Abstract
Background:
Hepatitis C virus (HCV) has been known as a major cause of hepatocellular carcinoma (HCC) worldwide. However, the distinct molecular mechanisms underlying the effects of HCV proteins on the HCC progression have remained unclear.
Objectives:
In the present study, we studied the possible role of HCV in the HCC initiation and invasion using topological analysis of protein-protein interaction (PPI) networks.
Materials and Methods:
After analysis with GEO2R, a PPI network of differentially expressed genes (DEGs) was constructed for both chronic HCV and HCC samples. The STRING and GeneMANIA databases were used to determine the putative interactions between DEGs. In parallel, the functional annotation of DEGs was performed using g: Profiler web tool. The topological analysis and network visualization was carried outperformed using Cytoscape software and the top hub genes were identified. We determined the hub genes-related miRNAs using miRTarBase server and reconstructed a miRNA-Hubgene network.
Results:
Based on the topological analysis of miRNA-Hubgene network, we identified the key hub miRNAs. In order to identify the most important common sub-network, we aligned two PPI networks using NETAL tool. The c-Jun gene was identified as the most important hub gene in both HCV and HCC networks. Furthermore, the hsa-miR-34a, hsa-miR-155, hsa-miR-24, hsa-miR-744 and hsa-miR-92a were recognized as the most important hub miRNAs with positive correlation in the chronic HCV and HCC samples. Functional annotation of differentially expressed miRNAs (DEMs) using the tool for annotations of human miRNAs (TAM) revealed that there is a considerable overlap between miRNA gene expression profiles of HCV-infected and HCC cells.
Conclusions:
Our results revealed the possible crucial genes and miRNAs involved in the initiation and progression of HCC cells infected with HCV.
Keywords: HCV, HCC, Topological Analysis, PPI Networks, miRNA
1. Background
Hepatitis C virus (HCV) is the most common cause of chronic liver disease and also an important factor in liver transplantation (CDC, www.cdc.gov/hepatitis/hcv/hcvfaq.htm). The world health organization (WHO) has estimated that 3% of the world’s population has been infected with hepatitis C virus (HCV) and currently more than 170 million carry chronic HCV (WHO, www.who.int). HCV is considered the cause of approximately one-third of HCC cases in the USA. According to various reports, it is estimated that the risk of development of HCC when liver cirrhosis has progressed is 0 to 3% per year. The risk of HCC in chronic infections of HCV is highly associated with fibrosis stage. The incidence of HCC is relatively high in patients with cirrhosis (1% - 7% per year) (1, 2).
Experimental observations have shown that HCV viral proteins interact with cell proliferation and survival pathways, which lead to the increased risk of HCC. The over-expression of HCV protein enhances cell proliferation, transformation, and tumor development in mice, which suggests that viral proteins lead to the activation of oncogenic pathways (3). Several studies have shown that core protein can participate in liver cancer caused by HCV. It has been shown that the core protein expression can modulate cellular mRNA transcription through the interaction with hnRNP K (4). It also has been shown that the core protein can affect p53 protein through various mechanisms such as physical connection (5, 6). Studies have suggested that non-structural HCV proteins may also play a role in carcinogenesis (7). In parallel with this hypothesis, it has been reported that the expression of three HCV genes, including NS3, NS5A and NS5B can affect cell proliferation. NS3 and NS5A proteins can inhibit transcription of p21 (8-10) and NS5A protein impairs the cell cycle pathway by targeting cdk1/2-cyclin complex (11). NS5A protein also activates the Wnt pathway and stimulates the β-catenin-responsive transcription by activating the phosphoinositide 3-kinase (12). Finally, expression of non-structural NS5B protein, by targeting proteasomal degradation of pRB, leads to up-regulation of E2F-responsive promoter and induction of cell entry into the S phase of the cell cycle (13).
Although the molecular mechanisms of HCV-induced HCC remain to be discovered, studies have suggested that HCV is the important factor in providing an appropriate microenvironment for progression and development of cancer (14). In addition, gene expression alterations of chronic liver disease and molecular mechanisms involved in the HCV-induced HCC are not well understood.
2. Objectives
Nowadays, the network-based analysis of high-throughput gene expression profiles is emerging as a reliable tool in the perception of biological processes. In the present study, we studied the possible role of HCV in HCC initiation and invasion using topological analysis of PPI networks.
3. Materials and Methods
3.1. Microarray Data
The gene expression profile GSE38226 was obtained from the public functional genomics data repository gene expression omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/). There are two Illumina platforms in this transcription profile, including GPL6947 and GPL8179 used as mRNA and miRNA expression beadchips, respectively. Before the mRNA and miRNA expression profiling, the liver biopsies were collected from chronic HCV patients (24 samples), patients with hepatocellular carcinoma (12 samples) and normal subjects (8 samples). The samples were used to identify the differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs) using GEO2R (15).
3.2. Functional Annotation of DEGs and DEMs
Gene ontology (GO) annotation of all DEGs was performed using g: Profiler public web server (http://biit.cs.ut.ee/gprofiler/). g: Profiler is a powerful web tool for characterizing and manipulating the gene lists obtained from analysis of the high-throughput genomic data. The overrepresented GO terms with significant P value filtered from the primary results.
