Abstract
AMPA-type glutamate receptors (AMPARs), central mediators of rapid neurotransmission and synaptic plasticity, predominantly exist as heteromers of the GluA1-4 subunits. Here we report first AMPAR heteromer structures, which deviate substantially from existing GluA2 homomers. Crystal structures of the GluA2/3 and GluA2/4 N-terminal domains reveal a novel compact conformation with an alternating arrangement of the four subunits around a central axis. This organization is confirmed by cysteine crosslinking in full-length receptors and permitted us to determine the structure of an intact GluA2/3 receptor by cryo-EM. Two models in the ligand-free state, at 8.25 Å and 10.3 Å resolution, exhibit a substantial vertical compression and close associations between domain layers, reminiscent of NMDA receptors. Model 1 resembles a resting state, model 2 a desensitized state, providing snapshots of gating transitions in the nominal absence of ligand. Our data reveal organizational features of heteromeric AMPARs and provide a framework to decipher AMPAR architecture and signaling.
Ionotropic glutamate receptors (iGluRs) are tetrameric cation channels that mediate fast excitatory signal transmission upon binding presynaptically released glutamate (1). They are essential for brain development and experience-dependent synaptic plasticity, which underlies learning. iGluR dysfunction is implicated in a number of neurological disorders including dementia, mood disorders and epilepsy (2). Three major subtypes, the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid, N-methyl-D-aspartate and kainate receptors (AMPARs, NMDARs and KARs), each contribute a different component to the synaptic signal (1). AMPARs mediate the initial depolarization of the postsynaptic membrane, triggering NMDAR activation and the generation of an excitatory postsynaptic potential. The rapid kinetics of AMPARs permit moment-to-moment signaling and their trafficking to synapses is central to synaptic plasticity (3).
Gating kinetics, ion permeation and trafficking are set by the subunit stoichiometry and ultimately shape the synaptic response. AMPAR tetramers are composed of the GluA1-GluA4 subunits with the vast majority containing GluA2, which renders the channel Ca2+-impermeable, lowers its conductance and alters its voltage dependence (1, 4).
Structures of GluA2 homomers have been instrumental in clarifying AMPAR modular architecture (5). The receptor is arranged in domain layers (Fig. 1A) - a two-fold symmetrical extracellular region (ECR), composed of the N-terminal domain (NTD) and ligand-binding domain (LBD), is attached to the transmembrane ion channel domain (TMD) of approximately four-fold symmetry (Fig. 1A), while a cytoplasmic tail mediates trafficking and anchorage at synapses. The four chains (A-D) in a tetramer are conformationally non-equivalent and likely contribute differently to gating: the ‘AC’ pair is positioned closer to the ion conduction pore axis (at the level of the LBD and TMD; pore-proximal) than the ‘BD’ chains (pore-distal). In NMDARs, obligatory heteromers, the GluN2 subunits occupy the BD position (6, 7) and exert a greater pulling force on the gate (8). The domain architecture of heteromeric AMPARs is currently unknown, and their intersubunit arrangement has only been inferred from modeling studies (9).
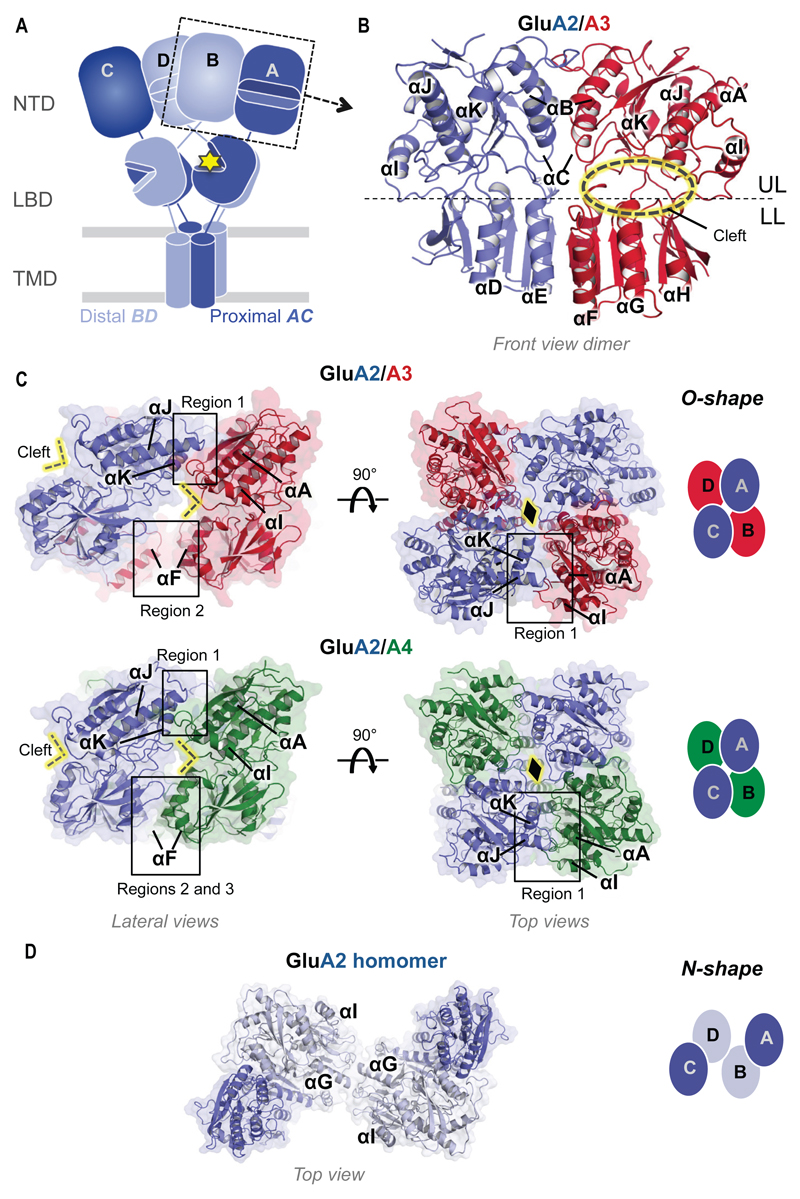
Fig. 1.
