Abstract
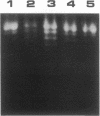
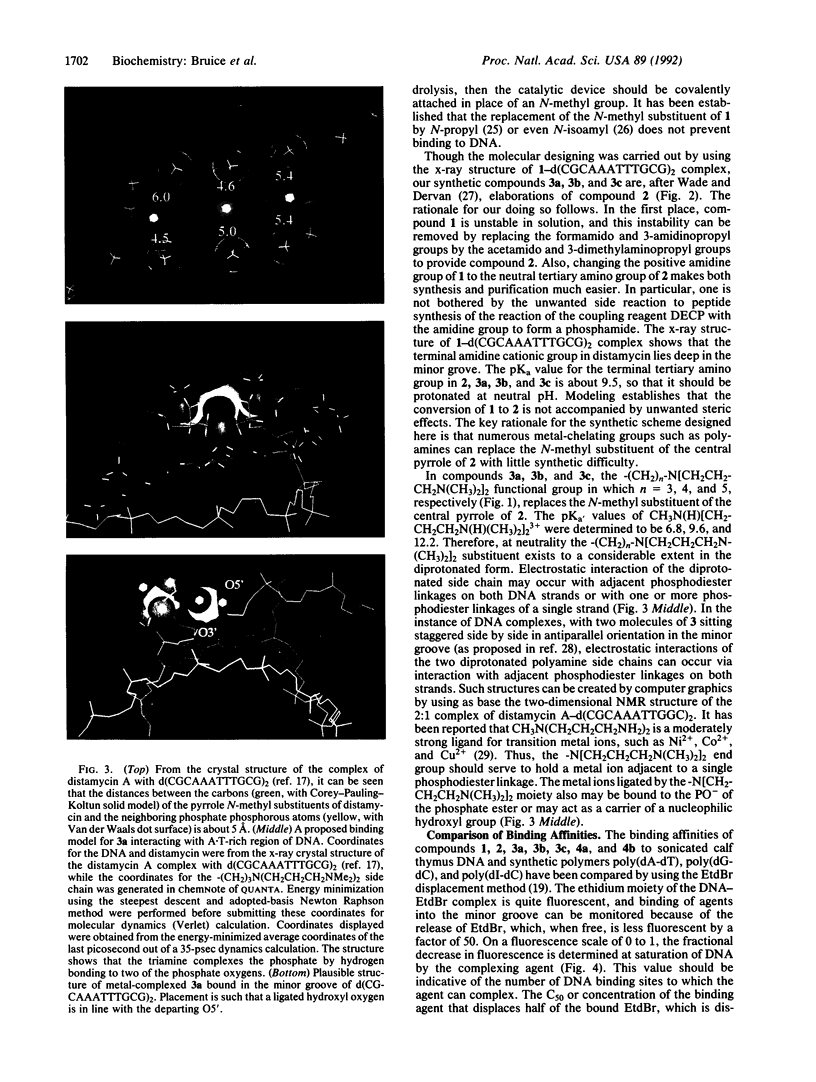
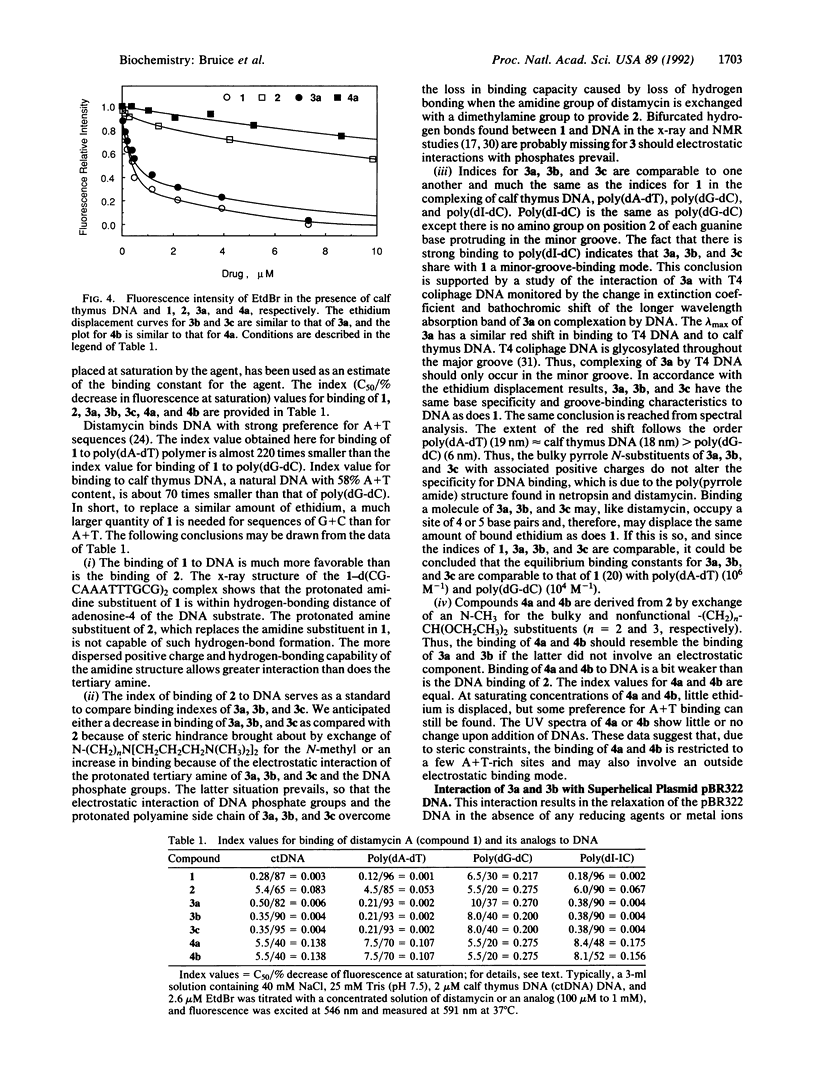
The structures of the compounds we call 3a, 3b, and 3c-compounds that incorporate (i) the tripyrrole peptide of the minor-groove-binding distamycin class of compounds and (ii) polyamine ligands that extend from the minor groove and can interact with phosphodiester bonds--were arrived at by computer-graphics designing by using the x-ray structure of distamycin A complexed in the minor groove of d(CGCAAATTTGCG)2. Compounds 3a, 3b, and 3c are elaborations of distamycin analog 2, designed for improved stability in solution and easier synthesis and purification, which itself binds weakly to DNA. Compounds 3a, 3b, and 3c have been synthesized, and the interaction of distamycin A, 2, 3a, 3b, and 3c with calf thymus DNA, poly(dA-dT), poly(dG-dC), poly(dI-dC), pBR322 superhelical plasmid DNA, and, in the case of 3b, T4 coliphage DNA have been studied. The following pertinent conclusions can be drawn. Binding of 3a, 3b, and 3c occurs in the minor groove of DNA and, because of favorable electrostatic interaction of diprotonated polyamine side chains and DNA phosphodiester linkages, the tenacity of DNA binding and site specificity of 3a, 3b, and 3c are comparable to that of native distamycin A. 3b has been found to induce changes in the superhelical density of pBR322 plasmid DNA. The study establishes that the central pyrrole N-CH3 substituent of 2 can be replaced by bulky polyamine metal ligands to create any number of compounds that bind into the minor groove at A + T-rich sites and are putative catalysts for the hydrolysis of DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barton J. K. Metals and DNA: molecular left-handed complements. Science. 1986 Aug 15;233(4765):727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton F. A., Day V. W., Hazen E. E., Jr, Larsen S., Wong S. T. Structure of bis(methylguanidinium) monohydrogen orthophosphate. A model for the arginine-phosphate interactions at the active site of staphylococcal nuclease and other phosphohydrolytic enzymes. J Am Chem Soc. 1974 Jul 10;96(14):4471–4478. doi: 10.1021/ja00821a020. [DOI] [PubMed] [Google Scholar]
