Abstract
Investigation into intratumoral heterogeneity (ITH) of the epigenome is in a formative stage. The patterns of tumor evolution inferred from epigenetic ITH and genetic ITH are remarkably similar, suggesting widespread co-dependency of these disparate mechanisms. The biological and clinical relevance of epigenetic ITH are becoming more apparent. Rare tumor cells with unique and reversible epigenetic states may drive drug resistance, and the degree of epigenetic ITH at diagnosis may predict patient outcome. This perspective presents these current concepts and clinical implications of epigenetic ITH, and the experimental and computational techniques at the forefront of ITH exploration.
At the time a patient is first diagnosed with cancer, the tumor may be composed of tens of millions of cells. These cell populations have already diversified, producing a tumor that can be highly heterogeneous. Such intratumoral heterogeneity (ITH) has been observed among spatially distinct regions of solid tumors and among individual cells in solid tumors or leukemias (Alpar et al., 2015). Profiling ITH provides a powerful opportunity to trace back through the formation of the malignancy and reconstruct the tumor's evolution, from the tumor initiating events through subsequent stepwise development of malignant clones (Gerlinger et al., 2012; Merlo et al., 2006). The models of tumor evolution, or tumor phylogenies, derived from ITH have improved our understanding of tumorigenesis. Despite the increased understanding, a majority of cancer therapies fail to achieve durable responses, which is often attributed to ITH. Importantly, most clinical trials still do not assess ITH and miss an opportunity to examine the prognostic value of ITH in a controlled setting.
ITH at diagnosis may be altered by selective pressures of cytotoxic or targeted cancer therapies, promoting outgrowth of one or more therapy-resistant tumor cell clones (Sharma et al., 2010). Therapeutic interventions could lead to contraction of ITH in some cases or expansion in others, influencing subsequent response and outcome. In most ITH studies, however, only a small fraction of the tumor is available for analysis. Furthermore, tumor samples typically lack information on where within the heterogeneous tumor they were obtained. Using image guided biopsies instead of random samples, molecular ITH could be compared to ITH charted by advanced imaging of tumors in patients (Sottoriva et al., 2013a).
ITH and tumor evolution have historically been assessed with genetic alterations such as somatic mutation and copy number alteration (CNA). However, an increasing number of studies have shown that in cell lines with a high degree of genetic homogeneity, epigenetic heterogeneity leads to cell to cell variability in response to therapy (Kreso et al., 2013; Sharma et al., 2010). Epigenetic mechanisms that may contribute to ITH include DNA methylation, post-translational modifications of histones and chromatin remodeling, which are essential for genome organization, gene expression and cell function (Portela and Esteller, 2010).
Epigenomics and cancer
Alteration to the epigenome is a fundamental characteristic of nearly all human cancers. Pioneering studies focused on DNA methylation and identified decreased 5-methylcytosine content in tumors compared to normal tissue (Feinberg and Vogelstein, 1983; Gama-Sosa et al., 1983), further loss of 5-methylcytosine during tumor progression (Goelz et al., 1985), and increased methylation in normally unmethylated CpG islands and promoter regions of a wide variety of genes including tumor suppressors (Greger et al., 1994; Herman et al., 1994; Merlo et al., 1995; Myöhänen et al., 1998; Sakai et al., 1991; Stirzaker et al., 1997), metastasis genes (Graff et al., 2000; Graff et al., 1995), and DNA repair genes (Costello et al., 1994; Herman et al., 1998; Kane et al., 1997; Pieper et al., 1991). The changes were hypothesized to affect gene expression and chromosomal stability. Indeed, induction of genome wide hypomethylation via reduction in DNA methyltransferase levels was associated with chromosomal abnormalities and was sufficient to induce aggressive T cell lymphoma in mice (Gaudet et al., 2003), suggesting a potentially causal role.