Structure and organization of the GluA2/3 and GluA2/4 NTD layer. (A) Overall organization of an iGluR, showing the typical layered architecture (TMD, LBD, NTD) and quaternary arrangement (the pore-proximal, AC and pore-distal, BD chains are depicted). The cytoplasmic tails are omitted for simplicity. The yellow star indicates L-glutamate in the LBD cleft. (B) GluA2/3 NTD heterodimer in front view (GluA2, blue; GluA3, red). The upper lobe (UL) and lower lobe (LL) are separated by a stippled line; helices are labelled αA-αK. The subunits face opposite directions providing a view of the GluA2 back and the GluA3 cleft (indicated by yellow ellipse). (C) O-shape tetrameric arrangement of the GluA2/3 (upper panels) and GluA2/4 (lower panels) in lateral (left) and top views (right). A black diamond marks the GluA2-A2 contact point. A schematic of the O-shape in top view is shown on the extreme right, with the four chains (A-D) indicated. GluA2, GluA3 and GluA4 are colored blue, red and green respectively. Tetrameric assembly regions 1-3 are boxed. (D) Top view of the classical N-shape arrangement found in GluA2 homomers (PDB code 3KG2). Two shades of blue are used to distinguish the different positions, according to panel (A).
Here we use X-ray crystallography and single-particle electron cryo-microscopy (cryo-EM), in combination with electrophysiology, biochemistry and coarse-grained simulations, to decipher the organization of GluA2-containing AMPAR heteromers.
Crystal structures and organization of the GluA2/3 and GluA2/4 NTD layers
Towards resolving the organization of AMPAR heteromers we first targeted the NTD layer for structural analysis. This membrane-distal domain encompasses 50% of the receptor mass and reaches midway into the synaptic cleft, where it forms a key platform for protein interactions and mediates AMPAR clustering (10, 11) (Fig. 1A). The bi-lobate NTD ‘clamshell’ contributes powerful allosteric modulation in NMDARs and thus represents a key drug target (12).
This highly sequence-diverse domain also initiates iGluR assembly (13–15). Although structures for the obligatory heteromeric NMDAR and KAR NTDs exist (16, 17), AMPAR NTD heteromers have remained elusive due to their preferential rather than obligatory heteromeric nature (14, 18). AMPAR NTDs form tight homodimers (nanomolar Kd) (18), which have been crystallized previously (14, 15). Here we developed a strategy to crystallize AMPAR NTD heteromers from purified homomers (supplementary methods) and tested all combinations harboring the critical GluA2 subunit.
We obtained crystals of the GluA2/3 and GluA2/4 NTDs and determined their structures at 2.1 Å and 2.5 Å resolution, respectively (Table S1). The overall structure of both heterodimers is similar to most AMPAR NTD homodimers (root mean square deviation [RMSD] ~ 1 Å) with upper lobe (UL) and lower lobe (LL) engaging a dimer interface of ~ 1400 Å2 (Fig. 1B and fig. S1). By contrast, GluA3 NTD homodimers deviate substantially (average RMSD ~ 2.5 Å), being destabilized by like-charge repulsion between Arg163 in the LLs (fig. S1, B and C) (19). In GluA2/3, salt bridges between GluA3-Arg163 and GluA2-Asp145 and H-bonds between GluA3-Arg184 and GluA2-Ala148 and GluA2-Glu149 zip up the heterodimer, readily explaining its higher affinity than GluA3 homodimers (fig. S1, A to C) (18). The structures underscore that differences in the LL interface are key determinants of selective heteromerization; UL interactions are mostly conserved between AMPAR paralogs.
Both NTD heterodimers assemble into a novel tetrameric arrangement (Fig. 1C). While GluA2 homomers adopt a loose ‘N’-shape when viewed from the top (Fig. 1D) (5), a compact ‘O’-shape with the four subunits alternating around a central axis is seen in the heteromers (Fig. 1C), an arrangement reminiscent of NMDARs (6, 7). In both tetramers, GluA2 comes to lie at specific positions, resulting in a diagonal alignment of the GluA2 loops that cap the dimer interface (Fig. 1C, yellow diamond).
The tetrameric interface harbors three major contact regions with most of the interactions mediated by the ULs (Fig. 1C and fig. S2). In region 1, the GluA2 back (helices J and K) engages the front face of the GluA3 and GluA4 NTDs (helix A, beta strand 2, loop after helix I), predominantly via polar and ionic contacts (Fig. 1C and fig. S2B). These are more extensive for GluA2/4 due to a more compact, near-parallel alignment of the two heterodimers along their vertical axis; in GluA2/3 the two dimers have separated at their LLs, resulting in a tilt angle of ~30° relative to GluA2/4 (fig. S3A). A key interface element is a loop emanating from helix I (‘Loop I’) in the non-GluA2 subunits, where an arginine (Arg265 in GluA3, Arg264 in GluA4) contacts a highly conserved glutamate (Glu288) on GluA2 helix J (Figs. 1C, fig. S2B). GluA2 helix K and the following hinge loop form additional ionic interactions with helix A and beta strand 2 in GluA3 and GluA4, which have been proposed to interact with N-cadherin in GluA2 (20). In region 2, diagonal contacts are mediated between the LLs of the non-GluA2 subunits via their helices F (fig. S2C), reminiscent of the GluN2B interactions in NMDARs (6, 7). Finally, in the more compact GluA2/4 heteromer, helix F interacts with GluA2 LL helix E (region 3). This alternative arrangement is expected to impact NTD interaction with synaptic proteins that mediate AMPAR clustering (10).
Mapping NTD interface contacts in full-length AMPAR heteromers
To validate this organization in intact AMPARs we introduced cysteines to crosslink region 1. The mutations Asn292Cys in GluA2myc and Arg265Cys in GluA3flag (Arg264Cys in GluA4flag) are expected to spontaneously crosslink the O-shape (Fig. 2, A and B). Isolation of the heteromers by Flag-immunoprecipitation followed by Western blotting of the partner subunit on non-reducing SDS-PAGE revealed crosslinked dimers only when both cysteine mutant subunits were present (Fig. 2, C and D; lane 3 versus lane 2). Similarly, diagonal crosslinking of GluA4 helices F in region 2 (Ser172Cys, Gln175Cys) produced spontaneous crosslinks of purified GluA2/4 heteromers (fig. S3B). These results support formation of the compact O-arrangement in intact AMPAR heteromers.
