000).
The effects are even worse when introduced species become invasive, alter the biodiversity and structure of native assemblages (Ruiz et al. 1997; Naylor et al. 2000; Grosholz 2002; Coles and Bolick 2007; Molnar et al. 2008) and modify the evolutionary rates of native species (Mooney and Cleland 2001; Grosholz 2002; Shine 2012). Indeed, marine invasions are currently considered the second most important cause of diversity loss in the world oceans (Blakeslee et al. 2010).
The genetic traits of alien populations may provide invaluable information on their capacity of proliferation and resilience in their introduction area (Holland 2000; Grosberg and Cunningham 2001; Féral 2002; Turon et al. 2003; Rius et al. 2008; Geller et al. 2010; Pineda et al. 2011). The study of the genetic structure of introduced populations allows understanding dispersal patterns, whether natural or man-produced, identifying colonization events, and predicting the species invasive potential (e.g. Rius et al. 2012).
Phylogeography studies of marine organisms have been traditionally based on mitochondrial genes, in particular, the COI barcode marker (Avise 2009). However, the several partitions of COI used up to now for phylogeography studies seem to be extraordinarily conserved in the Phylum Porifera (Duran et al. 2004a; Wörheide et al. 2005; Uriz and Turon 2012; but see León-Pech et al. 2015) and thus are poorly informative for analyses of population genetics at ecological time scales. Conversely, hypervariable molecular markers such as microsatellites have proved suitable for studying recent historical events in demography and population genetics of sponges (see Uriz and Turon 2012 for a review; Chaves-Fonnegra et al. 2015) and may help in tracking the expansion patterns of recently introduced species.
Only few sponges introduced to foreign habitats have been reported up to now (Calcinai et al. 2004; Pérez et al. 2006; Longo et al. 2007; Van Soest et al. 2007; Avila and Carballo 2009; Henkel and Janussen 2011) and none of them have been studied genetically at large geographical scales. The calcareous sponge Paraleucilla magna Klautau, Monteiro, and Borojevic 2004 is the first known alien sponge established in native Mediterranean assemblages (Longo et al. 2004, 2007) and emerges as an exceptional model for studying the invasive capacities of introduced, sessile marine species with poor natural dispersal capacity. It was first described from Rio de Janeiro, where it was considered an alien (Klautau et al. 2004) and has proliferated in the Atlantic and Mediterranean during the last years, with records in the central (Zammit et al. 2009) and western Mediterranean (Longo et al. 2007; Guardiola et al. 2012; this study), Adriatic (Cvitkovic et al. 2013), South of Portugal, and Madeira and Azores Archipelagos (this study). Adult individuals recorded on native Northwestern Mediterranean assemblages, disappeared in July after larval release (Guardiola et al. 2012, authors pers. obs.). However, presence along the whole year has been reported in South Italy (Longo et al. 2012) and Brazil (Padua et al. 2012; Lanna et al. 2014). Populations of P. magna appear to be genetically structured across time, and at very short distances (i.e., less than one hundred meters), which was attributed to the genetic drift associated with small population sizes (Guardiola et al. 2012).
Studies on population genetics of introduced species rarely have considered phenotypic and life history traits in parallel, although this type of information may help to foresee their success as invaders. Here, we analyzed the genetic features of ten populations of P. magna along the species introduction range and monitored the species life cycle and population dynamics on a native assemblage of Northwestern Mediterranean.
We did not attempt to establish the species origin since only introduced populations are currently known. To determine the species introduction pathway is also challenging because the species was simultaneously detected in several distant locations, which suggests parallel introduction events (Longo et al. 2012). Moreover, if several introductions have occurred in a short period of time, the phylogeographic signal might be weakened or even lost, which will difficult tracking the introduction pathway (Pineda et al. 2011). Rather, the study aimed at investigating the genetic makeup and connectivity of the introduced populations, exploring signs of local phenotypic adaptation in parallel to gain insight on the species invasive potential.
Materials and methods
Sampling
Population genetics
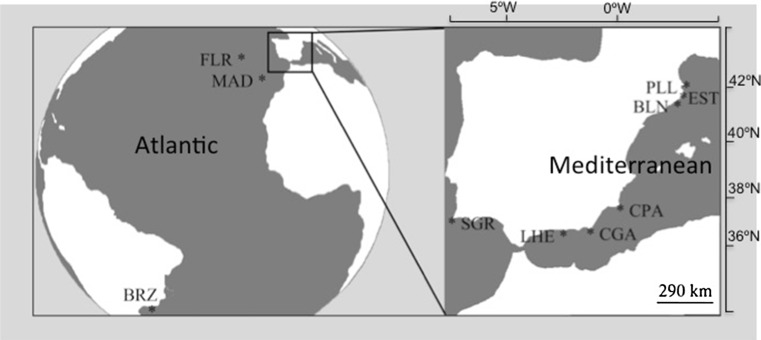
Samples of P. magna were collected from March to April 2008–2009 from ten localities: six from the Mediterranean Sea: Port Lligat (PLL) 42°17′37″N, 3°17′24″E; Estartit (EST), 42°3′1″N, 3°11′30″E; Blanes (BLN) 41°40′27″N, 2°47′25″ E; Cabo de Palos (CPA) 37°38′55.3″N, 0°47′11.7″W; Gabo de Gata (CGA) 36°56′6″N, 1°56′33″W; La Herradura (LHE) 36°44′9.5″N, 3°44′43.3″E—Iberian Peninsula; and four from the Atlantic Ocean: Azores-Flores (FLR) 39°26′50″N, 31°11′38″W; Madeira (M) 32°39′4″N, 16°54′35″W; Sagres (SGR)—Portugal—37°00′31″N, 8°56′35″W; and Rio de Janeiro—Brazil—(BRZ) 22°54′10″N, 43°12′27″W (Fig. 1). These locations were chosen to cover most of the species known range and include sites with biogeographic interest. All specimens were collected from rocky, vertical and sub-horizontal native assemblages at depths that ranged between 6 and 15 m, and dwelt on fleshy seaweeds, mussels, or directly on the rock.
Fig. 1.
Location of the sampled populations: PLL port Lligat, EST Estartit, BLN Blanes, CPA Cabo de Palos, CGA Cabo de Gata, LHE La Herradura, SGR Sagres, FLR Azores-Flores, MAD Madeira, BRZ Brazil
Most population samples consisted of ca. 30 individuals, although one population (La Herradura), which was sampled exhaustively, only provided 14 individuals (Table 1). A fragment of 0.5 cm3 was removed from the apical part of every individual to ensure not to take embryo-harboring tissue since the zone close to the oscula is oocyte free (Guardiola et al. 2012). A maximum of two alleles were obtained from each sample, confirming that we succeeded at obtaining parental tissue only. The samples were preserved in absolute ethanol and stored at −20 °C until DNA extraction.
Table 1.
P. magna: population genetic descriptors
| Population | N | Mean Na | Mean H E | Mean H O | F IS | HWE (p value) | Mean Na/pop |
|---|---|---|---|---|---|---|---|
| BLN | 30 | 6.8 | 0.69 | 0.57 | 0.25 | 0.00 | 61 |
| EST | 27 | 6.8 | 0.55 | 0.38 | 0.49 | 0.00 | 61 |
| PLL | 27 | 8.3 | 0.68 | 0.53 | 0.29 | 0.00 | 75 |
| CGA | 30 | 10.0 | 0.75 | 0.47 | 0.429 | 0.00 | 90 |
| CPA | 30 | 9.1 | 0.71 | 0.51 | 0.34 | 0.00 | 82 |
| LHE | 14 | 5.0 | 0.61 | 0.40 | 0.43 | 0.00 | 45 |
| FLR | 30 | 9.1 | 0.65 | 0.46 | 0.43 | 0.00 | 82 |
| SGR | 30 | 10.1 | 0.73 | 0.52 | 0.37 | 0.00 | 91 |
| MAD | 30 | 6.4 | 0.55 | 0.41 | 0.44 | 0.00 | 58 |
| BRZ | 18 | 5.4 | 0.59 | 0.58 | 0.19 | 0.01 | 49 |
N number of individuals, Na number of alleles, H E expected heterozygosity, H O observed heterozygosity, F IS inbreeding coefficient (estimated as a function of the heterozygote deficiency), HWE probability of departure from Hardy–Weinberg equilibrium
DNA extraction, amplification and microsatellite genotyping
DNA was extracted using PureLink Extraction Kit (Real Pure) following the manufacturer’s indications and the protocol described in Pascual et al. (1997). The nine polymorphic microsatellites were amplified using the following primers (cal_a-, cal_b-, cal_c-, cal_d-, cal_e-, cal_f-, cal_g-, cal_h- and cal_j forward and reverse) previously designed for the species (Agell et al. 2012). Forward primers were labeled with a fluorescent dye from Applied Biosystems: PET red (cal_a, cal_h), NED yellow (cal_b, cal_g), VIC green (cal_c, cal_e, cal_j) and FAM blue (cal_d, cal_f). The length of PCR products was estimated relative to the internal size standard GeneScan 500LIZ (Applied Biosystems) and determined using GeneMapper® and PeakScanner® software from Applied Biosystems. Three independent readers checked the results visually to avoid scoring errors.
Data analysis
The presence of null alleles was checked with Micro-Checker v.2.2.3 (Van Oosterhout et al. 2004). Departure from Hardy–Weinberg equilibrium, heterozygote deficit, linkage disequilibrium, allele frequencies, number of alleles per loci, and number and frequency of private alleles were calculated with GENEPOP web version 4.0 (Raymond and Rousset 1995; Rousset 2008). The p values of all analyses involving multiple comparisons were corrected using the False Discovery Rate (FDR) correction, which is less stringent in controlling the expected proportion of incorrectly rejected null hypothesis than Bonferroni correction and thus increases the analysis statistical power (Benjamini and Yekutieli 2001).