Several of these epigenetic defects may be linked to genetic mutations. Genes encoding regulators of the epigenome, including the writers, readers and erasers of epigenetic marks, are among the most commonly mutated genes observed across cancer types (Mack et al., 2015; Shen and Laird, 2013). Thus, epigenetic alterations may be a common mechanism linking genetic mutations to cancer phenotypes, although the details on how they are linked are just beginning to be explored. Indeed, recent work suggests that reprogramming of the epigenome to a progenitor-like state may create a cell state required for driver mutations to induce tumorigenesis (Kaufman et al., 2016). This work highlights the importance of studying premalignant cells and model systems to better understand when epigenomic changes arise and how stable they are over time. Naturally, clinical trials using new “epigenetic therapies” have been initiated to target the genetically mutant epigenetic regulators and their associated proteins (Rodriguez-Paredes and Esteller, 2011; Yoo and Jones, 2006).
In contrast to genetic alterations, epigenetic modifications are enzymatically reversible and their maintenance may have lower fidelity through DNA replication and mitosis. It had therefore been unclear whether epigenetic ITH (eITH) would be useful to infer tumor evolution. In normal adult cells, at least, epigenomic patterns in gene regulatory regions contain information related to their embryonic developmental history (Hon et al., 2013; Lowdon et al., 2014). Early ancestral information may therefore be encoded in epigenome patterns of subsequent cell generations.
Recurrent epigenetic alterations are well characterized in many tumor types, but only recently has eITH been profiled on a genome-wide scale. Intratumoral studies of the epigenome have unique advantages because they eliminate major variables that confound other study designs. For example, in contrast to inter-individual comparisons, intratumoral studies control for variables such as age, gender, germline differences and accumulated effects of dietary and environmental exposures, each of which can alter the epigenome. Moreover, interpretations of epigenetic differences that were identified by comparing tumor to normal tissues have been repeatedly questioned because the cell of origin of the tumor, representing the ideal normal tissue control, is often not known or hotly debated. Even when the cell of origin is known, the precise stage of differentiation at which a cell is first transformed impacts the tumor methylation state (Kulis et al., 2012; Kulis et al., 2015; Oakes et al., 2016). Thus investigation of eITH provides unique opportunities to identify epigenetic alterations associated with tumor evolution.
eITH can be examined at the level of histone modifications, chromatin conformation, or DNA methylation. To date, DNA methylation has been the main focus due to the quantitative nature of DNA methylation assays, and the relative ease of obtaining sufficient genomic DNA compared to chromatin. The following sections will thus focus on key issues surrounding ITH of DNA methylation (mITH).
Studies that examined the evolution of DNA methylation from initial tumors to recurrence found that overall DNA methylation levels can be lower at recurrence (Mazor et al., 2015), higher at recurrence (Hogan et al., 2011; Lin et al., 2013), or exhibit patterns that do not change across time (Cahill et al., 2013; Ju et al., 2011; Mazor et al., 2015; Schramm et al., 2015). These seemingly conflicting findings may be related to differences in the tumor type, the choice of the CpG sites analyzed, or to selective pressures of different therapies. Changes in methylation in samples obtained at different times in the course of a patient's disease could reflect clonal evolution or mITH. Here we focus specifically on studies that have explicitly examined mITH, either through multiple intratumoral samplings or quantification of mITH from a single sample per tumor.
ITH of DNA methylation at individual genes
Initial studies of mITH analyzed the promoters of individual tumor suppressor genes and other cancer related genes. Early studies used methylation-sensitive restriction enzyme digestion followed by southern blotting, which was sufficient to identify samples with both methylated and unmethylated signal, but not to determine the extent of mITH across the genome (Costello et al., 1996; Herman et al., 1994; Merlo et al., 1995). Many subsequent studies identified mITH using either the methylation-specific PCR method (MSP), a non-quantitative method with a binarized output (Herman et al., 1996; Klump et al., 1998; Nakamura et al., 2001a; Nakamura et al., 2001b; Watanabe et al., 2001), or a related quantitative assay (Eads et al., 2000a; Eads et al., 2000b). A common practice has been to classify a gene as methylated when both methylated and unmethylated alleles are detected. In many of these “methylated” cases, however, the evidence was also consistent with mITH. Indeed, while many studies examined multiple samples per tumor to identify mITH, few interpreted mixed methylated and unmethylated signal from a single sample as evidence of mITH. Mixed methylated and unmethylated signal was more often attributed to normal cells within the bulk tumor sample.