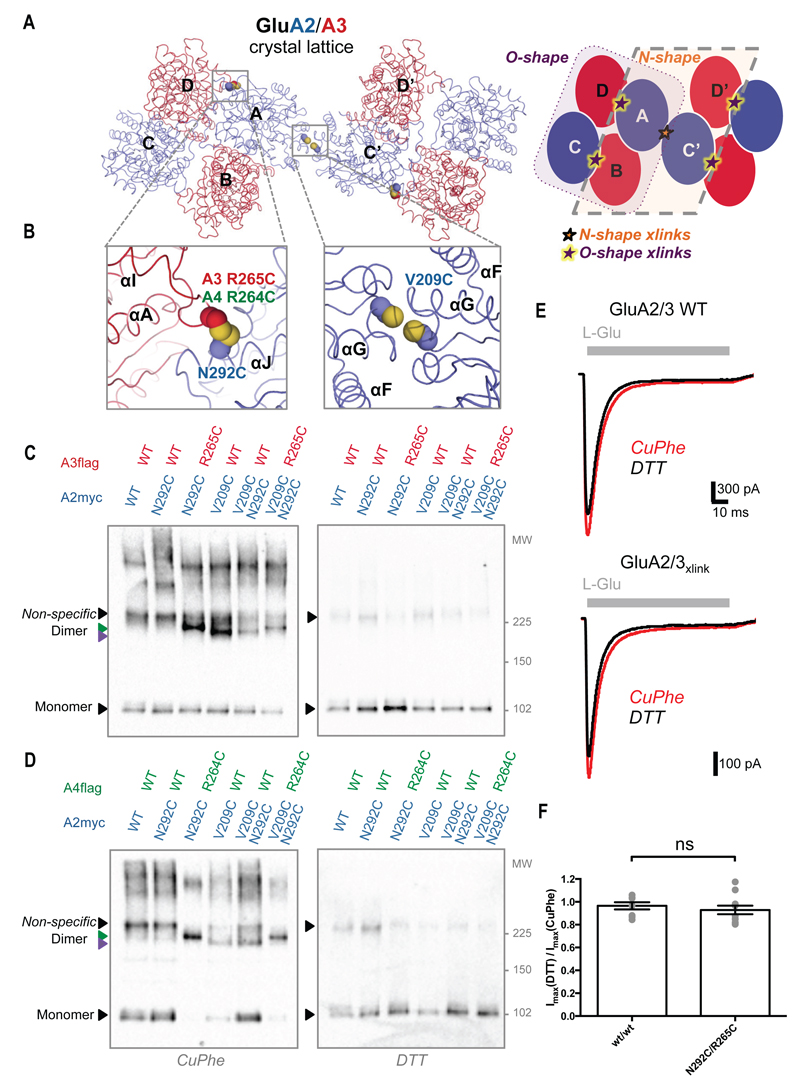
Fig. 2.
Probing the O-shape in full-length AMPARs with cysteine-crosslinking and electrophysiology. (A) The O- and N-shape arrangements coexist in the GluA2/3 crystal lattice with GluA2 in blue and GluA3 in red. N- and O-tetramers are highlighted in the schematic, their interfaces are depicted with stars. (B) Zooms of the boxed regions in A with cysteines introduced into the two tetrameric interfaces shown as spheres to mark the positions used for crosslinking. (C-D) Western blots showing crosslinked dimers for GluA2/3 (C) and GluA2/4 (D) on non-reducing SDS-PAGE (left panels). Green and purple arrowheads indicate N and O dimers. These disappear in the presence of DTT, non-specific dimers remain (right panels). Lanes 1 and 2: negative controls; lane 3: O-shape crosslinks; lane 4: N-shape crosslink, lane 5: N-shape crosslink control, and lane 6: combined N- and O-shape crosslinks. (E) Example traces showing AMPAR currents from HEK293T cells expressing GluA2/3 wt/wt (top) or N292C/R265C (bottom). Responses to 100 ms application of 10 mM glutamate were recorded in oxidizing (CuPhe; red) and reducing (DTT; black) conditions. (F) The peak current ratio between CuPhe and DTT for GluA2/3 wt/wt and N292C/R265C. No significant difference was found (P = 0.4883; unpaired t-test).
While GluA2/4 NTDs solely crystallized in the O-shape, the GluA2/3 crystal lattice also contains N-tetramers (Fig. 2A). N-shaped GluA2 homotetramers can be crosslinked via the Val209Cys mutation (Fig. 2B) (5). Crosslinking of GluA2-Val209Cys/GluA3 wild-type (wt) receptors suggests that the N-arrangement can also occur in heteromers (Fig. 2C; lane 4), with GluA2 at the crosslinked NTD central position and GluA3 at the periphery. This observation could be extended to the GluA2-V209Cys/GluA4-wt heteromer (Fig. 2D; lane 4). Two possible mechanisms may underlie the O-arrangement: i) lateral interactions between N-shape receptors or ii) an alternative NTD tetramer configuration within receptors. To resolve this question we generated a GluA2 double mutant with cysteines in both the N and O tetrameric interfaces. When combining GluA2-Val209Cys/Asn292Cys with GluA3-Arg265Cys, crosslinked dimers would appear if alternative conformers formed within receptors, whereas clustering of receptors through the O-shape arrangement would give rise to higher molecular weight aggregates. Consistent with the former, we observe crosslinked dimers, but not larger complexes, on SDS-PAGE (Fig. 2, C and D; lane 6) and tetramers on native-PAGE (fig. S3C), supporting the existence of alternative arrangements in the AMPAR NTD layer. Rearrangements of the NTD layer have been linked previously to the gating cycle (21–23).
