Abstract
Nitric oxide (NO) has a highly diverse range of biological functions from physiological signaling and maintenance of homeostasis to serving as an effector molecule in the immune system. Many of the dichotomous effects of NO and derivative reactive nitrogen species (RNS) can be explained by invoking precise interactions with different targets as a result of concentration and temporal constraints. Endogenous concentrations of NO span five orders of magnitude, with levels near the high picomolar range typically occurring in short bursts as compared to sustained production of low micromolar levels of NO during immune response. This article provides an overview of the redox landscape as it relates to increasing NO concentrations, which incrementally govern physiological signaling, nitrosative signaling and nitrosative stress-related signaling. Physiological signaling by NO primarily occurs upon interaction with the heme protein soluble guanylyl cyclase. As NO concentrations rise, interactions with nonheme iron complexes as well as indirect modification of thiols can stimulate additional signaling processes. At the highest levels of NO, production of a broader range of RNS, which subsequently interact with more diverse targets, can lead to chemical stress. However, even under such conditions, there is evidence that stress-related signaling mechanisms are triggered to protect cells or even resolve the stress. This review therefore also addresses the fundamental reactions and kinetics that initiate signaling through NO-dependent pathways, including the chemistry of RNS and their molecular targets.
Keywords: nitric oxide, nitric oxide synthase, breast cancer, cell signaling, cancer biology
Graphical Abstract
I. Introduction to the Principles of the Redox Landscape
The chemical biology of a number of elements such as oxygen, nitrogen and sulfur hinges on interconversion between redox-related species. Respiration for example depends on the controlled four-electron reduction of molecular oxygen to water through superoxo, peroxo (putatively) and oxo intermediates. Similarly, the nitrogen cycle, in which molecular nitrogen is converted into ammonia and nitrate to facilitate nitrogen assimilation, generates intermediate species both more oxidized and reduced than N2. Another example is thiol-disulfide cycling, which has long been known to impact protein structure and function and to maintain cellular redox homeostasis. Respiration, the nitrogen cycle and many other processes are dependent upon the well-known redox activity of metals. The functions of a number of organic cofactors, such as ascorbate, quinones and NADH, also rely on redox processes. The redox landscape can thus be defined as the sum of all redox active molecules in a specific environment such as the biosphere, the ocean or an individual organism, organ, cell or subcellular compartment.
Redox biology governs many aspects of prokaryotic and eukaryotic metabolism. In addition, modification of biological molecules by free radicals and other redox active species has long been considered to be integral to numerous pathological mechanisms. For instance, there is substantial evidence that under inflammatory conditions, abnormal production of redox active molecules can induce stress conditions, cell death and tissue injury [1]. However, redox active molecules also serve as signaling agents for control of a variety of physiological functions as well as coordination of inflammatory and tissue restoration mechanisms [2]. Furthermore, it is now apparent that redox signaling, particularly during inflammation, has significant potential to positively impact disease etiology. This review is focused on the signaling aspects of the redox landscape, particularly with respect to inflammatory conditions.
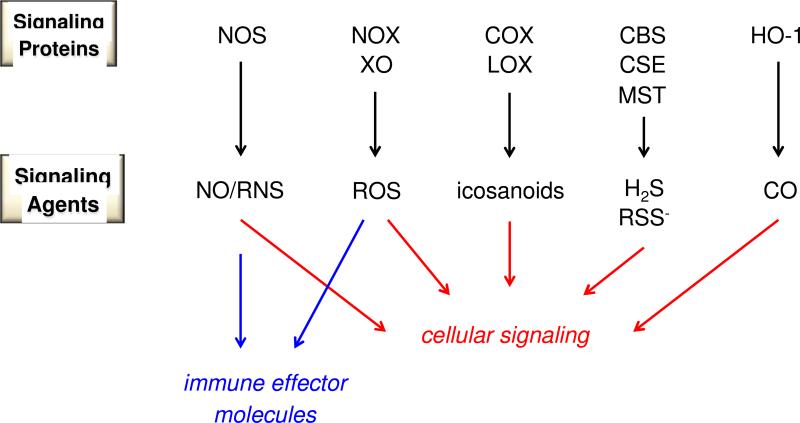
Intracellular communication and immune response are mediated by at least five classes of endogenously generated reactive small molecules (Figure 1): nitric oxide (NO) and reactive nitrogen oxide species (RNS), reactive oxygen species (ROS), eicosanoids and reactive lipid species (formed by oxidation of fatty acids), a variety of sulfur-based species such as hydrogen sulfide (H2S), thiols and disulfides and carbon monoxide (CO; formed from oxidation of heme [3]. Small molecules such as CO, H2S and eicosanoids are currently considered to primarily serve regulatory functions. In contrast, numerous studies describe deleterious as well as beneficial roles of NO, its related RNS and ROS. During pathogenic challenge or neoplastic development, ROS and NO/RNS serve as one arm of host defense and can be generated with high spatial focus. However, despite their anti-pathogen response, these species play vital roles in normal physiology, immunoregulation and coordination of tissue restoration. Electrophilic lipids, which serve as inhibitors of inflammation and initiators of apoptosis (see [4]), also can mediate pathophysiological conditions such as neurodegeneration [5].
Figure 1.
Components of the redox landscape. Cellular signaling includes physiological, stress-related and return to homeostasis signaling.
Within the context of the redox inflammatory landscape, both signaling and stress must be considered. While redox-induced stress is a result of extensive and often indiscriminate chemical modification of bioorganic molecules, redox signaling involves modification of individual targets that elicit specific biological cascades and result in precise phenotypic responses. Physiological signaling involves general maintenance of homeostasis and regulation of normal cellular function. On the other hand, stress-related signaling is initiated by sentinels that sense perturbations in normal homeostasis such as lack of oxygen or nutrients, the presence of pathogens, cellular modifications, mitochondria dysfunction and DNA damage. Stress signaling subsequently triggers mechanisms to protect the cell from such challenges, to regulate the inflammatory response or to resolve the stressful incident and restore homeostasis. Stress signaling can result either in cell protection or death and often involves the coordination of different types of cells.
To more fully define redox stress versus signaling, an understanding of the chemistry with respect to reaction conditions is necessary. Stress is traditionally defined by uncontrolled modifications of biomolecules, especially DNA and proteins, once natural cellular defenses, such as antioxidant systems, are overwhelmed [6]. Therefore, stress often results from exposure to very high levels of reactive species (in the stoichiometric sense), generally for prolonged periods of time, such that damage can accumulate to a point that results in cell dysfunction or death. However, lower fluxes of the same reactive species can instead initiate beneficial processes that lead to cell survival. The major differences between normal and stress signaling are the nature (or degree) of the targets that are modified as a function of the concentration of reactive species involved in the signaling event.
For any reaction, thermodynamic factors control chemical outcome. For large biological molecules such as enzymes that have evolved to serve specific purposes via conformational changes, entropy is often the predominant thermodynamic contributor. In contrast, enthalpic changes upon formation of covalent bonds with specific targets often dictate the effects of small signaling agents. These two processes are often interrelated, as exemplified by coordination of NO to the heme center of soluble guanylyl cyclase (sGC), which allosterically regulates the distant active site (for example, [7]). It is critical however to not discount the importance of kinetic factors. If the rate of a reaction is slow, thermodynamic favorability may be irrelevant, particularly in the complex environment of a living system. Reaction rates, and biological outcome, depend on concentration as well as spatial and temporal factors. These factors can directly impact the interaction of reaction partners, but also can influence the formation of such reacting species.
Many redox signaling agents are produced enzymatically and oxidatively in highly controlled fashion by enzymes including NO synthase (NOS), NADPH oxidase (NOX), cyclooxygenase (COX), lipoxygenase (LOX), cystathionine-β-synthase (CBS) and heme oxygenase (HO) (Figure 1). In addition to inherent spatial and temporal control, different isoforms also provide concentration gradients. For instance, the NOX2 isoform is generally associated with production of high levels of superoxide (O2−) for pathogen control, but is also critical for signaling in the inflammatory cascade in myeloid cells [8]. In contrast, NOX1, 3, 4, 5 and 6 all produce lower fluxes of O2− and are considered to serve more regulatory roles [9]. Isoforms can also produce different signaling agents as exemplified by the COX enzymes; COX2 is induced under inflammatory conditions to produce PGE2 and prostacyclin (PGI2) while COX1 leads to a signaling cascade primarily through thromboxane A2 (TXA2) [10]. In addition, that LOX, HO-1 and HO-2 systems have similar inflammatory redox and regulatory roles indicates that nature has harnessed parallel mechanisms to mediate varied and specific functions.
Under normal physiological conditions, redox active signaling agents are generally produced at low concentrations for short durations in specific locations. Under such conditions, rapid reactions between signaling agents and targets provide selective kinetic advantages, limiting other signaling processes that may be deleterious to cellular function. This type of kinetic specificity, allowing a single target to be modified at very low doses of a signaling agent, is again well exemplified by binding of NO to sGC. The rate constant for association of NO to heme centers is typically very high relative to other potential targets [11], such that low nanomolar levels of NO are sufficient to activate sGC. The resulting amplification of cGMP production then stimulates a biological cascade leading to vasodilation (for example, [12]).
While many physiological processes occur on the millisecond to minute timescale, inflammatory conditions are generally in effect for much longer time periods. Acute inflammation, such as results from response to pathogens, ischemia/reperfusion or trauma, is mediated over several days. In contrast, chronic inflammatory diseases such as Alzheimer's disease and cancer require months, years or even decades to fully develop. Such diseases are often described as creating amplified inflammatory loops [13]. Under acute and chronic inflammation, both the levels and duration of redox signaling agent production can reduce the kinetic specificity that exists under normal physiological conditions, such that there is an increased contribution from slower signaling processes.
Redox active signaling agents are highly interrelated, such that it is challenging to focus exclusively on a single class or species. One clear example of the complexity arising from the interactions of small redox active molecules is the classification of peroxynitrite formed from association of NO with O2− as both an RNS and ROS. There are numerous excellent reviews emphasizing various aspects of redox signaling including the effects of specific reactive species (e.g., H2O2, peroxynitrite) or classes (e.g., lipids, RNS), interactions with specific targets (e.g., post-translational modification of thiols) and impacts on specific conditions (e.g., cancer, heart failure). Here, we correlate NO concentration with corresponding physiological and stress-related signaling cascades, with an emphasis on inflammation, wound healing and oncogenesis.
The chemical biology of NO is critically dependent on localized chemistry [2, 14]. Furthermore, the three isoforms of NOS together produce NO over a wide concentration and temporal range. The constitutive isoforms, endothelial and neuronal NOS (eNOS/NOS3 and nNOS/NOS1), produce low cellular levels of NO, generally for regulation of cGMP and subsequent events (for example, [15]). Under hypoxia, reduction of nitrite (NO2−) can provide an alternative, oxygen-independent source of low levels of NO (for example, [16]). Inducible NOS (iNOS/NOS2) is also constitutively expressed for kidney and lung function, but upon induction can produce sustained NO fluxes in the micromolar range for days [14]. Among the NOS isoforms, NOS2 is therefore unique in being able to produce NO over four orders of magnitude, resulting in diverse signaling and immunotoxic effects [17-19]. Here, we highlight RNS formation and RNS-mediated modifications and their signaling ramifications as a function primarily of NOS2 activation. Discussion of control of NO levels and interplay of NO and RNS with other redox active species is also included.
II. Levels of NO and Cellular Signaling
Since the discovery of endogenous production of NO by NOS, there has been a debate as to whether regulation of cGMP formation or the immune response of macrophages is the more important function of NO. It remains a common practice to examine whether an observed NO-dependent process is also sGC-dependent, for example, by employing the sGC inhibitor ODQ. However, it is now understood that NO has more diverse functions, including regulation of iron and initiation of protective mechanisms during oxidative stress [20-24]. In correlation with the sGC/macrophage debate, NO levels have been historically thought to be bimodal: in the low nanomolar range to activate sCG or approaching low micromolar levels in localized regions as a function of immune response [25]. More recent data suggest instead a continuum in which specific NO fluxes correlate with specific signal transduction pathways.
Estimation of NO fluxes from NOS
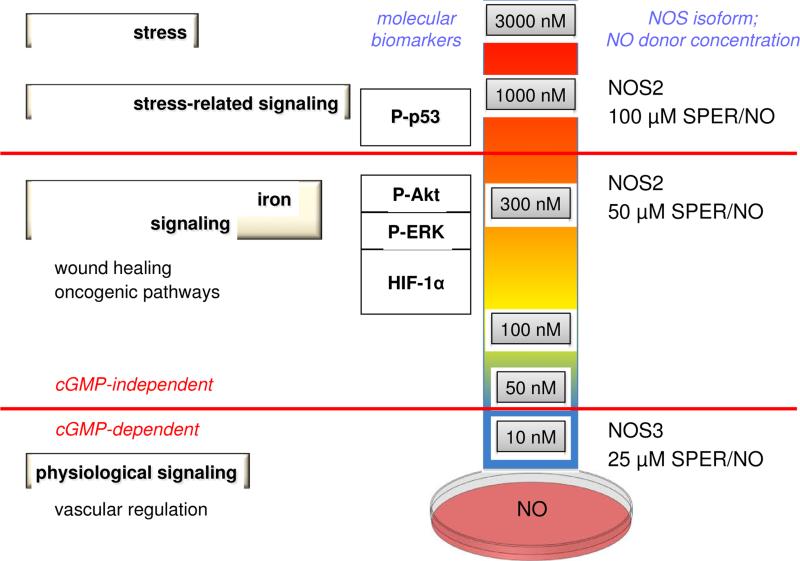
Despite substantial progress with respect to detection of NO, real-time analysis of NO production in cells and tissues continues to be challenging. In the absence of direct quantitative data, indirect methods have been developed to investigate NOS signaling from the perspective of both the concentration and temporal profile of NO production. In particular, NO donors that release NO in a controlled manner have proven to be useful mimics of NOS. For instance, the fluxes of NO produced upon decomposition of diazeniumdiolates (also commonly called NONOates) can be accurately measured in vessels of varied size and shape by use of an NO-selective electrode [26, 27]. Exposure of cultured cells to varied fluxes of NO from these donors, by changing the donor concentration or identity (for detailed discussions of NO donors, see [26, 28, 29]), has facilitated mapping of the large range of biological activities of NO as a function of donor concentration and the associated flux of NO (Figure 2) as well as time.
Figure 2.
Elucidation of NO levels and signaling mechanisms from NO donors in the Petri dish
That specific concentrations of NO donors are able to replicate the biological effects of activated murine macrophages (NOS2-specific) provides some quantitative insight about the NO fluxes that are either generated by these cells or are necessary to elicit a phenotypic response. For instance, co-culture of a 1:1 ratio of murine macrophages with other cell types produces an environment equivalent to the ~100 nM NO flux produced by 10-20 μM SPER/NO or 100 μM DETA/NO. On the other hand, a 4:1 ratio provides fluxes similar to 100-300 μM SPER/NO or 1-2 mM DETA/NO (~0.5-1 μM NO) [27, 30-32]. A similar comparison can be made with other cell types that express NOS2, including cancer cells. In such systems, NOS2 can be upregulated by a variety of conditions including cytokine exposure, hypoxia or serum withdrawal [33]. Responses attributed to generation of NO by NOS2 can be observed and then replicated with NO donors. Such systems also allow verification that the observed effect is NO-dependent by addition of specific NOS inhibitors or NO scavengers.
Exposure of cells to different fluxes of NO in this manner provides an important method to tease out in the Petri dish potential biological effects or signatures that can be extrapolated to animal models or even patients via immunohistochemical or gene expression analysis [33]. Although further studies are required to determine if NO donors reproduce the levels of NO generated in the cellular microenvironment, this method provides a first approximation toward deciphering the signaling and related chemical biology as a function of NO level.
It is important to note that significant differences may exist in the amount of NO generated by rodent and human NOS, as a function of both cellular and systemic dissimilarities. Early studies demonstrated that immune activators such as LPS and IL-2 clearly drive increases in plasma nitrate/nitrite in mice and in humans [34-42]. While there are similar resting levels of nitrate (25 μM) and nitrite (1 μM) in both species, administration of LPS to mice led to as much as 1 mM nitrate/nitrite while in humans peak levels were only 50-115 μM [34, 39-42]). Activation with IFNγ and LPS of murine macrophages leads to significant and rapid increases in nitrate/nitrite, reaching >10 μM in media within hours of NOS2 induction [42]. In contrast, human monocytes and peritoneal macrophages show virtually no accumulation of nitrate/nitrite and exhibit 50-fold lower NOS2 activity than stimulated murine macrophages [43]. This study suggested that reduced bioavailability of tetrahydrobiopterin was critical in limiting NOS2 activity.
Polymorphonuclear leukocytes showed little NOS activity compared to endothelial cells, which upon LPS-stimulation exhibit 400 times higher activity than resting cells [44]. Surprisingly, activation with IL-12 of natural killer cells leads to accumulation of >60 μM nitrate/nitrite during 72 h culture [45]. Moreover, human hepatocytes can generate similar levels of nitrate/nitrite (40-60 μM) in 24 h compared to murine models [46, 47]. Induction of NOS2 in breast cancer cells with IFNγ, PGE2, IL-6 or LPS results in 1 μM nitrate/nitrite [33]. In human fetal astrocytes, levels on the order of 40-60 μM were achieved over 72 h stimulation with IL-1/IFNγ or TNFα, indicating that astrocytes provide the major flux of NO in the human brain. However, in cytokine stimulated breast cancer cells, downstream NO-mediated signaling pathways correlated with an ~300 nM NO flux, in comparison to ~1-10 μM in activated murine macrophages [2]. At this point the basis and ramifications of the difference in NO levels between species and cell types has not been fully realized, but clearly there are important functional differences in NOS2 performance and biology between rodents and humans.
Use of NONOates has suggested that there are characteristic biomarkers and specific signal transduction pathways that correlate to specific fluxes of NO [26, 27] (Figure 2). It is useful to categorize NO signaling into three distinct tiers of steady state NO concentrations: low nanomolar (<100 nM), intermediate nanomolar (~100-500 nM) and above (approaching or even exceeding 1 μM) (Figure 2).
At the lowest levels, NO reacts primarily with ferrous hemes to mediate cGMP-dependent signaling as well as with reactive radicals [25, 48], such as those produced in lipid peroxidation. The direct interactions of NO with heme and free radical targets have been well documented previously [11, 25]. Although cGMP-dependent signaling such as in vascular regulation is a complex process, its initiation by binding of NO to sGC is relatively straightforward. Furthermore, such reactions, although important to signaling, are not generally categorized as redox reactions, although NO is produced through an oxidative mechanism.
When NO levels rise, other, more kinetically constrained reactions begin to be consequential along with radical scavenging and interactions with heme proteins. For instance, NO can interact with cytosolic, nonheme iron targets. In addition, conversion of NO into RNS can lead to indirect signaling through nitrosative, nitrative and oxidative modifications. Such modifications involve redox reactions and depend significantly on the redox landscape with respect to nitrogen, oxygen and sulfur-based species, demonstrating the interrelated nature of redox mediators (see section IV for a more detailed discussion).
Intermediate NO levels
Higher levels of NO are generally considered to be a component of abnormal conditions, including wound healing and stress-related responses, rather than normal physiology. Inflammation or injury stimulates numerous events. Early responses prevent blood loss, sterilize the area against pathogens and activate the innate and adaptive immune responses. In immune cells, NOS2 is generally induced, along with COX2, within 24-36 h of the stimulus [49]. This corresponds to the transition of the innate immune response to either an adaptive response if necessary or to the initiation of wound healing [50, 51]. NOS2 is involved in sterilization, immunosuppression and in a variety of wound healing responses and thus is critical to initiation and resolution of inflammation [14].
NO levels higher than 100 nM have been shown to initiate multiple pathways that are associated with wound healing and tissue restoration responses [20, 21, 32] (Figure 2). For example, inhibition of the nonheme iron protein prolyl hydroxylase by nitrosylation stabilizes hypoxia-inducible factor (HIF)-1α, a transcription factor that can in turn increase proliferation and migration of endothelial cells by stimulating release of vascular endothelial growth factor (VEGF) [20, 52]. Such studies build on earlier work by Drapier and coworkers who showed that regulation of the transferrin receptor and ferritin also occurred at higher levels of NO [53].
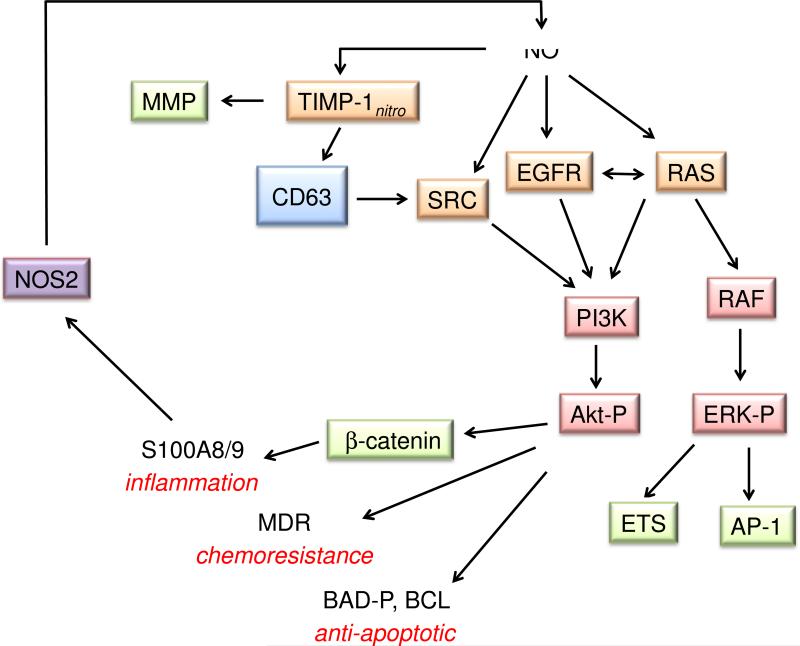
Thiol modifications are also involved in promotion of cell growth. For instance thiols in the membrane proteins transforming protein p21 (H-Ras), epidermal growth factor receptor (EGFR) and proto-oncogene tyrosine-protein kinase (Src) regulate activation of the classic growth factors and downstream targets phosphoinositide 3-kinase (PI3K) and extracellular-signal-regulated kinases (ERK) [54-56]. Recent studies have shown that levels of NO around 300 nM (300 μM DETA/NO) lead to increased S-nitrosation and activation of these targets [57, 58].