Quantitative methods for measuring mITH highlighted a high frequency of mITH across cancer types (Aggerholm et al., 1999; Korshunova et al., 2008; Moelans et al., 2014; Stirzaker et al., 1997; Varley et al., 2009). However, it remained difficult to compare the relative degree of mITH due to the lack of standardized methods and thresholds for calling mITH. For example, both Korshunova et al. (2008) and Moelans et al. (2014) examined mITH in breast cancer using valid measures, but methodological differences complicate comparison. Moreover, the methods implemented in these studies were unable to distinguish mITH from normal cell contamination.
Siegmund et al. (2009) took a different approach to mITH in colorectal cancer by analyzing DNA methylation patterns at two genomic loci that were assumed to have no role in gene regulation. In contrast to driver methylation changes (De Carvalho et al., 2012), methylation at such neutral loci were unlikely to be under selective pressure, and therefore could serve as a “molecular clock” to measure mitotic divisions based on the higher error rate of DNA methylation maintenance relative to the error rate of DNA polymerase. In this initial study Siegmund et al. (2009) profiled 8 alleles/cells per sample and 10 to 14 samples per patient, while follow-up studies used an average of 1,500 cells per sample in colorectal cancer (Sottoriva et al., 2013b) and glioblastoma (Sottoriva et al., 2013a). This molecular clock analysis revealed highly heterogeneous tumors, suggesting that the tumors had not undergone any recent clonal expansion or selective sweep (Siegmund et al., 2011; Sottoriva et al., 2013a; Sottoriva et al., 2013b). This conclusion based on neutral sites of DNA methylation, similar to the common practice of including silent somatic mutations in genomic evolutionary analyses, suggests an increase in power to differentiate evolutionary branch points by including non-regulatory domains in epigenomic analyses.
Inferring tumor evolution from genetic ITH
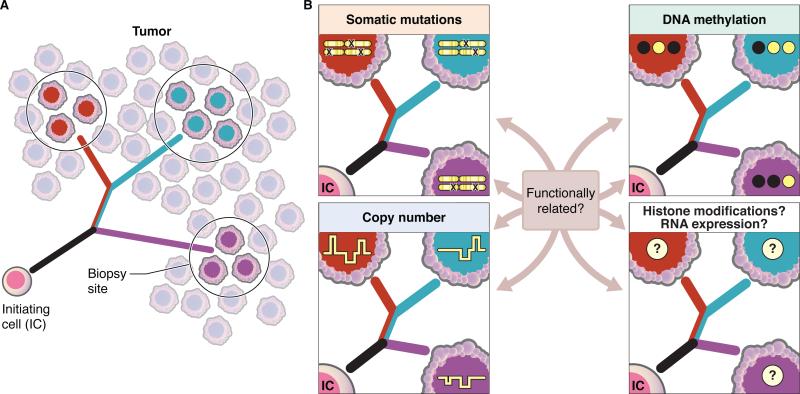
Clonal evolutionary theory (Greaves and Maley, 2012; Merlo et al., 2006; Nowell, 1976) provides a basis to infer the order in which molecular alterations were acquired. Inference of “evolutionary history” of a tumor from somatic mutations relies on the pattern of shared mutations across multiple samples of a tumor: mutations present in all samples of a tumor are inferred to be acquired by early precursor cells which clonally expanded (clonal mutations); in contrast, mutations present in only a subset of samples are inferred to be later events, acquired at some point during or after the initial clonal expansion (subclonal mutations). A seminal publication (Gerlinger et al., 2012) integrated the next-generation sequencing of tumors with principles from the clonal evolution theory of tumors. Gerlinger et al. (2012) performed exome-sequencing of 14 and 10 spatially distinct biopsies from two individuals with metastatic renal cell carcinoma (RCC). Taking advantage of the genetic ITH (gITH) delineated by the multiple samplings per tumor and analyzing the patterns of shared and unique mutations, early and late events were distinguished. Together the events revealed a branched evolutionary history with several instances of convergent evolution in which the same gene was mutated independently in multiple subclones within a single tumor. For each patient, these findings were presented with a phylogenetic tree, a graphical representation of the evolutionary history of a patient's tumor, as inferred from somatic mutations (Figure 1A). This approach to tracing tumor evolutionary history has since been applied across a wide range of malignancies (Cooper et al., 2015; Gerlinger et al., 2014; Gundem et al., 2015; Johnson et al., 2014; Nik-Zainal et al., 2012; Okosun et al., 2014; Schramm et al., 2015).