N-crosslinked dimers migrate faster on SDS-PAGE than O dimers, permitting us to distinguish between the two (Fig. 2, C and D; lanes 3 and 4, purple and green arrowheads, respectively). The heteromer containing both mutations in GluA2 (permitting the N- or O-crosslinks) migrated at the level of the O and not the N dimer, in both GluA2/3 and GluA2/4 (Fig. 2, C and D; lane 6); this behavior may indicate that the O-arrangement forms preferentially.
Characterization of the O-shaped GluA2/3 receptor
Negative-stain single particle EM
To further validate this new conformation in intact GluA2/3, O-crosslinked heteromers (GluA2/3xlink) were isolated via tandem affinity purifications and subjected to negative-stain single-particle EM (fig. S4; supplementary methods). 3D-reconstruction yielded a map at ~ 20 Å resolution with a compact O-shaped NTD layer; the GluA2/3 tetrameric crystal structure could be fitted into the EM density unambiguously (fig. S4C). Of note, a similar NTD organization was observed in 3D reconstructions of GluA2 homomers, although the rest of the receptor differed (24).
Agonist-induced desensitization triggers rearrangements in the LBD region (25). Recent AMPAR structures reveal complete rupture of both LBD dimers upon desensitization and imply that this is coupled to a rearrangement of the NTD layer (21–23). We likewise observe heterogeneity and rupture of the LBD layer in response to saturating L-glutamate, despite trapping the NTDs in the O-shape (fig. S5, A to C). Hence a compact NTD layer permits LBD dynamics and is compatible with gating transitions. Reducing the crosslinks with 10 mM DTT prior to the glutamate pulse resulted in additional heterogeneity in the NTD layer as seen in 2D averages (fig. S5D), revealing that the receptor can transit from the O-shape.
Coarse-grained simulations
Transitions of the NTD layer between N and O conformations were also evident in coarse-grained simulations using the anisotropic network model (ANM). The ANM calculates accessible conformational changes or global modes of motion, with the first modes describing the energetically most favorable motions (26) (supplementary methods). One low energy mode of a GluA2 homomer (PDB 3KG2), previously shown to vertically compress the AMPAR to an NMDAR-like conformation (mode 4) (27), also enables a transition from the N-shaped to an O-like NTD arrangement (fig. S6, left, and Movie 1). Moreover, ANM analysis of the GluA2/3 NTD crystal structure revealed a pathway for the reverse transition from O to N (fig S6, right; Movie S1), further suggesting that AMPARs can populate both conformations.
Movie 1.
ANM mode 4 of a GluA2 homomer showing an N to O transition.
Formation of a compact NTD arrangement resembling the GluA2/3 heteromer is observed in a simulation of a GluA2 homomer (PDB 3KG2); this is accompanied by a separation of the LBD dimers and a vertical compression of the receptor towards an c (27). The anisotropic network model creates springs between interacting residues (Cα within 15 Å) and therefore the lower lobe inter-dimer interface contacts are maintained. Strain within this interface is apparent and rearrangement of this region likely follows (see Movie S1).
Functional characterization
To test the functional impact of the O-shape we expressed GluA2/3 or GluA2/4 heteromers in HEK293T cells and recorded whole cell current responses to rapid glutamate application in oxidizing (copper phenanthroline; CuPhe) or reducing (dithiothreitol; DTT) conditions. Peak currents were similarly redox-sensitive in O-crosslink mutants and wt heteromers (Fig. 2, E and F, and fig. S7, A, B and E). The near equality in peak amplitude ratio (ImaxDTT/ImaxCuPhe) of mutants (crosslinked at either region 1 or region 2; Fig. 1C and fig. S3B) to wt heteromers suggests that the receptors trapped in this conformation remain functional.
Desensitization kinetics and recovery from desensitization showed no greater effect of the oxidizing/reducing treatment in the two O-crosslink heteromers compared to the respective wild types (Fig. 2E and figs. S7, C, D, F and G, and S8 and data not shown); together with the negative stain data (fig. S5), this suggests that rupture of the NTD layer (21–23) is not a prerequisite for desensitization.
Cryo-EM structure of a GluA2/3 AMPAR heteromer
Structure determination
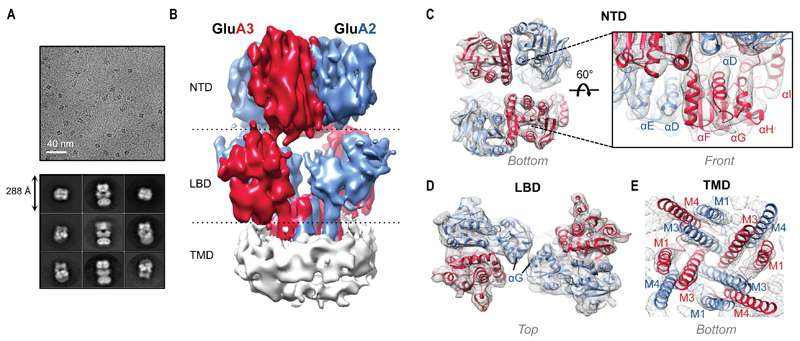
To determine the molecular organization of GluA2/3, we recorded a cryo-EM dataset of GluA2/3xlink in a ligand-free (apo) state using a Titan Krios microscope equipped with a Gatan K2 Summit® electron detector (supplementary methods). The direct electron detector allowed us to collect individual sub-frames and correct for beam-induced motion, while 3D classification using maximum likelihood methods was instrumental for isolating different conformations of the sample (28, 29). 3D classification of 107,939 motion-corrected particles using RELION (30) resulted in three classes with O-shaped NTDs (73,885 particles), one class (16,697 particles) resembling the classical Y-shaped receptor seen in GluA2 homomers (N-shaped when viewed from the top), and one class without AMPAR-like features (fig. S9). While processing of Y-shape data did not yield a model of sufficient quality, additional steps of 3D classification improved the O-shaped classes, especially at the heterogeneous LBD layer, resulting in two final models: M1 and M2. These models differed in both the orientation of the NTD tetramer and the arrangement of the LBDs (Figs. 3 and 4 and figs. S10 and S11). Refinement led to improved maps at 8.25 Å resolution for M1 and 10.3 Å for M2, allowing us to identify individual domains and secondary structure elements, with clear density within for the NTD, LBD and most TMD helices (Fig. 3 and fig. S10B). Our initial discussion focuses on M1.