Within the context of this review, we categorize such processes as nonheme iron nitrosylation or nitrosative signaling, signifying both initiating reactions as well as the level of NO required. Stabilized downstream products, such as HIF-1α or phosphorylated ERK, are often observable, indirect markers of these signaling pathways. Given the difficulty in quantifying NO levels in cells or tissue and the complexities of signaling pathways, it is important to emphasize that many studies provide only correlative evidence linking a specific signaling effect to a thiol modification (for a detailed discussion, see [59]). Furthermore, post-translational modification of thiols is by no means limited to S-nitrosation. Thiols can be converted into numerous oxidized products, of varying degrees of stability (see [60]). Thiol oxidation or nitrosation may also facilitate S-glutathionylation (see [61]). Given the complexity of the interchange reactions, both spontaneous and regulated, and that specific thiols may be susceptible to diverse modifications, identification of an individual signaling event and full elucidation of a signaling specific signaling pathway can be extremely challenging.
At present, our model is dependent on the correlation of NOS2 expression with elevation of a downstream marker and on detection of an S-nitrosothiol footprint as an indication of initiation of a signaling event by nitrosative chemistry. Use of specific scavengers to abate a signaling mechanism [57] is beginning to offer some experimental causation support for this model. Nonetheless, this model may be too reliant on detection of stable S-nitrosothiols, and future analyses may lead to expansions of the model to include other chemical modifications and more complex thiol-dependent pathways.
In addition to promotion of proliferation, the intermediate range of NO can lead to conversion of the immune suppressive factor transforming growth factor (TGF)-β from the latent to active form [62], presumably through S-nitrosation. NO also increases levels of the anti-inflammatory cytokine interleukin-10 (IL-10) in Treg cells as well as in some cancer cells [63-65]. An additional mechanism for IL-10 production is through NO-dependent increases of COX2 [3, 57]]. Taken together, this level of NO provides immunosuppression with respect to Th1 type responses, which is essential for tissue restoration.
Induction of COX2 by NO [57] can also facilitate angiogenesis. Furthermore, NO may play a critical role in increased angiogenesis and organization of the matrix for wound healing responses by suppressing tissue inhibitor of metallopeptidase 1 (TIMP1) while increasing the activity of matrix metalloproteinase (MMP)-9 [66], which degrades components of the extracellular matrix. NO mediated increases in MMP9 secretion and activity as well as reduced TIMP1 expression in microglial cells is critical for the elimination of plaques in murine models of Alzheimer's disease and in humans [67].
Together, such studies indicate that the intermediate level of NO produces a pro-growth, proliferative, immunosuppressed, pro-angiogenic and enhanced mobility environment that is critical to wound healing and tissue restoration through specific post-translational modifications.
High NO levels
At the high end of the spectrum, proteins that are related to stress response may be stabilized and activated. For example, high levels of NO lead to phosphorylation and stabilization of p53 [31], suggesting a role for NO in cell cycle regulation, DNA repair and apoptosis, particularly during inflammation. Interestingly, NOS2 and P-p53 are co-localized in tissues from patients with ulcerative colitis or Crohn's disease. S-Nitrosation of cysteine 258 of the phosphatase MKP-1 also increases its stability and activity [68], which decreases ERK signaling, causing cell cycle delay. The levels of S-nitrosated MKP-1 observed in tissues from head or neck cancer patients post radiation was reproduced by NO donor concentrations that produced high, prolonged fluxes of NO.
High levels of NO also induce the cellular growth transcription factor nuclear respiratory factor (NrF)-2A and an increase in proteins that have an antioxidant response element (ARE) [69, 70]. In addition, very high fluxes of NO, such as lead to nitrosative stress, can nitrosate and inhibit of caspases and poly(ADP-ribose) polymerase (PARP) [71, 72]. Under these conditions, apoptosis may instead be inhibited, for example as a protective measure in the vicinity of activated macrophages. However, chemical stress, especially within organelles such as the nucleus, mitochondrion or endoplasmic reticulum, may overwhelm a cell, particularly when proteins or nucleic acids are modified indiscriminately.
In chronic disease, involving extended exposure to pathogens or particulates such as plaques in the brain or following a critical mutation, wound healing processes persist and can lead to fibrosis or tumor growth [3]. In cancer, such pathways would then be considered to be pro-oncogenic and may lead to poor outcome [33]. Thus, the same NO signaling mechanism may lead to different biomedical outcomes, depending on the disease or the extent of the inflammation process. The seemingly conflicting roles of NO, for example in apoptosis, are often a result of both the level and timing of exposure to NO.
Although there are numerous examples of distinct responses to nitrosated proteins in vitro, questions remain as to whether NO concentrations necessary to induce nitrosative stress exist in vivo. Again, one indicator that high NO fluxes are possible in specific areas is co-localization of P-p53 with NOS2 [31]. In murine models of leukemia, NOS2 and NO have tumoricidal properties and induce the stabilization of p53 leading to apoptosis [17]. NOS2 and p53 double knockout mice develop leukemia and tumors, indicating that these proteins may serve as critical control switches for cancer development [73]. Xie et al. have suggested that NOS2 produces high levels of NO under these conditions and leads to increased tumoricidal activity [74]. In addition, fully activated murine macrophages or high levels of NO from donors can inhibit different pathogens such as Listeria, limiting their proteolysis [75]. Taken, together it has become clear that murine macrophages have the potential to sustain low micromolar NO fluxes at the cellular level.
In summary, the lowest levels of NO induce normal physiological signaling while intermediate levels of NO stimulate wound-healing responses, and high levels anti-pathogen and antitumor processes are activated. Each specific regulatory, tissue restoration or stress response pathway is mediated by interactions with unique molecular targets that are both spatially and kinetically accessible. The lowest levels of NO lead to activation of sGC and antioxidant reactions. At the intermediate level, activation of critical nonheme iron and thiol targets elicit increased anti-apoptotic and pro-growth responses, as well as increased immunosuppressive and angiogenic factors that lead to tissue restoration. At the highest NO or stress alert level, p53 is induced and proteins such as MKP-1 antagonize pathways that lead to NOS2 induction. This level of NO is anti-proliferative and induces cell cycle delay. In addition, Nrf2 is activated, which induces many antioxidant mechanisms. Furthermore, nuclear factor kappa-light-chain-enhancer of activated B-cells (NFκB) is down regulated, which may be important in leukocyte regulation. Prolonged NO can induce apoptosis, and the highest levels of NO lead to a build up of RNS and damaged proteins, resulting in chemical stress. The chemical interactions involved in nitrosative stress signaling are described in more detail in Sections IV and V, following a more detailed discussion of regulation of NO levels.
III. Control of NO Levels
As described in the previous section, NO-dependent signaling is highly sensitive to the level of NO, primarily as a function of kinetic constraints. Signaling will therefore depend on regulation of NO fluxes by modulation of production and consumption rates. Both physical and chemical processes are involved in shaping the NO profile, defined by concentration, duration and spatial orientation. Biosynthesis of NO by NOS is controlled by transcription, translation, post-transcriptional modification and substrate/cofactor availability. NOS-independent pathways to NO production also exist, for instance by chemical reduction of dietary nitrite. Such reactions are also regulated, particularly by oxygen tension. This section describes NOS regulation, while other mechanisms of NO formation are presented in Section IV.
Selective reactivity is an inherent characteristic of signaling molecules. In the case of NO, especially at low concentrations, there are few reactions that have the pseudo-first order kinetics, which depends on the rate constant and concentration of the target, to compete with simple diffusion away from the site of production. This low reactivity is particularly remarkable given the free radical nature of NO. That NO freely diffuses across cell membranes enables signaling in targets relatively distant from the point of origin, even within neighboring cells. The distance and rate of NO diffusion has been modeled extensively since an early report by Lancaster [76]. It has been long known that diffusion of NO to the lumen of blood vessels leads to rapid reaction with oxyhemoglobin. The resulting conversion of NO to nitrate (NO3−) is a major consumption pathway, limiting the intravascular lifetime of NO to approximately 2 ms [77]. Furthermore, consumption of NO by red blood cells leads to effectively unidirectional diffusion away from blood vessels, thereby localizing signaling to a specific region.
Although, hemoglobin is an effective scavenger of NO there is mounting evidence that hemoglobin as well as erythrocytes are involved in controlling the generation as well as the bioavailability of NO. Over a decade ago, nitrite was shown to be reduced to NO in the human vasculature with vasodilatory effects [78]. The presumption that deoxyhemoglobin was the primary erythrocytic nitrite reductase under physiological conditions was recently confirmed [79]. Interestingly, there are now reports characterizing an active and functional endothelial-like nitric oxide synthase within erythrocytes [80]. This enzyme is located in the plasma membrane and the cytoplasm of red blood cells and controls NO-mediated pathways including activation of platelets. Hemoglobin has also been shown to control NO bioavailability at the myoendothelial junction. An elegant study has demonstrated that the magnitude of NO signaling is a function of the redox state of a specific hemoglobin, Hbα, which is expressed in human arterial endothelial cells [81]. Such findings have important implications about the influence of NO-mediated signaling by non-erythrocytic hemoglobin.
Lancaster et al. have also demonstrated that non-erythroid cells consume NO in an oxygen-dependent fashion [82]. The biological lifetime of NO with respect to cellular consumption pathways depends on a variety of factors, including NO and O2 concentrations and metabolic rate. Although the dominant mechanism(s) by which cells metabolize NO is not known, modeling approximations indicate that the half-life of NO in tissue is between 0.01-2 s.
Another consumptive mechanism for NO is through reaction with ROS, which encompasses a large variety of species including O2− metal-oxo complexes and lipid radicals (NO does not directly react with H2O2) [25]. Due to the dismutation rate, O2− does not diffuse far from the source, for instance NOX. Similarly, metal complexes tend to be localized, for example in red blood cells or mitochondria. This localization provides spatial, temporal and concentration control that cooperatively leads to site-directed signaling processes. See Section IV for further discussion of the chemical reactions involved.
Regulation of NOS
The constitutive NOS isoforms are generally described as producing low levels of NO in short bursts in a calcium-dependent manner and therefore to be primarily associated with intercellular signaling through cGMP. However, regulation of NOS3 and NOS1 extends well beyond binding of calcium to calmodulin (for example, [83]). These two isoforms are also regulated through phosphorylation, myristoylation and palmitoylation, all of which impact location and calcium-dependence [84]. Phosphorylation of NOS3, for example, leads to calcium-independent production of NO. Although this prolongs NO production, the turnover rate is considerably slower than that of NOS2 such that NO is produced by NOS3 at a lower flux.
Although post-translational modification may extend the temporal impact of NOS3 and NOS1, these isoforms are unlikely to produce high enough NO fluxes to elicit nitrosative stress levels. Even though cytotoxic levels of NO are most likely not produced at the tissue level, spatial control may nonetheless lead to higher NO fluxes in specific locations. Therefore, nitrosation in the vicinity of NOS3 and NOS1 cannot be completely discounted but would be expected to be localized and a function of translocation. Nonetheless, it is important to consider that these isoforms may contribute to inflammation, especially in regulation of immune cells, platelet function and in epithelial and endothelial barrier function.
NOS2 is one of most regulated proteins at the translational and transcription level as well as through proteasome degradation and substrate availability [85]. Control of NOS2 transcription and translation has been the subject of extensive analysis, and a number of mechanisms are known to regulate NOS2 expression and activity. In murine leukocytes, activation and enhanced NO production is classically derived by cytokine and/or LPS exposure (e.g., IFNγ and TNFα/IL1β/LPS) [86]. LPS-mediated Toll-like receptor (TLR) 4 activation leads to approximately 10-fold higher NO levels than produced by cytokines [17]. Thus, different stimuli impact local NO concentrations, which influence signaling and stress.
Changes in metabolism provide another regulatory factor in redox signaling, as oxygen and cellular fuel levels are redox sensitive. Indeed, production of redox signaling molecules is tied directly to local oxygen and nutrient levels, which provide a tissue status readout. These molecules can act as sensors to steer signaling cascades during inflammation, for example from a T helper (Th) 1 to a Th2 cytokine response [3, 87].
Substrate and cofactor bioavailability are also important factors in the regulation of NO output and downstream signaling. Both NOS and arginase use arginine as a substrate and can be co-induced in macrophages by LPS [88]. Arginase can function as a substrate competitor in activated cells, and thus may serve a regulatory role in NO biosynthesis [88-90].
NOS cofactors such as tetrahydrobiopterin and NADPH are also influenced by the metabolic state of the cell. Reduction of tetrahydrobiopterin requires a reductase similar to folate reductase while NADPH is in part dependent on glucose-6-phosphate dehydrogenase (G6PD) and the hexose monophosphate shunt from glycolysis [91]. Thus, cofactor regulation by NOS2 can modulate the general metabolic state of the cell. In addition, limited cofactor levels can lead to production of species other than NO (e.g., O2− or HNO) [92-94].
The oxygen-dependence of NOS activity is another important factor. Interestingly, the three major NOS isoforms have widely varying KM values for oxygen, indicating a hierarchy of functionality under conditions of limiting oxygen availability. Purified NOS2 has a KM ~ 150 μM, but its activity is highly variable and depends on a number of factors including scavenging of NO, the environment of the enzyme (e.g., in cell lysate), the presence of nitrite and oxygen tension [95, 96]. NOS1 has a KM > 400 μM, suggesting it may also have oxygen sensing capabilities [97]. NOS3 has a low KM for oxygen, indicating that it retains activity even under low oxygen, which may be necessary to maintain blood flow [98].
These results suggest that the relationship between NO and O2 is a critical determinant of NO flux. Activated, NO-producing murine macrophages generate equivalent steady-state fluxes of NO under either 21 or 1% O2 [99]. However, maximal NO flux generated from NOS2 as well as downstream signaling effects have been reported to occur at 5-8% O2. This observation highlights the importance of both NO consumptive pathways and NO biosynthesis in determining the overall concentration of NO. At lower oxygen concentrations the KM (O2) for NO consumption in a cell is nearly identical to the KM (O2) for NO production by NOS2. Therefore, the steady-state concentration of NO is a function of the relative contributions of numerous oxygen-dependent processes that contribute to its synthesis as well as degradation.
In addition to control of physical flux of NO from NOS, there are negative feedback mechanisms for control of NO-dependent signaling. For example, thrombospondin-1 (TSP-1) can attenuate cGMP-dependent signaling that occurs at the lowest level of NO [100]. The physiological relevance of this NO/TSP-1 crosstalk relationship has been demonstrated in models of ischemia reperfusion as well as radiation injury, where suppression of TSP-1 or its receptor CD47 abates injury associated with these conditions [20, 101-106]. TSP-1 also affects the immune system where regulation of macrophage behavior inhibits NO/cGMP-dependent downregulation of TIMP1 [66]. In addition to direct inhibition of NOS3 signaling, TSP-1 indirectly modulates NO levels by production of ROS [107-110].
NOS2 activation of p53 leads to down regulation of NOS2 [111, 112]. Direct binding of NOS2 to p53 provides an additional inhibitory mechanism [111] as well as increased MKP-1, which abates ERK signaling of cell growth [100, 113] (see section II). In addition, regulation of nitrosative signaling may be provided by systems like thioredoxin (TRX) that can reverse S-nitrosation [114, 115]. By influencing the lifetime and rate of accumulation of RNS-mediated modifications, such proteins may thereby modulate the amplitude of cellular signaling.
Spatial orientation also influences NO flux, which can be tuned to impact cells differently, thus setting in motion specific phenotypic responses. An example is a fully activated murine macrophage that induces a Th1 type cytokine profile during the eradication of pathogens. Under these conditions, NO levels are high in the immediate vicinity of the macrophage; however several cell lengths away, diffusion has diluted NO such that neighboring unaffected regions experience levels that induce an anti-apoptotic response to protect normal tissue from the potential deleterious effects of the anti-pathogen response. As NO levels recede and other pro-growth mechanisms take effect, angiogenic factors induce immunosuppressive signaling mechanisms during wound healing and tissue restoration. Thus, NOS2 regulation is an important mediator of signaling mechanisms that initiate immunosuppression. In this context, NO flux profiles and tuning play critical roles in determining the course of disease or injury.
IV. Sources of RNS Signaling Agents
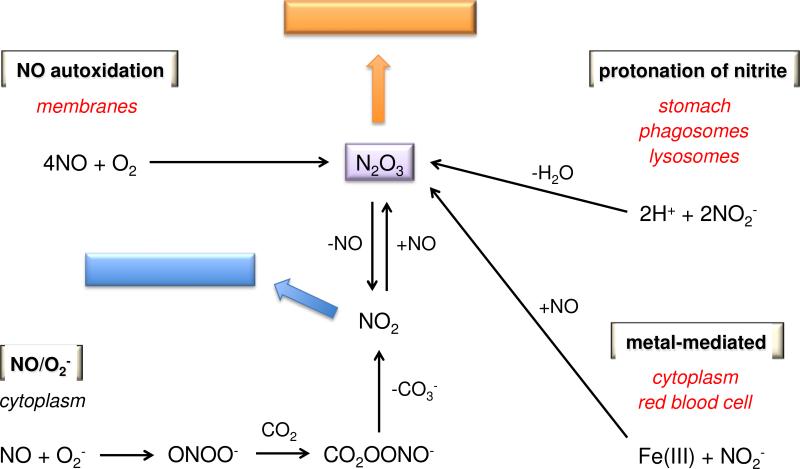
At the intermediate level of NO that promotes cell proliferation (mid-nanomolar), kinetics determines key targets in thiol signaling as well as metal binding. While nitrosylation of nonheme iron complexes occurs directly, nitrosation, nitration and oxidation can be initiated by several different mechanisms. These include NO autoxidation, protonation of nitrite, reaction of NO with O2− and metal mediated processes (Figure 3), all of which play critical roles in different compartments in cells and tissue [2]. Furthermore, these processes all lead to alterations in the redox landscape.
Figure 3.
Chemical sources of N2O3
Autoxidation
Autoxidation is a fundamental reaction type that occurs upon exposure of a chemical to air. Autoxidation of NO has been studied extensively for decades, but questions still remain about the mechanism. In water, it is clear that the reaction produces nitrite (Eq. 1) [116, 117].
| (1) |
It is also well understood that the RNS intermediates of this reaction can induce oxidative, nitrosative and nitrative modifications, all of which modulate cell signaling.
In open vessel experiments in which exogenous NO is added to an aerated, aqueous solution, autoxidation of NO is the primary determinant of NO flux. It is therefore important to understand the fundamental kinetic factors that affect the lifetime of NO under such conditions. Compared to the very common situation of bimolecular reactivity (e.g., A + B), especially in biological systems, autoxidation of NO is somewhat unusual because it is third order overall (Eq. 2) [116, 117].
| (2) |
Due to the second order dependence on NO concentration, the half-life of NO is inversely dependent on the initial concentration of NO. In other words, for every 10-fold increase in NO concentration, the half-life decreases by an order of magnitude. High concentrations of NO will thus be quickly consumed, while low concentrations can persist for minutes or even hours [116, 118]. In such experiments, it is also important to factor in the diffusion of NO from the liquid surface. Loss of NO from diffusion may be significant from high surface area to volume containers such as Petri dishes.
The situation is considerably more complicated when considering the lifetime of NO toward autoxidation within biological systems. First, O2 may no longer be an excess reagent, as is the case in open vessels, and the O2 concentration will vary depending on location. Thus, the biological outcome of NO autoxidation will depend on the redox landscape, in terms of the relative concentrations on NO and O2. Given the wide range of NO fluxes necessary to elicit biological responses, the concentration-dependence of consumption by autoxidation provides exquisite control of the location and timing of NO signaling. For example, the lowest levels of NO, produced for example by endothelial cells (NOS3), will persist long enough with respect to autoxidation to diffuse multiple cell lengths. In contrast, higher levels of NO will be more susceptible to autoxidation.
Small, neutral gases tend to be more soluble in organic solvents than in water. This translates to a higher solubility within cellular membranes than in the cytoplasm. The rate constant in Eq. 2 is relatively unaffected by solvent identity, pH or small temperature changes [116]. Thus, it is fairly straightforward to extrapolate higher concentrations of NO and O2 to higher rates of autoxidation. Lancaster and colleagues have suggested that NO autoxidation is 30-fold faster in membranes and thus that this compartment accounts for 90% of the total reaction [119]. They have also modeled the gradients of NO and O2 as a function of distance from blood vessels and the impact of these gradients on NO lifetime.
In addition to the solubility-dependence of NO autoxidation providing a lens-like focus to the membrane [120], the influence of RNS formed from NO autoxidation is likely to be further spatially localized by the existence of scavengers such as glutathione (GSH) in high concentration in the cytoplasm but not the cell membrane. As such, translocation of proteins to the membrane may facilitate RNS-driven regulation [121].
To better understand the potential role of NO autoxidation in signaling, a mathematical evaluation with temporal perspective in inflammation is useful. Activation of a growth factor receptor requires 20-50 ng/mL or picomolar levels of the ligand. Modification, such as S-nitrosation, of a single residue can also activate a receptor. If the NO flux in the region of the receptor is 500 nM, then the half-life of NO with respect to autoxidation at ambient O2-pressure is on the order of 500 s [116]. Assuming consumption of 250 nM NO per 500 s, 1.8 μM RNS would be produced in an hour. This is obviously a rough approximation, assuming both a constant NO flux and ambient O2 levels. The membrane effect is also not included, nor is conversion of the RNS intermediates to nitrite according to Eq. 1. Furthermore, RNS production rather than accumulation is estimated. Rapid reaction with targets would instead lead to accumulation of molecular modifications, which in turn stimulate signaling. Nonetheless, this approximation suggests that over the span of hours, NO autoxidation as derived from chronic upregulation of NOS2 can form sufficient RNS to induce long term signaling processes, since the estimated value is at least several orders of magnitude higher than required for activation of EGFR and other receptors.