Figure 1. Inferring the evolutionary history of a tumor from intratumoral heterogeneity.
(A) Analysis of spatially distinct biopsies can be used to build a phylogeny which represents the evolutionary history of a tumor. Phylogeny tree branches are colored according to the contributions of each cell population: the black branch contains alterations that are shared amongst all biopsies, the red/blue branch contains alterations shared between the red and blue biopsies and the red, blue and purple branches represents those alterations which are uniquely present in a single biopsy.
(B) Phylogenies have traditionally been built from genetic alterations, including somatic mutations and copy number alterations. Phyloepigenetic trees have been built from genome-wide DNA methylation data and could be similarly derived from other epigenetic marks, including histone modification patterns, open chromatin or RNA expression levels. Representative patterns of somatic mutations, copy number alterations and DNA methylation (black = methylated CpG site; white = unmethylated CpG site) are shown. To date, tumor histories derived from genetic alterations and DNA methylation show similar evolutionary patterns, raising the question of the extent to which these alterations are functionally related. Further work is still required to determine if phylogenies derived from histone modifications and RNA expression also reflect similar evolutionary histories.
While multiple samplings from a tumor is a powerful approach to profiling spatial heterogeneity and inferring tumor populations based on their spatial distributions, new analytical methods (Ding et al., 2014) have made it possible to infer clonal and subclonal structures from exome or whole genome sequencing of a single sample (Nik-Zainal et al., 2012; Shah et al., 2012). Moreover, by profiling a patient's tumor over time, it is possible to monitor clonal dynamics and infer the evolutionary history of the malignancy (Ding et al., 2012; Landau et al., 2013; Walter et al., 2012).
Inferring tumor evolution from DNA methylation profiles of intratumoral samples
Recent work in solid tumors has extended genome-wide profiling of multiple intratumoral samples from genomics into epigenomics. Given that DNA methylation is reversible and more error prone than DNA replication, the evolutionary history of a tumor might appear different when inferred from genetic versus epigenetic data from the same intratumoral samples. ITH analysis in prostate cancer (Aryee et al., 2013; Brocks et al., 2014) and glioma (Mazor et al., 2015) have shown somewhat surprisingly that the inferred histories from DNA methylation are highly similar to those from CNA or somatic mutation profiles (Figure 1B).
Aryee et al. (2013) and Brocks et al. (2014) used arrays to simultaneously profile genome-wide DNA methylation and CNA in prostate cancer. Brocks et al. (2014) examined multiple samples from the primary tumor site as well as premalignant lesions and metastases from five individuals, while Aryee et al. (2013) examined metastases from thirteen subjects. These studies revealed that while prostate-relevant enhancers frequently demonstrated mITH (Brocks et al., 2014), the sites of promoter mITH and the expression of target genes did not correlate well (Aryee et al., 2013). This may indicate that DNA methylation changes that alter gene expression are more likely to be selected for and become relatively homogenously present across the tumor (Aryee et al., 2013). In both studies, parallel analysis of the genome-wide DNA methylation and CNA produced highly similar tumor evolutionary histories.