The inbreeding coefficient (FIS, Weir and Cockerman 1984) for each locus individually as well as for all loci combined was calculated with ARLEQUIN v.3.11 (1000 permutations, Excoffier et al. 2005). The probability of recent effective population size reductions from allele data frequencies was tested under three mutation models of evolution: infinite allele mutation (IAM), two-phase mutation (TPM), and stepwise mutation model (SMM), using BOTTLENECK software. The method is based on the assumption that in non-bottlenecked populations (close to mutation drift equilibrium), the value of expected heterozygosity at Hardy–Weinberg equilibrium (He) is equal to the expected heterozygosity at mutation drift equilibrium (Heq). The excess of He over Heq is the evidence of severe reduction in population effective size that is compatible with a bottleneck or founder event (Cornuet and Luikart 1996).
The genetic differentiation among populations was determined by means of the statistics Fst (Weir and Cockerman 1984) with ARLEQUIN v.3.11 (1000 permutations, Excoffier et al. 2005) and Dest (Jost 2008), calculated with RStudio (2012) using the package DEMEtics v.0.8-3 (Gerlach et al. 2010). The latter software estimates the p values (FDR corrected) and the confidence intervals according to Manly (1997) with a bootstrap method that distributes the alleles for a specific locus randomly when the populations are in Hardy–Weinberg equilibrium for this locus. If the populations are not in Hardy–Weinberg equilibrium, the genotypes instead of the alleles are distributed randomly among populations (Goudet et al. 1996). The p values indicate how different the allele distribution in the studied sample is from a randomly obtained one. A hierarchical analysis of molecular variance (AMOVA) was performed with ARLEQUIN (Excoffier et al. 2005). The sources of variation considered in the AMOVA were between regions (Atlantic and Mediterranean basins), among groups (Iberian Mediterranean localities plus Sagres, NE Atlantic localities plus La Herradura, and Brazil), among populations, among individuals within populations, and within individuals. The AMOVA was run twice, either clustering populations into basins according to their geographical location or according to the nature of the water masses bathing the populations (Bryden et al. 1989) (i.e., the Atlantic Sagres was included in the South Mediterranean group and the Mediterranean La Herradura formed part of the North Atlantic group).
The number of genetically homogeneous groups was inferred using a Bayesian algorithm with STRUCTURE software v.2.3.3. (Pritchard et al. 2000; Falush et al. 2003, 2007; Hubisz et al. 2009) with the parameters: range of K = 1 to 4, MCMC repetitions = 50,000, burn-in range = 100,000, runs = 20) and the ad hoc statistic ΔK, based on the rate of change in the log probability of data between successive K values (Evanno et al. 2005). The results from STRUCTURE were merged across the 20 runs for the best K with CLUMPP v.1.1.2b (Jakobsson and Rosenberg 2007), while DISTRUCT v.1.1 (Rosenberg 2004) was used to graphically represent the results.
The genetic distance Ds (Nei’s standard distance 1972) between the populations was calculated with GENDIST, a program of the package PHYLIP v. 3.69 (Felsestein 1993). The matrix generated was then used in the program NEIGHBOR from PHYLIP to obtain the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) tree.
Isolation by distance was tested with a Mantel test using the correlation between pairwise genetic differentiation (Dest values) and the logarithm of the geographical distances between the populations (XLSTAT v. 2012 5.02 Microsoft Excel add-in, Addinsoft’s Core Software).
Population dynamics
A population of P. magna was monitored from July 2007 to July 2008. The population was established on a semi-sciophilous seaweed assemblage, dominated by Halopteris scoparia, from 8 to 12 m of depth, at the Blanes litoral (NW Mediterranean, 41°40′27″N, 2°47′25″E). Sampling periodicity was monthly, except on two occasions when the sea conditions prevented diving (i.e., no sampling was conducted in February 2008, and the sampling periodicity was 6 weeks instead of 4 from January to March). For the population dynamics’ study, randomly selected, 20 × 20 cm surfaces (three replicates) were scraped to reveal the rocky substrate monthly from July 2007 to April 2008. The samples were taken to the laboratory and examined under a stereomicroscope. Calcareous sponges from 1.5 mm in size on, purportedly belonging to P. magna, were isolated and their identity confirmed by amplifying three to five species-specific microsatellites (Guardiola et al. 2012). The height of those individuals unambiguously ascribed to P. magna was measured. In May–June, when the 20 × 20 cm2 quadrats proved inappropriate to represent adequately the size classes present in the population because of insufficient sponge density, the monitoring was conducted in situ by randomly placing three 1 m2 quadrats on the vertical rocky wall where the population was established. Individuals of P. magna within the quadrats were counted, and their maximum height measured. Then, counts were scaled to the 20 × 20 cm2 area used in the previous months to make data comparable.
From July to September (2007), no individual in the scrapped quadrats could be ascribed to P. magna recruits. These months without recruits are not included in the graphic representation of the population dynamics, which started in October 2007 (Fig. 6).
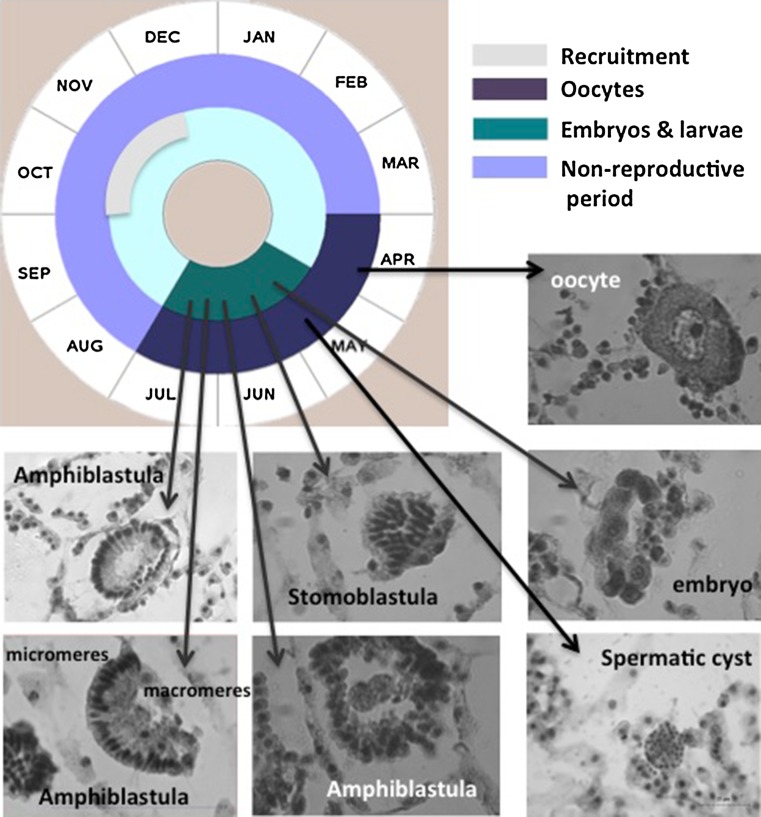
Fig. 6.
Life cycle of Paraleucilla magna at the Blanes (Northwestern Mediterranean) population. Oocytes were present during 4 months. Spermatogenesis was extremely short since our monthly sampling only allowed us to detect one spermatic cyst. Embryos and larvae were recorded from May to July. Larvae were abundant m specimens higher than 3 cm in height
Measuring individual height across the whole sponge cycle as an estimator of growth was a compromise solution. Height was a good proxy of increase in size for recruits and juveniles, which mainly grew up with a cylindrical shape. Although this measure was less appropriated for measuring larger irregular individuals, it also captured some growth, as irregular individuals also increased in high. Moreover, the study focused on uncovering the sponge growth during the first months after recruitment and on assessing how long the individuals took to reach their maximum size in the population, and these two goals were acceptably achieved with the measure used.
Life cycle
The sponge life cycle was analyzed from December 2007 (when some individuals reached 16 mm in height) to July 2008. Five, ≥16 mm high, randomly selected specimens were collected monthly for detecting the presence of reproductive elements by means of histological sections and ultrastructure observations. From April 2008 (when oocytes were first detected in the sponge tissues), sampling was conducted every 15 days to capture more accurately gamete formation, embryo hatching, and larval release.
Sponge samples for light microscopy were preserved in 10 % formaldehyde in seawater for 5 h and then changed to 4 % formaldehyde in seawater. Once fixed, the samples were decalcified in 5 % EDTA for 2 h and subsequently rinsed in distilled water, dehydrated in an ethanol series (increased concentrations from 70 to 100 %, 30 min each), cleared in toluene/ethanol 1:1 (30 min) and then in pure toluene (15 min) and embedded in paraffin before being cut in 5-µm thick sections using a Autocut 2040 Reichert-Jung microtome. After deparaffining with xylene, the sections were stained with hematoxylin and Schiff reagent. Photographs were taken with a digital camera mounted on a Zeiss Axioplan II microscope.
Samples were also processed for transmission electronic microscopy (TEM), to confirm the presence and the nature of the reproductive elements detected through light microscope. Samples of ca. 2 mm3 in size were fixed in 1 % OsO4 and 2 % glutaraldehyde in 0.45 M sodium acetate buffer (pH 6.4) with 10 % sucrose (1:3) (Leys and Reiswig 1998) for 12 h at 4 °C. Samples were repeatedly rinsed in the same buffer, dehydrated in an ethanol series and embedded in Spurr’s resin. Ultrathin sections were stained with uranyl acetate and lead citrate, and observed with a TEM (JEOL 1010), implemented with a Bioscan system (Gatan) for image digitalization (Microscopy Unit of the Scientific and Technical Services of the Universitat de Barcelona).