To determine functional categories of DEMs obtained from the previous step, the list of DEMs of both chronic HCV and HCC expression profiles were introduced to the TAM tool (16). The list of DEMs was functionally annotated and summarized according to the integrated miRNA categorical data. The important functional categories were chosen for further analysis. There was a considerable overlap between the enrichment results of chronic HCV and HCC in the given DEMs lists.
3.3. Protein-Protein Interaction (PPI) Network Construction
Based on the STRING (a protein-protein interaction database) (17) and GenMANIA (a web service used for prediction of gene function) regulation databases, we constructed and visualized the corresponding PPI networks using Cytoscape software (18). We considered directed interactions as well as important undirected interactions such as genetic interactions for building PPI networks. The number of nodes-edges in the PPI networks constructed from chronic HCV and HCC gene expression profiles were 171 - 1090 and 175 - 1161, respectively.
3.4. Topological Analysis of PPI Networks
Topological network analysis was performed for PPI networks and the most important hub genes were identified. The network analyzer plugin of cytoscape was used to analyze topological properties of the networks. Among nine evaluated methods, degree, betweenness centrality and closeness centrality had better outcomes and used for topological analysis of the PPI networks. Degree measure refers to the number of neighbors (edges) of a node and the nodes with higher degree centrality are considered biologically important hub genes within biological networks. Betweenness centrality is another centrality measure that represents the number of times each node is visited during traversing all shortest paths and is computed as follows:
| (1) |
The s and t are nodes in the network different from n, σst indicates the number of shortest paths from s to t and σst (n) is the number of shortest paths from s to t that n lies on.
The closeness centrality of a node is a measure of how fast information spreads from a given node to other reachable nodes in the given network. This measure is computed as follows:
| (2) |
Where L (n, m) is the length of the shortest path between two n and m nodes. The closeness centrality of each node is a number between 0 and 1.
We selected the top 50 genes according to the ranked results of three mentioned methods. Furthermore, the putative miRNAs regulating hub genes were predicted in silico using mirTarBase server (19) and a new miRNA-Hubgene network was reconstructed for both chronic HCV and HCC gene expression profiles. Topological analysis of the new miRNA-Hubgene networks was also performed using the degree measure to identify the hub miRNAs in each network. Interestingly, most hub miRNAs had been determined as DEMs after miRNA expression analysis of chronic HCV and HCC samples with GEO2R.
3.5. PPI Networks Alignment
A network alignment allows us to find the conserved functional complexes between two or more given networks. The alignment between chronic HCV and HCC networks was performed by NETAL tool (20). The NETAL is a new method for global alignment of PPI networks based on both biological and topological information of input networks. This method is highly capable of finding the conserved functional complexes and evolutionary relationships. The edge correctness (EC) measure was considered an important parameter to determine topological similarity between chronic HCV and HCC derived PPI networks. EC is the percentage of interactions aligned between the given networks. After comparative analysis of chronic HCV and HCC networks, the most important common sub-network was determined and visualized.
4. Results
4.1. Functional Annotation Analysis
Analysis of the functional annotation of chronic HCV and HCC related DEGs specifically indicated that most genes, in both chronic HCV and HCC related DEGs, are involved in important cellular processes such as cell signaling, cell proliferation, cell death, cell migration, etc. (Table 1). These functional categories are obviously involved in the cancer progression and invasion.
Table 1. Overrepresented gProfiler Terms in the DEGs From HCC and Chronic HCV.
| DEGs Source | Number of Genes | P Value |
|---|---|---|
| HCC | ||
| Localization | 85 | 1.58e - 03 |
| Signaling | 87 | 2.68e - 02 |
| Cell proliferation | 38 | 2.39e - 02 |
| Cell death | 37 | 3.14e - 02 |
| Cell migration | 36 | 5.18e - 06 |
| Extracellular matrix | 75 | 3.15e - 04 |
| Chronic HCV | ||
| Signaling | 101 | 1.66e - 07 |
| Localization | 81 | 2.12e - 02 |
| Cell adhesion | 56 | 3.19e - 17 |
| Immune response | 59 | 3.68e - 08 |
| Cell death | 40 | 7.57e - 03 |
| Angiogenesis | 21 | 1.05e- 06 |
Furthermore, we enriched the list of DEMs obtained from miRNA expression analysis of both HCC and chronic HCV samples. Interestingly, there was a considerable overlap between the functional categories of chronic HCV and HCC related DEMs (Figure 1). Similarly to DEGs, a high number of miRNAs predicted as regulators of several cancer-related pathways such as cell cycle, cell proliferation, cell death, apoptosis, epithelial-mesenchymal transition and angiogenesis. We found that the four important onco-miRNA (hsa-miR-24, hsa-miR-92a, hsa-miR-194 and hsa-miR-191) were up-regulated in both chronic HCV and HCC samples compared to the control liver cells. Furthermore, several miRNA tumor suppressors, including hsa-miR-181b, hsa-miR-451, hsa-miR-34a, hsa-miR-34b and hsa-miR-34c were significantly down-regulated in chronic HCV and HCC samples compared to the control samples.