Independent genetic and epigenetic approaches were also applied to brain tumors (Mazor et al., 2015). Genome-wide patterns of DNA methylation across six individuals, including multiple samplings of paired initial and recurrent gliomas, were compared to somatic mutations derived from exome-sequencing. Construction of phylogenies independently from the DNA methylation (phyloepigenetic tree) and somatic mutations (phylogenetic tree) yielded highly concordant as well as complementary evolutionary histories.
Together, these studies support the concept of co-dependency of aberrant DNA methylation and genetic alterations, including either CNAs or somatic point mutation. Moreover, evolutionary histories could potentially be inferred from a range of additional data types, allowing future research to address the question of the extent to which other types of epigenetic marks evolve along similar evolutionary paths. Other datasets that could be used to construct phylogenies and infer a tumor's evolutionary history include, but are not limited to, histone modifications or gene expression (Figure 1B). Additional research is still needed to determine the extent to which gITH or eITH reflects differences in regional or clone-specific driver or passenger events, and to what extent the alterations to genetics and epigenetics may be functionally related. It is also not yet known whether the patterns in prostate and glioma data will be widely generalizable to other cancer types.
Linking epigenetic changes to specific genetic alterations
Aberrant epigenetic states may promote genetic instability or may arise from specific genetic alterations (Aumann and Abdel-Wahab, 2014; Brena and Costello, 2007). As an example, using single samples per patient from a large cohort of patients, comparison of samples with and without particular somatic mutations have identified associations between mutations and DNA methylation patterns. These associations reflect both mutations that drove altered DNA methylation, as with IDH1 mutation in glioma (Noushmehr et al., 2010; Turcan et al., 2012) and altered DNA methylation landscapes which promoted the acquisition of particular mutations, as with BRAF mutation in colorectal cancer (Hinoue et al., 2009; Weisenberger et al., 2006).
Despite the success of previous studies identifying the relationship between a mutation and specific methylation changes, these statistical analyses required large cohorts of cases with and without each mutation to overcome the inherent inter-individual variability of DNA methylation arising from germline epigenetic differences, age, gender and other covariates. One approach to identify genetic-epigenetic associations in a smaller cohort is to use intratumoral heterogeneity of mutations and DNA methylation. For example, chromatin modifier genes, including SMARCA4 and BAP1, are often mutated as late events in tumorigenesis and therefore are present heterogeneously within a tumor (Gerlinger et al., 2014; Johnson et al., 2014; Suzuki et al., 2015). By profiling mITH and contrasting the samples with and without mutation in a particular gene, and then extending the analysis across a cohort of patients with similarly heterogeneous mutations in the same gene, substantial inter-patient heterogeneity can be excluded to then identify DNA methylation changes that result from the specific mutation.
mITH from single samples of tumor
In contrast to the spatial heterogeneity of solid tumors, samples from distinct locations (peripheral blood and lymph node) in blood cell malignancies share similar epigenomic patterns (Cahill et al., 2013), suggesting that a single sample represents the full diversity of tumor cell populations. Using an array-based approach, Oakes et al. (2013) calculated the mITH levels of 68 chronic lymphocytic leukemia (CLL) and found that overall CLL had low mITH relative to several solid tumors. As with the analysis of multiple samplings, Oakes et al. (2013) noted that the level of mITH within an individual's leukemia was positively correlated with the level of gITH. Landau et al. (2014) used reduced representation bisulfite sequencing (RRBS), also in CLL, and found a similar correlation between high numbers of subclonal mutations and high mITH. They also discovered a different kind of correlation between genetic and epigenetic heterogeneity. Looking at the genomic locations exhibiting mITH, they noted that CpG sites with high mITH were found in regions of high genetic variation, such as regions of late DNA replication and gene-poor regions. Interestingly, De et al. (2013) also investigated genomic locations with high mITH and found lymphomas to have high mITH in gene-rich regions, suggesting that the association of genomic features and levels of mITH might be variable between different types of malignancies.