Results
Demographic estimators
A total of 266 individuals of P. magna belonging to ten populations were genotyped for nine microsatellite primers previously designed (Agell et al. 2012). All loci were polymorphic, with a total number of alleles per locus ranging from 4 to 45: 18 (cal_a), 13 (cal_b), 4 (cal_c), 21 (cal_d), 8 (cal_e), 15 (cal_f), 28 (cal_g), 45 (cal _h), and 13 (cal_i). No loci showed linkage disequilibrium after false discovery rate (FDR) correction. Populations showed between 1 (EST) to 13 (PLL) private alleles (Table S1).
The inbreeding coefficient values were positive and significant (p < 0.01) for all populations (Table 1). The exact tests for Hardy–Weinberg equilibrium confirmed these results showing significant deviations for each population (Table 1). Analysis with Micro-Checker indicated the presence of null alleles for all loci in all populations (Table 2a), which might be the cause of the deviation from Hardy–Weinberg equilibrium detected. The percentage of failed amplifications is reported in Table 2b. No evidence of scoring errors due to stuttering or large allele drop out resulted from the Micro-Checker analysis. No identical multilocus genotypes were found in any of the populations analyzed. Hence, asexual reproduction is rare or inexistent in the introduced populations.
Table 2.
P. magna: frequencies of null alleles (a) and percentage of failed amplifications (b) across loci and populations
| Population | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BLN | EST | PLL | CGA | CPA | LHE | FLR | SGR | MAD | BRZ | |
| a | ||||||||||
| cal_a | – | 0.168 | – | – | 0.133 | – | – | – | – | – |
| cal_b | – | 0.241 | 0.167 | 0.197 | – | – | 0.100 | 0.356 | 0.249 | – |
| cal_c | – | – | – | – | – | – | – | – | – | – |
| cal_d | 0.369 | 0.407 | 0.412 | 0.342 | 0.404 | 0.396 | 0.293 | 0.242 | 0.261 | – |
| cal_e | – | – | – | – | – | – | – | – | – | – |
| cal_f | 0.195 | – | 0.232 | 0.355 | 0.346 | 0.409 | 0.349 | 0.134 | – | – |
| cal_g | 0.204 | 0.125 | 0.116 | 0.194 | – | – | 0.170 | 0.200 | – | – |
| cal_h | – | 0.233 | 0.182 | 0.297 | 0.205 | – | 0.289 | 0.365 | 0.200 | 0.206 |
| cal_j | – | 0.205 | – | 0.103 | – | 0.224 | 0.100 | 0.128 | 0.140 | – |
| b | ||||||||||
| cal_a | 6.67 | 25.92 | 2.00 | – | 3.33 | 7.14 | 6.67 | 10.00 | 13.33 | 44.44 |
| cal_b | 6.67 | 29.63 | – | 3.33 | 3.33 | 7.14 | 26.67 | 3.33 | 6.67 | – |
| cal_c | 6.67 | 3.70 | – | 3.33 | 3.33 | – | 6.67 | – | 16.67 | – |
| cal_d | 23.33 | 29.63 | 29.63 | 23.33 | 6.67 | 28.57 | 23.33 | 23.33 | 13.33 | 16.67 |
| cal_e | – | 3.70 | – | – | – | 7.14 | 10.00 | – | 13.33 | – |
| cal_f | – | 22.22 | – | 6.67 | 23.33 | 7.14 | 13.33 | 16.67 | 20.00 | 11.11 |
| cal_g | 3.33 | 14.81 | 3.70 | – | – | 7.14 | 10.00 | – | 20.00 | 5.56 |
| cal_h | 3.33 | 25.92 | 7.40 | 10.00 | 10.00 | 7.14 | 43.33 | 26.67 | 20.00 | 22.22 |
| cal_j | – | 14.81 | – | 10.00 | – | 7.14 | 3.33 | 3.33 | 13.33 | 11.11 |
Bottleneck analysis showed that from three to seven populations, depending on the mutation model considered, differed significantly from mutation drift equilibrium (Table 3) and thus underwent a recent founder effect. However, only Blanes, Cabo de Gata and Sagres showed bottleneck under the TPM model, while Estartit, Flores, and Brazil did it under the SMM model. The populations from Port Lligat and Madeira did not show bottleneck independently of the mutation model considered (Table 3).
Table 3.
Bottleneck analysis of populations of P. magna under three mutation models
| Population | Mutation model | ||
|---|---|---|---|
| IAM | TPM | SMM | |
| Blanes | 0.004 | 0.037 | 0.652 |
| Estartit | 0.049 | 0.910 | 0.014 |
| Port Lligat | 0.652 | 0.910 | 0.203 |
| Cabo de Gata | 0.002 | 0.037 | 0.359 |
| Cabo de Palos | 0.010 | 0.426 | 0.426 |
| La Herradura | 0.020 | 0.250 | 0.820 |
| Flores | 0.129 | 0.426 | 0.014 |
| Sagres | 0.002 | 0.020 | 0.426 |
| Madeira | 0.203 | 1.000 | 0.164 |
| Brazil | 0.037 | 0.910 | 0.049 |
IAM infinite allele mutation, TPM two-phase mutation, SMM stepwise mutation model, p values in bold indicate significant deviation from mutation drift equilibrium and thus bottleneck
Genetic differentiation and structure
The genetic differentiation values (Fst and Dest) were significant for all pairwise comparisons (Dest) or for all the comparisons but three (Fst), after the FDR correction (Table 4). This indicates that the genetic diversity was spatially structured in most of the introduced populations studied. The values of Dest ranged from moderate to high in most cases being the highest between Brazil and the remaining populations but Port Lligat, and between Madeira and the Mediterranean populations. The Fst values were low in general being the highest between Madeira and most of the Mediterranean populations.
Table 4.
P. magna: pairwise genetic differentiation between populations
| BLN | EST | PLL | CGA | CPA | LHE | FLR | SGR | MAD | BRZ | |
|---|---|---|---|---|---|---|---|---|---|---|
| BLN | 0.291 | 0.384 | 0.266 | 0.225 | 0.311 | 0.343 | 0.263 | 0.344 | 0.304 | |
| EST | 0.078 | 0.284 | 0.213 | 0.216 | 0.216 | 0.279 | 0.294 | 0.377 | 0.357 | |
| PLL | 0.094 | 0.079 | 0.315 | 0.345 | 0.413 | 0.375 | 0.374 | 0.486 | 0.224 | |
| CGA | 0.050 | 0.047 | 0.061 | 0.177 | 0.266 | 0.143 | 0.255 | 0.397 | 0.306 | |
| CPA | 0.057 | 0.064 | 0.091 | 0.038 | 0.324 | 0.203 | 0.262 | 0.417 | 0.333 | |
| LHE | 0.077 | 0.061 | 0.105 | 0.048 | 0.085 | 0.359 | 0.276 | 0.283 | 0.448 | |
| FLR | 0.075 | 0.064 | 0.092 | 0.045 | 0.064 | 0.085 | 0.202 | 0.296 | 0.368 | |
| SGR | 0.041 | 0.026 | 0.064 | 0.018 | 0.022 | 0.060 | 0.033 | 0.163 | 0.376 | |
| MAD | 0.102 | 0.103 | 0.146 | 0.093 | 0.108 | 0.106 | 0.046 | 0.073 | 0.410 | |
| BRZ | 0.088 | 0.101 | 0.073 | 0.078 | 0.091 | 0.132 | 0.097 | 0.078 | 0.132 |
D est values above the diagonal and F ST values below the diagonal. All comparisons were significant after FDR correction (p < 0.001), except the ones in bold (p > 0.05)
The hierarchical AMOVA results (Table 5) revealed that most of the genetic diversity was due to variation within individuals (55.34 %) and among individuals within populations (34.52 %). However, differentiation among populations, which explained 6.51 % of the variance, was also significant (p < 0.001). No significant differences were found between the Mediterranean and Atlantic basins or among population clusters formed according to their geographical proximity (data not shown). However, between basins and among groups differences turned out to be significant and explained 1.18 and 2.5 % of the variance, respectively (p < 0.05 in both cases), when the groups were established according to the nature of the water masses bathing the coasts of the sampling sites (i.e., placing Sagres within the Southwestern Mediterranean group, and La Herradura in the North Atlantic group) (Table 5).
Table 5.
Paraleucilla magna: Summary of AMOVA results
| Source of variation | df | Sum of squares | Variance components | Percentage of variation | p value |
|---|---|---|---|---|---|
| Between basins | 1 | 26.86 | 0.044 | 1.18 | p < 0.05 |
| Among groups | 2 | 51.04 | 0.092 | 2.45 | p < 0.01 |
| Among populations | 9 | 157.46 | 0.243 | 6.51 | p < 0.001 |
| Among individuals within populations | 254 | 1210.17 | 1.285 | 34.52 | p < 0.001 |
| Within individuals | 264 | 579.50 | 2.105 | 55.34 | p < 0.001 |
Basins: (1) Cabo de Palos–Cabo de Gata–Blanes–Estartit–Port Lligat–Sagres and (2) Flores–Madeira–Brazil–La Herradura. Groups: (1) Blanes–Estartit–Port Lligat–Cabo de Gata–Cabo de Palos–Sagres. (2) La Herradura–Flores–Madeira. (3) Brazil
na not addressed issue
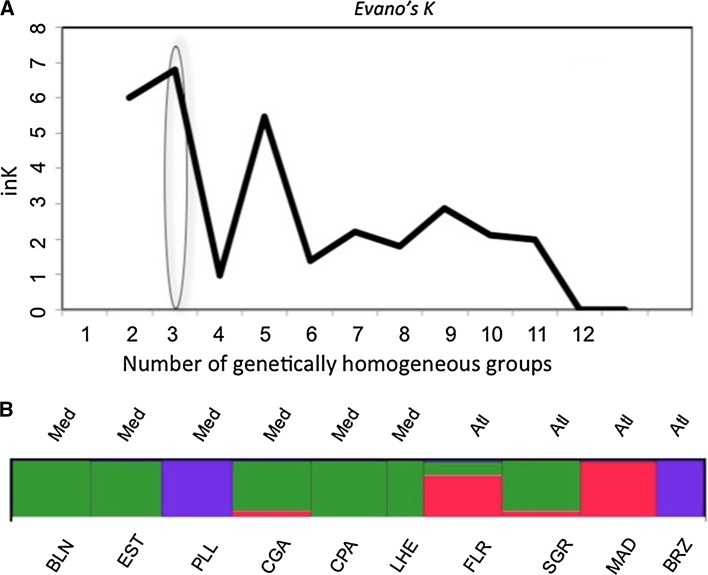
The Bayesian clustering method (STRUCTURE) indicated the highest likelihood for the model with three genetically homogeneous groups of individuals (k) according to the highest increment of k between the models with two and three groups (Fig. 2a). The graphical representation of the Structure’s coefficient memberships for the populations indicated that these three groups corresponded to the populations from the Mediterranean basin, the North Atlantic, and the South Atlantic (Brazil) plus Port Lligat (Mediterranean) regions and showed some allele sharing between the Atlantic Flores and the Mediterranean populations (Fig. 2b). The Atlantic population of Sagres (continental Portugal) shared more alleles with the Mediterranean populations than with the other Atlantic populations (Fig. 2b).