Fig. 3.
Cryo-EM structure of GluA2/3x-link. (A) Sample micrograph at 3.5 μm defocus and a nominal magnification of 28,409x with representative 2D class averages shown below. (B) 8.25 Å resolution map of GluA2/3 M1 with GluA3 red, GluA2 blue, and micelle white. (C) Sample fitting of the NTD bottom (left) showing individual density for secondary structure elements. The front view (right) shows a zoom onto the GluA3 NTD lower lobe front face (αF-H). Individual protomers were fitted (GluA2: PDB 3HSY chain B; GluA3: PDB 3O21 chain D). (D) Sample fitting of the individual LBD protomers (GluA2: PDB 1FTO chain B; GluA3: PDB 3UA8). (E) Fitting of the TMD from a GluA2 homomer (PDB 3KG2). Good density was obtained for the gate helices but not for the re-entrant pore loops, which were omitted.
Fig. 4.
Ligand-free GluA2/3 in ‘resting’ and ‘desensitized’ conformations. (A) Model 1 (M1) of GluA2/3xlink with a view onto an LBD dimer and the NTD inter-dimer interface (left panel). The LBD dimer interface is intact and is mediated by the backs of the LBD upper lobes (boxed). In an equivalent view onto M2 (right panel), one LBD dimer has ruptured (chains B and C; boxed), adopting a state seen in desensitized structures (21). (B) Top views onto the LBD layers of M1 (left) and M2 (right). In M1 both LBD dimers are intact, ‘resting’ (R). In M2, chains A and D form an R dimer and chains B and C are ‘desensitized’ (D).
Overall architecture of GluA2/3 (M1)
Contrary to GluA2 homomers, the heteromer is strikingly contracted along its vertical axis (Fig. 3B and 5). A compact NTD assembly closely nests between widely separated LBD dimers, deviating from the loosely arranged (Y-shaped) GluA2 homomer (5) and approaching an NMDAR-like architecture (6, 7) (Fig. 5B). We performed rigid-body fitting of nine independent crystal structures into the M1 density, resulting in a model composed of two GluA2 NTD protomers, two GluA3 NTD protomers, four LBD protomers and the TMD of GluA2 (PDB 3KG2) (Fig. 3, C to E, and fig. S10). Although the NTD protomers were fitted independently, the resulting tetramer closely resembles our NTD crystal structure (RMSD ~ 3 Å). The distance between the Cβ of GluA2 Asn292 and GluA3 Arg265 is ~ 6 Å, thus marking the position of the introduced disulfide bond. Importantly, we observed clear density for the GluA3-specific N-glycosylation site at Asn35 (fig. S10, A and B), confirming the positioning of the GluA2 and GluA3 subunits in the receptor in agreement with the NTD crystal structure (Fig. 1C).
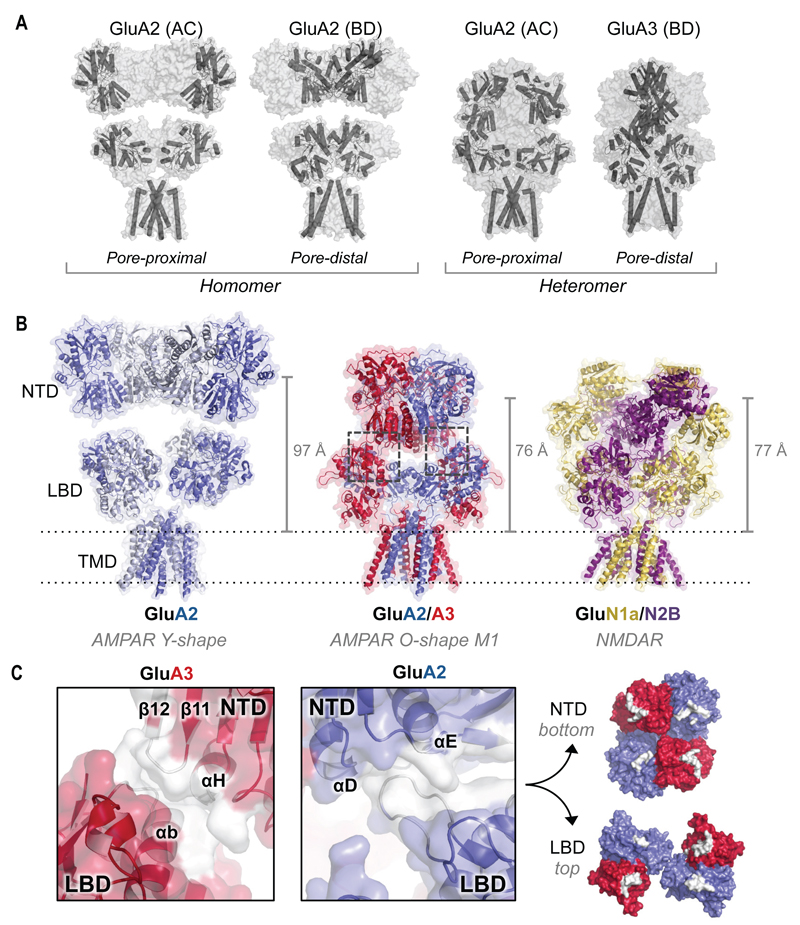
Fig. 5.
Architecture of GluA2/3 M1. (A) Distinct conformations of the non-equivalent chain pairs (AC and BD) in the Y-shaped GluA2 homomer (PDB 4UQJ; left) and the O-shaped GluA2/3 heteromer (right). (B) The GluA2/3 heteromer (middle) exhibits a substantial vertical compression relative to GluA2 homomers (PDB 4UQJ; left), akin to NMDARs (PDB 4PE5; right). Compression of the GluA2/3 ECR is indicated by the center of mass distance between the NTD and Thr625 (vertical lines on the right). (C) NTD-LBD contact regions for GluA2 and GluA3 are shown in white (boxed in panel B). The protein backbone is depicted in cartoon representation within a transparent surface. Right panel: surface view of the inter-layer interface; contacts formed between the NTD and LBD are shown in white.