Protonation of nitrite
Autoxidation of NO produces nitrite as an end product (Eq. 1). Nitrite is also ingested in the diet and is generated by bacterial nitrate reductases, for example in oral flora. Not long ago, nitrite was considered to be a relatively stable species indicative of biological NO synthesis and was thought to be primarily excreted either directly or after oxidation to nitrate by oxyheme proteins such as hemoglobin. It is now clear, however, that nitrite serves as both a NOS-dependent and exogenous source of NO and RNS [122-124]. For example, under hypoxic conditions, deoxyheme systems can reduce nitrite to NO to elicit vasodilation [122]. In such circumstances, nitrite can be envisioned as a redox buffer and storage pool for endogenous NO.
In acidic environments such as the stomach, the chemistry of nitrite is more complex, and can lead to RNS and NO production [125], independent of both NOS and oxygen. Protonation of nitrite can induce an interchange between nitrous acid and N2O3 (Eq. 3). Dissociation of N2O3 (Eq. 4) completes the disproportionation reaction, although it is reversible.
| (3) |
| (4) |
Since neutral, small molecules such as N2O3, NO and NO2 can readily diffuse across cellular membranes, within biological systems, an equilibrium condition as portrayed in Eqs. 3 and 4 is unlikely to exist. Furthermore, each of these nitrogen oxides reacts with different biological targets. Of the three, NO is the least reactive, and thus has the longest lifetime. NO2 is much less discriminate, interacting readily with a wide range of biomolecules including the common antioxidants GSH, ascorbate and urate. Given that nitrite will primarily be found in aqueous media where these antioxidants also exist in high concentration, NO2 is expected to be primarily scavenged by the antioxidant defense system. Under such conditions, acidification of nitrite would then serve primarily as an NO-production pathway.
Since N2O3 can also be scavenged, for example by ascorbate, the yield of NO from this pathway will be dependent on location within the redox landscape. In fact, Tannenbaum and coworkers suggested that ascorbate serves as an important protective mechanism against N-nitrosation of amines to carcinogenic nitrosamines in the digestive system [126]. Nonetheless, conversion of nitrite to NO in the stomach has been reported to be important to immune defense and mucosal integrity (see [127]). Dietary nitrite may in fact be an important alternative source of NO and RNS [128]. Foods rich in nitrite/nitrate have also been shown to protect against non-steroidal anti-inflammatory drug (NSAID) toxicity that results in perforation of the mucosal lining [129, 130].
There are numerous acidic environments in vivo, but the pH varies widely from the high acidity of the stomach to the only slightly acidic conditions that transiently arise in hypoxic cells. Intermediate acidities exist in phagosomes and at the surface of macrophages (pH 4.5) [131] as well as in areas of the small intestine (pH 5.7) [132]. The pKa value of 3.7 for HNO2 [133] suggests that relatively low pH conditions are required for Eq. 3 to progress significantly. However, since non-equilibrium conditions can drive the reaction forward, a kinetic comparison can be more informative.
The rate of the acidic nitrite reaction is dependent on both pH and nitrite concentration as follows (Eq. 5) [134, 135]:
| (5) |
The second-order nature on each reactant dictates that at pH 7.4, the reaction rate will be a million times slower than at pH 4.5. Thus, acidity will dictate the location of the reaction.
The concentration of nitrite in blood is 0.2-1 μM [136]. Feelisch and coworkers have determined that nitrite and nitrate levels in the micromolar range exist in different tissues [137-139]. Assuming a nitrite concentration of 1 μM, the half-life is on the order of years at pH 7, but reduces to hours at pH 4.5. A tenfold increase in nitrite would increase the reaction rate by 100 times. Thus, in localized areas of high nitrite levels, this reaction may have increased significance.
Reaction with superoxide
Compared to NO autoxidation, the bimolecular reaction of NO with O2− to form peroxynitrite is straightforward (Eq. 6).
| (6) |
This nearly diffusion-controlled reaction provides protection against oxidative damage by O2−. However, in the process, NO levels are also reduced. It is well known that O2− production will decrease cGMP-dependent signaling, but it also can impact nitrosative signaling by scavenging of NO.
Modulation of NO and O2− levels is only one aspect of Eq. 6. The product, peroxynitrite, is itself an RNS, and interaction with excess NO or O2− can produce still other RNS [25]. The complexities of these interactions have been described in detail previously [140-142]. The discussion can be much simplified by solely considering the chemistry that occurs upon reaction of peroxynitrite with CO2. In carbonate buffer, the lifetime of peroxynitrite is on the order of 15 ms [143] (Eq. 7).
| (7) |
The resulting adduct (nitrosoperoxycarbonate) can decompose through a caged radical pair to NO2 and the carbonate radical anion (Eq. 8).
| (8) |
These radicals can also recombine to give nitrocarbonate (O2NOCO2−), which ultimately reacts with H2O to give to nitrate and carbonate in less than 1 ms. The short lifetime significantly limits the spatial influence of the chemistry of peroxynitrite. Furthermore, as with nitrite, the anionic character of peroxynitrite and ONOOCO2− is expected to limit their chemistry to aqueous environments. Scavenging of NO2 may also be significant as described in the above section.
In regions where NO levels exceed those of O2−, NO2 may instead react significantly with NO to produce N2O3 (comproportionation direction of Eq. 4). Thus, the relative levels of NO and O2− may also modulate signaling mechanisms, with nitration yielding to nitrosation as NO fluxes increase. That the reaction of NO2 with NO will be in direct competition with cellular thiols suggests an alternate route to nitrosated thiols involving oxidation by NO2 to the thiyl radical (RS•) followed by radical combination with NO. Based on kinetic analysis, Lancaster [59] has predicted that this latter pathway will predominate.
In summary, NO autoxidation, protonation of nitrite and association of NO and O2− can all produce both NO2 and N2O3, albeit by unique mechanisms. The dependence on NO of the formation of N2O3 varies as does the order of production of NO2 and N2O3. Furthermore, while NO autoxidation is expected to primarily occur in hydrophobic environments, the latter two processes will principally take place in aqueous media. Thus, although each process is spatially limited, in sum, RNS formation is possible throughout cellular systems.
Metal-mediated processes
As ROS levels rise, reactive metal-oxo and -peroxo complexes can accumulate (Eq. 9; Fenton-type chemistry) and lead to cellular damage.
| (9) |
Rapid reaction of NO with such hypervalent metal complexes can reduce the metal to a normal oxidation state (Eq. 10) [144-146].
| (10) |
The impacts of such reactions will parallel those described above for reaction of NO with O2−. Furthermore, such metal-based reactions can lead to oxidation of NO directly to nitrite without the intermediacy of RNS such as NO2 and N2O3.
Under highly oxidative conditions, nitrite can be further oxidized to NO2, which can contribute to chemical sterilization of tissue [147]. Nitrite can also be formally oxidized to NO2 by ferric systems in the presence of NO [148]. The mechanism is currently a matter of debate, for instance whether NO or nitrite is initially bound to the metal, as is the driving force (e.g., a second NO binding to Fe2+ to form a stable nitrosyl complex), but ultimately, this interaction may serve as a source of N2O3 (Eq. 11)
| (11) |
Hypervalent species (e.g., Eq. 9) can also oxidize nitrite to NO2 [149].
Interestingly, nitrogen oxides such as hydroxylamine (NH2OH) can be oxidized by hypervalent metal complexes to produce RNS with lower oxidation states than NO including nitroxyl (HNO) (Eq. 12) [150].
| (12) |
The NO/NO2− couple
This section highlights not only the diverse reactions of NO that are important to signaling, but also the interrelated nature of NO and nitrite. There are certain parallels to be drawn to the thiol/disulfide system, which has critical roles in protein structure, signaling and redox homeostasis. Although the NO/NO2− couple is not directly involved in structural modifications, as is the case for thiol/disulfide couples, the RNS that are formed from these nitrogen oxides certainly influence structure through post-translational modifications. That cysteine residues are often the targets of these modifications demonstrates the depth of the interconnected nature of redox mediators. The NO/NO2− couple is more directly involved in oxygen-dependent signaling. For example, when NO is produced under normoxic conditions in endothelial cells, it partitions between the signaling pathway involving binding to sGC and consumptive pathways in which NO is converted in an oxygen-dependent manner to nitrite or nitrate (e.g., by reaction with oxyhemoglobin, autoxidation or by other as yet poorly defined cellular processes). Under hypoxic conditions, nitrite can in turn be reduced by hemoglobin back to NO. In this manner, NO-mediated vasodilation can occur over a wide range of O2 concentrations.
From the perspective that NO is the initial signaling agent that can function directly or through the intermediacy of RNS, it is clear that nitrite could be labeled as an RNS. It is interesting to reverse the bias and consider instead nitrite signaling. In such a scenario, ROS/NO reactivity would serve as routes to focused formation of nitrite deposits. Nitrite signaling would then lead to various effects under hypoxic, acidic and oxidative conditions. Under hypoxia, nitrite would be directly converted to NO, while acidification would produce NO and NO2 through the intermediacy of N2O3. Under oxidative conditions, NO2 would be the initial species, and association with NO could then produce N2O3.
The antimicrobial response provides a strong case for a nitrite-centric viewpoint. NOX2 and NOS2 are translocated to phagosomes in neutrophils through Ras-related C3 botulinum toxin substrate 2 (Rac2) [151-153]. Reaction of O2− with NO may then rapidly and site-specifically form a nitrite pool through the reactions outlined above. In this iron-rich, oxidative environment, nitrite may also be formed from reaction of NO with hypervalent metal complexes. Nitrite may then be oxidized to the antimicrobial agent NO2 directly or through protonation, in the acidic environment of the phagosome. Together, the interplay between NO, nitrite and RNS such as NO2 creates a destructive cauldron to combat pathogens or digest proteins.
Interestingly, some pathogens have developed adaptive/evasive mechanisms that target proton pumps, suggesting the importance of acidic nitrite in pathogen defense [3] Tuberculosis is a long-term pathogen that can live in macrophages, where acidic nitrite can induce cytotoxicity [154, 155]. However, diffusion of NO from the macrophage may induce additional signaling mechanisms that coordinate supporting responses by the immune system. In fact, the facile diffusion of NO, as well as its highly regulated biosynthesis by NOS, argues for retaining NO as the central signaling agent relative to other nitrogen oxides.
V. RNS-derived Modifications
As a class, RNS are capable of nitrosative, nitrative and oxidative chemistry. The interactions of RNS with antioxidants often fall in the oxidative category. However, this aspect of RNS chemistry has received the least attention. In similarity to NO and related RNS, consideration of sulfur-containing compounds within the redox signaling landscape may reasonably be focused on thiols and their conversion into other related species, both reactive and not, and primarily more oxidized than the parent thiol. The biological significance of the thiol/disulfide redox couple cannot be overstated. However, a variety of oxidized sulfur species, such as sulfenic, sulfinic and sulfonic acids, have been detected following exposure to thiols to RNS. The mechanisms of formation and the impacts of such modifications are not well resolved. Sulfenic acid may in part be produced through oxygen insertion during the reaction of NO with O2− [156].
Nitration
Conversely, the mechanisms and effects of nitration, particularly of tyrosine and lipids, have been analyzed extensively. In both cases, oxidative chemistry may actually be a first step in nitration [2, 157]. For example, lipid oxidation or peroxidation by NO2 can allow subsequent addition of NO2 to form the nitrolipid. These modified lipids have known physiological properties. For instance, Freeman and coworkers have published a series of papers suggesting that nitrolipids have anti-inflammatory capacity [158]. At present, nitration is considered to be a permanent modification, which can impact versatility in signaling. However, the search for denitrases is an active research area (e.g., [159, 160]).
Although NO2 can be produced by multiple pathways (see [149, 157] and section IV), the reaction of NO with O2− is often assumed to be the primary source of NO2 and thus of nitrative modifications. Exposure of tyrosine to a bolus dose of synthetic peroxynitrite leads to significant nitration [161, 162]. That approximately equal fluxes of NO with O2− produce considerably lower yields of 3-nitrotyrosine [149] highlight the common finding of condition-dependent outcomes, especially with exogenous donor systems.
Formation of 3-nitrotyrosine can inhibit protein function, for example by preventing tyrosine phosphorylation. In addition, 3-nitrotyrosine has also been proposed as a hapten, stimulating an immune response. Improvements in detection have led to demonstration that protein nitration is site specific but occurs at low levels. Observation of internal nitrosation preferentially over more exposed tyrosine residues [163] supports the discussion in section IV that NO2-mediated signaling may primarily occur in regions of low water content, including hydrophobic pockets.
Measurement of enhanced protein nitration in diseased tissue has led to significant interest in the role of this post-translational modification in disease onset and progression, particularly involving neurodegeneration such as occurs during Alzheimer's disease and Parkinson's disease (see [157, 164]). Although correlative, demonstrations of causation are limited. Recently, nitration of a single tyrosine residue on 90-kDa heat-shock protein (Hsp90) by exogenous peroxynitrite was shown to induce motor neuron death [165]. Additional demonstrations of the impacts of site-specific nitration are expected.
Nitrosation and S-nitrosothiols
Within the RNS signaling arena, nitrosation, which can be reversed by several pathways, has received significant attention. Nitrosation indicates a chemical reaction in which NO+ is added to a nucleophile. Given the very short lifetime of free NO+ in biological systems, nitrosation occurs through a transfer reaction, particularly to a thiol or amine (Eqs. 13 and 14).
| (13) |
| (14) |
Here, we emphasize the S-nitrosation of thiols, or in all likelihood the more nucleophilic anions, by N2O3 (Eq. 15).
| (15) |
In this reaction, N2O3 serves as an NO+ transfer agent rather than an NO+ donor. In other words, the reaction is concerted with NO+ directly moving from N2O3 to the thiolate, rather than stepwise following heterolytic dissociation of N2O3 to NO+ and NO2−. This latter reaction is not favorable compared to homolytic dissociation of N2O3 (Eq. 4). The impact of Eq. 15 is often discussed with respect to the S-nitrosothiol product, but it is important to note that direct transfer of NO+ from N2O3 to a nucleophile serves as yet another route for interconversion of nitrogen oxides (compare to Eq. 3).
Nitrosation is often referred to as nitrosylation to draw analogy to other well-known signaling processes such as phosphorylation or methylation. However, from a chemical perspective the term nitrosylation refers to addition of NO, for example to a metal center. Although NO can associate directly with both amines and thiols, such reactions require non-physiological conditions such as high pressures of NO. It is chemically incorrect to use the terms nitrosylation and nitrosation synonymously (for further details, see [166]). In fact these distinct, although related, chemical modifications lead to unique signaling processes as illustrated throughout this review.
N-Nitrosation of amines (Eq. 14) has been implicated in formation of potentially carcinogenic N-nitrosamines [126, 167], for example when meats are preserved by addition of nitrite or nitrate salts [168]. N-Nitrosation can also be mutagenic [169], which may play a role in inflammation-associated carcinogenesis. Although N-nitrosamines have been observed in vivo, especially during chronic inflammation, knowledge of their signaling capacity is rather limited. See [21] for a more detailed discussion of N-nitrosation.
In contrast, S-nitrosation (Eq. 13) has been implicated as a major player in signal transduction [170-173]. Nitrosative signaling involves eliciting a specific response by targeting specific thiols that have been evolutionarily engineered to be kinetically accessible under specific biological conditions. Important determinants for kinetic control are hydrophobicity of the thiol environment and low thiol pKa to enhance nucleophilicity. As is the case with binding of NO to sGC, the small modification of a thiol to an S-nitrosothiol can translate to large structural changes. In membrane receptors, for example, such structural alterations may impact docking or recruitment of proteins.
In biological systems, there are three primary sources of NO+: N2O3, S-nitrosothiols and iron nitrosyls with FeIINO+ character [25]. As illustrated in Section IV, accumulation of N2O3 requires relatively high levels of NO, and at least three different mechanisms lead to its formation in a site-specific manner. Such site-specificity is especially important with respect to signaling given the high cytoplasmic levels of GSH.
In analogy to the reaction of NO and O2− (Eq. 6), association of N2O3 with thiols (Eq. 13) can modulate the effects of both reactants and can produce a species with its own signaling capacity. S-Nitrosothiols are stable in aqueous media under biological conditions, and both low and high molecular weight S-nitrosothiols have been detected in biological samples. S-Nitrosothiols can participate in a variety of reactions, both enzymatic and non-enzymatic [2]. Perhaps the central reaction in terms of signaling is the ability to serve as a nitrosating agent through transnitrosation (Eq. 16) [174].
| (16) |
The term transnitrosation is generally reserved for the specific transfer of NO+ from an S-nitrosothiol to a thiol (e.g., XNO in Eq. 13 is an S-nitrosothiol). This process represents a pathway by which a mobile species such as S-nitrosoglutathione (GSNO) mediates signaling through protein thiol nitrosation [175, 176].
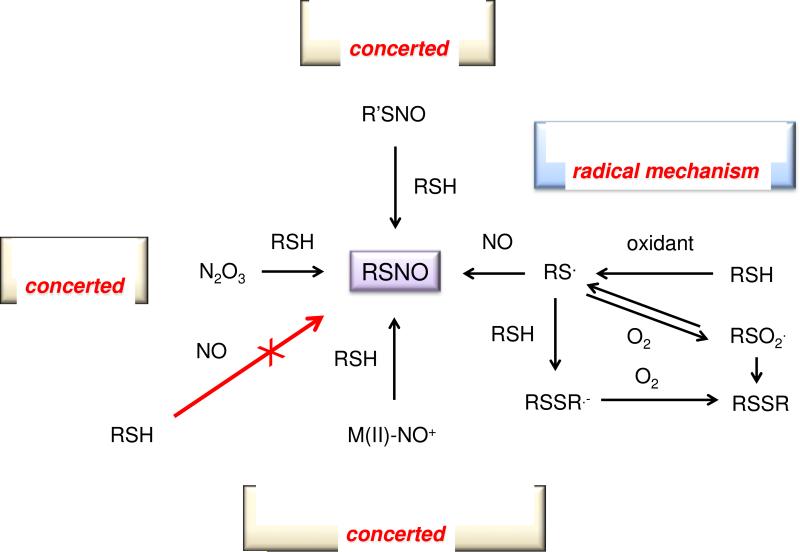
S-Nitrosothiols may be formed by routes other than reaction with an NO+ transfer agent (summarized [175]) (Figure 4). Perhaps the most viable of the alternative pathways is oxidative nitrosylation in which a thiol is first oxidized, for example by an ROS or NO2, to a thiyl radical, which then associates directly with NO. However, the thiyl intermediate has a complex reaction pattern due to its free radical nature. For example, thiyl radicals can associate with other thiolates to form RSSR·− radical anions. These species are very good reductants and can in turn be further oxidized by oxidants such as O2 (e.g., producing O2− and RSSR') [173].
Figure 4.
Three major mechanisms for formation of RSNOs
Since S-nitrosothiols can be homolytically cleaved by light or traces metals, particularly Cu+ (Eq. 17), they are often used as NO donors.
| (17) |
This reaction presents the possibility that S-nitrosothiols may serve as a storage pool for NO, in analogy to nitrite. In fact, nitrite is also a product of the reaction of thiols with N2O3 (Eq. 13). It is important to draw attention to the potential for competition between homolysis to produce NO and other processes such as NO+ transfer. As such, S-nitrosothiols cannot be unequivocally labeled as NO donors, and care should be given to interpretation of biological results obtained with S-nitrosothiols [177].
Interestingly, in addition to transnitrosation, the reaction of S-nitrosothiols with other thiols can follow an alternative pathway (Eq. 18) [178, 179].
| (18) |
As with homolytic cleavage of S-nitrosothiols, this reaction provides an RSNO-dependent mechanism to alter the thiol:disulfide ratio. These processes are in addition to the oxidative mechanisms involving thiyl radicals (Figure 4) and together, contribute to regulation of the redox landscape. Furthermore, this reaction highlights the potential to endogenously form RNS more reduced than NO (Figure 5).
Figure 5.
Nitroxyl (HNO) and thiols as novel signaling species
HNO and nitroxylation
Detailed discussion of low oxidation state RNS has been the subject of numerous reviews and is beyond the scope of this article. However, it is clear that the importance of the NO/HNO redox couple is just beginning to unfold. Although direct interconversion is suggested to be limited [180], NOS itself has been suggested to produce HNO under hypoxic or low cofactor conditions or by oxidative degradation of the intermediate Nω-hydroxy-L-arginine [92, 93, 181-187]. The possibility of biosynthesis of HNO by NOS offers intriguing oxygen-dependent response pathways, in analogy to those previously described for the NO/nitrite couple. For instance Colton and colleagues have demonstrated that HNO and NO uniquely impact glycine-independent desensitization of the NMDA receptor [188, 189]. While both NO and HNO potentiate glutamate-mediated channel activation, hypoxia augments the NO-mediated response, resulting in enhanced neuronal toxicity. In contrast, hypoxia attenuates the effect of HNO on calcium influx, potentially offering a degree of protection toward ischemia/reperfusion injury in the brain.