Fig. 2.
a Number of genetically homogeneous groups inferred using a Bayesian algorithm with STRUCTURE and the ad hoc statistic ΔK, based on the rate of change in the log probability of data between successive K values. b Graphic representation of the results from STRUCTURE merged across the 20 runs for the best K with CLUMPP
Genetic distances
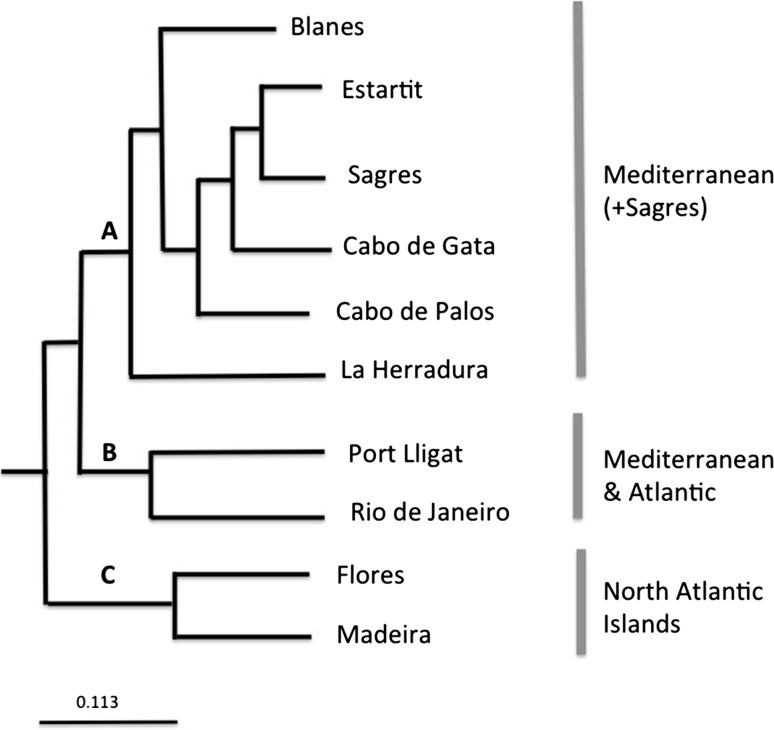
The UPGMA clustering method based on genetic mean distances between populations, recovered three main clades corresponding to: (A) Mediterranean populations plus that of Sagres (South Portugal) (B) the Brazil population plus that of Port Lligat, and (C) North Atlantic insular populations (Flores and Madeira) (Fig. 3). The Flores and Madeira populations appear as the sister clade of the remaining populations.
Fig. 3.
Dendrogram of the sampled populations using the UPGMA algorithm on a pairwise matrix of genetic distances (D)
The Mantel test indicated a low but significant correlation between population genetic differentiation and geographical distance (Fig. S1). When the Port Lligat population was removed from the analysis, the correlation value increased as well as the level of significance (Mantel test, r = 0.44, p < 0.01).
Population dynamics and reproductive cycle
The population monitored dwelt on a seaweed assemblage dominated by Halopteris scoparia, placed on a vertical rocky wall exposed to water flow, at 5–10 m depth. Individuals settled directly on the rock or on the seaweeds, and the latter were then subjected to continuous movement. Individuals <2 cm in height mainly formed single tubes (Fig. 4a–c), while larger individuals tended to be roughly massive with short osculifer processes (Fig. 4d–h).
Fig. 4.
P. magna growth habits in Northwestern Mediterranean assemblages. a–c 1 cm individuals. d–h Adult individuals. Bar scales = 0.5 cm
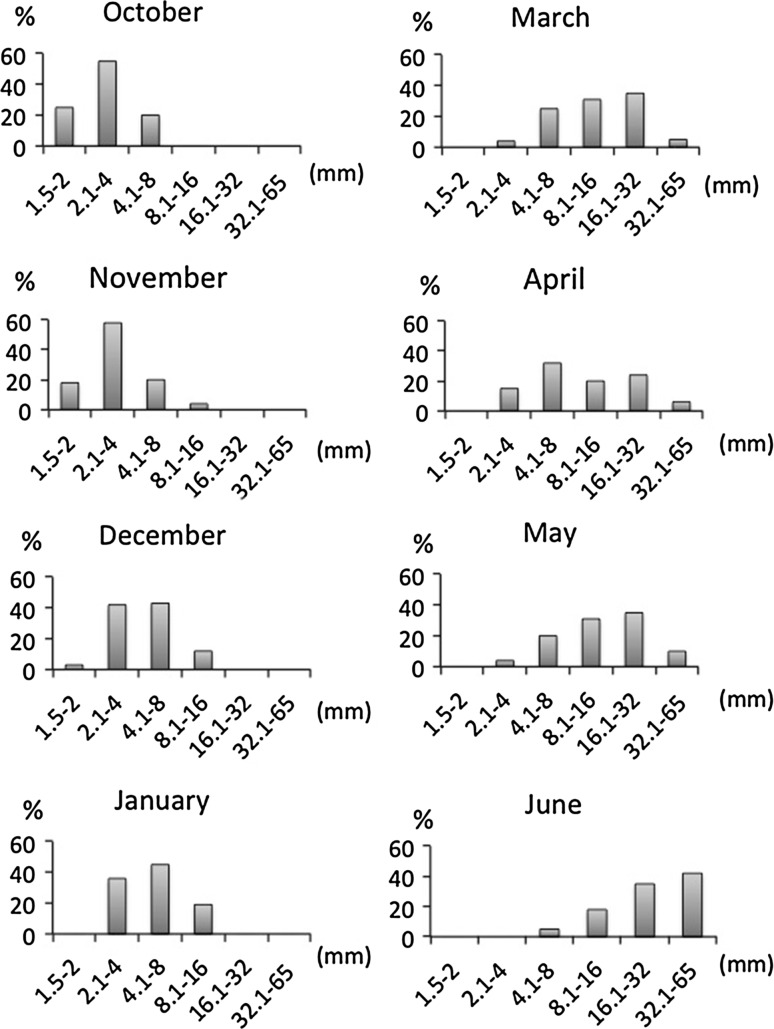
Early recruits, 1.5 mm ± 0.8 mm in height (mean ± SE), were first recorded in October (2007). Recruitment pursued for 3 months as recruits of the 1.5–2 mm size class were observed until December (2007) (Fig. 5). The individuals grew to higher size classes with time and some representatives of the largest size class (32.1–65 mm in height) were already recorded in March 2008. Individuals reached the maximum size 8 months after the first recruits were observed. From March to June, the frequencies of the individuals belonging to the three largest size-classed increased progressively (Fig. 4) but growth stopped in April 2008 for members of the largest size class, concomitantly with the beginning of the reproductive cycle (Fig. 5).
Fig. 5.
Population dynamics of Paraleucilla magna on a native assemblage of the Northwestern Mediterranean (Blanes, NE Spain) bars represent % of individuals belonging to each of the six arbitrarily selected size classes. Only one cohort was detected per year despite asynchronous recruitment. The maximum individual size (32.1–65 mm) was reached 8 months after the adults disappeared
Oocytes (15–20 µm in size) were recorded in the sponge tissue during ca. 4.5 months (from April to July 2008), and one 15 µm in diameter spermatic cyst was observed in April (Fig. 6). Oocyte formation was synchronous within individuals but asynchronous between individuals. From May to July, embryos and larvae at different developmental stages were present in the population sharing the sponge tissue with oocytes (Fig. 6). Ready-to-release larvae were observed within the sponge tissue in June–July (Fig. 6). Sponge size was highly variable within the population, which indicates asynchronous larval release. Nonetheless, only one three-month long recruitment event was recorded (Fig. 5), suggesting one reproductive cycle per year. In July 2008, most larvae had already been released and adult individuals started to collapse.
Larval characteristics were the typical of Calcarea. Cleavage occurred inside a follicle “placental membrane” and gave rise to a stomoblastula (Figs. 6, 7). Micromeres produced nucleolate ciliated cells with the cilia inwards (Fig. 7a, b) and continued to cleave forming a single layered epithelium (Fig. 7d, e) before the inversion of layers formed a hollow amphiblastula with the cilia outwards (Fig. 8). Larvae remained within the “placental membrane” until release (Figs. 6, 8a, b). A couple of granulose cells, purportedly of maternal origin, were observed in the larval central zone together with numerous bacteria (Figs. 6, 8b). Four symmetrically placed cells (cells in croix) were present between the larval ciliated cells (Fig. 8b). The cells in croix were packed with multi-vesicular dark bodies (Fig. 8c). These and the ciliated cells enclosed yolk reserves and abundant collagen-containing vesicles. Some of them open to the extracellular space between adjacent cells, purportedly corresponded to spicule formation centers (Fig. 8a, d).