The diagonal subunit pairs in GluA2 homomers, AC and BD, are conformationally distinct (Figs. 1A and 5A), which is expected to be of functional consequence (5). In particular, the linker connecting the LBD to TM3 in the BD pair is predicted to have a greater influence on gating (5, 31). In our structure, GluA2 occupies the AC and GluA3 the BD position. As a consequence, GluA3 is placed pore-distal at the level of the LBD and TMD while GluA2 is pore-proximal (Fig. 5A). This positioning appears not to be strict in AMPARs as GluA2 can also locate to the BD position according to the GluA2/3 crystal lattice (Fig. 2A) and the crosslinking data (Fig. 2C). Hence, rules underlying subunit placement in preferential iGluR heteromers may be relaxed (14), contrary to obligatory heteromers (6, 7, 17).
NTD-LBD contacts are apparent in the compact heteromer
One of the most striking features of the heteromer is the vertical compression relative to existing AMPAR homomer structures (Fig. 5, A and B). To describe this difference in M1 we used the center of mass (COM) distance between the NTD layer and the gate residue Thr625 within TM3 as a metric (21). This distance was reduced by 21 Å when comparing GluA2/3 M1 (76 Å) to the antagonist-bound GluA2 cryo-EM structure (97 Å; Fig. 5B) (22). The two LBD dimers of the heteromer are splayed apart to accommodate the compact NTD assembly, which partially fills the inter-layer cavity characteristic of GluA2 homomers.
The compression gives rise to novel interfaces between the NTD and LBD layers (Fig. 5C), a region pivotal to allosteric signal transmission in NMDARs, triggered by NTD ligands (6, 7, 12). The inter-layer contact points are approximately equidistant in all chains of M1 (Fig. 5, B and C) but are more extensive in the BD chains (GluA3). In GluA2, a loop (Ile444-Gly462) in the LBD projects towards helices D and E at the backside of the NTD. The GluA3 NTDs wedge into the LBD inter-dimer interface, with NTD helix H contacting LBD helix B. These GluA3 contacts are also seen in M2, where GluA2 interactions are lost (Fig. 4A and fig. S11). By contrast, complete separation of the ECR layers is apparent in GluA2 cryo-EM structures (22) (Fig. 5B) and in a crystal structure of a GluA2-snail toxin complex, where the toxin pushes the NTD away (31). Such a separation between the NTD and LBD layers may also occur in the O-shaped receptors as revealed by ANM mode 6 of GluA2/3 M1 (fig. S12 and Movie S2), further highlighting the flexible nature of the AMPAR ECR. These interlayer dynamics may alter AMPAR interactions at synapses and enable allosteric modulation by the NTD akin to NMDARs.
GluA2/3 in a desensitized state (M2)
Glutamate binding to the LBD (Fig. 1A, star) triggers conformational changes that result in activation and desensitization. Individual LBDs can gate the receptor in response to agonist and occupation of a single subunit triggers desensitization (32, 33). The two loosely associated, back-to-back dimers rupture upon desensitization in AMPARs and KARs (22, 25, 34). This dynamic architecture underlies the speed of AMPAR gating (25, 35).
Heterogeneity in our O-shape classes largely results from mobility of the LBD layer. Dramatic rearrangements of the LBD layer are apparent in M2; the BC dimer has completely ruptured whereas the AD dimer remains intact, characteristic of the resting/non-desensitized state (Fig. 4 and fig. S11). The structure mimics agonist-bound, desensitized GluA2 (21) and provides structural evidence that the receptor can transit into desensitized-like states in the nominal absence of agonist (32, 35, 36). Consistent with this, M1 could be fitted best with agonist-free, open-cleft LBD conformations (PDB 1FTO and 3UA8; fig. S10C).
Rearrangements of the LBD gating machinery in M1
Rearrangements in the GluA2/3 LBD layer are also seen in M1, which exhibits intact dimers (Fig. 6, A and B, and fig. S11D). The interaction between LBD dimers changes during gating and their tilting apart has been linked to activation (Fig. 6A) (21, 22). In GluA2/3 M1, the angle between the LBD dimers has opened, substantially deviating from the GluA2 ligand-free receptor and surpassing the activated-state conformation (by 15°) (Fig. 6A). ANM-based simulations suggest that this separation is associated with ‘NTD sinking’ (M1 mode 6) (fig. S12 and Movie S2). Together, these data raise the possibility that NTD-LBD interlayer dynamics impact the gating machinery.
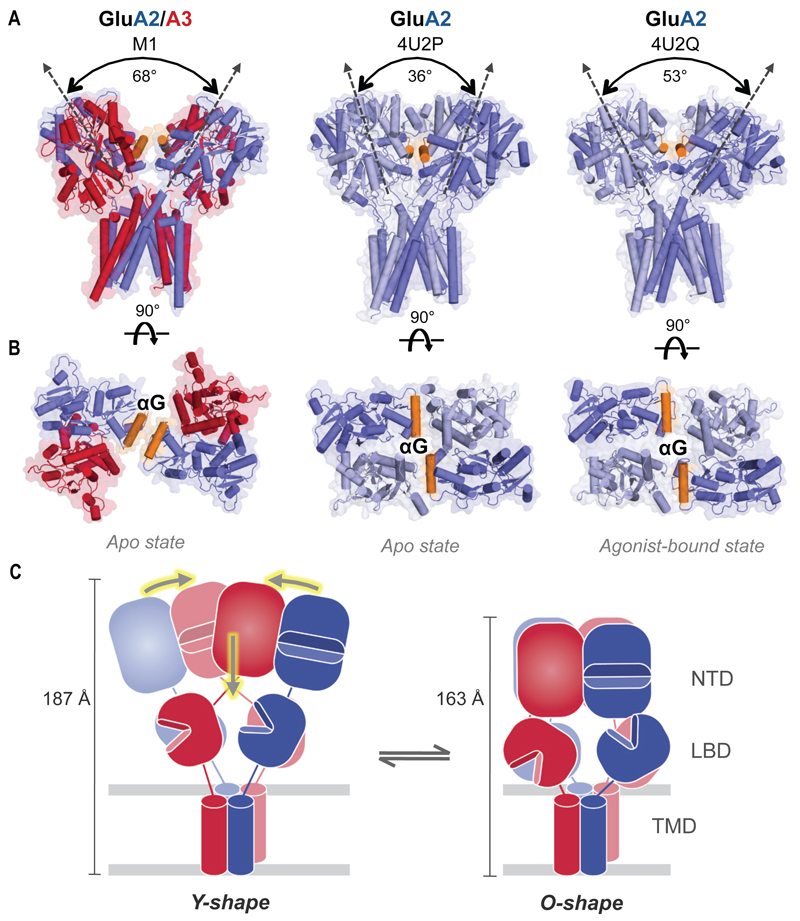
Fig. 6.