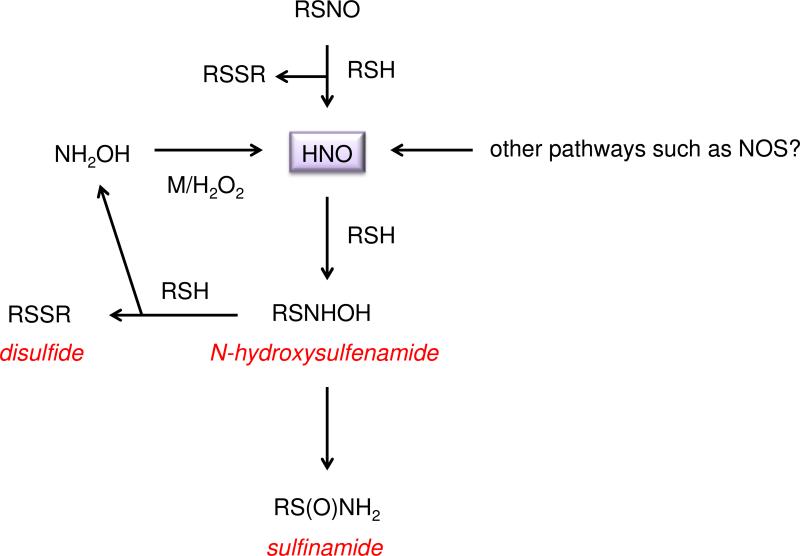
HNO has its own unique signaling mechanisms that are distinct from NO and other RNS [180, 190]. A principal modification mediated by HNO is the conversion of a thiol to the corresponding sulfinamide [178] (Eq. 19; Figure 5).
| (19) |
Since the reaction between HNO and GSH is facile (2 × 106 M−1 s−1, pH 7.4, 37 °C) [190] in comparison to other biomolecules, and the concentration of GSH in cells (0.5-10 mM) [191] is higher than many other cellular components, glutathione sulfinamide (GS(O)NH2) has been proposed to be a selective, endogenous intracellular biomarker for HNO [178, 192].
Sulfinamides may be hydrolyzed to the respective sulfinic acid (RSO2H) or react further with excess thiols to form thiosulfinates (RS(O)SR) [178]. Nonetheless, synthetic GS(O)NH2 has been detected with a reasonable recovery percentage in cell lysate and has been observed after exposure of cells to an HNO donor [193].
As the ratio of thiol to HNO increases, a second reaction pathway, in which the disulfide and hydroxylamine are formed (Eq. 20) [178], becomes progressively competitive such that a mixture of RS(O)NH2 and RSSR is produced. The sulfinamide is indicated to be the major product however [193, 194].
| (20) |
Interestingly NH2OH can be reoxidized to HNO forming a shunt system for HNO that is initiated by RSNOs [150].
Within the context of the redox landscape, it is currently unclear what factors control the outcome of the interactions of S-nitrosothiols with thiols (e.g., Eqs. 17 and 18). However, this reaction may provide a nitrosative/nitroxylative switch. In the vascular system, this could be a unique mechanism of vasodilation. S-Nitrosothiol activation has been well known since Ignarro and coworkers described metabolism of nitroglycerin [195]. Kemp-Harper et al. have suggested that HNO is a mediator of dilation through activation of Kca and Kv channels in vascular smooth muscle [196, 197]. As such HNO may be an endothelial-derived hyperpolarizing factor (EDHF) biosynthesized by NOS3. Taken together, both NO and HNO are suggested to play important roles in maintenance of blood flow.
The potential for oxygen-sensitive signaling by NOS may also indicate the existence of a nitrosative/nitroxylative switch as a function of thiol modification. For example, under normoxic conditions, conversion of NO to N2O3 will produce S-nitrosothiols. Hypoxia could instead lead to HNO production and HNO-mediated signaling. Both modifications, S-nitrosothiol via nitrosation chemistry and sulfinamide via HNO chemistry, involve electrophilic reactions, rather than the intermediacy of radicals. This may provide higher specificity for signaling compared to for example NO2, which can lead to both oxidative and nitrative modifications.
Several proteomic studies have been performed to examine the modifications derived by exposure of thiol-containing proteins to NO or HNO [198-202]. In addition to S-nitrosothiol and sulfinamide products, disulfides and other oxidized species have been observed, demonstrating the diversity of species that may be important in protein activation or regulation. In addition, protein S-nitrosothiols, for example on alkyltransferases and H-Ras [203], can be converted to a sulfinamide by GSNO reductase/alcohol dehydrogenase V [204]. It is not at present clear whether this reaction is concerted or involves formation of free HNO. Nonetheless, GSNO reductase represents a pathway to covert an NO signature into a marker of HNO. As mentioned above, sulfinamides are susceptible to hydrolysis to the corresponding sulfinic acid [178, 205]. In specific proteins, such oxidized products can be reduced by sulfiredoxin (SRX) [206] revealing the potential of enzymatic cycling.
NO and mitochondria
Over the past several decades, NO has been shown to have various effects on mitochondrial function and metabolism by interacting at specific sites in the respiratory chain. Although peroxynitrite has been shown to inhibit complex III [207], NO only affects Complexes I and IV. This is perhaps due to the relatively high levels of MnSOD, which scavenge O2− prior to reaction with NO.
At Complex IV, cytochrome c oxidase, subnanomolar amounts of NO can competitively inhibit respiration. NO interacts with either the ferrous heme iron or oxidized copper, but not both simultaneously at the heme iron:copper binuclear center of cytochrome c oxidase (a(3)/Cu(B)) [208]. The Ki, however, is dependent on the respiration rate. For example in cardiac muscle, which has a high respiration rate, the Ki is 10-fold lower than epithelial or endothelial cells [209].
Higher NO levels have been shown to persistently inhibit Complex I [210, 211] in macrophages, via oxidation or S-nitrosation of specific thiols. Murphy and coworkers used a mitochondria-selective S-nitrosating agent to demonstrate that nitrosation of Complex I serves a protective role during ischemia/reperfusion by reducing oxidative damage and tissue necrosis [212, 213]. In porcine endothelial cells, higher NO levels have also been shown to lead to inhibition of Complex IV via S-nitrosation of two cysteine residues [214].
A comparative study in endothelial cells found different mechanisms of action by CysNO (an NO+ transfer agent) and DETA/NO (an NO donor) [215]. DETA/NO (0.5 mM) reduced mitochondrial reserve capacity and increased glycolysis but did not impact basal respiration. These effects were reversible, likely due to coordination of NO at the Complex IV heme rather than perturbation of the glycolytic protein GAPDH. In contrast, treatment with CysNO led to inhibition of basal respiration, ATP-dependent oxygen consumption rate and reserve capacity. These results indicate that the actions and targets of NO can be significantly different from those of transnitrosation agents. In fact, profound differences in the types and amounts of NO-derived cellular adducts have recently been shown in experiments conducted with a nitrosating agent (CysNO) vs. an authentic NO donors (DETA/NO) [177].
In addition to the direct effects of NO on mitochondrial inhibition, there is a subsequent shift from oxidative to glycolytic metabolism, which can influence other systems by perturbing the balance of metabolic cofactors. For example NO-mediated mitochondrial inhibition in dendritic cells leads to a switch from oxidative to glycolic metabolism similar to the Warburg effect that is observed in hypoxic cancer cells [216]. Other studies indicate that stabilization of HIF-1α by NO leads to an increase in glycolytic pathways [217] as well as activation of AMPK [218]. These metabolic shifts can have significant downstream effects, specifically on epigenetic regulatory mechanisms [219]. For example, many of the enzymes necessary to modify histones or to methylate/demethylate DNA require metabolites as substrates or cofactors. Thus, their enzymatic activities can be influenced by the metabolic state of the cell.
Histone acetyltransferases require acetyl-CoA as the source of acetyl equivalents to acetylate histones. The Km values of most acetyltransferases are within the range that can be effected by changes in the intracellular acetyl-CoA pool, suggesting that NO could indirectly affect histone acetylation by altering acetyl-CoA levels [220]. During glycolysis, there is a shift from NAD+ to NADH. A change in the NAD+/NADH ratio could be important in the case of sirtuins, which are deacetylases. Since their enzymatic activity depends on NAD+, reduced sirtuin activity, by alterations in the NAD+/NADH ratio, could also shift the degree of histone acetylation, which directly influences chromatin structure and gene transcription [221].
Methylation and demethylation of histones and DNA is also accomplished by enzymes that utilize a variety of cofactors. LSD histone demethylases require FAD while KDM demethylases require α-ketoglutarate (α-KG) [222]. Thus, depletion of these cofactors via alterations in cellular metabolism could have significant effects on their activities and the overall histone methylation status. This could affect cell phenotype as histone methylation can either positively or negatively impact gene expression depending on the degree of methylation as well as the location.
TET enzymes oxidatively demethylate cytosine DNA bases. The activities of these α-KG-dependent enzymes also hinge on cofactor availability. DNA methylation, especially at promoter regions, is associated with gene silencing and therefore demethylation of cytosines by TET enzymes can result in gene activation. Therefore, fluctuations in α-KG could have an influence on gene expression via regulation of TET activity. Another important implication of NO-mediated mitochondrial inhibition is associated with changes in oxygen concentration. As inhibition of respiration prevents oxygen consumption, this may lead to significant alterations in the local oxygen concentration. Many of the epigenetic regulatory enzymes mentioned above are oxygen-dependent dioxygenases, and their activities will be affected by oxygen changes depending on their relative Km values for oxygen. These examples highlight the numerous mechanisms whereby NO can influence gene expression and cell phenotype through direct mitochondrial interactions as well as indirectly by dynamic changes in metabolic intermediates and cofactors.
H2S/persulfide interactions with NO
The last few years have led to an emergence in interest in endogenous H2S formation and its role in physiology and pathophysiology. The observation of biosynthesis of H2S from CBS and cystathionine gamma-lyase (CSE) as well as mercaptopyruvate sulfur transferase (MST) has led to a new field of study [223, 224]. The CBS/CSE system is linked to conversion of methionine to cysteine, and as such, is a critical source of GSH (a cysteine-containing tripeptide). Very recently, the presence of H2S has been suggested to be a marker for a larger persulfide (RSS−) pool. Ida and colleagues [225] showed that CSE, and to a lesser extent CBS, can catalyze conversion of cystine (CysSSCys) to cysteine persulfide (CysSS−), which leads to high levels of glutathione persulfide (GSS−) and protein persulfides in mammalian tissues and plasma. Polysulfide proteins were also observed, as a consequence of CysSS−, formation. In addition, H2S is consumed to protect the mitochondria by sulfide:quinone oxidoreductase (SQR) at a significant rate. This process generates GSS−, suggesting that there is a biological and chemical equilibrium between H2S and persulfides [223, 226]. Since persulfides are estimated to exist at as much as 1000 times higher levels than H2S, it has been hypothesized that persulfides may be a major factor/player in thiol-based signaling [225], potentially of even higher significance than H2S. The TSP-1/CD47 pathway also inhibits H2S signaling in T cells and inhibits activation-induced up-regulation of CBS and CSE expression [227].
The ubiquitous presence of persulfides changes the current perspective with respect to the interaction of RNS and thiols and may add a unique layer of complexity to the redox signaling landscape (Figure 6). Persulfides and H2S may provide a new pathway for the interchange of thiol and nitrosative signaling. Several recent articles suggest unique chemistry associated with RSS− (for instance, [228]). Since RSS− is a better nucleophile than free thiols, persulfide formation may represent a mechanism to increase nucleophilicity. Nitrosation of persulfides could result in RSSNO species, which can decompose through RSH attack to RSSR and HSNO [229]. Thionitrite (ONS−) has been proposed as an intermediate in NO and H2S chemistry, presumably through a nitrosative mechanism [230]. Interestingly, Feelisch and coworkers have shown that ONS− is a stable species that activates sGC [231]. Decomposition of these species, which are relatively stable, can even lead to HNO. Ida and colleagues [225] detected polysulfides (RSSnR, n ≥ 2), which suggests the possibility that species like RSSNO or RSSOH can lead to RSSSR for example. The possible physiological roles of polysulfide formation have yet to be elucidated.
Figure 6.
The interplay between H2S/persulfides and RNS
The realization that H2S can be an important physiological mediator led to the proposal that, as mentioned above, ONS− and related/derived species may be significant signaling molecules [231]. As discussed below, these species may be part of a new signaling paradigm linking H2S/persulfides with NO and RNS. That RSNOs have the potential to release or degrade to a variety of RNS and/or reactive sulfur species may explain why there is so little RSNO in circulating blood [232-234].
VI. Nonheme Iron Signaling
At physiological levels of NO (<100 nM), heme proteins, in particular sGC, are the primary targets of NO. As NO levels rise, the kinetic accessibility of other targets, especially nonheme iron complexes, increases. In biological systems, iron is typically bound by a variety of sulfur, oxygen and nitrogen containing ligands. Perhaps the most well-known nonheme complexes are iron-sulfur clusters, which play diverse roles from electron transport to gene expression. A prototypical example of an iron-sulfur cluster protein is the TCA cycle enzyme aconitase. Another classic example that highlights the diversity and importance of nonheme iron complexes in metabolism [235, 236] is the prolyl hydroxylases, which are involved in collagen formation and stabilization of HIF-1α [237-239]. The iron-binding site in these proteins consists of two histidines and an aspartate residue in a facial triad.
With respect to coordination of NO, nonheme iron complexes tend to have slower on rates 103-106 [238] than heme systems (>107; [11]). The off rates also differ, and the impact on stability can be simplified by considering KNO [240]. To provide an estimate of stability, consider simply that 1/KNO approximates the concentration of NO needed to result in 50% complexation. For example, sGC has a KNO of 108 suggesting that a 10 nM NO flux is required to lead to occupation of half of the binding sites. The KNO for nonheme iron systems is typically lower (~106) suggesting that a higher NO flux, in the 300 nM range, is required to significantly bind NO [2]. In analogy to protein environments enhancing thiol nucleophilicity, as indicated by a reduced pKa value, tuning of the KNO by the ligand field of an iron complex may be a key determinant to enzymatic activity and signaling functionality.
Many studies of nonheme protein-mediated signaling have focused on iron metabolism, in particular iron binding and the regulation of transferrin and ferritin in iron homeostasis [53]. The interrelated nature of NO, iron and O2 in regulation of the uptake and storage of cellular iron has been reviewed extensively elsewhere (see [24, 241, 242]). Briefly, binding of NO to key iron-sulfur clusters can lead to regulation of translation of proteins containing iron responsive element (IRE) motifs in their mRNA such as transferrin receptor and ferritin [243-245]. Interestingly, a similar coordinative mechanism for the regulation of ferrochelatase leads to inhibition of heme synthesis [246], potentially serving as a negative feedback mechanism for NO production from NOS. Nitrosylation of the nonheme iron sites in aconitase [238] and prolyl hydroxylase [247] also lead to enzyme inhibition and thus can serve a regulatory role that impacts metabolism and response to low oxygen levels.
In the phagosome, iron deposition facilitates the sterilization process in conjunction with ROS and nitrite [3]. In addition, as described in Section IV, dysregulation of iron can lead to generation of highly oxidizing and deleterious species, such as by serving as catalytic sources for Fenton reactivity [235]. Importantly, NO can serve a protective, antioxidant role in this regard since it can either react rapidly with deleterious radical intermediates associated with oxidative stress or bind catalytic metals responsible for oxidant generation.
Nonheme iron complexes have recently been shown to play a vital role in regulating critical epigenetic modifications in the cell by modulating both DNA and histone modifications. Many of the demethylases that act on histone or CpG islands in DNA contain nonheme iron centers that oxidize the methyl group in a series of reactions ultimately releasing CO2 and the demethylated product [248]. For example, the JmjC class of lysine specific demethylases (KDMs) are mononuclear iron dioxygenases that use α-KG and O2 as co-substrates to oxidatively demethylate specific lysine residues. The iron-binding motif of this family of demethylases is similar to that of prolyl hydroxylase. NO, iron chelators and divalent metals such as Co2+ and Ni2+ have all been shown to inhibit dioxygenases that contain this structural motif, by binding, removing or replacing iron. Binding of NO to JmjC domain-containing histone demethylases orchestrates remodeling of global histone methylation patterns through three distinct mechanisms: direct inhibition, dinitrosyliron complex-mediated reduction in iron cofactor availability, and regulation of gene expression of methyl-modifying enzymes [249].
Lysine methylation status is maintained by lysine methyltransferases (KMT) and KDMs, respectively. Methylation status is therefore a dynamic process that represents a balance of KMT and KDM activities. Since the methylation status of histone lysine residues can positively or negatively influence gene transcription, the KMT/KDM-driven balance can have both acute and long-term impacts on chromatin structure and mRNA expression.
Another critical epigenetic event that has profound effects on gene expression is methylation of DNA cytosine bases. Specifically, 5-methylcytosine (5-mC) is associated with transcriptional repression and is an important determinant of the phenotypic outcome of numerous cellular events. Although 5-mC is the most studied epigenetic DNA modification, at least three other methylcytosine derivatives with gene regulatory functions exist: 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Conversion of cytosine to 5-mC is accomplished by DNA methyltransferases while the sequential oxidation of 5mC to 5fC and 5caC is catalyzed by the ten-eleven-translocation (TET) family of proteins. These TET enzymes belong to the same class of Fe(II)-dependent mononuclear nonheme dioxygenases as the demethylases. TET proteins should be candidates for inhibition by NO under normoxic conditions resulting in alterations in 5-hmC patterns. The hypothesis that NO acts as an epigenetic regulator by modulating TET pathways is compelling and could account for a significant proportion of the phenotypic responses to NO.
The chelatable iron pool (CIP) is another important cellular target for NO. Reaction of excess NO with the CIP leads to quantitative conversion to paramagnetic dinitrosyliron complexes (DNICs) with thiol containing ligands [250, 251]. Although DNICs have been detected under both normal and pathological conditions, the functional consequences attributed to their formation have remained obscure. Hickok et al. examined regulation of the tumor suppressor protein N-Myc downstream-regulated gene 1 (NDRG1) by NO via its reaction with the CIP [252]. Previous reports indicated that NDRG1 is strongly upregulated by iron chelators [253], and therefore NDRG1 was hypothesized to be similarly regulated by iron sequestration through DNIC formation. DNIC-mediated sequestration of chelatable iron in tumor cells did in fact result in an iron-starved phenotype [254]. The appearance of DNICs was also found to be a key determinant for NDRG1 upregulation in triple negative breast cancer cells [252]. These studies showed that NO-mediated suppression of tumor cell migration was mediated through NDRG1 expression, indicating a distinct functional consequence of the reaction of NO with chelatable iron.
Nonheme iron may be at the crossroads for formation of intermediates that lead to species like, NO, NO2 or RSNO, thus directing/changing the redox signaling landscape [238, 240]. The oxidation of reduced species such as NH2OH (Eq. 12) has been investigated extensively with heme proteins [150]. However, Vanin originally proposed that DNICs could lead to a series of reactions that result in RSNO and possibly HNO formation [255, 256]. The potential importance of nonheme iron in the redox landscape has likely been underappreciated.
VII. RNS-derived Signaling in Cancer
The role of chronic inflammation in oncogenesis and the contribution of NOS2 and NO/RNS signaling in tumor growth and metastasis have become areas of increased interest. Expression of NOS2 has been reported in malignancies of the breast, prostate, lung, brain and colon [257-266]. Increased NOS2 expression predicts poor survival in estrogen receptor α-negative (ER(−)) breast cancer patients [267]. In contrast, NO derived from NOS2 in macrophages can have cytostatic and cytotoxic effects on cancer cells [268, 269].
The considerable disparity of outcome relative to NOS2 expression in tumor cells compared to immune cells may be a consequence of the location of NO production. It can be envisioned that production of NO by cancer cells may either modify unique targets or highjack an existing signaling modification. As such inhibition of NOS2 within cancer cells may be therapeutic [270] while delivery of exogenous NO may initiate tumor regression in simulation of the immune system. In fact, NO donors have been shown to lead to chemo- [271, 272] and radiosensitization [273-275] and to overcome drug resistance in tumor cells [276].
As introduced in Section II, S-nitrosation of the membrane proteins H-Ras, EGFR, and TIMP1 regulates activation of the classic growth factors PI3K and ERK [54-56] (Figure 7). H-Ras, PI3K and ERK are regulated by TSP-1/CD47 signaling via cGMP-independent pathways [227, 277, 278]. In some tissues, TSP-1 may also regulate S-nitrosation of these proteins by controlling NOS2 expression [279]. The tumor suppressor protein phosphatase 2A (PP2A) also plays an important inhibitory role in these pathways [32]. PP2A is a threonine serine phosphatase that interacts with Akt, ERK and other downstream targets. PP2A is inhibited by EGFR-mediated phosphorylation or binding to SET nuclear oncogene (SET) and cancerous inhibitor of protein phosphatase 2 (CIP2) [280]. Interestingly, apolipoprotein E (ApoE)-3 also displaces SET and increases PP2A activity, suggesting a potential role of ApoE at nitrosative levels of NO [281].
Figure 7.
Examples of non-cGMP signaling by NO
In chronic disease, increases in ROS and oxidative stress can inhibit NO signaling in wound healing processes, leading to perpetual inflammation and tissue restoration. Exposure of NO to breast cancer cells increased HIF-1α and P-p53 levels and activated EGFR and Src via nitrosation [27, 57, 282]. However, NO-induced HIF-1α stabilization and p53 phosphorylation levels are decreased by O2− [57, 283]. Addition of SOD proved to be protective toward nitrosative signaling [284]. These results suggest that NO levels and subsequent downstream signaling events are regulated by O2−. The antagonistic relationship between NO and ROS is therefore an important aspect of NO signaling and is a factor in tailoring the redox landscape.
In addition to nitrosation, protein nitration has been shown to modulate signaling pathways and disease outcome [67, 84, 163]. Increases in nitration of specific tyrosine residues can impact signaling, for example in mitochondria with dependence on fluctuations in O2 tension [285]. Nitration of p53 and TIMP1 has been demonstrated to induce interesting changes in cellular function [84, 163, 286]. In the case of p53, nitration of tyrosine 327 by low doses of NO specifically leads to stabilization of p53 without ataxia telangiectasia and Rad3-related protein/ataxia telangiectasia mutated (ATR/ATM)-dependent phosphorylation [286]. At such doses, DNA damage was not sufficient for the ATR system to sense and initiate cell cycle arrest.
TIMP1 nitration has recently been reported in human macrophages treated with LPS, which reduced the ability to bind and inhibit active MMP-9 [84]. In support of this observation, mass spectrometry has recently been employed to identify NO donor-mediated tyrosine nitration of TIMP-1 [163]. Internal tyrosines were specifically nitrated (Y95 and Y143), while more exposed residues were not modified. These tyrosine residues reside near key disulfide bonds that form knot structures, which are critical for TIMP-1 binding and inhibition of active MMPs [287]. As MMPs are involved in breakdown and remodeling of the extracellular matrix, they are a critical component of the metastatic process. NO-mediated activation of MMP activity could therefore increase cancer cell invasion and migration.