Landau et al. (2014) further correlated mITH at promoters with gene expression data from single-cell RNA sequencing. They noted that promoters with high mITH showed high cell-to-cell expression heterogeneity of the corresponding gene. This discrepancy with the finding by Aryee et al. (2013) in prostate cancer may reflect a functional distinction between solid and liquid malignancies, differences in genomic regions profiled, or the power of single-cell RNA sequencing to identify signals that are obscured in bulk sampling.
Evolution of mITH
mITH can be further tracked for evolution over time. Intriguingly, mITH can increase or decrease at relapse. Landau et al. (2014) classified 14 cases of CLL into those that did or did not undergo genetic evolution and found that those patients with genetic evolution showed higher mITH at relapse. Looking in diffuse large B-cell lymphoma (DLBCL), Pan et al. (2015) also calculated mITH from RRBS data in 11 paired diagnostic and relapse samples, but found that the level of mITH decreased at relapse in all but one case. They interpreted this result as reflecting a series of clonal outgrowths from more diverse cell populations, as diagnostic samples showed lower mITH than the normal B cell population from which they arose, and a further decrease in mITH was apparent at relapse.
Computational techniques for inferring epigenetic heterogeneity and evolution
The biological conclusions in these studies are strongly influenced by the computational methods used in the analysis. Thus, it is necessary to understand not just the differences between the analytical methods, but also the advantages and drawbacks in each experimental design.
Methods to infer evolution from multiple intratumoral samples
The availability of multiple samples from individual tumors leads to a natural application of clustering to infer the evolutionary history of tumors. While mutation information is often simplified to a binary readout, DNA methylation data, which measures the fraction of methylated alleles at a given CpG site, is typically represented on a continuous scale from 0 to 1, and often requires heuristic choices in the data analysis that could alter the biological interpretation of the results. Moreover, the few published phyloepigenetic trees from DNA methylation data are built by a distance-based clustering method. In these instances, the choice of distance metric not only dictates the topology and branch lengths, but also determines the relative importance of each CpG site. In contrast, building a distance matrix from binary mutation data weighs clonal and subclonal mutations equally. Thus it is important to understand the biological implications of each distance metric and data type.
A major benefit of understanding the phylogeny of a tumor is better knowledge of the genetic and epigenetic signatures underlying subclones of a tumor. In evolutionary analysis, simple set-difference calculation can often be used to identify the specific mutations responsible for tree branch points (Merlo et al., 2006). However, identifying the CpG sites responsible for a bifurcation in a phyloepigenetic tree is not as straightforward. Mazor et al. (2015) calculated the singular vectors along the samples and determined the weighting of CpG sites when projected onto those directions. A heuristic method was then applied to determine how many CpG sites were most important for forming that bifurcation. Additional work is still required to identify the most appropriate computational techniques for uncovering the epigenetic signature underlying branch points in a phyloepigenetic tree.
Methods to infer heterogeneity from a single sample of a tumor
Many computational methods have been developed to quantify tissue heterogeneity or remove the influence of mixed cell populations when conducting hypothesis testing in both microarray and high-throughput sequencing data. McGregor et al. (2015) and Yadav and De (2015) provide a comprehensive review and simulation results on this topic. In brief, methods to infer ITH can be generally divided into two categories: reference-based approaches that separate an observed heterogeneous sample into proportions of previously profiled cell types and any remaining signal of unknown origin; and reference-free methods that attempt to identify unmeasured confounding variables, such as ITH, with an unsupervised approach.
In a reference-free approach applicable to sequencing data, Landan et al. (2012) used the methylation status of CpG sites across a single read from RRBS to calculate the epipolymorphism, a measure of mITH. For any four CpG sites that are close enough to be covered by a single sequencing read, the epipolymorphism is defined as the probability of any two reads having a different pattern of methylation across those sites, given the overall methylation level. While heavily methylated or unmethylated loci always have a low level of epipolymorphism, because most alleles are identical, intermediately methylated loci can have low (e.g. an imprinted locus) or high values, indicating the heterogeneity of methylation patterns (Elliott et al., 2015; Lister et al., 2009).