Figure 1. Functional Enrichment Analysis of DEMs Resulted From miRNA Expression Analysis of Chronic HCV and HCC Samples.
4.2. Topological Analysis of PPI Networks
The PPI networks for DEGs resulted from analysis of chronic HCV and HCC gene expression profiles were constructed with 171 - 1090 and 175 - 1161 nodes-edges, respectively. We used the degree, betweenness centrality and closeness centrality measures to analyze topology of resulted PPI networks. The top 50 hub genes of these three measures were selected for further analysis (Tables 2 and 3). After filtering and removing duplicate values, we obtained 61 and 64 unique hub genes from the topological analysis of chronic HCV and HCC related PPIs, respectively. There were 32 common hub genes between chronic HCV and HCC unique hub genes. Interestingly, the c-Jun gene was identified as the most important hub gene in both chronic HCV and HCC related PPI networks. In parallel, validated corresponding miRNAs for each hub gene were determined and a new miRNAs-Hubgenes network was reconstructed with 217 and 319 nodes and edges, respectively. Topological analysis of miRNAs-Hubgenes regulatory network was performed and the hub miRNAs were identified and visualized along with their putative targets (Figure 2). We compared the hub miRNAs with DEMs obtained from the miRNA expression profile of chronic HCV and HCC samples. The comparison indicated a positive correlation between chronic HCV and HCC resulted DEMs. Two hub miRNAs, including hsa-miR-34a and hsa-miR-155 were significantly down-regulated in both chronic HCV and HCC samples compared to normal liver samples. Moreover, three other hub miRNAs (hsa-miR-24, hsa-miR-744 and hsa-miR-92a) were up-regulated in chronic HCV and HCC samples compared to the control samples.
Table 2. The Top 50 Hub Genes Obtained From Analysis of the Chronic HCV Network.
| Betweenness | Value | Degree | Value | Closeness | Value |
|---|---|---|---|---|---|
| JUN | 0.088456 | JUN | 60 | JUN | 0.563107 |
| KCNMA1 | 0.045263 | KCNMA1 | 38 | KCNMA1 | 0.537037 |
| SLC1A1 | 0.044697 | COL4A1 | 36 | SLC1A1 | 0.52568 |
| BCL2 | 0.035565 | SLC1A1 | 35 | LEF1 | 0.517857 |
| COL4A1 | 0.035263 | LEF1 | 33 | COL4A1 | 0.514793 |
| PODXL | 0.031336 | BCL2 | 32 | ACACB | 0.513274 |
| BIRC3 | 0.030732 | VEGFC | 31 | ENAH | 0.511765 |
| VEGFC | 0.029737 | COL1A2 | 31 | BCL2 | 0.508772 |
| WNT4 | 0.029547 | BIRC3 | 30 | VEGFC | 0.507289 |
| ENAH | 0.02707 | ENAH | 30 | COL1A2 | 0.501441 |
| POLQ | 0.026244 | SOX9 | 29 | PID1 | 0.501441 |
| ACACB | 0.025908 | COL5A1 | 29 | PODXL | 0.5 |
| LEF1 | 0.025767 | COL15A1 | 29 | WNT4 | 0.5 |
| CELF4 | 0.024233 | PODXL | 28 | BIRC3 | 0.498567 |
| FANCC | 0.022376 | WNT4 | 28 | COL15A1 | 0.497143 |
| SLCO3A1 | 0.022328 | ACACB | 28 | CELF4 | 0.494318 |
| TSPAN8 | 0.021671 | SLCO3A1 | 28 | SLCO3A1 | 0.492918 |
| SMOC2 | 0.020669 | MYOF | 28 | CD34 | 0.492918 |