Novel conformation of the LBD layer in GluA2/3 M1. (A) Front view onto the GluA2/3 LBD and TMD layers (left), which is more open and reaches down towards the TMD as compared to the ligand-free and agonist-bound GluA2 crystal structures (PDB 4U2P, middle and 4U2Q, right). The roll angle, measured between the two-fold pseudosymmetry axes of the two LBD dimers, is indicated. (B) Top view onto the two LBD dimers reveals a novel, parallel approximation of helices G (orange) in chains A and C of GluA2 (blue subunit). In contrast, the ligand-free (apo) homomer (PDB 4U2P) exhibits a head-to-head placement of these helices forming a ‘gating ring’, which opens further in the agonist-bound structure (PDB 4U2Q). This ring is closed in GluA2/3. (C) Models of an AMPAR Y-shape (left) characteristic of GluA2 homomers and an O-shape (right) as seen for GluA2/3xlink. Double, grey arrows indicate transition between the Y-shape and O-shape receptors, as also suggested by the ANM simulations. Yellow arrows show potential movements permitting this interconversion: horizontal arrows compact the NTD layer towards an O-shape, while vertical arrows indicate vertical compression of the receptor tetramer. Vertical heights of the receptors are indicated on their right sides.
When viewing the LBD layer from the top we see an entirely new arrangement. The channel-active state has been suggested to be accompanied by an iris-like opening of the LBD tetramer interface, formed between helices G of chains A and C (21, 22). These helices, which are arranged ‘head-on’ in the activated (and resting) states of GluA2 homomers, come to lie parallel in GluA2/3 M1, essentially closing the ‘LBD gating ring’ (Fig. 6B and Movie 2). This suggests that the receptor can access a multitude of conformations with both a separation of the LBD dimers and an open gating ring required for activation.
Movie 2.
Morph showing conformational changes in the LBD layer revealed by GluA2/3 model 1.
The morph command in PyMOL was used to generate a series of intermediate conformations between the LBD layer of a GluA2 homomer (PDB 4U2P) and that of GluA2/3 model 1. A rotation of the two dimers about the inter-dimer interface is apparent resulting in an approximation of helices G in chains A and C (orange). This is accompanied by an opening of the roll angle between the LBDs.
Conclusion
Here we present the first view of AMPAR heteromers containing the GluA2 subunit, the prevailing variety in the brain. We delineate the most sequence-diverse subunit interface at atomic resolution and show that heterodimers associate to form a tetramer with the four subunits alternating around the pore axis. Although GluA2 occupies the pore-proximal (AC) position, subunit placement does not appear to follow strict rules, contrasting with obligatory iGluR heteromers (6, 7, 17). Whether and how these ‘relaxed’ assembly rules impact signaling in preferential heteromeric iGluRs (AMPARs and low-affinity KARs) is now an open question.
Ligand-free GluA2/3 was captured in resting (M1) and desensitized (M2) conformations, providing structural evidence that the apo receptor can transit between states. Overall, its architecture departs from the classic Y-shape and approximates NMDARs. In conjunction with ANM simulations we describe a conformational trajectory, with the NTDs transiting from an N-shape into a compact O-assembly, which is associated with a vertical compression (Fig. 6C and Movie 1). This receptor conformation could be of functional consequence. First, the interfaces resulting from approximation of the NTD and LBD could permit allosteric coupling within the ECR and provide a novel substrate for AMPAR therapeutics, analogous to NMDARs (12). Second, since the NTD mediates AMPAR clustering at synapses and dendritic spine dynamics (11, 20), transition from N to O will alter a strategic docking platform for synaptic interaction partners, including pentraxins (11) and AMPAR auxiliary subunits (37). The GluA2/3 structure opens avenues to further understanding of AMPAR signal transmission.
One sentence summary: 3D structures of a GluA2/3 AMPA receptor heteromer
Supplementary Material
Materials and Methods
Figures S1 to S12
Tables S1 and S2
Movies S1 to S2
References 38 to 68
Acknowledgements
We thank Ole Paulsen, Andrew Penn and Mark Farrant for comments on the manuscript. We also acknowledge Paula da Fonseca and Sjors Scheres for helpful comments regarding cryo-EM, Nigel Unwin for carbon-coated grids and Christos Savva for help with the EM. We thank the staff at the Diamond Light Source (beamline I04-1) for provision of synchrotron facilities. BH, J G-N, OC, R F-L, JK, HH and IHG were supported by the Medical Research Council (MC_U105174197). Coordinates and structure factors for the NTD have been deposited in the protein data bank, GluA2/4 has PDB ID 5FWX and GluA2/3 has PDB ID 5FWY. The cryo-EM density maps representing GluA2/3 Model 1 and GluA2/3 Model 2 have been deposited in the EM-databank with accession numbers EMD-8090 and EMD-8091 respectively and the corresponding models in the protein data bank are PDB 5IDE and PDB 5IDF.