Fig. 7.
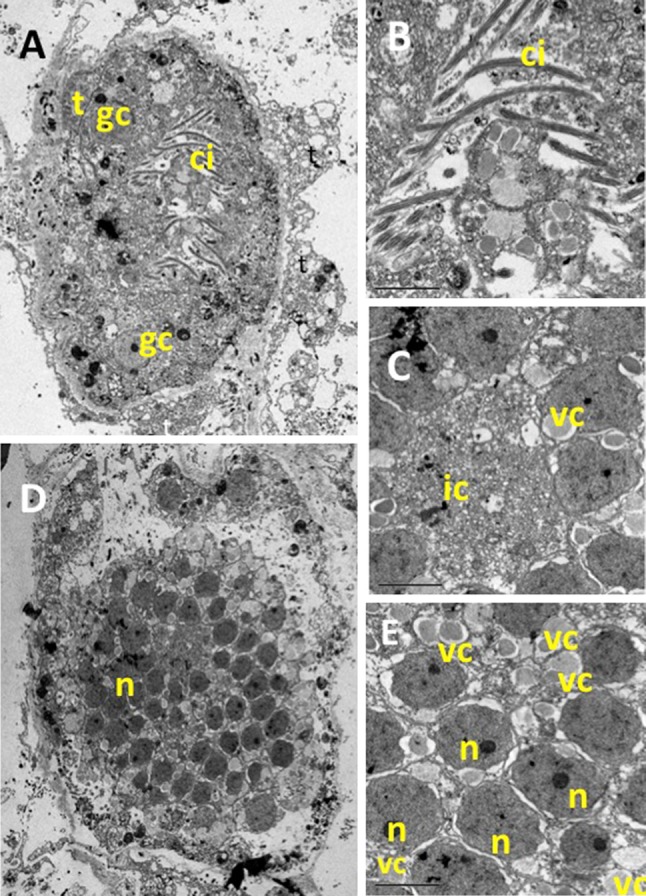
TEM pictures of a P. magna early larvae (stomoblastula). a Section surrounded by maternal trophocytes (t) showing macromeres and nucleolate micromeres and the two granular nucleolate cells (gc) that will move to the internal space of the amphiblastula. b Detail of the inner zone showing the cilia of the epithelial layer already differentiated but still inwards. c Detail of the anterior zone of the stomoblastula with numerous vacuoles containing collagen filaments (vc) and multivesicular inner cell (ic). d, e Larval section at the nuclei level (n)
Fig. 8.
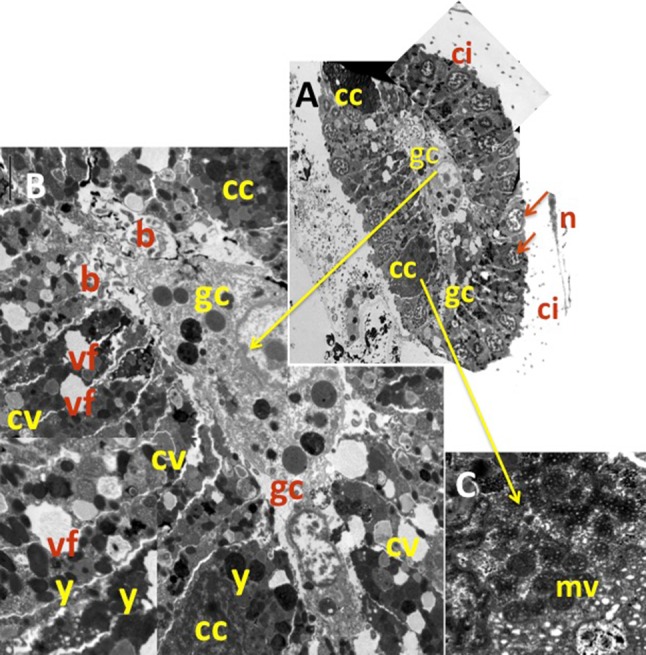
a Cross section of the anterior zone of amphiblastula with the ciliated cells showing their nucleus (n) at a distal position, and the cilia (ci) outwards. Two of the four cells en croix (cc) are also visible. b Enlarged TEM picture of the amphiblastula section with details of the two granulate central cells (gc) inner abundant bacteria (b). The cells en croix, (cc), abundant yolk reserves (y) and numerous vacuoles containing collagen (cv) within the ciliated cells, with instances of vacuole fusion (vf) between adjacent cells. c Detail of a cell en croix showing abundant multivesicular dark bodies (mv)
Some relevant differences in the species biological cycle were evident between the monitored population and those from other localities previously studied (Table 6). Populations forming part of fouling assemblages subjected to eutrophic conditions reproduced continuously along the year and remain in the same place for years (Table 6), while those established on native assemblages under non-eutrophic conditions were clearly seasonal and disappeared after reproduction (Table 6; Fig. 6).
Table 6.
Comparisons of phenotypic characteristics of Paraleucilla magna in several localities with contrasting environmental conditions
| Author | Lanna et al. (2007) | Longo et al. (2012) | Lanna et al. (2014) | Padua et al. (2012) | Current study |
|---|---|---|---|---|---|
| Site | Brazil: Punta Vermelha | Italy: Mar Piccolo Taranto | Brazil: Punta Vermelha | Brazil: Rio de Janeiro | Spain: Blanes |
| Sea | SW Atlantic | Ionian sea (Central Mediterranean | SW Atlantic | SW Atlantic | NW Mediterranean |
| Assemblage | Rock/calcareous algae | Fouling assemblage | Fouling assemblage | Fouling assemblage | Native seaweeds assemblages |
| Water conditions | Locally variable. Close to an eutrophic bay | Eutrophic/sheltered | Locally variable: close to an eutrophic Bay | Locally variable: close to an eutrophic Bay | Clean/exposed |
| Substrate | Rock | Mussel ropes/artificial substrates | n.a. | Concrete blocks | Halopteris scoparia/Corallina sp |
| Shape/max. size | Massive/n.a. | Massive-tubular/10 cma | Massive-tubular/n.a. | n.a. | Massive/7 cm |
| Reproduction/recruitment period | Seasonal: once a year | All year round (picks in warm months) | All year round | All year round | Seasonal: 4 months/6 months |
| Cycle duration (months) | n.a. | 6 months | n.a. | n.a. | 6 months |
| Population resilience | n.a. | >10 years | n.a. | n.a. | 10 months |
Only the literature sources containing information on the species cycle or population resilience have been considered
n.a. not addressed issue
aApproximate height derived from the size in volume supplied by the authors
Discussion
Genetic features
Although several sponge species had been studied using microsatellites at geographical scales (Duran et al. 2004b; Blanquer and Uriz 2010; Dailianis et al. 2011; Chaves-Fonnegra et al. 2015), none of them belonged in the Calcarea class or was allochthonous in its known distribution range. Calcareous sponges have particularly short-living, short dispersal larvae (Lanna and Klautau 2012) and thus expectations would point to genetic differentiation among geographically distant populations where only natural dispersal was acting. However, man-mediated dispersal modifies natural gene flow and the introduced populations of P. magna might be connected.
The microsatellite markers designed for P. magna (Agell et al. 2012) proved informative to establish the genetic structure and relationships of its introduced populations, which were located from tens to thousands of kilometers apart. These markers were more polymorphic (mean Na per locus = 18.5) in the populations analyzed than microsatellites previously studied for native demosponges (mean Na = 3.9, Na = 5.9, Na = 15.1, for populations of Scopalina lophyropoda, Crambe crambe, and Spongia officinalis, respectively), covering similar geographical ranges (Duran et al. 2004b; Blanquer and Uriz 2010; Dailianis et al. 2011). Heterozygote deficit and, thus, inbreeding was shown in all the populations studied. The presence of null alleles for all of the loci in all populations may have contributed to the deviation from Hardy–Weinberg equilibrium observed, but the homozygote excess may also be the result of mating among relatives and the formation of pedigree structures subsequently to recurrent introductions of related (non-randomly selected) individuals from the native sources.
The genetic diversity of these populations, with a mean number of alleles per population (all loci included) ranging from 45 to 90, was higher than generally expected for a recent introduction (Longo et al. 2004). Recently introduced species are classically reported to suffer from reduced within-population genetic variation due to founder effects and population bottleneck (Grosberg 1987; Carlon 1999). However, empirical studies have challenged the classical genetic scenario of invasive species by reporting higher than expected genetic variation in introduced populations, due to multiple introductions from native-range sources (Kolbe et al. 2007), which could be the case for P. magna and may explain the high genetic within-population diversity and the mutation drift equilibrium found in most populations under the TPM and SMM models.
Moreover, most populations were strongly structured as proved by significant Dest and Fst values, significant differences among populations (AMOVA), and the presence of private alleles, which may be caused by both the genetic drift and recurrent introductions from genetically differentiated populations within the native range. Nevertheless, the Mantel test showed significant isolation by distance, which became more significant when the Port Lligat population, genetically close to the distant population of Brazil, was removed. This isolation by distance pattern suggests that either the introductions followed a stepping stone model, which is not likely, or that the species distribution was not only man-mediated but also produced by some natural dispersal.
The AMOVA analysis showed significant genetic differences between the Mediterranean and Atlantic basins only when they were defined as a function of the water masses (either Atlantic or Mediterranean) bathing the populations: Sagres (South of Portugal) is under the influence of Mediterranean water masses entering the Atlantic trough the Gibraltar Strait, while La Herradura (Mediterranean) is bathed by large, tidally induced pulses of Atlantic water entering the Mediterranean (Bryden et al. 1989; Estrada 1996). This suggests some gene exchange between populations at both sides of the Strait of Gibraltar favored by the water circulation in the area.