| SOX9 | 0.020065 | TSPAN8 | 27 | BCAS4 | 0.491525 |
| TMEM200A | 0.019163 | SOX4 | 26 | MYOF | 0.490141 |
| SOX4 | 0.01913 | BCAS4 | 26 | TSPAN8 | 0.490141 |
| STK39 | 0.018437 | THBS2 | 26 | AGTRAP | 0.490141 |
| COL5A1 | 0.017963 | CD44 | 26 | BRCA1 | 0.488764 |
| CBLN1 | 0.017321 | CELF4 | 25 | KCNN2 | 0.487395 |
| ADCY1 | 0.016641 | PID1 | 25 | SOX9 | 0.486034 |
| OSBPL3 | 0.016441 | COL4A2 | 25 | SOX4 | 0.486034 |
| BCAS4 | 0.01626 | ADRA1A | 24 | SPP1 | 0.486034 |
| MYOF | 0.015722 | CD34 | 24 | STK39 | 0.483333 |
| COL1A2 | 0.014797 | STK39 | 23 | COL5A1 | 0.481994 |
| THBS2 | 0.014385 | BRCA1 | 23 | PDGFRB | 0.481994 |
| CCL20 | 0.014217 | SPP1 | 23 | FRZB | 0.481994 |
| TGFBI | 0.014096 | CHRM3 | 23 | CHRM3 | 0.480663 |
| FRZB | 0.014042 | KCNN2 | 23 | SMOC2 | 0.480663 |
| COL15A1 | 0.014024 | POLQ | 22 | CBLN1 | 0.480663 |
| CD44 | 0.013973 | FANCC | 22 | ERBB2 | 0.479339 |
| FST | 0.013728 | SMOC2 | 22 | THBS2 | 0.476712 |
| STAG3 | 0.01337 | TMEM200A | 22 | ADRA1A | 0.476712 |
| S100A13 | 0.013157 | CBLN1 | 22 | POLQ | 0.476712 |
| ACSL1 | 0.012823 | TGFBI | 22 | VWF | 0.476712 |
| DACT2 | 0.012597 | ST3GAL6 | 22 | PARM1 | 0.476712 |
| BRCA1 | 0.01237 | COL1A1 | 22 | NOS1AP | 0.476712 |
| RRM2 | 0.012267 | OSBPL3 | 21 | ST3GAL6 | 0.47541 |
| ADRA1A | 0.012258 | PDGFRB | 21 | COL1A1 | 0.47541 |
| PLVAP | 0.011888 | B3GALNT1 | 20 | CMIP | 0.47541 |
| SPP1 | 0.011585 | ERBB2 | 20 | TGFBI | 0.472826 |
| B3GALNT1 | 0.011486 | AGTRAP | 20 | TMEM200A | 0.471545 |
| SFRP5 | 0.011395 | ADCY1 | 19 | ASPH | 0.47027 |
| CHRM3 | 0.011097 | STAG3 | 19 | CD44 | 0.469003 |
| KCNN2 | 0.01109 | DCDC2 | 19 | SERPINE2 | 0.469003 |
| PID1 | 0.011078 | VWF | 19 | COL4A2 | 0.467742 |
Table 3. The Top 50 Hub Genes Obtained From Analysis of the HCC Network.
| Betweenness | Value | Degree | Value | Closeness | Value |
|---|---|---|---|---|---|
| JUN | 0.152054 | JUN | 79 | JUN | 0.602837 |
| PAPPA | 0.064493 | BCL2 | 39 | BCL2 | 0.532915 |
| BCL2 | 0.04056 | KCNMA1 | 36 | COL1A2 | 0.519878 |
| KCNMA1 | 0.040488 | COL1A2 | 36 | PAPPA | 0.519878 |
| VEGFC | 0.038859 | PAPPA | 35 | VEGFC | 0.519878 |
| PDGFD | 0.037164 | SOX9 | 33 | KCNMA1 | 0.516717 |
| COL4A1 | 0.033355 | COL4A1 | 32 | COL4A1 | 0.515152 |
| ERBB2 | 0.033017 | CD44 | 32 | ERBB2 | 0.515152 |
| COL1A2 | 0.031187 | EGR1 | 32 | SOX9 | 0.505952 |
| SOX9 | 0.029953 | VEGFC | 31 | EGR1 | 0.505952 |
| IL8 | 0.028423 | ERBB2 | 29 | FMOD | 0.504451 |
| CCR6 | 0.02778 | IL8 | 28 | SOD3 | 0.501475 |
| FMOD | 0.024919 | FMOD | 27 | PODXL | 0.5 |
| PODXL | 0.023075 | PODXL | 27 | CD34 | 0.5 |
| CD44 | 0.022533 | WNT4 | 27 | PDGFRB | 0.5 |
| EGR1 | 0.021941 | IL6 | 27 | CD44 | 0.498534 |
| MOXD1 | 0.02144 | COL5A1 | 27 | WNT4 | 0.498534 |
| SPP1 | 0.020341 | MOXD1 | 26 | SPP1 | 0.498534 |
| CD34 | 0.019815 | CD34 | 26 | IL8 | 0.497076 |
| PDLIM3 | 0.018409 | SPP1 | 25 | COL1A1 | 0.497076 |