Nitration of TIMP1 prevents inhibition of active MMP9. Concurrently, a similar level of NO activates latent MMP9 [66, 288, 289]. Nitration of TIMP1 is also associated with enhanced CD63 binding, which promotes pro-survival signaling through PI3K/Akt/BAD phosphorylation, as well as increased drug resistance, cellular proliferation and brain metastasis [33, 163]. CD63 is associated with induction of autophagy [290] and may be a trigger for prosurvival autophagy. Thus, selective nitration leads to specific phenotypes. NO donor-mediated nitration events occurred in the range of 100-700 nM steady state NO fluxes and correlated with NO-mediated Akt pathway activation and reduced survival in ER(−) breast cancer patients with high tumor NOS2 expression [163].
TIMP1 modulates cellular functions via MMP-dependent and -independent mechanisms. MMP-independent pathways can occur through TIMP-1 interaction with the cell surface tetraspanin CD63/integrin complex, which initiates PI3K/Akt-dependent pro-survival signaling [163, 291-293]. Interestingly, immunoprecipitation studies have demonstrated that mutated TIMP1, which lacks MMP inhibitory function, binds CD63 to a higher degree when compared to wild type protein [294]. Similarly, TIMP-1 from NO donor treated cells showed enhanced nitration and co-immunoprecipitated with CD63 to a higher degree than that of untreated controls [163]. Collectively, these results suggest that elevated NO flux in high NOS2-expressing breast tumors may shift TIMP-1 function away from MMP-inhibition to favor TIMP1/CD63-mediated pro-survival signaling while maintaining MMP activity, which further promotes disease progression through the selection of invasive phenotypes.
Conclusion
As described here, the interactions between nitrogen oxides, oxygen and reactive oxygen species, thiol and thiol-derived species and nonheme iron complexes are vital to regulation of cellular metabolism and temporal response to the cellular environment. Due to this interconnectivity, it is often difficult to study the mechanisms of isolated species. That is, focusing on a single signaling species may greatly over-simplify the overall effect and role of this species in an integrated system. Nonetheless, it is clear that NO and related species provide a multitude of reactions that shape the redox landscape. The reactions mediated by NO provide a chemical approach to signaling that is independent of normal ligand activation but dependent on NO flux and NOS2 activity. Additionally, nitrosative and non-heme iron signaling is key to both normal biological functions as well as disease mechanisms. In particular, a central role for NOS2 in wound healing and cancer progression is emerging.
Highlights.
NO concentration is correlated with physiological and stress-related signaling
redox active signaling agents are highly interrelated
nitrosative modifications are involved in signaling and stress processes
stress-related signaling mechanisms are triggered to protect cells
Acknowledgments
This work was supported in part by a National Institutes of Health grant (R01GM076247 to KMM; R01GM085232 to DDT), the Intramural Research Program of the National Institutes of Health, National Cancer Institute and Center for Cancer Research (DAW, DDR) and the NSF (CHE-0645818 to KMM).
Abbreviations
- 5-caC
5-carboxylcytosine
- 5-fC
5-formylcytosine
- 5-hmc
5-hydroxymethylcytosine
- 5-mC
5- methylcytosine
- α-KG
α-ketoglutarate
- Akt
v-akt murine thymoma viral oncogene homolog
- ApoE
apolipoprotein E
- ARE
antioxidant response element
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related protein
- BAD
BCL2-associated agonist of cell death
- CBS
cystathionine β-synthase
- cGMP
cyclic guanosine monophosphate
- CIP
chelatable iron pool
- CIP2
cancerous inhibitor of protein phosphatase 2
- CO
carbon monoxide
- COX
cyclooxygenase
- CSE
cystathionine gamma-lyase
- DETA/NO
diethylenetriamine NONOate
- DNIC
dinitrosyliron complexes
- EGFR
epidermal growth factor receptor
- EDHF
endothelial-derived hyperpolarizing factor
- eNOS
endothelial nitric oxide synthase (aka NOS3)
- ER
estrogen receptor
- ERK
extracellular-signal-regulated kinases
- G6PD
glucose-6-phosphate dehydrogenase
- GSH
glutathione
- GSNO
S-nitrosoglutathione
- H2S
hydrogen sulfide
- HIF
hypoxia-inducible factor
- HNO
nitroxyl
- H-Ras
transforming protein p21 (GTPase)
- HO
heme oxygenase
- IFNγ
interferon gamma
- IL1β
interleukin 1 beta
- IL10
interleukin 10
- iNOS
inducible nitric oxide synthase (aka NOS2)
- IRE
iron responsive element
- JmjC
Jumonji C
- KDM
histone lysine demethylase
- KMT
lysine methyltransferases
- LOX
lipoxygenase
- LPS
lipopolysaccharides
- MKP
mitogen-activated protein kinase phosphatase
- MMP
matrix metalloproteinase
- MST
mercaptopyruvate sulfur transferase
- N2O3
dinitrogen trioxide
- NADPH
nicotinamide adenine dinucleotide phosphate
- NDRG1
N-Myc downstream-regulated gene 1
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B-cells
- NH2OH
hydroxylamine
- nNOS
neuronal nitric oxide synthase (aka NOS1)
- NRF
nuclear respiratory factor
- NO
nitric oxide
- NO2−
nitrite
- NO3−
nitrate
- NONOate
diazeniumdiolate
- NOS
nitric oxide synthase
- NOX
NADPH oxidase
- NSAID
non-steroidal anti-inflammatory drug
- O2−
superoxide
- O2NOCO2−
nitrocarbonate
- ODQ
1H-[1, 2, 4]oxadiazolo[4, 3-a]quinoxalin-1-one
- ONOO−
peroxynitrite
- ONOOCO2−
nitrosoperoxycarbonate
- ONS−
thionitrite
- PARP
poly(ADP-ribose) polymerase
- PGE2
prostaglandin E2
- PGI2
prostacyclin
- PI3K
phosphoinositide 3-kinase
- PP2A
protein phosphatase 2A
- Rac2
Ras-related C3 botulinum toxin substrate 2
- Ras
small GTPase
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RSH
thiol
- RSNO
S-nitrosothiol
- RS(O)NH2
sulfinamide
- RS(O)OH
sulfinic acid
- RSS−
persulfide
- RSSR
disulfide
- SET
SET nuclear oncogene
- sGC
soluble guanylyl cyclase
- SQR
sulfide:quinone oxidoreductase
- Src
proto-oncogene tyrosine-protein kinase
- SRX
sulfiredoxin
- SPER/NO
spermine NONOate
- TCA
tricarboxylic acid
- TET
ten-eleven-translocation
- TGF
transforming growth factor
- Th
T helper
- TIMP1
tissue inhibitor of metallopeptidase 1
- TLR
toll like receptor
- TNFα
tumor necrosis factor alpha
- Treg
regulatory T cell
- TRX
thioredoxin
- TSP1
thrombospondin-1
- TXA2
thromboxane A2
- VEGF
vascular endothelial growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sies H, Cadenas E. Oxidative stress: damage to intact cells and organs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1985;311:617–631. doi: 10.1098/rstb.1985.0168. [DOI] [PubMed] [Google Scholar]
- 2.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: implications in cellular signaling. Free Radic. Biol. Med. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wink DA, Hines HB, Cheng RY, Switzer CH, Flores-Santana W, Vitek MP, Ridnour LA, Colton CA. Nitric oxide and redox mechanisms in the immune response. J. Leukoc. Biol. 2011;89:873–891. doi: 10.1189/jlb.1010550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higdon A, Diers A, Oh J, Landar A, Darley-Usmar V. Cell signalling by reactive lipid species: new concepts and molecular mechanisms. Biochem. J. 2012;442:453–464. doi: 10.1042/BJ20111752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musiek ES, Breeding RS, Milne GL, Zanoni G, Morrow JD, McLaughlin B. Cyclopentenone isoprostanes are novel bioactive products of lipid oxidation which enhance neurodegeneration. J. Neurochem. 2006;97:1301–1313. doi: 10.1111/j.1471-4159.2006.03797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadenas E, Sies H. Oxidative stress: excited oxygen species and enzyme activity. Adv. Enzyme Regul. 1985;23:217–237. doi: 10.1016/0065-2571(85)90049-4. [DOI] [PubMed] [Google Scholar]
- 7.Garthwaite J. New insight into the functioning of nitric oxide-receptive guanylyl cyclase: physiological and pharmacological implications. Mol. Cell. Biochem. 2010;334:221–232. doi: 10.1007/s11010-009-0318-8. [DOI] [PubMed] [Google Scholar]
- 8.Reddy MM, Fernandes MS, Salgia R, Levine RL, Griffin JD, Sattler M. NADPH oxidases regulate cell growth and migration in myeloid cells transformed by oncogenic tyrosine kinases. Leukemia. 2011;25:281–289. doi: 10.1038/leu.2010.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leto TL, Geiszt M. Role of Nox family NADPH oxidases in host defense. Antioxid. Redox Signal. 2006;8:1549–1561. doi: 10.1089/ars.2006.8.1549. [DOI] [PubMed] [Google Scholar]
- 10.Caughey GE, Cleland LG, Penglis PS, Gamble JR, James MJ. Roles of cyclooxygenase (COX)-1 and COX-2 in prostanoid production by human endothelial cells: selective up-regulation of prostacyclin synthesis by COX-2. J. Immunol. 2001;167:2831–2838. doi: 10.4049/jimmunol.167.5.2831. [DOI] [PubMed] [Google Scholar]
- 11.Ford PC, Lorkovic IM. Mechanistic aspects of the reactions of nitric oxide with transition-metal complexes. Chem. Rev. 2002;102:993–1018. doi: 10.1021/cr0000271. [DOI] [PubMed] [Google Scholar]
- 12.Ignarro LJ. Nitric oxide: a unique endogenous signaling molecule in vascular biology. Biosci. Rep. 1999;19:51–71. doi: 10.1023/a:1020150124721. [DOI] [PubMed] [Google Scholar]
- 13.Ridnour LA, Cheng RY, Switzer CH, Heinecke JL, Ambs S, Glynn S, Young HA, Trinchieri G, Wink DA. Molecular pathways: toll-like receptors in the tumor microenvironment-poor prognosis or new therapeutic opportunity. Clin. Cancer Res. 2013;19:1340–1346. doi: 10.1158/1078-0432.CCR-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridnour LA, Thomas DD, Switzer C, Flores-Santana W, Isenberg JS, Ambs S, Roberts DD, Wink DA. Molecular mechanisms for discrete nitric oxide levels in cancer. Nitric Oxide. 2008;19:73–76. doi: 10.1016/j.niox.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murad F. Nitric oxide and cyclic GMP in cell signaling and drug development. N. Engl. J. Med. 2006;355:2003–2011. doi: 10.1056/NEJMsa063904. [DOI] [PubMed] [Google Scholar]
- 16.Modin A, Bjorne H, Herulf M, Alving K, Weitzberg E, Lundberg JON. Nitrite-derived nitric oxide: A possible mediator of ‘acidic-metabolic’ vasodilation. Acta Physiol. Scand. 2001;171:9–16. doi: 10.1046/j.1365-201X.2001.00771.x. [DOI] [PubMed] [Google Scholar]
- 17.Stuehr DJ, Nathan CF. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J. Exp. Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuehr DJ. Mammalian nitric oxide synthases. Biochim. Biophys. Acta. 1999;1411:217–230. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 19.Stuehr DJ. Enzymes of the L-arginine to nitric oxide pathway. J. Nutr. 2004;134:2748S–2751S. doi: 10.1093/jn/134.10.2748S. [DOI] [PubMed] [Google Scholar]
- 20.Ridnour LA, Thomas DD, Donzelli S, Espey MG, Roberts DD, Wink DA, Isenberg JS. The biphasic nature of nitric oxide responses in tumor biology. Antioxid. Redox Signal. 2006;8:1329–1337. doi: 10.1089/ars.2006.8.1329. [DOI] [PubMed] [Google Scholar]
- 21.Ridnour LA, Thomas DD, Mancardi D, Paolocci N, Espey MG, Miranda KM, Feelisch M, Fukuto J, Wink DA. The chemistry of nitrosative stress by nitric oxide and reactive nitrogen oxide species: putting perspective on stressful biological situations. Biol. Chem. 2004;385:1–10. doi: 10.1515/BC.2004.001. [DOI] [PubMed] [Google Scholar]
- 22.Pantopoulos K, Hentze MW. Nitric oxide signaling to iron-regulatory protein: direct control of ferritin mRNA translation and transferrin receptor mRNA stability in transfected fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1267–1271. doi: 10.1073/pnas.92.5.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalyanaraman B. Iron signaling and oxidant damage. Cardiovasc. Toxicol. 2007;7:92–94. doi: 10.1007/s12012-007-0025-1. [DOI] [PubMed] [Google Scholar]
- 24.Kotamraju S, Tampo Y, Kalivendi SV, Joseph J, Chitambar CR, Kalyanaraman B. Nitric oxide mitigates peroxide-induced iron-signaling, oxidative damage, and apoptosis in endothelial cells: role of proteasomal function? Arch. Biochem. Biophys. 2004;423:74–80. doi: 10.1016/j.abb.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 25.Wink DA, Mitchell JB. Chemical biology of nitric oxide: insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic. Biol. Med. 1998;25:434–456. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 26.Thomas DD, Miranda KM, Espey MG, Citrin D, Jourd'heuil D, Paolocci N, Hewett SJ, Colton CA, Grisham MB, Feelisch M, Wink DA. Guide for the use of nitric oxide (NO) donors as probes of the chemistry of NO and related redox species in biological systems. Methods Enzymol. 2002;359:84–105. doi: 10.1016/s0076-6879(02)59174-6. [DOI] [PubMed] [Google Scholar]
- 27.Thomas DD, Espey MG, Ridnour LA, Hofseth LJ, Mancardi D, Harris CC, Wink DA. Hypoxic inducible factor 1alpha, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8894–8899. doi: 10.1073/pnas.0400453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang PG, Xian M, Tang X, Wu X, Wen Z, Cai T, Janczuk AJ. Nitric oxide donors: chemical activities and biological applications. Chem. Rev. 2002;102:1091–1134. doi: 10.1021/cr000040l. [DOI] [PubMed] [Google Scholar]
- 29.Feelisch M, Stamler JS. Donors of Nitrogen Oxides. In: Feelisch M, Stamler JS, editors. Methods in Nitric Oxide Research. John Wiley & Sons; New York: 1996. pp. 71–115. [Google Scholar]
- 30.Brüne B, von Knethen A, Sandau KB. Transcription factors p53 and HIF-1alpha as targets of nitric oxide. Cell. Signal. 2001;13:525–533. doi: 10.1016/s0898-6568(01)00175-9. [DOI] [PubMed] [Google Scholar]
- 31.Hofseth LJ, Saito S, Hussain SP, Espey MG, Miranda KM, Araki Y, Jhappan C, Higashimoto Y, He P, Linke SP, Quezadol MM, Zurer I, Rotter V, Wink DA, Appella E, Harris CC. Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:143–148. doi: 10.1073/pnas.0237083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Switzer CH, Glynn SA, Ridnour LA, Cheng RY, Vitek MP, Ambs S, Wink DA. Nitric oxide and protein phosphatase 2A provide novel therapeutic opportunities in ER-negative breast cancer. Trends Pharmacol. Sci. 2011;32:644–651. doi: 10.1016/j.tips.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinecke JL, Ridnour LA, Cheng RY, Switzer CH, Lizardo MM, Khanna C, Glynn SA, Hussain SP, Young HA, Ambs S, Wink DA. Tumor microenvironment-based feed-forward regulation of NOS2 in breast cancer progression. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6323–6328. doi: 10.1073/pnas.1401799111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochoa JB, Udekwu AO, Billiar TR, Curran RD, Cerra FB, Simmons RL, Peitzman AB. Nitrogen oxide levels in patients after trauma and during sepsis. Ann. Surg. 1991;214:621–626. doi: 10.1097/00000658-199111000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guarner C, Soriano G, Tomas A, Bulbena O, Novella M, Balanzo J, Vilardell F, Mourelle M, Moncada S. Increased serum nitrite and nitrate levels in patients with cirrhosis: relationship to endotoxemia. Hepatology. 1993;18:1139–1143. [PubMed] [Google Scholar]
- 36.Jacob TD, Ochoa JB, Udekwu AO, Wilkinson J, Murray T, Billiar TR, Simmons RL, Marion DW, Peitzman AB. Nitric oxide production is inhibited in trauma patients. J. Trauma. 1993;35:590–596. doi: 10.1097/00005373-199310000-00015. [DOI] [PubMed] [Google Scholar]
- 37.Miles D, Thomsen L, Balkwill F, Thavasu P, Moncada S. Association between biosynthesis of nitric oxide and changes in immunological and vascular parameters in patients treated with interleukin-2. Eur. J. Clin. Invest. 1994;24:287–290. doi: 10.1111/j.1365-2362.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 38.Citterio G, Pellegatta F, Lucca GD, Fragasso G, Scaglietti U, Pini D, Fortis C, Tresoldi M, Rugarli C. Plasma nitrate plus nitrite changes during continuous intravenous infusion interleukin 2. Br. J. Cancer. 1996;74:1297–1301. doi: 10.1038/bjc.1996.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Billiar TR, Curran RD, Harbrecht BG, Stuehr DJ, Demetris AJ, Simmons RL. Modulation of nitrogen oxide synthesis in vivo: NG-monomethyl-L-arginine inhibits endotoxin-induced nitrate/nitrate biosynthesis while promoting hepatic damage. J. Leukoc. Biol. 1990;48:565–569. doi: 10.1002/jlb.48.6.565. [DOI] [PubMed] [Google Scholar]
- 40.Tracey WR, Tse J, Carter G. Lipopolysaccharide-induced changes in plasma nitrite and nitrate concentrations in rats and mice: pharmacological evaluation of nitric oxide synthase inhibitors. J. Pharmacol. Exp. Ther. 1995;272:1011–1015. [PubMed] [Google Scholar]
- 41.Rees DD, Monkhouse JE, Cambridge D, Moncada S. Nitric oxide and the haemodynamic profile of endotoxin shock in the conscious mouse. Br. J. Pharmacol. 1998;124:540–546. doi: 10.1038/sj.bjp.0701815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuehr DJ, Marletta MA. Mammalian nitrate biosynthesis: mouse macrophages produce nitrite and nitrate in response to Escherichia coli lipopolysaccharide. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7738–7742. doi: 10.1073/pnas.82.22.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinberg JB, Misukonis MA, Shami PJ, Mason SN, Sauls DL, Dittman WA, Wood ER, Smith GK, McDonald B, Bachus KE. Human mononuclear phagocyte inducible nitric oxide synthase (iNOS): analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood. 1995;86:1184–1195. [PubMed] [Google Scholar]
- 44.Yan L, Vandivier RW, Suffredini AF, Danner RL. Human polymorphonuclear leukocytes lack detectable nitric oxide synthase activity. J. Immunol. 1994;153:1825–1834. [PubMed] [Google Scholar]
- 45.Salvucci O, Kolb J, Dugas B, Dugas N, Chouaib S. The induction of nitric oxide by interleukin-12 and tumor necrosis factor-alpha in human natural killer cells: relationship with the regulation of lytic activity. Blood. 1998;92:2093–2102. [PubMed] [Google Scholar]
- 46.Nüssler AK, Di Silvio M, Billiar TR, Hoffman RA, Geller DA, Selby R, Madariaga J, Simmons RL. Stimulation of the nitric oxide synthase pathway in human hepatocytes by cytokines and endotoxin. J. Exp. Med. 1992;176:261–264. doi: 10.1084/jem.176.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nüssler AK, Geller DA, Sweetland MA, Di Silvio M, Billiar TR, Madariaga JB, Simmons RL, Lancaster JR. Induction of nitric oxide synthesis and its reactions in cultured human and rat hepatocytes stimulated with cytokines plus LPS. Biochem. Biophys. Res. Commun. 1993;194:826–835. doi: 10.1006/bbrc.1993.1896. [DOI] [PubMed] [Google Scholar]
- 48.Wink DA, Miranda KM, Espey MG. Cytotoxicity related to oxidative and nitrosative stress by nitric oxide. Exp. Biol. Med. 2001;226:621–623. doi: 10.1177/153537020222600704. [DOI] [PubMed] [Google Scholar]
- 49.Weinberg JB. Nitric oxide synthase 2 and cyclooxygenase 2 interactions in inflammation. Immunol. Res. 2000;22:319–341. doi: 10.1385/IR:22:2-3:319. [DOI] [PubMed] [Google Scholar]
- 50.Schwentker A, Vodovotz Y, Weller R, Billiar TR. Nitric oxide and wound repair: role of cytokines? Nitric Oxide. 2002;7:1–10. doi: 10.1016/s1089-8603(02)00002-2. [DOI] [PubMed] [Google Scholar]
- 51.Schwentker A, Billiar TR. Nitric oxide and wound repair. Surg. Clin. North Am. 2003;83:521–530. doi: 10.1016/S0039-6109(02)00207-4. [DOI] [PubMed] [Google Scholar]
- 52.Sandau KB, Fandrey J, Brüne B. Accumulation of HIF-1alpha under the influence of nitric oxide. Blood. 2001;97:1009–1015. doi: 10.1182/blood.v97.4.1009. [DOI] [PubMed] [Google Scholar]
- 53.Bouton C, Drapier JC. Iron regulatory proteins as NO signal transducers. Sci STKE. 2003;2003:pe17. doi: 10.1126/stke.2003.182.pe17. [DOI] [PubMed] [Google Scholar]
- 54.Lander HM, Ogiste JS, Pearce SF, Levi R, Novogrodsky A. Nitric oxide-stimulated guanine nucleotide exchange on p21ras. J. Biol. Chem. 1995;270:7017–7020. doi: 10.1074/jbc.270.13.7017. [DOI] [PubMed] [Google Scholar]