The analyzed populations belonged to three genetically homogeneous groups according to the Bayesian algorithm of Structure and the UPGMA dendrogram, which corresponded to the North Atlantic (including La Herradura), Mediterranean (including Sagres), and Port Lligat Brazil populations. The Sagres population shared alleles with those from the Mediterranean, and the same occurred between La Herradura and the North Atlantic populations.
Phenotypic adaptation traits
Several phenotypic characteristics of the species vary across the colonized habitats. Shape ranges from long tubes in sheltered eutrophic habitats (Klautau et al. 2004; Longo et al. 2012; Cvitkovic et al. 2013) to massive clumps with a series of oscula on the top in exposed clean habitats (current study). Reproduction is almost continuous along the year in eutrophic areas of Brazil (Padua et al. 2012; Lanna et al. 2014) and South Italy (Pierri et al. 2010; Longo et al. 2012), while just one reproduction event per year was recorded in clean environments of Northwestern Mediterranean (current study).
Investment in reproduction also varied as a function of the habitat conditions. High temperatures seem to enhance the species reproductive effort in South Italy where reproduction extended along the whole year and the maximum gamete production occurred in August–September with water temperatures peaking above 25 °C (Longo et al. 2012). However, food supply has also been proposed to influence the species reproduction in Brazil, where water temperature undergoes minor changes between summer and winter and P. magna reproduced in summer (South Hemisphere) concomitantly with the runoff of nutrients into the seawater produced during the raining season (Lanna et al. 2007). The species reproduces in the Northwestern Mediterranean in winter–spring (this study), when temperature range between 14 °C (March) and 21 °C (June) (Blanquer et al. 2008) and primary production is high due to river runoff (Estrada 1996). Moreover, recruitment occurred in autumn (this study) alongside with the second peak of primary production in the area (Estrada 1996), after thermocline breakage and the arrival of bottom nutrients to the photic zone. On the other hand, TEM studies showed that, in both eutrophic (Lanna and Klautau 2012) and non-eutrophic (this study) habitats, larvae are well equipped with reserves and collagen (Lanna and Klautau 2012; Figs. 6, 7), which confer them strength and might facilitate fast spicule formation at settlement.
Population span also seems to vary according to food supply in the introduction habitats. The adult populations completely disappeared after larval release in August in native assemblages of Northwestern Mediterranean (this study) when most filter-feeders experience food depletion (Coma et al. 2000), while overlapping of generations gave rise to stable populations in eutrophic habitats such as the Mar Piccolo de Taranto—South Italy—(Longo et al. 2012), which is heavily exploited for mussel farming (Caroppo et al. 2012).
Likewise, the species growth rates also seem to vary according to the trophic conditions of the colonized habitat. Early, 1.5–2 mm high recruits employ six months to reach the maximum size (32.1–65 cm high size class) in well-preserved assemblages of Northwestern Mediterranean (this study), while 5 mm3 in volume (ca. 2 × 1.75 × 1.5 mm in size) specimens (approximated estimate) spend only three months to reach the highest size class of 100 ml (ca. 10.5 cm × 3.5 cm × 3 cm) in the eutrophic Mar Piccolo de Taranto (Longo et al. 2012).
General conclusions
The massive proliferation of P. magna around shellfish farms, in particular Mytilus cultures (Longo et al. 2007, 2012; current study) points to shellfish fouling as the most likely introduction pathway (Longo et al. 2007), although introduction by vessels cannot be totally discarded. Epibiont removal, previously to placing mussels into depuration tanks (Longo et al. 2012; authors’ obs.), may have caused the arrival of brooding individuals/fragments of P. magna to native assemblages, where larval release and settlement could have occurred. Moreover, given the species fragility, rafting of brooding fragments from established populations may also be produced by natural causes such as storms.
The species origin remains undetermined, as it has been identified as an alien in its currently known distribution range. The genus Paraleucilla was unknown out of the Indo-Pacific (http://www.marinespecies.org/porifera/) before P. magna was recorded in the Mediterranean (Longo et al. 2004) and Atlantic (Klautau et al. 2004), which points to its Indo-Pacific origin. High temperatures seem to favor the species reproductive effort (Longo et al. 2012), suggesting a warm ocean as its native source. Moreover, there are several Indo-Pacific countries with an intensive production of Mytilus and Crassostrea, which export around the globe (Chew 1990) and have been responsible for the distribution of many unwanted species (Haupt et al. 2009) including some of the few introduced sponges known up to now (Henkel and Janussen 2011).
Adaptation to disturbance, wide environmental tolerance, rapid growth, and high reproductive capacity are characteristics of foulers (Murray et al. 2012) shared by P. magna (Lanna et al. 2007; Longo et al. 2007, 2012; Zammit et al. 2009; current study). Thus, the species might be an opportunistic fouler also in its native range. A fouler nature and the scarce taxonomical studies of fouling sponges in the Indo-Pacific might explain why the species has not been recorded in its purported native habitat yet.
Although very small populations of P. magna in native communities were unstable across time because of the genetic drift, lack of natural gene flow, and stochastic recruitment failure (Guardiola et al. 2012), the genetic traits of the introduced populations indicate that the species is not in risk of extinction, and constrain on adaptation does not seem to hinder the spread of the species in the Atlanto-Mediterranean region.
The genetic makeup and the high levels of phenotypic plasticity of P. magna altogether allow predicting expansion of its introduced populations (Brown et al. 2011) by adopting the best-suited phenotypes to local conditions. Moreover, long-living populations such as those of eutrophic habitats (Longo et al. 2012) may operate as reservoir for dispersal and fast colonization of additional locations (Ridley and Ellstrand 2010).
To summarize, P. magna shows both, tolerance to contrasting local conditions and phenotypic plasticity, which are typical traits of invasive species (Matesanz et al. 2012). The species is expanding in the colonized regions, in particular under favorable trophic circumstances, where it may overgrow native seaweeds and filter-feeder invertebrates, and compete for trophic resources with shellfish cultures, lowering culture yields and hampering culture manipulation and maintenance. The future spread of P. magna in its introduced range is likely to be fueled by phenotypic adaptation to local conditions and high fitness across diverse habitats, rather than by local evolution of adapted genotypes.
Additional introductions can be expected in the near future reinforcing population stability and expansion since no adequate policies can be implemented until the introduction vector is unambiguously identified.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank M Klautau from the Universidade Federal do Rio de Janeiro, for collecting the Brazil population of P. magna. This study has been partially funded by MARSYMBIOMICS project ref. CTM2013-43287-P (Spanish Ministry of Economy and Competitiveness (MINECO)—FECYT agency, and Consolidate Group Award 2009-SGR655 (Generalitat of Catalonian) to MJU. MG has benefited from a FPU fellowship from the Spanish MINECO and JF from a Marie Curie (EU) fellowship, associated, respectively, with the BENTHOMICS and BIOCAPITAL projects to MJU.
Footnotes
Reviewed by M.-C. Pineda and an undisclosed expert.
This article is part of the Topical Collection on Invasive Species.
References
- Agell G, Frotscher J, Guardiola M, Pascual M, Uriz MJ, et al. Characterization of nine microsatellite loci for the calcarean sponge Paraleucilla magna Klautau et al. 2004 introduced to the Mediterranean Sea. Conserv Genet. 2012;4(2):403–405. doi: 10.1007/s12686-011-9560-y. [DOI] [Google Scholar]
- Avila E, Carballo JL. A preliminary assessment of the invasiveness of the Indo-Pacific sponge Chalinula nematifera on coral communities from the tropical Eastern Pacific. Biol Invasions. 2009;11(2):257–264. doi: 10.1007/s10530-008-9230-5. [DOI] [Google Scholar]
- Avise JC. Phylogeography: retrospect and prospect. J Biogeogr. 2009;36:3–15. doi: 10.1111/j.1365-2699.2008.02032.x. [DOI] [Google Scholar]
- Benjamini BY, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- Blackburn TM, Pyšek P, Bacher S, Carlton JT, Duncan RP, Jarošík V, Wilson JRU, Richardson DM. A proposed unified framework for biological invasions. Trends Ecol Evol. 2011;26(7):333–339. doi: 10.1016/j.tree.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Blakeslee AMH, McKenzie CH, Darling JA, Byers JE, Pringle JM, et al. A hitchhiker’s guide to the Maritimes: anthropogenic transport facilitates long-distance dispersal of an invasive marine crab to Newfoundland. Divers Distrib. 2010;16:879–891. doi: 10.1111/j.1472-4642.2010.00703.x. [DOI] [Google Scholar]