| WNT4 | 0.017109 | COL4A2 | 25 | MOXD1 | 0.494186 |
| SLCO3A1 | 0.017079 | COL15A1 | 25 | IL6 | 0.492754 |
| IL6 | 0.016092 | COL1A1 | 24 | UCHL1 | 0.489914 |
| TGFBI | 0.015425 | SLCO3A1 | 23 | FRZB | 0.489914 |
| LYVE1 | 0.015167 | SOD3 | 23 | VWF | 0.487106 |
| HDC | 0.014886 | PDGFRB | 23 | COL5A1 | 0.48433 |
| SOD3 | 0.014787 | COL6A3 | 23 | SLCO3A1 | 0.482955 |
| MYOF | 0.014711 | PDGFD | 22 | AGTRAP | 0.481586 |
| PTGS2 | 0.014547 | MYOF | 22 | PID1 | 0.478873 |
| SMOC2 | 0.014195 | TGFBI | 21 | PTGS2 | 0.478873 |
| COL4A2 | 0.013321 | SMOC2 | 21 | COL6A3 | 0.477528 |
| UCHL1 | 0.013228 | UCHL1 | 21 | COL3A1 | 0.477528 |
| COL1A1 | 0.013075 | VWF | 21 | COL4A2 | 0.47619 |
| VWF | 0.01304 | PID1 | 21 | TGFBI | 0.47619 |
| COL5A1 | 0.013023 | FRZB | 21 | SMOC2 | 0.47619 |
| CBLN1 | 0.012941 | HDC | 20 | SGCD | 0.47619 |
| PID1 | 0.012849 | PTGS2 | 20 | NAB2 | 0.47619 |
| MYL9 | 0.012038 | CCR6 | 19 | BRCA1 | 0.47486 |
| FAP | 0.011566 | SGCD | 19 | CBLN1 | 0.47486 |
| FRZB | 0.011414 | NAB2 | 19 | COL15A1 | 0.473538 |
| ENO2 | 0.010941 | COL3A1 | 19 | MYOF | 0.473538 |
| TMEM200A | 0.01088 | TMEM200A | 18 | HDC | 0.472222 |
| SGCD | 0.009918 | BRCA1 | 18 | CMIP | 0.470914 |
| CD3E | 0.009637 | FOSB | 17 | ACTA2 | 0.469613 |
| PDGFRB | 0.009418 | THBS2 | 17 | PDGFD | 0.467033 |
| COL15A1 | 0.00936 | PDLIM3 | 16 | CCR6 | 0.465753 |
| PLAT | 0.009294 | CBLN1 | 16 | DPYSL3 | 0.465753 |
| FBLN1 | 0.009258 | FAP | 16 | FOSB | 0.464481 |
| BRCA1 | 0.008361 | CD3E | 16 | SERPINE2 | 0.464481 |
| CLEC2B | 0.008261 | SAA1 | 16 | PLAT | 0.464481 |
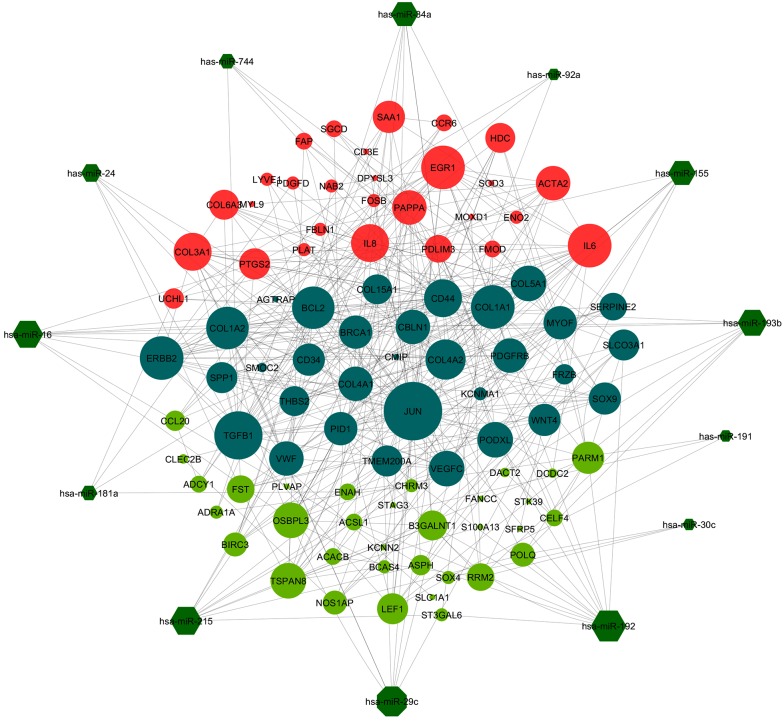
Figure 2. The Regulatory Network of Hub miRNAs (Dark Green Hexagons) Along With Hub Genes of Chronic HCV (Red Circles) and HCC (Pale Green Circles) PPI Networks.
Nodes with higher degrees are shown in the bigger circles. The common hub genes between chronic HCV and HCC are shown with dark cyan circles.
4.3. PPI Networks Alignment
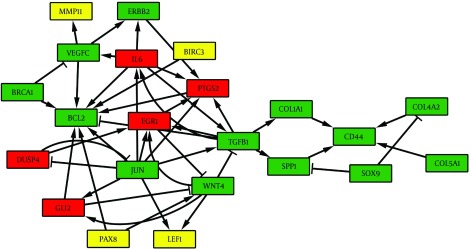
After networks alignment, several sub-networks were identified as common complexes between chronic HCV and HCC networks. The EC measure was equal to 0.45 indicating approximately 45% of topological similarity between the given networks. The connected component between the two PPI networks had 156 nodes and 471 edges. We selected a 22-node sub-network as the most important functional complex, which consists of 13 common genes with positive correlation (Figure 3). The functional annotation analysis demonstrated that this sub-network is actively involved in regulation of cell proliferation, cell migration and apoptosis.
Figure 3. The Most Important Functional Sub-Network Obtained From Two PPI Network Alignments.
The common genes between chronic HCV and HCC networks are shown in the green color boxes. The red and yellow boxes represent chronic HCV and HCC specific genes, respectively.