- Dervan P. B. Design of sequence-specific DNA-binding molecules. Science. 1986 Apr 25;232(4749):464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. P., Benkovic S. J. Stereochemical course of the 3'----5'-exonuclease activity of DNA polymerase I. Biochemistry. 1984 Nov 20;23(24):5874–5881. doi: 10.1021/bi00319a029. [DOI] [PubMed] [Google Scholar]
- Gursky G. V., Zasedatelev A. S., Zhuze A. L., Khorlin A. A., Grokhovsky S. L., Streltsov S. A., Surovaya A. N., Nikitin S. M., Krylov A. S., Retchinsky V. O. Synthetic sequence-specific ligands. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):367–378. doi: 10.1101/sqb.1983.047.01.043. [DOI] [PubMed] [Google Scholar]
- Hough E., Hansen L. K., Birknes B., Jynge K., Hansen S., Hordvik A., Little C., Dodson E., Derewenda Z. High-resolution (1.5 A) crystal structure of phospholipase C from Bacillus cereus. Nature. 1989 Mar 23;338(6213):357–360. doi: 10.1038/338357a0. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. E., Wyckoff H. W. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J Mol Biol. 1991 Mar 20;218(2):449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- LePecq J. B., Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol. 1967 Jul 14;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- Mokul'skii M. A., Kapitonova K. A., Mokul'skaya T. D. Secondary structure of phage T2 DNA. Mol Biol. 1972 Nov-Dec;6(6):716–731. [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural characterization of a 2:1 distamycin A.d(CGCAAATTGGC) complex by two-dimensional NMR. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5723–5727. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural modeling of the distamycin A-d(CGCGAATTCGCG)2 complex using 2D NMR and molecular mechanics. Biochemistry. 1988 Oct 18;27(21):8088–8096. doi: 10.1021/bi00421a018. [DOI] [PubMed] [Google Scholar]
- Snounou G., Malcom A. D. Production of positively supercoiled DNA by netropsin. J Mol Biol. 1983 Jun 15;167(1):211–216. doi: 10.1016/s0022-2836(83)80043-6. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]