- Blanquer A, Uriz MJ. Population genetics at three spatial scales of a rare sponge living in fragmented habitats. BMC Evol Biol. 2010;10:13. doi: 10.1186/1471-2148-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquer A, Uriz MJ, Agell G. Hidden diversity in sympatric sponges: adjusting life-history dynamics to share substrate. Mar Ecol Progr Ser. 2008;371:109–115. doi: 10.3354/meps07679. [DOI] [Google Scholar]
- Brown GP, Kelehear C, Shine R. Effects of seasonal aridity on the ecology and behaviour of invasive cane toads (Rhinella marina) in the Australian wet-dry tropics. Funct Ecol. 2011 [Google Scholar]
- Bryden HL, Candela J, Kinder TH. Exchange through the Strait of Gibraltar. Progr Oceanog. 1989;33:201–248. doi: 10.1016/0079-6611(94)90028-0. [DOI] [Google Scholar]
- Bulleri F, Chapman MG. The introduction of coastal infrastructure as a driver of change in marine environments. J Appl Ecol. 2010;47:26–35. doi: 10.1111/j.1365-2664.2009.01751.x. [DOI] [Google Scholar]
- Calcinai B, Bavestrello G, Cerrano C. Dispersal and association of two alien species in the Indonesian coral reefs: the octocoral Carijoariisei and the demosponge Desmapsamma anchorata. J Mar Biol Assoc UK. 2004;84:937–941. doi: 10.1017/S0025315404010227h. [DOI] [Google Scholar]
- Carlon DB. The evolution of mating systems in tropical reef corals. Trends Ecol Evol. 1999;14:491–495. doi: 10.1016/S0169-5347(99)01709-7. [DOI] [PubMed] [Google Scholar]
- Carlton JT (2009) Deep invasion ecology and the assembly of communities in historical time. In: Rilov G, Crooks JA (eds) Biological invasions in marine ecosystems: ecological, management, and geographic perspectives: ecological studies, vol 204. Springer, Berlin, Heidelberg, pp 13–56
- Caroppo C, Giordano L, Palmieri N, Bellio G, Paride Bisci A, Portacci G, Sclafani P, Sawyer Hopkins T. Progress towards sustainable mussel aquaculture in Mar Piccolo, Italy. Ecol Soc. 2012;17(3):10. [Google Scholar]
- Chaves-Fonnegra A, Feldheinm KA, Secord J, Lopez JV. Population structure and dispersal of the coral-excavating sponge Cliona delitrix. Mol Ecol. 2015;24:1447–1466. doi: 10.1111/mec.13134. [DOI] [PubMed] [Google Scholar]
- Chew KK. Global bivalve shellfish introductions. J World Aquac. 1990;21:9–22. [Google Scholar]
- Colautti RI, MacIsaac HJ. A neutral terminology to define invasive species. Divers Distrib. 2004;10:135–141. doi: 10.1111/j.1366-9516.2004.00061.x. [DOI] [Google Scholar]
- Coles S, Bolick H. Invasive introduced sponge Mycale grandis overgrows reef corals in Kane’ohe Bay, O’ahu, Hawai’i. Coral Reefs. 2007;26(4):911–911. doi: 10.1007/s00338-007-0295-x. [DOI] [Google Scholar]
- Coma R, Ribes M, Gili JM, Zabala M. Seasonality in coastal benthic ecosystems. Trends Ecol Evol. 2000;15:448–453. doi: 10.1016/S0169-5347(00)01970-4. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts ADM, Dodgshun TJ. The nature and extent of organisms in vessel sea-chests: a protected mechanism for marine bioinvasions. Mar Poll Bull. 2007;54:875–886. doi: 10.1016/j.marpolbul.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Cvitković I, Despalatović M, Grubelić I, Nikolić V, Pleše B, Žuljecić A. Occurrence of Paraleucilla magna (Porifera: Calcarea) in the Eastern Adriatic Sea. Acta Adriat. 2013;54(1):93–99. [Google Scholar]
- Dafforn KA, Johnston EL, Glasby TM. Shallow moving structures promote marine invader dominance. Biofouling. 2009;25:277–287. doi: 10.1080/08927010802710618. [DOI] [PubMed] [Google Scholar]
- Dailianis T, Tsigenopoulos CS, Dounas C, Voultsiadou E. Genetic diversity of the imperiled bath sponge Spongia officinalis Linnaeus, 1759 across the Mediterranean Sea: patterns of population differentiation and implications for taxonomy and conservation. Mol Ecol. 2011;20:3757–3772. doi: 10.1111/j.1365-294X.2011.05222.x. [DOI] [PubMed] [Google Scholar]
- Duran S, Pascual M, Turon X. Low levels of genetic variation in mtDNA sequences over the western Mediterranean and Atlantic range of the sponge Crambe crambe (Poecilosclerida) Mar Biol. 2004;144:31–35. doi: 10.1007/s00227-003-1178-5. [DOI] [Google Scholar]
- Duran S, Pascual M, Estoup A, Turon X. Strong population structure in the marine sponge Crambe crambe (Poecilosclerida) as revealed by microsatellite markers. Mol Ecol. 2004;13:511–522. doi: 10.1046/j.1365-294X.2004.2080.x. [DOI] [PubMed] [Google Scholar]
- Estrada M. Primary production in Northwestern Mediterranean. Sci Mar. 1996;60:55–64. [Google Scholar]
- Evanno GS, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsestein J (1993) PHYLIP-phylogenetic inference package. Version 3.69 University of Washington, Seattle. http://evolution.genetics.washington.edu/phylip/getme.html
- Féral JP. How useful are the genetic markers in attempts to understand and manage marine biodiversity? J Exp Mar Biol Ecol. 2002;268:121–145. doi: 10.1016/S0022-0981(01)00382-3. [DOI] [Google Scholar]
- Galil BS. Loss or gain? Invasive aliens and biodiversity in the Mediterranean Sea. Mar Poll Bull. 2007;55:314–322. doi: 10.1016/j.marpolbul.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Geller JB, Darling JA, Carlton JT. Genetic perspectives on marine biological invasions. Annu Rev Mar Sci. 2010;2:367–393. doi: 10.1146/annurev.marine.010908.163745. [DOI] [PubMed] [Google Scholar]
- Gerlach G, Jueterbock A, Kraemer P, Deppermann J, Harmand P. Calculations of population differentiation based on GST and D: forget GST but not all of statistics! Mol Ecol. 2010;19:3845–3852. doi: 10.1111/j.1365-294X.2010.04784.x. [DOI] [PubMed] [Google Scholar]
- Glasby TM, Connell SD, Holloway MG, Hewitt CL. Nonindigenous biota on artificial structures: could habitat creation facilitate biological invasions? Mar Biol. 2007;151:887–895. doi: 10.1007/s00227-006-0552-5. [DOI] [Google Scholar]
- Goudet J, Raymond M, de Meeus T, Rousset F. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg RK. Limited dispersal and proximity-dependent mating success in the colonial ascidian Botryllus schlosseri. Evolution. 1987;41:372–384. doi: 10.2307/2409145. [DOI] [PubMed] [Google Scholar]
- Grosberg R, Cunningham CW. Genetic structure in the sea. From populations to communities. In: Bertness MD, Gaines SD, Hay ME, editors. Marine community ecology. Sunderland: Sinauer Associates Inc; 2001. pp. 61–84. [Google Scholar]
- Grosholz E. Ecological and evolutionary consequences of coastal invasions. Trends Ecol Evol. 2002;17:22–27. doi: 10.1016/S0169-5347(01)02358-8. [DOI] [Google Scholar]
- Guardiola M, Frotscher J, Uriz MJ. Genetic structure and differentiation at a short-time scale of the introduced calcareous sponge Paraleucilla magna to the western Mediterranean. Hydrobiologia. 2012;687:71–84. doi: 10.1007/s10750-011-0948-1. [DOI] [Google Scholar]
- Haupt TM, Griffiths CL, Robinson TB, Tonin AFG. Oysters as vectors of marine aliens, with notes on four introduced species associated with oyster farming in South Africa. Afr Zool. 2009;45(1):52–62. doi: 10.3377/004.045.0101. [DOI] [Google Scholar]
- Henkel D, Janussen D. Redescription and new records of Celtodoryx ciocalyptoides (Demospongiae: Poecilosclerida)—a sponge invader in the north east Atlantic Ocean of Asian origin? J Mar Biol Assoc UK. 2011;91(2):347–355. doi: 10.1017/S0025315410001487. [DOI] [Google Scholar]
- Holland BS. Genetics of marine bioinvasions. Hydrobiologia. 2000;420:63–71. doi: 10.1023/A:1003929519809. [DOI] [Google Scholar]
- Hubisz M, Falush D, Stephens M, Pritchard J. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Mol Ecol. 2008;17(18):40. doi: 10.1111/j.1365-294X.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Klautau M, Monteiro L, Borojevic R. First occurrence of the genus Paraleucilla (Calcarea, Porifera) in the Atlantic Ocean: P. magna sp. nov. Zootaxa. 2004;710:1–8. [Google Scholar]
- Kolbe JJ, Glor RE, Schetino LR, Lara AC, Larson A, Losos JB. Multiple sources, admixture, and genetic variation in introduced Anolis lizard populations. Conserv Biol. 2007;221:1612–1625. doi: 10.1111/j.1523-1739.2007.00826.x. [DOI] [PubMed] [Google Scholar]
- Lanna E, Klautau M. Embryogenesis and larval ultrastructure in Paraleucilla magna (Calcarea, Calcaronea) with remarks on the epilarval trophocyte epithelium (“placental” membrane) Zoomorphology. 2012;131:277–292. doi: 10.1007/s00435-012-0160-5. [DOI] [Google Scholar]
- Lanna E, Monteiro LC, Klautau M (2007) Life cycle of Paraleucilla magna Klautau, Monteiro and Borojevic, 2004 (Porifera, Calcarea). In: Custodio MR, Lôbo-Hajdu G, Hajdu E, Muricy G (eds) Porifera research: biodiversity, innovation, and sustainability. Museu Nacional, Rio de Janeiro, pp 413-441
- Lanna E, Paranhos R, Paiva PC, Klautau M. Environmental effects on the reproduction and fecundity of the introduced calcareous sponge Paraleucilla magna in Rio de Janeiro, Brazil. Mar Ecol. 2014 [Google Scholar]
- León-Pech MG, Cruz-Barraza JA, Carballo JL, Calderon-Aguilera LE, Rocha-Olivares A. Pervasive genetic structure at different geographic scales in the coral-excavating sponge Cliona vermifera (Hancock, 1867) in the Mexican Pacific. Coral Reefs. 2015;34:1316. doi: 10.1007/s00338-015-1316-9. [DOI] [Google Scholar]