- 55.Peranovich TM, da Silva AM, Fries DM, Stern A, Monteiro HP. Nitric oxide stimulates tyrosine phosphorylation in murine fibroblasts in the absence and presence of epidermal growth factor. Biochem. J. 1995;305:613–619. doi: 10.1042/bj3050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahman MA, Senga T, Ito S, Hyodo T, Hasegawa H, Hamaguchi M. S-nitrosylation at cysteine 498 of c-Src tyrosine kinase regulates nitric oxide-mediated cell invasion. J. Biol. Chem. 2010;285:3806–3814. doi: 10.1074/jbc.M109.059782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Switzer CH, Glynn SA, Cheng RY, Ridnour LA, Green JE, Ambs S, Wink DA. S-nitrosylation of EGFR and Src activates an oncogenic signaling network in human basal-like breast cancer. Mol. Cancer Res. 2012;10:1203–1215. doi: 10.1158/1541-7786.MCR-12-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Switzer CH, Cheng RY, Ridnour LA, Glynn SA, Ambs S, Wink DA. Ets-1 is a transcriptional mediator of oncogenic nitric oxide signaling in estrogen receptor-negative breast cancer. Breast Cancer Res. 2012;14:R125. doi: 10.1186/bcr3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lancaster JR. Protein cysteine thiol nitrosation: maker or marker of reactive nitrogen species-induced nonerythroid cellular signaling? Nitric Oxide. 2008;19:68–72. doi: 10.1016/j.niox.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 60.Poole LB. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80C:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vodovotz Y, Chesler L, Chong HY, Kim SJ, Simpson JT, DeGraff W, Cox GW, Roberts AB, Wink DA, Barcellos-Hoff MH. Regulation of transforming growth factor beta 1 by nitric oxide. Cancer Res. 1999;59:2142–2149. [PubMed] [Google Scholar]
- 63.Niedbala W, Cai B, Liu H, Pitman N, Chang L, Liew FY. Nitric oxide induces CD4+CD25+ Foxp3 regulatory T cells from CD4+CD25 T cells via p53, IL-2, and OX40. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15478–15483. doi: 10.1073/pnas.0703725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts AB, Vodovotz Y, Roche NS, Sporn MB, Nathan CF. Role of nitric oxide in antagonistic effects of transforming growth factor-beta and interleukin-1 beta on the beating rate of cultured cardiac myocytes. Mol. Endocrinol. 1992;6:1921–1930. doi: 10.1210/mend.6.11.1282674. [DOI] [PubMed] [Google Scholar]
- 65.Cunha FQ, Moncada S, Liew FY. Interleukin-10 (IL-10) inhibits the induction of nitric oxide synthase by interferon-gamma in murine macrophages. Biochem. Biophys. Res. Commun. 1992;182:1155–1159. doi: 10.1016/0006-291x(92)91852-h. [DOI] [PubMed] [Google Scholar]
- 66.Ridnour LA, Windhausen AN, Isenberg JS, Yeung N, Thomas DD, Vitek MP, Roberts DD, Wink DA. Nitric oxide regulates matrix metalloproteinase-9 activity by guanylyl-cyclase-dependent and -independent pathways. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16898–16903. doi: 10.1073/pnas.0702761104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ridnour LA, Dhanapal S, Hoos M, Wilson J, Lee J, Cheng RY, Brueggemann EE, Hines HB, Wilcock DM, Vitek MP, Wink DA, Colton CA. Nitric oxide-mediated regulation of -amyloid clearance via alterations of MMP-9/TIMP-1. J. Neurochem. 2012;123:736–479. doi: 10.1111/jnc.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guan W, Sha J, Chen X, Xing Y, Yan J, Wang Z. S-Nitrosylation of mitogen activated protein kinase phosphatase-1 suppresses radiation-induced apoptosis. Cancer Lett. 2012;314:137–146. doi: 10.1016/j.canlet.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 69.Um HC, Jang JH, Kim DH, Lee C, Surh YJ. Nitric oxide activates Nrf2 through S-nitrosylation of Keap1 in PC12 cells. Nitric Oxide. 2011;25:161–168. doi: 10.1016/j.niox.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Li CQ, Kim MY, Godoy LC, Thiantanawat A, Trudel LJ, Wogan GN. Nitric oxide activation of Keap1/Nrf2 signaling in human colon carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14547–14551. doi: 10.1073/pnas.0907539106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sidorkina O, Espey MG, Miranda KM, Wink DA, Laval J. Inhibition of poly(ADP-ribose) polymerase (PARP) by nitric oxide and reactive nitrogen oxide species. Free Radic. Biol. Med. 2003;35:1431–1438. doi: 10.1016/j.freeradbiomed.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 72.Kim YM, Talanian RV, Billiar TR. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J. Biol. Chem. 1997;272:31138–31134. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- 73.Hussain SP, He P, Subleski J, Hofseth LJ, Trivers GE, Mechanic L, Hofseth AB, Bernard M, Schwank J, Nguyen G, Mathe E, Djurickovic D, Haines D, Weiss J, Back T, Gruys E, Laubach VE, Wiltrout RH, Harris CC. Nitric oxide is a key component in inflammation-accelerated tumorigenesis. Cancer Res. 2008;68:7130–7136. doi: 10.1158/0008-5472.CAN-08-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie K, Fidler IJ. Therapy of cancer metastasis by activation of the inducible nitric oxide synthase. Cancer Metastasis Rev. 1998;17:55–75. doi: 10.1023/a:1005956721457. [DOI] [PubMed] [Google Scholar]
- 75.Ogawa R, Pacelli R, Espey MG, Miranda KM, Friedman N, Kim SM, Cox G, Mitchell JB, Wink DA, Russo A. Comparison of control of Listeria by nitric oxide redox chemistry from murine macrophages and no donors: insights into listeriocidal activity of oxidative and nitrosative stress. Free Radic. Biol. Med. 2001;30:268–276. doi: 10.1016/s0891-5849(00)00470-6. [DOI] [PubMed] [Google Scholar]
- 76.Lancaster JRJ. Simulation of the diffusion and reaction of endogenously produced nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8137–8141. doi: 10.1073/pnas.91.17.8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu X, Miller MJ, Joshi MS, Sadowska-Krowicka H, Clark DA, Lancaster JR. Diffusion-limited reaction of free nitric oxide with erythrocytes. J. Biol. Chem. 1998;273:18709–18713. doi: 10.1074/jbc.273.30.18709. [DOI] [PubMed] [Google Scholar]
- 78.Cosby K, Partovi KS, Crawford JH, Patel RP, Reiter CD, Martyr S, Yang BK, Waclawiw MA, Zalos G, Xu X, Huang KT, Shields H, Kim-Shapiro DB, Schechter AN, Cannon RO, Gladwin MT. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nature Med. 2003;9:1498–1505. doi: 10.1038/nm954. [DOI] [PubMed] [Google Scholar]
- 79.Liu C, Wajih N, Liu X, Basu S, Janes J, Marvel M, Keggi C, Helms CC, Lee AN, Belanger AM, Diz DI, Laurienti PJ, Caudell DL, Wang J, Gladwin MT, Kim-Shapiro DB. Mechanisms of human erythrocytic bioactivation of nitrite. J. Biol. Chem. 2015;290:1281–1294. doi: 10.1074/jbc.M114.609222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kleinbongard P, Schulz R, Rassaf T, Lauer T, Dejam A, Jax T, Kumara I, Gharini P, Kabanova S, Ozüyaman B, Schnürch H, Gödecke A, Weber A, Robenek M, Robenek H, Bloch W, Rösen P, Kelm M. Red blood cells express a functional endothelial nitric oxide synthase. Blood. 2006;107:2943–2951. doi: 10.1182/blood-2005-10-3992. [DOI] [PubMed] [Google Scholar]
- 81.Straub AC, Lohman AW, Billaud M, Johnstone SR, Dwyer ST, Lee MY, Bortz PS, Best AK, Columbus L, Gaston B, Isakson BE. Endothelial cell expression of haemoglobin regulates nitric oxide signalling. Nature. 2012;491:473–477. doi: 10.1038/nature11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomas DD, Liu X, Kantrow SP, Lancaster JRJ. The biological lifetime of nitric oxide: implications for the perivascular dynamics of NO and O2. Proc. Natl. Acad. Sci. U.S.A. 2001;98:355–360. doi: 10.1073/pnas.011379598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Forstermann U, Sessa WC. Nitric oxide synthases: Regulation and function. Eur. Heart J. 2012;33:829–837. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patruno A, Pesce M, Marrone A, Speranza L, Grilli A, De Lutiis MA, Felaco M, Reale M. Activity of matrix metallo proteinases (MMPs) and the tissue inhibitor of MMP (TIMP)-1 in electromagnetic field-exposed THP-1 cells. J. Cell. Physiol. 2012;227:2767–2774. doi: 10.1002/jcp.23024. [DOI] [PubMed] [Google Scholar]
- 85.Foster MW, Thompson JW, Forrester MT, Sha Y, McMahon TJ, Bowles DE, Moseley MA, Marshall HE. Proteomic analysis of the NOS2 interactome in human airway epithelial cells. Nitric Oxide. 2013;34:37–46. doi: 10.1016/j.niox.2013.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Espey MG, Miranda KM, Pluta RM, Wink DA. Nitrosative capacity of macrophages is dependent on nitric oxide synthase induction signals. J. Biol. Chem. 2000;275:11341–11347. doi: 10.1074/jbc.275.15.11341. [DOI] [PubMed] [Google Scholar]
- 87.Colton CA. Microglial-neuronal interactions during neurodegenerative diseases. J. Neuroimmune Pharmacol. 2013;8:4–6. doi: 10.1007/s11481-013-9437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kepka-Lenhart D, Ash DE, Morris SM. Determination of mammalian arginase activity. Methods Enzymol. 2008;440:221–230. doi: 10.1016/S0076-6879(07)00813-0. [DOI] [PubMed] [Google Scholar]
- 89.Christianson DW. Arginase: Structure, mechanism, and physiological role in male and female sexual arousal. Acc. Chem. Res. 2005;38:91–201. doi: 10.1021/ar040183k. [DOI] [PubMed] [Google Scholar]
- 90.Boucher JL, Moali C, Tenu JP. Nitric oxide biosynthesis, nitric oxide synthase inhibitors and arginase competition for L-arginine utilization. Cell Mol. Life Sci. 1999;55:1015–1028. doi: 10.1007/s000180050352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Werner ER, Blau N, Thöny B. Tetrahydrobiopterin: biochemistry and pathophysiology. Biochem. J. 2011;438:397–414. doi: 10.1042/BJ20110293. [DOI] [PubMed] [Google Scholar]
- 92.Schmidt HHHW, Hofmann H, Schindler U, Shutenko ZS, Cunningham DD, Feelisch M. No NO from NO synthase. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14492–14497. doi: 10.1073/pnas.93.25.14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adak S, Wang Q, Stuehr DJ. Arginine conversion to nitroxide by tetrahydrobiopterin-free neuronal nitric-oxide synthase - implications for mechanism. J. Biol. Chem. 2000;275:33554–33561. doi: 10.1074/jbc.M004337200. [DOI] [PubMed] [Google Scholar]
- 94.Pou S, Pou WS, Bredt DS, Snyder SH, Rosen GM. Generation of superoxide by purified brain nitric oxide synthase. J. Biol. Chem. 1992;267:24173–24176. [PubMed] [Google Scholar]
- 95.Dweik RA, Laskowski D, Abu-Soud HM, Kaneko F, Hutte R, Stuehr DJ, Erzurum SC. Nitric oxide synthesis in the lung. Regulation by oxygen through a kinetic mechanism. J. Clin. Invest. 1998;101:660–666. doi: 10.1172/JCI1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson MA, Baumgardner JE, Otto CM. Oxygen-dependent regulation of nitric oxide production by inducible nitric oxide synthase. Free Radic. Biol. Med. 2011;51:1952–1965. doi: 10.1016/j.freeradbiomed.2011.08.034. [DOI] [PubMed] [Google Scholar]
- 97.Haque MM, Tejero J, Bayachou M, Wang ZQ, Fadlalla M, Stuehr DJ. Thermodynamic characterization of five key kinetic parameters that define neuronal nitric oxide synthase catalysis. FEBS Journal. 2013;280:4439–4453. doi: 10.1111/febs.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abu-Soud HM, Ichimori K, Presta A, Stuehr DJ. Electron transfer, oxygen binding, and nitric oxide feedback inhibition in endothelial nitric-oxide synthase. J. Biol. Chem. 2000;275:17349–17357. doi: 10.1074/jbc.M000050200. [DOI] [PubMed] [Google Scholar]
- 99.Hickok JR, Vasudevan D, Jablonski K, Thomas DD. Oxygen dependence of nitric oxide-mediated signaling. Redox Biol. 2013;1:203–209. doi: 10.1016/j.redox.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Isenberg JS, Ridnour LA, Perruccio EM, Espey MG, Wink DA, Roberts DD. Thrombospondin-1 inhibits endothelial cell responses to nitric oxide in a cGMP-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13141–13146. doi: 10.1073/pnas.0502977102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Isenberg JS, Maxhimer JB, Powers P, Tsokos M, Frazier WA, Roberts DD. Treatment of liver ischemia-reperfusion injury by limiting thrombospondin-1/CD47 signaling. Surgery. 2008;144:752–761. doi: 10.1016/j.surg.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Isenberg JS, Maxhimer JB, Hyodo F, Pendrak ML, Ridnour LA, DeGraff WG, Tsokos M, Wink DA, Roberts DD. Thrombospondin-1 and CD47 limit cell and tissue survival of radiation injury. Am. J. Pathol. 2008;173:1100–1112. doi: 10.2353/ajpath.2008.080237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maxhimer JB, Soto-Pantoja DR, Ridnour LA, Shih HB, Degraff WG, Tsokos M, Wink DA, Isenberg JS, Roberts DD. Radioprotection in normal tissue and delayed tumor growth by blockade of CD47 signaling. Sci. Transl. Med. 2009;1:3ra7. doi: 10.1126/scitranslmed.3000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Soto-Pantoja D, Isenberg J, Roberts D. Therapeutic targeting of CD47 to modulate tssue responses to ischemia and radiation. J. Genet. Syndr. Gene. Ther. 2011;2 doi: 10.4172/2157-7412.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soto-Pantoja DR, Miller TW, Pendrak ML, DeGraff WG, Sullivan C, Ridnour LA, Abu-Asab M, Wink DA, Tsokos M, Roberts DD. CD47 deficiency confers cell and tissue radioprotection by activation of autophagy. Autophagy. 2012;8:1628–1642. doi: 10.4161/auto.21562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soto-Pantoja DR, Ridnour LA, Wink DA, Roberts DD. Blockade of CD47 increases survival of mice exposed to lethal total body irradiation. Sci. Rep. 2013;3:1038. doi: 10.1038/srep01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martin-Manso G, Galli S, Ridnour LA, Tsokos M, Wink DA, Roberts DD. Thrombospondin 1 promotes tumor macrophage recruitment and enhances tumor cell cytotoxicity of differentiated U937 cells. Cancer Res. 2008;68:7090–7099. doi: 10.1158/0008-5472.CAN-08-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts DD, Isenberg JS, Ridnour LA, Wink DA. Nitric oxide and its gatekeeper thrombospondin-1 in tumor angiogenesis. Clin. Cancer Res. 2007;13:795–798. doi: 10.1158/1078-0432.CCR-06-1758. [DOI] [PubMed] [Google Scholar]
- 109.Csányi G, Yao M, Rodríguez AI, Al Ghouleh I, Sharifi-Sanjani M, Frazziano G, Huang X, Kelley EE, Isenberg JS, Pagano PJ. Thrombospondin-1 regulates blood flow via CD47 receptor-mediated activation of NADPH oxidase 1. Arterioscler. Thromb. Vasc. Biol. 2012;32:2966–2973. doi: 10.1161/ATVBAHA.112.300031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bauer PM, Bauer EM, Rogers NM, Yao M, Feijoo-Cuaresma M, Pilewski JM, Champion HC, Zuckerbraun BS, Calzada MJ, Isenberg JS. Activated CD47 promotes pulmonary arterial hypertension through targeting caveolin-1. Cardiovasc. Res. 2012;93:682–693. doi: 10.1093/cvr/cvr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ambs S, Bennett WP, Merriam WG, Ogunfusika MO, Oser SM, Harrington AM, Shields PG, Felley-Bosco E, Hussain SP, Harris CC. Relationship between p53 mutations and inducible nitric oxide synthase expression in human colorectal cancer. J. Natl. Cancer Inst. 1999;91:86–88. doi: 10.1093/jnci/91.1.86. [DOI] [PubMed] [Google Scholar]
- 112.Ambs S, Hussain SP, Harris CC. Interactive effects of nitric oxide and the p53 tumor suppressor gene in carcinogenesis and tumor progression. FASEB J. 1997;11:443–448. doi: 10.1096/fasebj.11.6.9194524. [DOI] [PubMed] [Google Scholar]
- 113.Ridnour LA, Isenberg JS, Espey MG, Thomas DD, Roberts DD, Wink DA. Nitric oxide regulates angiogenesis through a functional switch involving thrombospondin-1. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13147–13152. doi: 10.1073/pnas.0502979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sengupta R, Holmgren A. Thioredoxin and thioredoxin reductase in relation to reversible S-nitrosylation. Antioxid. Redox Signal. 2013;18:259–269. doi: 10.1089/ars.2012.4716. [DOI] [PubMed] [Google Scholar]
- 115.Sengupta R, Holmgren A. The role of thioredoxin in the regulation of cellular processes by S-nitrosylation. Biochim. Biophys. Acta. 2012;1820:689–700. doi: 10.1016/j.bbagen.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 116.Ford PC, Wink DA, Stanbury DM. Autoxidation kinetics of aqueous nitric oxide. FEBS Lett. 1993;326:1–3. doi: 10.1016/0014-5793(93)81748-o. [DOI] [PubMed] [Google Scholar]
- 117.Pogrebnaya VL, Usov AP, Baranov AV, Nesterenko AI, Bezyazychnyi PI. Oxidation of nitric-oxide by oxygen in liquid-phase. J. Appl. Chem. USSR (Engl. Transl.) 1975;48:1004–1007. [Google Scholar]
- 118.Wink DA, Darbyshire JF, Nims RW, Saavedra JE, Ford PC. Reactions of the bioregulatory agent nitric oxide in oxygenated aqueous media -determination of the kinetics for oxidation and nitrosation by intermediates generated in the NO/O2 reaction. Chem. Res. Toxicol. 1993;6:23–27. doi: 10.1021/tx00031a003. [DOI] [PubMed] [Google Scholar]
- 119.Liu XP, Miller MJS, Joshi MS, Thomas DD, Lancaster JR. Accelerated reaction of nitric oxide with O2- within the hydrophobic interior of biological membranes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2175–2179. doi: 10.1073/pnas.95.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Möller MN, Li Q, Lancaster JR, Denicola A. Acceleration of nitric oxide autoxidation and nitrosation by membranes. IUBMB Life. 2007;59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 121.Zhou XL, Espey MG, Chen JX, Hofseth LJ, Miranda KM, Hussain SP, Wink DA, Harris CC. Inhibitory effects of nitric oxide and nitrosative stress on dopamine-beta-hydroxylase. J. Biol. Chem. 2000;275:21241–21246. doi: 10.1074/jbc.M904498199. [DOI] [PubMed] [Google Scholar]
- 122.Gladwin MT. Nitrite as an intrinsic signaling molecule. Nat. Chem. Biol. 2005;1:245–246. doi: 10.1038/nchembio1005-245. [DOI] [PubMed] [Google Scholar]
- 123.Williams DL, Aldred SE. Inhibition of nitrosation of amines by thiols, alcohols and carbohydrates. Food Chem. Toxicol. 1982;20:79–81. doi: 10.1016/s0278-6915(82)80013-6. [DOI] [PubMed] [Google Scholar]
- 124.Lundberg JO, Gladwin MT, Ahluwalia A, Benjamin N, Bryan NS, Butler A, Cabrales P, Fago A, Feelisch M, Ford PC, Freeman BA, Frenneaux M, Friedman J, Kelm M, Kevil CG, Kim-Shapiro DB, Kozlov AV, Lancaster JR, Lefer D, McColl K, McCurry K, Patel R, Petersson J, Rassaf T, Reutov VP, Richter-Addo GB, Schechter A, Shiva S, Tsuchiya K, van Faassen EE, Webb AJ, Zuckerbraun BS, Zweier JL, Weitzberg E. Nitrate and nitrite in biology, nutrition and therapeutics. Nat. Chem. Biol. 2009;5:865–869. doi: 10.1038/nchembio.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Casella L, Monzani E, Roncone R, Nicolis S, Sala A, De Riso A. Formation of reactive nitrogen species at biologic heme centers: a potential mechanism of nitric oxide-dependent toxicity. Environ. Health Perspect. 2002;110:709–711. doi: 10.1289/ehp.02110s5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tannenbaum SR. Preventive action of vitamin C on nitrosamine formation. Int. J. Vitam. Nutr. Res. Suppl. 1989;30:109–113. [PubMed] [Google Scholar]
- 127.Sparacino-Watkins CE, Lai YC, Gladwin MT. Nitrate-nitrite-nitric oxide pathway in pulmonary arterial hypertension therapeutics. Circulation. 2012;125:2824–2826. doi: 10.1161/CIRCULATIONAHA.112.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lundberg JO, Weitzberg E. Biology of nitrogen oxides in the gastrointestinal tract. Gut. 2013;62:616–629. doi: 10.1136/gutjnl-2011-301649. [DOI] [PubMed] [Google Scholar]
- 129.Lundberg JO, Weitzberg E. NO generation from inorganic nitrate and nitrite: Role in physiology, nutrition and therapeutics. Arch. Pharm. Res. 2009;32:1119–1126. doi: 10.1007/s12272-009-1803-z. [DOI] [PubMed] [Google Scholar]