5. Discussion
Strong epidemiological evidence links hepatitis C infection with hepatocellular carcinoma (3). Liver cancer usually develops decades after HCV infection and liver cirrhosis. However, the molecular mechanisms involved in cancer development have not been fully understood. HCV is classified as a positive-strand RNA virus that replicates in the cytoplasm and does not integrate into the host DNA; however, its carcinogenic features are remarkable. It is possible that indirect mechanisms such as chronic liver inflammation, oxidative damages, presence of cirrhosis and DNA damage are involved in cancer development (21, 22). Cirrhosis as the result of fibrosis due to hepatitis C infection is the indication of preneoplastic stage in development of liver cancer, which can cause rapid proliferation of dysplastic liver cells (21). Studies have shown that HCV proteins, especially core protein, are inherently oncogenic and their expression in the cell can be involved in the development of HCC directly and in the absence of immune responses or cirrhosis. Several HCV proteins can interact with host cell proteins and induce cell proliferation. Evidence has shown that replication of HCV replicon RNAs is strongly dependent on cell proliferation (23), while the HCV RNA synthesis is stimulated in the S phase of the cell cycle (24). Hepatocytes usually have low proliferation rate and it seems that some viral proteins expressed during HCV infection trigger cell division and are involved in carcinogenic effects of chronic HCV infection (22, 25).
A positive correlation between chronic HCV infection and HCC progression has been frequently reported. However, the main mechanisms underlying pathogenesis of HCV-induced HCC has remained unclear. We reported here molecular mechanisms regulated by HCV, which seem to be involved in HCC progression and invasion. We found that the c-Jun gene is the most important hub gene obtained from topological analysis of both HCV and HCC networks. The c-Jun has a key role in enhanced cell proliferation of cancer cells by regulating the expression of CCND1 and Cdc2 genes (26). In addition, c‐Jun can prevent TNFα-mediated apoptosis through cooperation with NFKB1. Erhadt and colleagues reported that HCV can protect various cells from TNFα-induced apoptosis via activating NFKB signaling pathway (27). Taken together, we suggest that the role of HCV in the inhibition of apoptosis is mediated by c‐Jun function. Two proto-oncogene, BCL2 and ERBB2, were identified as the common hub genes of HCV and HCC-derived PPI networks. The BCL2 overexpressing cells tend to have a prolonged survival and reduced apoptosis in a variety of cancer cells. This gene is highly capable of suppressing apoptosis by either inhibiting the release of cytochrome c from the mitochondria or blocking the function of APAF1 (28). It is now well established that HCV triggers cancer cell apoptosis and survival through up-regulation of BCL2 expression, resulting in reduced CASP3 activity. The nonstructural protein 4B (NS4B) is the key gene responsible for stimulating BCL2 gene through inhibition of the function of SOCS3 gene (29). ERBB2 is a protein tyrosine kinase that can bind to other growth factor receptors to form a heterodimer, stabilizing ligand-receptor complex and enhancing several downstream signaling pathways such as mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3 kinase (PI3K/Akt1) (10, 30). This gene was identified as an important hub gene in both chronic HCV and HCC networks. The statistically significant correlation between NS3, an HCV serine protease, and ERBB2 has been reported in HCC cases (31). VEGFC is an important gene involved in promoting angiogenesis. This hub gene was up-regulated in HCV and HCC samples compared to normal liver cells. It is now widely accepted that HCV is of central importance for vascular development and angiogenesis through enhancing the expression of VEGFC (32).
Extracellular matrix proteins play a pivotal role in tumor progression and their alterations are usually associated with an aggressive phenotype of various cancer types. Our results revealed that expression of COL4A2, COL1A1 and COL5A1 hub genes were significantly altered in the HCV-infected and HCC cells compared to the normal cell samples. These genes are necessary for spreading HCC cells through remodeling of extracellular matrix (33). Recently, it has been determined that HCV infection can lead to an increased invasive phenotype by regulating the matrix metalloproteinases (MMPs), a family of extracellular matrix components (34). However, the role of HCV in regulation of collagen genes is poorly understood.
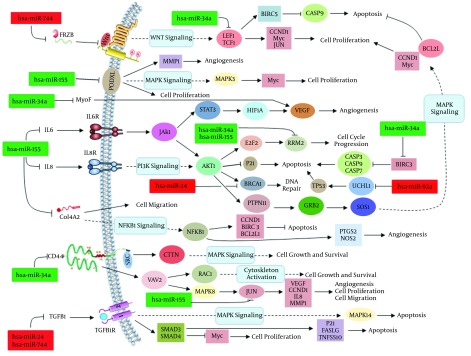
Amongst non-coding RNAs, the role of miRNAs in carcinogenesis has been extensively investigated. Varnholt and colleagues examined the miRNA gene expression profile of HCV-infected HCC cells and identified several important miRNAs that seem to be involved in the pathogenesis of HCV-associated HCC (26). We found that the miRNA gene expression profile of HCV-infected cells is clearly similar to HCC cells. The hsa-miR-155 and hsa-miR-34a were down-regulated in both HCV-infected and HCC cells compared to the control samples. The HCV core and NS5A proteins have been implicated in pathogenesis of HCC through induction of Wnt signaling pathway (28). HCV-associated activation of Wnt signaling pathway may be mediated by expression alterations of hsa-miR-34a and has-miR-744. Additionally, the HCV can actively induce cell proliferation and MAPK signaling pathway in vitro (27). However, the distinct mechanism defining the effect of HCV on the cell proliferation has remained unknown. Here, we show that HCV can positively impair cell cycle progression through inhibition of hsa-miR-34a. The expression of two cell proliferation-related genes, RRM2 and BIRC3, is regulated by hsa-miR-34a (Figure 4). This miRNA can activate the MAPK signaling pathway.