References
- 1.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62(3):405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowie D. Ionotropic glutamate receptors & CNS disorders. CNS Neurol Disord Drug Targets. 2008;7(2):129–143. doi: 10.2174/187152708784083821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: the last 25 years. Neuron. 2013;80(3):704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isaac JT, Ashby M, McBain CJ. The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron. 2007;54:859–871. doi: 10.1016/j.neuron.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344(6187):992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CH, et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511(7508):191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat Neurosci. 2014;17(7):914–922. doi: 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansour M, Nagarajan N, Nehring RB, Clements JD, Rosenmund C. Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron. 2001;32(5):841–853. doi: 10.1016/s0896-6273(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Nafria J, Herguedas B, Watson JF, Greger IH. The dynamic AMPA receptor extracellular region: a platform for synaptic protein interactions. J Physiol. 2016 doi: 10.1113/JP271844. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sia GM, et al. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55(1):87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Zhu S, Paoletti P. Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Curr Opin Pharmacol. 2015;20C:14–23. doi: 10.1016/j.coph.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Hansen KB, Furukawa H, Traynelis SF. Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol. 2010;78(4):535–549. doi: 10.1124/mol.110.067157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herguedas B, Krieger J, Greger IH. Receptor heteromeric assembly-how it works and why it matters: the case of ionotropic glutamate receptors. Prog Mol Biol Transl Sci. 2013;117:361–386. doi: 10.1016/B978-0-12-386931-9.00013-1. [DOI] [PubMed] [Google Scholar]
- 15.Kumar J, Mayer ML. Functional Insights from Glutamate Receptor Ion Channel Structures. Annu Rev Physiol. 2013 doi: 10.1146/annurev-physiol-030212-183711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475(7355):249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar J, Schuck P, Mayer ML. Structure and assembly mechanism for heteromeric kainate receptors. Neuron. 2011;71(2):319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossmann M, et al. Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers. EMBO J. 2011;30:959–971. doi: 10.1038/emboj.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sukumaran M, et al. Dynamics and allosteric potential of the AMPA receptor N- terminal domain. EMBO Journal. 2011;30(5):972–982. doi: 10.1038/emboj.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saglietti L, et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54(3):461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Durr KL, et al. Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States. Cell. 2014;158(4):778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyerson JR, et al. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514(7522):328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433(7025):545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 24.Midgett CR, Madden DR. The quaternary structure of a calcium-permeable AMPA receptor: conservation of shape and symmetry across functionally distinct subunit assemblies. J Mol Biol. 2008;382(3):578–584. doi: 10.1016/j.jmb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 26.Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: bridging between structure and function. Annu Rev Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutta A, et al. Cooperative dynamics in intact AMPA and NMDA glutamate receptors – similarities and subfamily-specific differences. Structure. 2015;23(9):1692. doi: 10.1016/j.str.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40(1):49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Liao M, Cao E, Julius D, Cheng Y. Single particle electron cryo-microscopy of a mammalian ion channel. Curr Opin Struct Biol. 2014;27:1–7. doi: 10.1016/j.sbi.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Durr KL, Gouaux E. X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science. 2014;345(6200):1021–1026. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robert A, Howe JR. How AMPA receptor desensitization depends on receptor occupancy. J Neurosci. 2003;23:847–858. doi: 10.1523/JNEUROSCI.23-03-00847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- 34.Schauder DM, et al. Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proc Natl Acad Sci U S A. 2013;110(15):5921–5926. doi: 10.1073/pnas.1217549110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plested AJ, Mayer ML. AMPA receptor ligand binding domain mobility revealed by functional cross linking. J Neurosci. 2009;29(38):11912–11923. doi: 10.1523/JNEUROSCI.2971-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez J, Du M, Parameshwaran K, Suppiramaniam V, Jayaraman V. Role of dimer interface in activation and desensitization in AMPA receptors. Proc Natl Acad Sci U S A. 2010;107(21):9891–9896. doi: 10.1073/pnas.0911854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cais O, et al. Mapping the interaction sites between AMPA receptors and TARPs reveals a role for the receptor N-terminal domain in channel gating. Cell Rep. 2014;9:1–13. doi: 10.1016/j.celrep.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reeves PJ, Kim JM, Khorana HG. Structure and function in rhodopsin: a tetracycline-inducible system in stable mammalian cell lines for high-level expression of opsin mutants. Proc Natl Acad Sci U S A. 2002;99(21):13413–13418. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longo PA, Kavran JM, Kim MS, Leahy DJ. Transient mammalian cell transfection with polyethylenimine (PEI) Methods Enzymol. 2013;529:227–240. doi: 10.1016/B978-0-12-418687-3.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seiradake E, Zhao Y, Lu W, Aricescu AR, Jones EY. Production of cell surface and secreted glycoproteins in mammalian cells. Methods Mol Biol. 2015;1261:115–127. doi: 10.1007/978-1-4939-2230-7_6. [DOI] [PubMed] [Google Scholar]
- 41.Winter G, Lobley CM, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 45.Dutta A, Shrivastava IH, Sukumaran M, Greger IH, Bahar I. Comparative Dynamics of NMDA- and AMPA-Glutamate Receptor N-Terminal Domains. Structure. 2012;20(11):1838–1849. doi: 10.1016/j.str.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K. Gctf: real-time CTF determination and correction. J Struct Biol. 2015 doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3(6):977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Suhre K, Navaza J, Sanejouand YH. NORMA: a tool for flexible fitting of high-resolution protein structures into low-resolution electron-microscopy-derived density maps. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):1098–1100. doi: 10.1107/S090744490602244X. [DOI] [PubMed] [Google Scholar]
- 59.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 60.Orain D, et al. 6-Amino quinazolinedione sulfonamides as orally active competitive AMPA receptor antagonists. Bioorg Med Chem Lett. 2013;22(2):996–999. doi: 10.1016/j.bmcl.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 61.DeLano WL. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 62.Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 63.Robert A, Armstrong N, Gouaux JE, Howe JR. AMPA receptor binding cleft mutations that alter affinity, efficacy, and recovery from desensitization. J Neurosci. 2005;25:3752–3762. doi: 10.1523/JNEUROSCI.0188-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atilgan AR, et al. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80(1):505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu C, Tobi D, Bahar I. Allosteric changes in protein structure computed by a simple mechanical model: hemoglobin T<-->R2 transition. J Mol Biol. 2003;333(1):153–168. doi: 10.1016/j.jmb.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 66.Eyal E, Lum G, Bahar I. The anisotropic network model web server at 2015 (ANM 2.0) Bioinformatics. 2015;31(9):1487–1489. doi: 10.1093/bioinformatics/btu847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27(11):1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.