- 130.Björne HH, Petersson J, Phillipson M, Weitzberg E, Holm L, Lundberg JO. Nitrite in saliva increases gastric mucosal blood flow and mucus thickness. J. Clin. Invest. 2004;113:106–114. doi: 10.1172/JCI200419019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vergne I, Constant P, Lanéelle G. Phagosomal pH determination by dual fluorescence flow cytometry. Anal. Biochem. 1998;255:127–132. doi: 10.1006/abio.1997.2466. [DOI] [PubMed] [Google Scholar]
- 132.Fallingborg J. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 1999;46:183–196. [PubMed] [Google Scholar]
- 133.The Handbook of Chemistry and Physics. CRC Press; Boca Raton, FL: 1999. [Google Scholar]
- 134.Williams DLH. Nitrosation. Cambridge University Press; Cambridge, UK: 1988. [Google Scholar]
- 135.Williams DLH. Nitrosation Reactions and the Chemistry of Nitric Oxide. Elsevier; Amsterdam: 2004. [Google Scholar]
- 136.van Faassen EE, Bahrami S, Feelisch M, Hogg N, Kelm M, Kim-Shapiro DB, Kozlov AV, Li H, Lundberg JO, Mason R, Nohl H, Rassaf T, Samouilov A, Slama-Schwok A, Shiva S, Vanin AF, Weitzberg E, Zweier J, Gladwin MT. Nitrite as regulator of hypoxic signaling in mammalian physiology. Med. Res. Rev. 2009;29:683–741. doi: 10.1002/med.20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rassaf T, Feelisch M, Kelm M. Circulating NO pool: assessment of nitrite and nitroso species in blood and tissues. Free Radic. Biol. Med. 2004;36:413–422. doi: 10.1016/j.freeradbiomed.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 138.Kleinbongard P, Dejam A, Lauer T, Rassaf T, Schindler A, Picker O, Scheeren T, Gödecke A, Schrader J, Schulz R, Heusch G, Schaub GA, Bryan NS, Feelisch M, Kelm M. Plasma nitrite reflects constitutive nitric oxide synthase activity in mammals. Free Radic. Biol. Med. 2003;35:790–796. doi: 10.1016/s0891-5849(03)00406-4. [DOI] [PubMed] [Google Scholar]
- 139.Pelletier MM, Kleinbongard P, Ringwood L, Hito R, Hunter CJ, Schechter AN, Gladwin MT, Dejam A. The measurement of blood and plasma nitrite by chemiluminescence: pitfalls and solutions. Free Radic. Biol. Med. 2006;41:541–548. doi: 10.1016/j.freeradbiomed.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 140.Jourd'heuil D, Jourd'heuil FL, Kutchukian PS, Musah RA, Wink DA, Grisham MB. Reaction of superoxide and nitric oxide with peroxynitrite - implications for peroxynitrite-mediated oxidation reactions in vivo. J. Biol. Chem. 2001;276:28799–28805. doi: 10.1074/jbc.M102341200. [DOI] [PubMed] [Google Scholar]
- 141.Lancaster JR. Nitroxidative, nitrosative, and nitrative stress: kinetic predictions of reactive nitrogen species chemistry under biological conditions. Chem. Res. Toxicol. 2006;19:1160–1174. doi: 10.1021/tx060061w. [DOI] [PubMed] [Google Scholar]
- 142.Gunaydin H, Houk KN. Molecular dynamics simulation of the HOONO decomposition and the HO*/NO2* caged radical pair in water. J. Am. Chem. Soc. 2008;130:10036–10037. doi: 10.1021/ja711365e. [DOI] [PubMed] [Google Scholar]
- 143.Lymar SV, Jiang Q, Hurst JK. Mechanism of carbon dioxide-catalyzed oxidation of tyrosine by peroxynitrite. Biochemistry. 1996;35:7855–7861. doi: 10.1021/bi960331h. [DOI] [PubMed] [Google Scholar]
- 144.Gorbunov NV, Osipov AN, Day BW, Zayas-Rivera B, Kagan VE, Elsayed NM. Reduction of ferrylmyoglobin and ferrylhemoglobin by nitric oxide: a protective mechanism against ferryl hemoprotein-induced oxidations. Biochemistry. 1995;34:6689–6699. doi: 10.1021/bi00020a014. [DOI] [PubMed] [Google Scholar]
- 145.Kanner J, Harel S, Granit R. Nitric oxide as an antioxidant. Arch. Biochem. Biophys. 1991;289:130–136. doi: 10.1016/0003-9861(91)90452-o. [DOI] [PubMed] [Google Scholar]
- 146.Wink DA, Nims RW, Saavedra JE, Utermahlen WE, Ford PC. The Fenton oxidation mechanism - reactivities of biologically relevant substrates with two oxidizing intermediates differ from those predicted for the hydroxyl radical. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6604–6608. doi: 10.1073/pnas.91.14.6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Takahama U, Hirota S. Nitrogen dioxide-dependent oxidation of uric acid in the human oral cavity under acidic conditions: implications for its occurrence in acidic dental plaque. Chem. Res. Toxicol. 2010;23:1067–1075. doi: 10.1021/tx1000464. [DOI] [PubMed] [Google Scholar]
- 148.Fernandez BO, Ford PC. Nitrite catalyzes ferriheme protein reductive nitrosylation. J. Am. Chem. Soc. 2003;125:10510–10511. doi: 10.1021/ja036693b. [DOI] [PubMed] [Google Scholar]
- 149.Thomas DD, Espey MG, Vitek MP, Miranda KM, Wink DA. Protein nitration is mediated by heme and free metals through Fenton-type chemistry: an alternative to the NO/O2- reaction. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12691–12696. doi: 10.1073/pnas.202312699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Donzelli S, Espey MG, Flores-Santana W, Switzer CH, Yeh GC, Huang J, Stuehr DJ, King SB, Miranda KM, Wink DA. Generation of nitroxyl by heme protein-mediated peroxidation of hydroxylamine but not N-hydroxy-L-arginine. Free Radic. Biol. Med. 2008;45:578–584. doi: 10.1016/j.freeradbiomed.2008.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kuncewicz T, Balakrishnan P, Snuggs MB, Kone BC. Specific association of nitric oxide synthase-2 with Rac isoforms in activated murine macrophages. Am. J. Physiol. Renal Physiol. 2001;281:F326–F336. doi: 10.1152/ajprenal.2001.281.2.F326. [DOI] [PubMed] [Google Scholar]
- 152.Jyoti A, Singh AK, Dubey M, Kumar S, Saluja R, Keshari RS, Verma A, Chandra T, Kumar A, Bajpai V, Barthwal MK, Dikshit M. Interaction of inducible nitric oxide synthase with rac2 regulates reactive oxygen and nitrogen species generation in the human neutrophil phagosomes: implication in microbial killing. Antioxid. Redox Signal. 2014;20:417–431. doi: 10.1089/ars.2012.4970. [DOI] [PubMed] [Google Scholar]
- 153.Hordijk PL. Regulation of NADPH oxidases: the role of Rac proteins. Circ. Res. 2006;98:453–462. doi: 10.1161/01.RES.0000204727.46710.5e. [DOI] [PubMed] [Google Scholar]
- 154.Harvey BH. Acid-dependent dismutation of nitrogen oxides may be a critical source of nitric oxide in human macrophages. Med. Hypotheses. 2000;54:829–831. doi: 10.1054/mehy.1999.0960. [DOI] [PubMed] [Google Scholar]
- 155.O'Brien L, Carmichael J, Lowrie DB, Andrew PW. Strains of Mycobacterium tuberculosis differ in susceptibility to reactive nitrogen intermediates in vitro. Infect. Immun. 1994;62:5187–5190. doi: 10.1128/iai.62.11.5187-5190.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J. Biol. Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- 157.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schopfer FJ, Baker PR, Giles G, Chumley P, Batthyany C, Crawford J, Patel RP, Hogg N, Branchaud BP, Lancaster JR, Freeman BA. Fatty acid transduction of nitric oxide signaling. Nitrolinoleic acid is a hydrophobically stabilized nitric oxide donor. J. Biol. Chem. 2005;280:19289–19297. doi: 10.1074/jbc.M414689200. [DOI] [PubMed] [Google Scholar]
- 159.Deeb RS, Nuriel T, Cheung C, Summers B, Lamon BD, Gross SS, Hajjar DP. Characterization of a cellular denitrase activity that reverses nitration of cyclooxygenase. Am. J. Physiol.-Heart Circul. Physiol. 2013;305:H687–H698. doi: 10.1152/ajpheart.00876.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kamisaki Y, Wada K, Bian K, Balabanli B, Davis K, Martin E, Behbod F, Lee YC, Murad F. An activity in rat tissues that modifies nitrotyrosine-containing proteins. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11584–11589. doi: 10.1073/pnas.95.20.11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Beckman JS, Ischiropoulos H, Zhu L, van der Woerd M, Smith C, Chen J, Harrison J, Martin JC, Tsai M. Kinetics of superoxide dismutase- and iron-catalyzed nitration of phenolics by peroxynitrite. Arch. Biochem. Biophys. 1992;298:438–445. doi: 10.1016/0003-9861(92)90432-v. [DOI] [PubMed] [Google Scholar]
- 162.Ischiropoulos H, Zhu L, Beckman JS. Peroxynitrite formation from macrophage-derived nitric oxide. Arch. Biochem. Biophys. 1992;298:446–451. doi: 10.1016/0003-9861(92)90433-w. [DOI] [PubMed] [Google Scholar]
- 163.Ridnour LA, Barasch KM, Windhausen AN, Dorsey TH, Lizardo MM, Yfantis HG, Lee DH, Switzer CH, Cheng RY, Heinecke JL, Brueggemann E, Hines HB, Khanna C, Glynn SA, Ambs S, Wink DA. Nitric oxide synthase and breast cancer: role of TIMP-1 in NO-mediated Akt activation. PLOS ONE. 2012;7:e44081. doi: 10.1371/journal.pone.0044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Reynolds MR, Berry RW, Binder LI. Nitration in neurodegeneration: deciphering the “Hows” “nYs”. Biochemistry. 2007;46:7325–7336. doi: 10.1021/bi700430y. [DOI] [PubMed] [Google Scholar]
- 165.Franco MC, Ye Y, Refakis CA, Feldman JL, Stokes AL, Basso M, Melero Fernández de Mera RM, Sparrow NA, Calingasan NY, Kiaei M, Rhoads TW, Ma TC, Grumet M, Barnes S, Beal MF, Beckman JS, Mehl R, Estévez AG. Nitration of Hsp90 induces cell death. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E1102–1111. doi: 10.1073/pnas.1215177110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Heinrich T, da Silva R, Miranda KM, Switzer CH, Wink DA, Fukuto JM. Biological nitric oxide signaling: chemistry and terminology. Br. J. Pharmacol. 2013;169:1417–1429. doi: 10.1111/bph.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Issenberg P. Nitrite, nitrosamines, and cancer. Fed. Proc. 1976;35:1322–1326. [PubMed] [Google Scholar]
- 168.Leaf CD, Tannenbaum SR. The role of dietary nitrate and nitrite in human cancer. In: Watson RR, editor. Nutrition and Cancer Prevention. CRC Press; 1996. pp. 317–324. [Google Scholar]
- 169.Routledge MN, Wink DA, Keefer LK, Dipple A. Mutations induced by saturated aqueous nitric oxide in the Psp189 supf gene in human Ad293 and Escherichia coli Mbm7070 cells. Carcinogenesis. 1993;14:1251–1254. doi: 10.1093/carcin/14.7.1251. [DOI] [PubMed] [Google Scholar]
- 170.Stamler JS. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 171.Go YM, Jones DP. The redox proteome. J. Biol. Chem. 2013;288:26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Go YM, Jones DP. Thiol/disulfide redox states in signaling and sensing. Crit. Rev. Biochem. Mol. Biol. 2013;48:173–181. doi: 10.3109/10409238.2013.764840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Forman HJ, Maiorino M, Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Barnett DJ, Mcaninly J, Williams DLH. Transnitrosation between nitrosothiols and thiols. J. Chem. Soc. Perkin Trans. 1. 1994;2:1131–1133. [Google Scholar]
- 175.Broniowska KA, Hogg N. The chemical biology of S-nitrosothiols. Antioxid. Redox Signal. 2012;17:969–980. doi: 10.1089/ars.2012.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hogg N. The kinetics of S-transnitrosation, a reversible second-order reaction. Anal. Biochem. 1999;272:257–262. doi: 10.1006/abio.1999.4199. [DOI] [PubMed] [Google Scholar]
- 177.Hickok JR, Vasudevan D, Thatcher GR, Thomas DD. Is S-nitrosocysteine a true surrogate for nitric oxide? Antioxid. Redox Signal. 2012;17:962–968. doi: 10.1089/ars.2012.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wong PSY, Hyun J, Fukuto JM, Shirota FN, DeMaster EG, Shoeman DW, Nagasawa HT. Reaction between S-nitrosothiols and thiols: generation of nitroxyl (HNO) and subsequent chemistry. Biochemistry. 1998;37:5362–5371. doi: 10.1021/bi973153g. [DOI] [PubMed] [Google Scholar]
- 179.Arnelle DR, Stamler JS. NO+, NO, and NO- donation by S-nitrosothiols -implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys. 1995;318:279–285. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 180.Miranda KM. The chemistry of nitroxyl (HNO) and implications in biology. Coord. Chem. Rev. 2005;249:433–455. [Google Scholar]
- 181.Fukuto JM, Chiang K, Hszieh R, Wong P, Chaudhuri G. The pharmacological activity of nitroxyl: a potent vasodilator with activity similar to nitric oxide and/or endothelium-derived relaxing factor. J. Pharmacol. Exp. Ther. 1992;263:546–551. [PubMed] [Google Scholar]
- 182.Fukuto JM, Wallace GC, Hszieh R, Chaudhuri G. Chemical oxidation of N-hydroxyguanidine compounds: release of nitric oxide, nitroxyl and possible relationship to the mechanism of biological nitric oxide generation. Biochem. Pharmacol. 1992;43:607–613. doi: 10.1016/0006-2952(92)90584-6. [DOI] [PubMed] [Google Scholar]
- 183.Fukuto JM, Hobbs AJ, Ignarro LJ. Conversion of nitroxyl (HNO) to nitric oxide (NO) in biological systems - the role of physiological oxidants and relevance to the biological activity of HNO. Biochem. Biophys. Res. Commun. 1993;196:707–713. doi: 10.1006/bbrc.1993.2307. [DOI] [PubMed] [Google Scholar]
- 184.Hobbs AJ, Fukuto JM, Ignarro LJ. Formation of free nitric oxide from L-arginine by nitric oxide synthase - direct enhancement of generation by superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10992–10996. doi: 10.1073/pnas.91.23.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rusche KM, Spiering MM, Marletta MA. Reactions catalyzed by tetrahydrobiopterin-free nitric oxide synthase. Biochemistry. 1998;37:15503–15512. doi: 10.1021/bi9813936. [DOI] [PubMed] [Google Scholar]
- 186.Clague MJ, Wishnok JS, Marletta MA. Formation of N-delta-cyanoornithine from NG-hydroxy-L-arginine and hydrogen peroxide by neuronal nitric oxide synthase: implications for mechanism. Biochemistry. 1997;36:14465–14473. doi: 10.1021/bi971024u. [DOI] [PubMed] [Google Scholar]
- 187.Pufahl RA, Wishnok JS, Marletta MA. Hydrogen peroxide-supported oxidation of N-G-hydroxy-L-arginine by nitric oxide synthase. Biochemistry. 1995;34:1930–1941. doi: 10.1021/bi00006a014. [DOI] [PubMed] [Google Scholar]
- 188.Colton CA, Gbadegesin M, Wink DA, Miranda KM, Espey MG, Vicini S. Nitroxyl anion regulation of the NMDA receptor. J. Neurochem. 2001;78:1126–1134. doi: 10.1046/j.1471-4159.2001.00509.x. [DOI] [PubMed] [Google Scholar]
- 189.Gbadegesin M, Vicini S, Hewett SJ, Wink DA, Espey M, Pluta RM, Colton CA. Hypoxia modulates nitric oxide-induced regulation of NMDA receptor currents and neuronal cell death. Am. J. Physiol.-Cell Physiol. 1999;277:C673–C683. doi: 10.1152/ajpcell.1999.277.4.C673. [DOI] [PubMed] [Google Scholar]
- 190.Miranda KM, Paolocci N, Katori T, Thomas DD, Ford E, Bartberger MD, Espey MG, Kass DA, Feelisch M, Fukuto JM, Wink DA. A biochemical rationale for the discrete behavior of nitroxyl and nitric oxide in the cardiovascular system. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9196–9201. doi: 10.1073/pnas.1430507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Forman HJ, Zhang H, Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol. Aspects Med. 2009;30:1–12. doi: 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Donzelli S, Espey MG, Thomas DD, Mancardi D, Tocchetti CG, Ridnour LA, Paolocci N, King SB, Miranda KM, Lazzarino G, Fukuto JM, Wink DA. Discriminating formation of HNO from other reactive nitrogen oxide species. Free Radic. Biol. Med. 2006;40:1056–1066. doi: 10.1016/j.freeradbiomed.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 193.Johnson GM, Chozinski TJ, Gallagher ES, Aspinwall CA, Miranda KM. Glutathione sulfinamide serves as a selective, endogenous biomarker for nitroxyl following exposure to therapeutic levels of donors. Free Radic. Biol. Med. 2014;76:299–307. doi: 10.1016/j.freeradbiomed.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Johnson GM, Chozinski TJ, Salmon DJ, Moghaddam AD, Chen HC, Miranda KM. Quantitative detection of nitroxyl upon trapping with glutathione and labeling with a specific fluorogenic reagent. Free Radic. Biol. Med. 2013;63:476–844. doi: 10.1016/j.freeradbiomed.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 195.Ignarro LJ, Gruetter CA. Requirement of thiols for activation of coronary arterial guanylate cyclase by glyceryl trinitrate and sodium nitrite: possible involvement of S-nitrosothiols. Biochim. Biophys. Acta. 1980;63:221–231. doi: 10.1016/0304-4165(80)90297-4. [DOI] [PubMed] [Google Scholar]
- 196.Irvine JC, Ritchie RH, Favaloro JL, Andrews KL, Widdop RE, Kemp-Harper BK. Nitroxyl (HNO): the Cinderella of the nitric oxide story. Trends Pharmacol. Sci. 2008;29:601–608. doi: 10.1016/j.tips.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 197.Kemp-Harper BK. Nitroxyl (HNO): a novel redox signaling molecule. Antioxid. Redox Signal. 2011;14:1609–1613. doi: 10.1089/ars.2011.3937. [DOI] [PubMed] [Google Scholar]
- 198.Shiva S, Crawford JH, Ramachandran A, Ceaser EK, Hillson T, Brookes PS, Patel RP, Darley-Usmar VM. Mechanisms of the interaction of nitroxyl with mitochondria. Biochem. J. 2004;379:359–366. doi: 10.1042/BJ20031758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang YT, Piyankarage SC, Williams DL, Thatcher GR. Proteomic profiling of nitrosative stress: protein S-oxidation accompanies S-nitrosylation. ACS Chem. Biol. 2014;9:821–830. doi: 10.1021/cb400547u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hoffman MD, Walsh GM, Rogalski JC, Kast J. Identification of nitroxyl-induced modifications in human platelet proteins using a novel mass spectrometric detection method. Mol. Cell. Proteomics. 2009;8:887–903. doi: 10.1074/mcp.M800230-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Agnetti G, Kaludercic N, Kane LA, Elliott ST, Guo Y, Chakir K, Samantapudi D, Paolocci N, Tomaselli GF, Kass DA, Van Eyk JE. Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circ. Cardiovasc. Genet. 2010;3:78–87. doi: 10.1161/CIRCGENETICS.109.871236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gould N, Doulias PT, Tenopoulou M, Raju K, Ischiropoulos H. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J. Biol. Chem. 2013;288:26473–26479. doi: 10.1074/jbc.R113.460261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Leung J, Wei W, Liu L. S-nitrosoglutathione reductase deficiency increases mutagenesis from alkylation in mouse liver. Carcinogenesis. 2013;34:984–989. doi: 10.1093/carcin/bgt031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Staab CA, Alander J, Brandt M, Lengqvist J, Morgenstern R, Grafström RC, Höög JO. Reduction of S-nitrosoglutathione by alcohol dehydrogenase 3 is facilitated by substrate alcohols via direct cofactor recycling and leads to GSH-controlled formation of glutathione transferase inhibitors. Biochem. J. 2008;413:493–504. doi: 10.1042/BJ20071666. [DOI] [PubMed] [Google Scholar]
- 205.Keceli G, Moore CD, Labonte JW, Toscano JP. NMR detection and study of hydrolysis of HNO-derived sulfinamides. Biochemistry. 2013;52:7387–7396. doi: 10.1021/bi401110f. [DOI] [PubMed] [Google Scholar]
- 206.Abbas K, Riquier S, Drapier JC. Peroxiredoxins and sulfiredoxin at the crossroads of the NO and H2O2 signaling pathways. Methods Enzymol. 2013;527:113–128. doi: 10.1016/B978-0-12-405882-8.00006-4. [DOI] [PubMed] [Google Scholar]
- 207.Riobó NA, Clementi E, Melani M, Boveris A, Cadenas E, Moncada S, Poderoso JJ. Nitric oxide inhibits mitochondrial NADH:ubiquinone reductase activity through peroxynitrite formation. Biochem. J. 2001;359:139–145. doi: 10.1042/0264-6021:3590139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mason MG, Nicholls P, Wilson MT, Cooper CE. Nitric oxide inhibition of respiration involves both competitive (heme) and noncompetitive (copper) binding to cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:708–713. doi: 10.1073/pnas.0506562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cooper CE, Mason MG, Nicholls P. A dynamic model of nitric oxide inhibition of mitochondrial cytochrome c oxidase. Biochim. Biophys. Acta. 2008;1777:867–786. doi: 10.1016/j.bbabio.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 210.Orsi A, Beltrán B, Clementi E, Hallén K, Feelisch M, Moncada S. Continuous exposure to high concentrations of nitric oxide leads to persistent inhibition of oxygen consumption by J774 cells as well as extraction of oxygen by the extracellular medium. Biochem. J. 2000;346:407–412. [PMC free article] [PubMed] [Google Scholar]