- Leys SP, Reiswig HM. Transport pathways in the neotropical sponge Aplysina. Biol Bull. 1998;195:30–42. doi: 10.2307/1542773. [DOI] [PubMed] [Google Scholar]
- Longo C, Scalera-Liaci L, Manuel M, Corriero G. Note sui poriferi del Mar Grande e del Mar Piccolo di Taranto (Mar Ionio) Biol Mar Med. 2004;11:444–447. [Google Scholar]
- Longo C, Mastrototaro F, Corriero G. Occurrence of Paraleucilla magna (Porifera, Calcarea) in the Mediterranean Sea. J Mar Biol Ass UK. 2007;87(6):1749–1755. doi: 10.1017/S0025315407057748. [DOI] [Google Scholar]
- Longo C, Pontassuglia C, Corriero G, Gaino E. Life-Cycle Traits of Paraleucilla magna, a Calcareous Sponge Invasive in a Coastal Mediterranean Basin. PLoS ONE. 2012;7(8):e42392. doi: 10.1371/journal.pone.0042392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack RN, Simberloff D, Lonsdale WM, et al. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol Appl. 2000;10:689–710. doi: 10.1890/1051-0761(2000)010[0689:BICEGC]2.0.CO;2. [DOI] [Google Scholar]
- Manly BFJ. Randomization, bootstrap 368 and Monte Carlo methods in biology. Comput Stat. 1997;24(2):371–372. [Google Scholar]
- Matesanz S, Horgan-Kobelski T, Sultan SE. Phenotypic plasticity and population differentiation in an ongoing species invasion. PLoS ONE. 2012;7(9):e44955. doi: 10.1371/journal.pone.0044955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar JL, Gamboa RL, Revenga C, Spalding MD. Assessing the global threat of invasive species to marine biodiversity. Front Ecol Environ. 2008;6(9):485–492. doi: 10.1890/070064. [DOI] [Google Scholar]
- Mooney HA, Cleland EE. The evolutionary impact of invasive species. Proc Natl Acad Sci. 2001;98:5446–5451. doi: 10.1073/pnas.091093398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CC, Thomas W, Therriault TW, Martone PT. Adapted for invasion? Comparing attachment, drag and dislodgment of native and nonindigenous hull fouling species. Biol Invasions. 2012;14(8):1651–1663. doi: 10.1007/s10530-012-0178-0. [DOI] [Google Scholar]
- Naylor RL, Goldburg RJ, Primavera JH, Kautsky N, Beveridge MCM, ClayJ Folke C, Lubchenco J, Mooney H, Troell M. Effect of aquaculture on world fish supplies. Nature. 2000;405:1017–1024. doi: 10.1038/35016500. [DOI] [PubMed] [Google Scholar]
- Naylor RL, Willians SL, Strong DR. Aquaculture – A gateway for exotic species. Science. 2001;294:1655–1656. doi: 10.1126/science.1064875. [DOI] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. Am Nat. 1972;106:283–292. doi: 10.1086/282771. [DOI] [Google Scholar]
- Orensanz JM, Schwindt E, Pastorino G, Bortolus A, Casas Darrigran G, Elias R, López-Gappa JJ, Obenat S, Pascual M, Penchaszadeh P, Piriz ML, Scarabino F, Spivak ED, Vallarino EA. No longer the pristine confines of the world ocean: a survey of exotic marine species in the southwestern Atlantic. Biol Invasions. 2002;4:115–143. doi: 10.1023/A:1020596916153. [DOI] [Google Scholar]
- Padua A, Lanna E, Zilberberg C, de Paiva PC, Klautau M (2012) Recruitment, habitat selection and larval photoresponse of Paraleucilla magna (Porifera, Calcarea) in Rio de Janeiro, Brazil. Mar Ecol:1–6
- Pascual M, Balanyà J, Latorre A, Serra L. Analysis of the variability of Drosophila azteca and Drosophila athabasca populations revealed by random amplified polymorphic DNA. JZSER J Zool Syst Evolut Res. 1997;35:159–164. doi: 10.1111/j.1439-0469.1997.tb00419.x. [DOI] [Google Scholar]
- Pérez T, Perrin B, Carteron S, Vacelet J, Boury-Esnault N (2006) Celtodoryx girardae gen.nov. sp. nov., a new sponge species (Poecilosclerida: Demospongiae) invading the Gulf of Morbihan (North East Atlantic, France). Cah Biol Mar 47:205-214
- Pierri C, Longo C, Giangrande A. Variability of fouling communities in the Mar Piccolo of Taranto (Northern Ionian Sea - Mediterranean Sea) J Mar Biol Ass UK. 2010;90:159–167. doi: 10.1017/S0025315409990798. [DOI] [Google Scholar]
- Pimentel D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the United States. Bioscience. 2000;50:53–65. doi: 10.1641/0006-3568(2000)050[0053:EAECON]2.3.CO;2. [DOI] [Google Scholar]
- Pineda MC, López-Legentil S, Turon X. The Whereabouts of an Ancient Wanderer: global Phylogeography of the Solitary Ascidian Styela plicata. PLoS ONE. 2011;6(9):e25495. doi: 10.1371/journal.pone.0025495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta P, Karakassis L, Zivanovic M, Occhipinti-Ambrogi A, Marchini A, Cantone G, Castelli A, Chimenz C, et al. Alien species along the Italian coasts: an overview. Biol Invasions. 2011;13:215–237. doi: 10.1007/s10530-010-9803-y. [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Ridley CE, Ellstrand NC. Rapid evolution of morphology and adaptive life history in the invasive California wild radish (Raphanus sativus) and the implications for management. Evol Appl. 2010;3:64–76. doi: 10.1111/j.1752-4571.2009.00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilov G, Croocks JA. Biological invasions in marine ecosystems: ecological, management, and geographic perspectives. Heilderberg: Springer; 2009. [Google Scholar]
- Rius M, Pascual M, Turon X. Phylogeography of the widespread marine invader Microcosmus squamiger (Ascidiacea) reveals high genetic diversity of introduced populations and non-independent colonizations. Divers Distrib. 2008;14:818–828. doi: 10.1111/j.1472-4642.2008.00485.x. [DOI] [Google Scholar]
- Rius M, Turon X, Ordóñez V, Pascual M. Tracking invasion histories in the sea: facing complex scenarios using multilocus data. PLoS ONE. 2012;7(4):e35815. doi: 10.1371/journal.pone.0035815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson TB, Griffiths CL, McQuaid CD, Rius M. Marine alien species from South Africa –Status and impacts. Afr J Mar Sci. 2005;27(1):297–306. doi: 10.2989/18142320509504088. [DOI] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- Rousset F. Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- RStudio (2012) RStudio: Integrated development environment for R (Version 0.96.122) [Computer software]. Boston, MA. Retrieved May 20, 2012
- Ruesink JL, Lenihan HS, Trimble AC, Heiman KW, Micheli F, Byers JE, Kay MC. Introduction of non-native oysters: ecosystem effects and restoration implications. Annu Rev Ecol Evol Syst. 2005;36:643–689. doi: 10.1146/annurev.ecolsys.36.102003.152638. [DOI] [Google Scholar]
- Ruiz GM, Carlton JT. Invasive species: vectors and management strategies. Washington: Island Press; 2003. [Google Scholar]
- Ruiz GM, Carlton JT, Grosholz ED, Hines AH. Global invasions of marine and estuarine habitats by non-indigenous species: mechanisms, extent, and consequences. Am Zool. 1997;37:621–632. doi: 10.1093/icb/37.6.621. [DOI] [Google Scholar]
- Shine R. Invasive species as drivers of evolutionary change: cane toads in tropical Australia. Evol Applications. 2012;5(2):107–116. doi: 10.1111/j.1752-4571.2011.00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen MS, Wernberg T, Olden JD, Griffin JN, Silliman BR. A framework to study the context-dependent impacts of marine invasions. J Exp Mar Biol Ecol. 2011;400:322–327. doi: 10.1016/j.jembe.2011.02.033. [DOI] [Google Scholar]
- Turon X, Tarjuelo I, Duran S, Pascual M. Characterising invasion processes with genetic data: an Atlantic clade of Clavelina lepadiformis (Ascidiacea) introduced into Mediterranean harbours. Hydrobiologia. 2003;503:29–35. doi: 10.1023/B:HYDR.0000008481.10705.c2. [DOI] [Google Scholar]
- Tyrrell MC, Byers JE. Do artificial substrates favor nonindigenous fouling species over native species? J Exp Mar Biol Ecol. 2007;342:54–60. doi: 10.1016/j.jembe.2006.10.014. [DOI] [Google Scholar]
- Uriz MJ, Turon X. Sponge ecology in the molecular era. Adv Mar Biol. 2012;61:345–410. doi: 10.1016/B978-0-12-387787-1.00006-4. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mole Ecol Notes. 2004;4(3):535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- Van Soest RWM, De Kluijver MJ, Van Bragt PH, Faasse M, Nijland R, et al. Sponge invaders in Dutch coastal waters. J Mar Biol Ass UK. 2007;87(6):1733–1748. [Google Scholar]
- Weir BS, Cockerman CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- Wörheide G, Solé-Cava AM, Hooper JNA. Biodiversity, molecular ecology and phylogeography of marine sponges: patterns, implications and outlooks. Integr Comp Biol. 2005;45:377–385. doi: 10.1093/icb/45.2.377. [DOI] [PubMed] [Google Scholar]
- National Invasive Council (NISC) http://www.invasivespecies.gov/global/ISAC/ISAC_whitepapers.html
- XLSTAT v. 2012 5.02 Microsoft Excel add-in, Addinsoft’s Core Software
- Zammit PP, Longo C, Schembri PJ, et al. Occurrence of Paraleucilla magna Klautau, et al., 2004 (Porifera: Calcarea) in Malta. Medit Mar Sci. 2009;10(2):135–138. doi: 10.12681/mms.114. [DOI] [Google Scholar]
- Zenetos A, Gofas S, Verlaque M, Inar ME, García Raso JE, et al. (2010) Alien species in the Mediterranean Sea by 2010. A contribution to the application of European Union’s Marine Strategy Framework Directive (MSFD). Part I. Spatial distribution. Medit Mar Sci 11 (2):381-493
- Zibrowius H. Ongoing modification of the Mediterranean marine fauna and flora by the establishment of exotic species. Mésogée. 1991;51:83–107. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.