Figure 4. The Hub miRNAs Obtained From Topological Analysis of miRNA-Hubgene Network Along With Their Putative Targets.
Up-regulated and down-regulated hub miRNAs with positive correlation in both HCV-infected and HCC cells are shown in red and green boxes, respectively.
However, activation of MAPK, AKT1, NFKB1 and JAK-STAT signaling pathways is also mediated through down-regulation of hsa-miR-155. This hub miRNA was significantly down-regulated in both HCV-infected and HCC cells compared to normal cells. However, functional study of hsa-miR-155 targets revealed that down-regulation of this miRNA can induce cell proliferation and angiogenesis and inhibit apoptosis through activating aforementioned signaling pathways (Figure 4). The hsa-miR-24 acts as an anti-apoptotic miRNA by repressing the expression of Bim gene (35). We found that this hub miRNA was up-regulated in HCV-infected and HCC samples, suggesting its possible effect on the progression of HCC cells. Furthermore, the function of tumor suppressor BRCA1 is considerably modulated by hsa-miR-24. The hsa-miR-92a is a key regulator of angiogenesis and apoptosis. It has been shown that overexpression of hsa-miR-744 can moderately induce tube formation and reduce apoptosis in vitro (36).
5.1. Conclusion
This study indicated that characteristics of invasive phenotype HCC cells are associated with differential expressions of several genes and miRNAs. The graph-based information may give us a new insight into molecular mechanisms underlying the pathogenesis of HCV-induced HCC.
Acknowledgments
This work was supported by Tehran University of Medical Sciences. We also appreciate Hassan Askari’s collaboration at the early stage of this study.
Footnotes
Authors’ Contribution:Vahdat Poortahmasebi: literature search, writing and reviewing and bioinformatic analysis; Mansour Poorebrahim: study concept and design, main writing part and bioinformatic analysis; Saeideh Najafi: data collection and writing; Seyed Mohammad Jazayeri, Seyed Moayed Alavian, and Seyed Shahriar Arab: participated in the design of study and reviewed manuscript; Saeid Ghavami: critical revision of the manuscript for important intellectual content; Seyed Ehsan Alavian: Adel Rezaei Moghadam, and Mehdi Amiri: contributed equally to this paper. All authors read and approved the final manuscript.
References
- 1.Yoshida H, Shiratori Y, Moriyama M, Arakawa Y, Ide T, Sata M, et al. Interferon therapy reduces the risk for hepatocellular carcinoma: national surveillance program of cirrhotic and noncirrhotic patients with chronic hepatitis C in Japan. IHIT Study Group. Inhibition of Hepatocarcinogenesis by Interferon Therapy. Ann Intern Med. 1999;131(3):174–81. doi: 10.7326/0003-4819-131-3-199908030-00003. [DOI] [PubMed] [Google Scholar]
- 2.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):448–58. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goossens N, Hoshida Y. Hepatitis C virus-induced hepatocellular carcinoma. Clin Mol Hepatol. 2015;21(2):105–14. doi: 10.3350/cmh.2015.21.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh TY, Matsumoto M, Chou HC, Schneider R, Hwang SB, Lee AS, et al. Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. J Biol Chem. 1998;273(28):17651–9. doi: 10.1074/jbc.273.28.17651. [DOI] [PubMed] [Google Scholar]
- 5.Kao CF, Chen SY, Chen JY, Wu Lee YH. Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein. Oncogene. 2004;23(14):2472–83. doi: 10.1038/sj.onc.1207368. [DOI] [PubMed] [Google Scholar]
- 6.Otsuka M, Kato N, Lan K, Yoshida H, Kato J, Goto T, et al. Hepatitis C virus core protein enhances p53 function through augmentation of DNA binding affinity and transcriptional ability. J Biol Chem. 2000;275(44):34122–30. doi: 10.1074/jbc.M000578200. [DOI] [PubMed] [Google Scholar]
- 7.Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, et al. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122(2):352–65. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 8.Kwun HJ, Jung EY, Ahn JY, Lee MN, Jang KL. p53-dependent transcriptional repression of p21(waf1) by hepatitis C virus NS3. J Gen Virol. 2001;82(Pt 9):2235–41. doi: 10.1099/0022-1317-82-9-2235. [DOI] [PubMed] [Google Scholar]
- 9.Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC, et al. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21(31):4801–11. doi: 10.1038/sj.onc.1205589. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh R, Narasanna A, Wang SE, Liu S, Chakrabarty A, Balko JM, et al. Trastuzumab has preferential activity against breast cancers driven by HER2 homodimers. Cancer Res. 2011;71(5):1871–82. doi: 10.1158/0008-5472.CAN-10-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arima N, Kao CY, Licht T, Padmanabhan R, Sasaguri Y, Padmanabhan R. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J Biol Chem. 2001;276(16):12675–84. doi: 10.1074/jbc.M008329200. [DOI] [PubMed] [Google Scholar]