- 211.Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dahm CC, Moore K, Murphy MP. Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: implications for the interaction of nitric oxide with mitochondria. J. Biol. Chem. 2006;281:10056–10065. doi: 10.1074/jbc.M512203200. [DOI] [PubMed] [Google Scholar]
- 213.Chouchani ET, Methner C, Nadtochiy SM, Logan A, Pell VR, Ding S, James AM, Cochemé HM, Reinhold J, Lilley KS, Partridge L, Fearnley IM, Robinson AJ, Hartley RC, Smith RA, Krieg T, Brookes PS, Murphy MP. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nature Med. 2013:753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang J, Jin B, Li L, Block ER, Patel JM. Nitric oxide-induced persistent inhibition and nitrosylation of active site cysteine residues of mitochondrial cytochrome-c oxidase in lung endothelial cells. Am. J. Physiol.-Cell Physiol. 2005;288:C840–C849. doi: 10.1152/ajpcell.00325.2004. [DOI] [PubMed] [Google Scholar]
- 215.Diers AR, Broniowska KA, Darley-Usmar VM, Hogg N. Differential regulation of metabolism by nitric oxide and S-nitrosothiols in endothelial cells. Am. J. Physiol.-Heart Circul. Physiol. 2011;301:H803–H812. doi: 10.1152/ajpheart.00210.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Everts B, Amiel E, van der Windt GJ, Freitas TC, Chott R, Yarasheski KE, Pearce EL, Pearce EJ. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Garedew A, Moncada S. Mitochondrial dysfunction and HIF1alpha stabilization in inflammation. J. Cell Sci. 2008;121:3468–3475. doi: 10.1242/jcs.034660. [DOI] [PubMed] [Google Scholar]
- 218.Erusalimsky JD, Moncada S. Nitric oxide and mitochondrial signaling: from physiology to pathophysiology. Arterioscler. Thromb. Vasc. Biol. 2007;27:2524–2531. doi: 10.1161/ATVBAHA.107.151167. [DOI] [PubMed] [Google Scholar]
- 219.Yun J, Johnson JL, Hanigan CL, Locasale JW. Interactions between epigenetics and metabolism in cancers. Front. Oncol. 2012;2:163. doi: 10.3389/fonc.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 221.Hitchler MJ, Domann FE. Metabolic defects provide a spark for the epigenetic switch in cancer. Free Radic. Biol. Med. 2009;47:115–127. doi: 10.1016/j.freeradbiomed.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kabil O, Motl N, Banerjee R. H2S and its role in redox signaling. Biochim. Biophys. Acta. 2014;1844:1355–1366. doi: 10.1016/j.bbapap.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kabil O, Banerjee R. Enzymology of H2S biogenesis, decay and signaling. Antioxid. Redox Signal. 2014;20:770–782. doi: 10.1089/ars.2013.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, Akaike T. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kolluru GK, Shen X, Bir SC, Kevil CG. Hydrogen sulfide chemical biology: pathophysiological roles and detection. Nitric Oxide. 2013;35:5–20. doi: 10.1016/j.niox.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Miller TW, Kaur S, Ivins-O'Keefe K, Roberts DD. Thrombospondin-1 is a CD47-dependent endogenous inhibitor of hydrogen sulfide signaling in T cell activation. Matrix Biol. 2013;32:316–324. doi: 10.1016/j.matbio.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Francoleon NE, Carrington SJ, Fukuto JM. The reaction of H(2)S with oxidized thiols: generation of persulfides and implications to H2S biology. Arch. Biochem. Biophys. 2011;516:146–153. doi: 10.1016/j.abb.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 229.King SB. Potential biological chemistry of hydrogen sulfide (H2S) with the nitrogen oxides. Free Radic. Biol. Med. 2013;55:1–7. doi: 10.1016/j.freeradbiomed.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Filipovic MR, Miljkovic JL, Nauser T, Royzen M, Klos K, Shubina T, Koppenol WH, Lippard SJ. Ivanovi -Burmazovi , I. Chemical characterization of the smallest S-nitrosothiol, HSNO; cellular cross-talk of H2S and S-nitrosothiols. J. Am. Chem. Soc. 2012;134:12016–12027. doi: 10.1021/ja3009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cortese-Krott MM, Fernandez BO, Santos JLT, Mergia E, Grman M, Nagy P, Kelm M, Butler AR, Feelisch M. Nitrosopersulfide (SSNO(−)) accounts for sustained NO bioactivity of S-nitrosothiols following reaction with sulfide. Redox Biol. 2014;2:234–244. doi: 10.1016/j.redox.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Gladwin MT, Shelhamer JH, Schechter AN, Pease-Fye ME, Waclawiw MA, Panza JA, Ognibene FP, Cannon RO. Role of circulating nitrite and S-nitrosohemoglobin in the regulation of regional blood flow in humans. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11482–11487. doi: 10.1073/pnas.97.21.11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Rassaf T, Kleinbongard P, Preik M, Dejam A, Gharin i. P., Lauer T, Erckenbrecht J, Duschin A, Schulz R, Heusch G, Feelisch M, Kelm M. Plasma nitrosothiols contribute to the systemic vasodilator effects of intravenously applied NO: experimental and clinical Study on the fate of NO in human blood. Circ. Res. 2002;91:470–477. doi: 10.1161/01.res.0000035038.41739.cb. [DOI] [PubMed] [Google Scholar]
- 234.Jourd'heuil D, Hallén K, Feelisch M, Grisham MB. Dynamic state of S-nitrosothiols in human plasma and whole blood. Free Radic. Biol. Med. 2000;28:409–417. doi: 10.1016/s0891-5849(99)00257-9. [DOI] [PubMed] [Google Scholar]
- 235.Halliwell B, Gutteridge JM. C. Free Radicals in Biology and Medicine. Clarendon Press; Oxford: 1985. [DOI] [PubMed] [Google Scholar]
- 236.Minotti G. The role of an endogenous nonheme iron in microsomal redox reactions. Arch. Biochem. Biophys. 1992;297:189–198. doi: 10.1016/0003-9861(92)90661-f. [DOI] [PubMed] [Google Scholar]
- 237.Butler AR, Megson IL. Non-heme iron nitrosyls in biology. Chem. Rev. 2002;102:1155–1165. doi: 10.1021/cr000076d. [DOI] [PubMed] [Google Scholar]
- 238.Cooper CE. Nitric oxide and iron proteins. Biochim. Biophys. Acta-Bioenerg. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 239.Fontecave M. Ribonucleotide reductases and radical reactions. Cell Mol. Life Sci. 1998;54:684–965. doi: 10.1007/s000180050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Flores-Santana W, Switzer C, Ridnour LA, Basudhar D, Mancardi D, Donzelli S, Thomas DD, Miranda KM, Fukuto JM, Wink DA. Comparing the chemical biology of NO and HNO. Arch. Pharm. Res. 2009;32:1139–1153. doi: 10.1007/s12272-009-1805-x. [DOI] [PubMed] [Google Scholar]
- 241.Koskenkorva-Frank TS, Weiss G, Koppenol WH, Burckhardt S. The complex interplay of iron metabolism, reactive oxygen species, and reactive nitrogen species: insights into the potential of various iron therapies to induce oxidative and nitrosative stress. Free Radic. Biol. Med. 2013;65:1174–1194. doi: 10.1016/j.freeradbiomed.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 242.Bouton C, Raveau M, Drapier JC. Modulation of iron regulatory protein functions. Further insights into the role of nitrogen- and oxygen-derived reactive species. J. Biol. Chem. 1996;271:2300–2306. doi: 10.1074/jbc.271.4.2300. [DOI] [PubMed] [Google Scholar]
- 243.Drapier JC, Bouton C. Modulation by nitric oxide of metalloprotein regulatory activities. Bioessays. 1996;18:549–556. doi: 10.1002/bies.950180706. [DOI] [PubMed] [Google Scholar]
- 244.Hentze MW, Kuhn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Klausner RD, Rouault TA, Harford JB. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 246.Kim YM, Bergonia HA, Muller C, Pitt BR, Watkins WD, Lancaster JRJ. Loss and degradation of enzyme-bound heme induced by cellular nitric oxide synthesis. J. Biol. Chem. 1995;270:5710–5713. doi: 10.1074/jbc.270.11.5710. [DOI] [PubMed] [Google Scholar]
- 247.Metzen E, Zhou J, Jelkmann W, Fandrey J, Brüne B. Nitric oxide impairs normoxic degradation of HIF-1alpha by inhibition of prolyl hydroxylases. Mol. Biol. Cell. 2003;14:3470–8341. doi: 10.1091/mbc.E02-12-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hickok JR, Vasudevan D, Antholine WE, Thomas DD. Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases. J. Biol. Chem. 2013;288:16004–16015. doi: 10.1074/jbc.M112.432294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hickok JR, Sahni S, Shen H, Arvind A, Antoniou C, Fung LW, Thomas DD. Dinitrosyliron complexes are the most abundant nitric oxide-derived cellular adduct: biological parameters of assembly and disappearance. Free Radic. Biol. Med. 2011;51:1558–1556. doi: 10.1016/j.freeradbiomed.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lancaster JRJ, Hibbs JBJ. EPR demonstration of iron-nitrosyl complex formation by cytotoxic activated macrophages. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1223–1227. doi: 10.1073/pnas.87.3.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hickok JR, Sahni S, Mikhed Y, Bonini MG, Thomas DD. Nitric oxide suppresses tumor cell migration through N-Myc downstream-regulated gene-1 (NDRG1) expression: role of chelatable iron. J. Biol. Chem. 2011;286:41413–41424. doi: 10.1074/jbc.M111.287052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sahni S, Bae DH, Lane DJ, Kovacevic Z, Kalinowski DS, Jansson PJ, Richardson DR. The metastasis suppressor, N-myc downstream-regulated gene 1 (NDRG1), inhibits stress-induced autophagy in cancer cells. J. Biol. Chem. 2014;289:9692–9709. doi: 10.1074/jbc.M113.529511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fang BA, Kovačevi Ž, Park KC, Kalinowski DS, Jansson PJ, Lane DJ, Sahni S, Richardson DR. Molecular functions of the iron-regulated metastasis suppressor, NDRG1, and its potential as a molecular target for cancer therapy. Biochim. Biophys. Acta. 2014;1845:1–19. doi: 10.1016/j.bbcan.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 255.Vanin AF. Endothelium-derived relaxing factor is a nitrosyl iron complex with thiol ligands. FEBS Lett. 1991;289:1–3. doi: 10.1016/0014-5793(91)80894-9. [DOI] [PubMed] [Google Scholar]
- 256.Ueno T, Suzuki Y, Fujii S, Vanin AF, Yoshimura T. In vivo nitric oxide transfer of a physiological NO carrier, dinitrosyl dithiolato iron complex, to target complex. Biochem. Pharmacol. 2002;63:485–493. doi: 10.1016/s0006-2952(01)00869-3. [DOI] [PubMed] [Google Scholar]
- 257.Thomsen LL, Miles DW, Happerfield L, Bobrow LG, Knowles RG, Moncada S. Nitric oxide synthase activity in human breast cancer. Br. J. Cancer. 1995;72:41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Cobbs CS, Brenman JE, Aldape KD, Bredt DS, Israel MA. Expression of nitric oxide synthase in human central nervous system tumors. Cancer Res. 1995;55:727–730. [PubMed] [Google Scholar]
- 259.Masri FA, Comhair SA, Koeck T, Xu W, Janocha A, Ghosh S, Dweik RA, Golish J, Kinter M, Stuehr DJ, Erzurum SC, Aulak KS. Abnormalities in nitric oxide and its derivatives in lung cancer. Am. J. Respir. Crit. Care Med. 2005;172:597–605. doi: 10.1164/rccm.200411-1523OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Klotz T, Bloch W, Volberg C, Engelmann U, Addicks K. Selective expression of inducible nitric oxide synthase in human prostate carcinoma. Cancer. 1998;82:1897–1903. [PubMed] [Google Scholar]
- 261.Lagares-Garcia JA, Moore RA, Collier B, Heggere M, Diaz F, Qian F. Nitric oxide synthase as a marker in colorectal carcinoma. Am. Surg. 2001;67:709–713. [PubMed] [Google Scholar]
- 262.Hajri A, Metzger E, Vallat F, Coffy S, Flatter E, Evrard S, Marescaux J, Aprahamian M. Role of nitric oxide in pancreatic tumour growth: in vivo and in vitro studies. Br. J. Cancer. 1998;78:841–849. doi: 10.1038/bjc.1998.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Weninger W, Rendl M, Pammer J, Mildner M, Tschugguel W, Schneeberger C, Stürzl M, Tschachler E. Nitric oxide synthases in Kaposi's sarcoma are expressed predominantly by vessels and tissue macrophages. Lab. Invest. 1998;78:949–955. [PubMed] [Google Scholar]
- 264.Ekmekcioglu S, Ellerhorst J, Smid CM, Prieto VG, Munsell M, Buzaid AC, Grimm EA. Inducible nitric oxide synthase and nitrotyrosine in human metastatic melanoma tumors correlate with poor survival. Clin. Cancer Res. 2000;6:4768–4775. [PubMed] [Google Scholar]
- 265.Ambs S, Merriam WG, Bennett WP, Felley-Bosco E, Ogunfusika MO, Oser SM, Klein S, Shields PG, Billiar TR, Harris CC. Frequent nitric oxide synthase-2 expression in human colon adenomas: implication for tumor angiogenesis and colon cancer progression. Cancer Res. 1998;58:334–341. [PubMed] [Google Scholar]
- 266.Gallo O, Masini E, Morbidelli L, Franchi A, Fini-Storchi I, Vergari WA, Ziche M. Role of nitric oxide in angiogenesis and tumor progression in head and neck cancer. J. Natl. Cancer Inst. 1998;90:587–589. doi: 10.1093/jnci/90.8.587. [DOI] [PubMed] [Google Scholar]
- 267.Glynn SA, Boersma BJ, Dorsey TH, Yi M, Yfantis HG, Ridnour LA, Martin DN, Switzer CH, Hudson RS, Wink DA, Lee DH, Stephens RM, Ambs S. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J. Clin. Invest. 2010;120:3843–3854. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lechner M, Lirk P, Rieder J. Inducible nitric oxide synthase (iNOS) in tumor biology: the two sides of the same coin. Semin. Cancer Biol. 2005;15:277–278. doi: 10.1016/j.semcancer.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 269.Menon C, Bauer TW, Kelley ST, Raz DJ, Bleier JI, Patel K, Steele K, Prabakaran I, Shifrin A, Buerk DG, Sehgal CM, Fraker DL. Tumoricidal activity of high-dose tumor necrosis factor-alpha is mediated by macrophage-derived nitric oxide burst and permanent blood flow shutdown. Int. J. Cancer. 2008;123:464–475. doi: 10.1002/ijc.23499. [DOI] [PubMed] [Google Scholar]
- 270.Singh S, Gupta AK. Nitric oxide: role in tumour biology and iNOS/NO-based anticancer therapies. Cancer Chemother. Pharmacol. 2011;67:1211–1224. doi: 10.1007/s00280-011-1654-4. [DOI] [PubMed] [Google Scholar]
- 271.Cook JA, Krishna MC, Pacelli R, DeGraff W, Liebmann J, Mitchell JB, Russo A, Wink DA. Nitric oxide enhancement of melphalan-induced cytotoxicity. Br. J. Cancer. 1997;76:325–334. doi: 10.1038/bjc.1997.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wink DA, Cook JA, Christodoulou D, Krishna MC, Pacelli R, Kim S, DeGraff W, Gamson J, Vodovotz Y, Russo A, Mitchell JB. Nitric oxide and some nitric oxide donor compounds enhance the cytotoxicity of cisplatin. Nitric Oxide. 1997;1:88–94. doi: 10.1006/niox.1996.0108. [DOI] [PubMed] [Google Scholar]
- 273.Gray LH, Green FO, Hawes CA. Effect of nitric oxide on the radiosensitivity of tumour cells. Nature. 1958;182:952–953. doi: 10.1038/182952a0. [DOI] [PubMed] [Google Scholar]
- 274.Mitchell JB, Wink DA, Degraff W, Gamson J, Keefer LK, Krishna MC. Hypoxic mammalian cell radiosensitization by nitric oxide. Cancer Res. 1993;53:5845–5848. [PubMed] [Google Scholar]
- 275.Mitchell JB, Cook JA, Krishna MC, DeGraff W, Gamson J, Fisher J, Christodoulou D, Wink DA. Radiation sensitisation by nitric oxide releasing agents. Br. J. Cancer. 1996;74:S181–S184. [PMC free article] [PubMed] [Google Scholar]
- 276.Bonavida B, Baritaki S. Dual role of NO donors in the reversal of tumor cell resistance and EMT: downregulation of the NF- B/Snail/YY1/RKIP circuitry. Nitric Oxide. 2011;24:1–7. doi: 10.1016/j.niox.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 277.Wilson KE, Li Z, Kara M, Gardner KL, Roberts DD. Beta 1 integrinand proteoglycan-mediated stimulation of T lymphoma cell adhesion and mitogen-activated protein kinase signaling by thrombospondin-1 and thrombospondin-1 peptides. J. Immunol. 1999;163:3621–3628. [PubMed] [Google Scholar]
- 278.Kaur S, Martin-Manso G, Pendrak ML, Garfield SH, Isenberg JS, Roberts DD. Thrombospondin-1 inhibits VEGF receptor-2 signaling by disrupting its association with CD47. J. Biol. Chem. 2010;285:38923–38932. doi: 10.1074/jbc.M110.172304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ng TF, Turpie B, Masli S. Thrombospondin-1-mediated regulation of microglia activation after retinal injury. Invest. Ophthalmol. Vis. Sci. 2009;50:5472–5478. doi: 10.1167/iovs.08-2877. [DOI] [PubMed] [Google Scholar]
- 280.Switzer CH, Cheng RY, Vitek TM, Christensen DJ, Wink DA, Vitek MP. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene. 2011;30:2504–2513. doi: 10.1038/onc.2010.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Christensen DJ, Ohkubo N, Oddo J, Van Kanegan MJ, Neil J, Li F, Colton CA, Vitek MP. Apolipoprotein E and peptide mimetics modulate inflammation by binding the SET protein and activating protein phosphatase 2A. J. Immunol. 2011;186:2535–2542. doi: 10.4049/jimmunol.1002847. [DOI] [PubMed] [Google Scholar]
- 282.Switzer CH, Ridnour LA, Cheng R, Heinecke J, Burke A, Glynn S, Ambs S, Wink DA. S-Nitrosation mediates multiple pathways that lead to tumor progression in estrogen receptor-negative breast cancer. For. Immunopathol. Dis. Therap. 2012;3:117–124. doi: 10.1615/ForumImmunDisTher.2012006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Thomas DD, Ridnour LA, Espey MG, Donzelli S, Ambs S, Hussain SP, Harris CC, DeGraff W, Roberts DD, Mitchell JB, Wink DA. Superoxide fluxes limit nitric oxide-induced signaling. J. Biol. Chem. 2006;281:25984–25993. doi: 10.1074/jbc.M602242200. [DOI] [PubMed] [Google Scholar]
- 284.Edirisinghe I, Arunachalam G, Wong C, Yao H, Rahman A, Phipps RP, Jin ZG, Rahman I. Cigarette-smoke-induced oxidative/nitrosative stress impairs VEGF- and fluid-shear-stress-mediated signaling in endothelial cells. Antioxid Redox Signal. 2010;12:1355–1369. doi: 10.1089/ars.2009.2874. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 285.Koeck T, Stuehr DJ, Aulak KS. Mitochondria and regulated tyrosine nitration. Biochem. Soc. Trans. 2005;33:1399–1403. doi: 10.1042/BST0331399. [DOI] [PubMed] [Google Scholar]
- 286.Yakovlev VA, Bayden AS, Graves PR, Kellogg GE, Mikkelsen RB. Nitration of the tumor suppressor protein p53 at tyrosine 327 promotes p53 oligomerization and activation. Biochemistry. 2010;49:5331–5339. doi: 10.1021/bi100564w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Gomis-Rüth FX, Maskos K, Betz M, Bergner A, Huber RS,K, Yoshida N, Nagase H, Brew K, Bourenkov GP, Bartunik H, Bode W. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature. 1997;389:77–81. doi: 10.1038/37995. [DOI] [PubMed] [Google Scholar]
- 288.Krishnatry AS, Kamei T, Wang H, Qu J, Fung HL. Identification of nitroglycerin-induced cysteine modifications of pro-matrix metalloproteinase-9. Rapid Commun, Mass Spectrom. 2011;25:2291–2298. doi: 10.1002/rcm.5118. [DOI] [PubMed] [Google Scholar]
- 289.Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 290.Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- 291.Chirco R, Liu XW, Jung KK, Kim HR. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006;25:99–113. doi: 10.1007/s10555-006-7893-x. [DOI] [PubMed] [Google Scholar]
- 292.Bommarito A, Richiusa P, Carissimi E, Pizzolanti G, Rodolico V, Zito G, Criscimanna A, Di Blasi F, Pitrone M, Zerilli M, Amato M, Spinelli G, Carina V, Modica G, Latteri M, Galluzzo A, Giordano C. BRAFV600E mutation, TIMP-1 upregulation, and NF- B activation: closing the loop on the papillary thyroid cancer trilogy. Endocr. Relat. Cancer. 2011;18:669–685. doi: 10.1530/ERC-11-0076. [DOI] [PubMed] [Google Scholar]
- 293.Lee S, Kim J, Cho S, Kim H, Shin H, Jeon J, Kausar R, Jeong S, Lee Y, Lee M. TIMP-1 modulates chemotaxis of human neural stem cells through CD63 and integrin signalling. Biochem. J. 2014;459:565–576. doi: 10.1042/BJ20131119. [DOI] [PubMed] [Google Scholar]
- 294.Stilley JA, Sharpe-Timms KL. TIMP1 contributes to ovarian anomalies in both an MMP-dependent and -independent manner in a rat model. Biol. Reprod. 2012;86:47. doi: 10.1095/biolreprod.111.094680. [DOI] [PMC free article] [PubMed] [Google Scholar]