- 12.Street A, Macdonald A, McCormick C, Harris M. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. J Virol. 2005;79(8):5006–16. doi: 10.1128/JVI.79.8.5006-5016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 2005;102(50):18159–64. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koike K. Molecular Basis of Hepatitis C Virus–Associated Hepatocarcinogenesis: Lessons From Animal Model Studies. Clin Gastroenterol Hepatol. 2005;3:S132–5. doi: 10.1016/s1542-3565(05)00700-7. [DOI] [PubMed] [Google Scholar]
- 15.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu M, Shi B, Wang J, Cao Q, Cui Q. TAM: a method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics. 2010;11:419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu S, Lin F, Wu W, Liang C, Huang W, Chan W, et al. miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic acids res. 2010:gkq1107. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neyshabur B, Khadem A, Hashemifar S, Arab SS. NETAL: a new graph-based method for global alignment of protein-protein interaction networks. Bioinformatics. 2013;29(13):1654–62. doi: 10.1093/bioinformatics/btt202. [DOI] [PubMed] [Google Scholar]
- 21.Hoshida Y, Fuchs BC, Bardeesy N, Baumert TF, Chung RT. Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma. J Hepatol. 2014;61(1 Suppl):S79–90. doi: 10.1016/j.jhep.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, et al. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122(2):366–75. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 23.Pietschmann T, Lohmann V, Kaul A, Krieger N, Rinck G, Rutter G, et al. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J Virol. 2002;76(8):4008–21. doi: 10.1128/JVI.76.8.4008-4021.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scholle F, Li K, Bodola F, Ikeda M, Luxon BA, Lemon SM. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J Virol. 2004;78(3):1513–24. doi: 10.1128/JVI.78.3.1513-1524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwer B, Ren S, Pietschmann T, Kartenbeck J, Kaehlcke K, Bartenschlager R, et al. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J Virol. 2004;78(15):7958–68. doi: 10.1128/JVI.78.15.7958-7968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47(4):1223–32. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 27.Erhardt A, Hassan M, Heintges T, Haussinger D. Hepatitis C virus core protein induces cell proliferation and activates ERK, JNK, and p38 MAP kinases together with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line. Virology. 2002;292(2):272–84. doi: 10.1006/viro.2001.1227. [DOI] [PubMed] [Google Scholar]
- 28.Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene. 2006;25(27):3834–47. doi: 10.1038/sj.onc.1209562. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Zhang Q, Liu Y, Luo Z, Kang L, Qu J, et al. Hepatitis C virus activates Bcl-2 and MMP-2 expression through multiple cellular signaling pathways. J Virol. 2012;86(23):12531–43. doi: 10.1128/JVI.01136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knuefermann C, Lu Y, Liu B, Jin W, Liang K, Wu L, et al. HER2/PI-3K/Akt activation leads to a multidrug resistance in human breast adenocarcinoma cells. Oncogene. 2003;22(21):3205–12. doi: 10.1038/sj.onc.1206394. [DOI] [PubMed] [Google Scholar]
- 31.Bahnassi AA, Zekri AR, El-Houssini S, Mokhtar NM, Abdel-Aziz AO, Sherif GM, et al. Hepatitis C virus-NS3P in relation to p53, p21waf, mdm2, p21-ras and c-erbB2 in hepatocarcinogenesis. J Gastroenterol Hepatol. 2005;20(11):1731–40. doi: 10.1111/j.1440-1746.2005.04002.x. [DOI] [PubMed] [Google Scholar]
- 32.Hassan M, Selimovic D, Ghozlan H, Abdel-kader O. Hepatitis C virus core protein triggers hepatic angiogenesis by a mechanism including multiple pathways. Hepatology. 2009;49(5):1469–82. doi: 10.1002/hep.22849. [DOI] [PubMed] [Google Scholar]
- 33.Lin ZY, Chuang WL. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother. 2012;66(6):454–8. doi: 10.1016/j.biopha.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Nunez O, Fernandez-Martinez A, Majano PL, Apolinario A, Gomez-Gonzalo M, Benedicto I, et al. Increased intrahepatic cyclooxygenase 2, matrix metalloproteinase 2, and matrix metalloproteinase 9 expression is associated with progressive liver disease in chronic hepatitis C virus infection: role of viral core and NS5A proteins. Gut. 2004;53(11):1665–72. doi: 10.1136/gut.2003.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med. 2011;208(3):549–60. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Zhou M, Qin G, Weintraub NL, Tang Y. MiR-92a regulates viability and angiogenesis of endothelial cells under oxidative stress. Biochem Biophys Res Commun. 2014;446(4):952–8. doi: 10.1016/j.bbrc.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]