An alternate reference-free approach applicable to array data uses the distribution of methylation proportions across all probes to impute the amount of heterogeneity (Oakes et al., 2013). This method takes into account the enrichment of probes centered at 0 and 1 in normal tissue and calculates the deviation from that enrichment in tumor samples. Within a single cell, or a homogenous population of cells, each CpG site can only exist in three states: methylated, unmethylated, or heterozygously methylated. Therefore any intermediate methylation in a tumor, excluding allele-specific methylation, may be considered mITH.
In contrast, reference-based approaches can be applied to deconvoluting normal contaminating cells from the tumor population, especially for those tumor samples for which tumor purity is low. Such an approach has already been developed for expression datasets (Yoshihara et al., 2013). Methylation data has been used to identify component populations of normal tissue (Koestler et al., 2013), suggesting that a reference-based approach may be successful in identifying contaminating normal cells within DNA methylation datasets from tumors. This approach is especially useful for identifying the amount of signal originating from non-tumor cells with well-characterized epigenetic profiles.
Future explorations combining genetic and epigenetic data from spatially separated samples
While important steps have been taken to understand heterogeneity from both multi-sample and single-sample data, additional work is still necessary to compute an overall measure of heterogeneity for a spatially distributed tumor. Thus an essential next step is to develop methods for inferring tumor-wide heterogeneity across multiple spatially distinct samples from the same individual. Lui et al. (2014) demonstrated the effectiveness of one approach for determining heterogeneity across multiple samples. The authors built a gene co-expression network from 96 serial samplings of normal brain tissue. They then identified modules of genes with similar expression profiles across the 96 samples. Finally, using the number of separate modules that the genes were separated into, the authors estimated the number of subtypes present within this tissue. This method has been applied to gene expression, but the underlying technique could be applied to data types including DNA methylation and other epigenetic marks. While the utility of this approach has been demonstrated for normal tissue, further work is required to extend this method into cancer. It is also important to note that even though accurately estimating a large correlation matrix from a small sample size is mathematically difficult, this analysis led to new insights that were independently validated
To better understand tumor heterogeneity between tumor subclones and to build a comprehensive evolutionary history of cancer progression, a novel analytical approach combining genetic and epigenetic data is required. Although several studies have found substantial agreement between tumor evolution traced from DNA methylation compared to somatic mutations or CNAs, it is not yet possible to create a theoretical mathematical model to understand how much co-dependency exists between the genetics and epigenetics, as the rate, timing, and location of exact DNA methylation changes is not well known.
Epigenetic heterogeneity and patient outcomes
One provocative question is the possibility that mITH may be tied to patient outcomes, and moreover that it may be used as a prognostic feature. Analysis of gITH has shown promise as a prognostic feature in a variety of malignancies. In the premalignant condition Barrett's esophagus, increased gITH of lesions correlated with progression to malignant esophageal adenocarcinoma (Maley et al., 2006; Merlo et al., 2010). Similarly, studies in hematopoietic malignancies including acute myeloid leukemia (AML) and CLL have shown that increased gITH, as measured by the number of subclones or subclonal mutations, is associated with worse patient outcomes (Bochtler et al., 2013; Landau et al., 2013).
Several groups have investigated a potential relationship between mITH and outcome (Figure 2A). In CLL, high mITH at diagnosis correlated with a shorter interval until treatment is required (Oakes et al., 2013) and, furthermore, higher mITH also correlated with shorter progression-free survival (PFS) (Landau et al., 2014). In diffuse large B-cell lymphoma (DLBCL), patients that had not relapsed in 5 years post treatment had lower mITH at diagnosis than those that did relapse within 5 years (Pan et al., 2015). Furthermore, lower mITH correlated with longer PFS following initial treatment. In a cohort including follicular lymphoma and DLBCL, De et al. (2013) similarly showed that higher mITH predicts poor PFS. While these initial results are promising, larger studies will be required to determine the value of mITH as a prognostic feature.
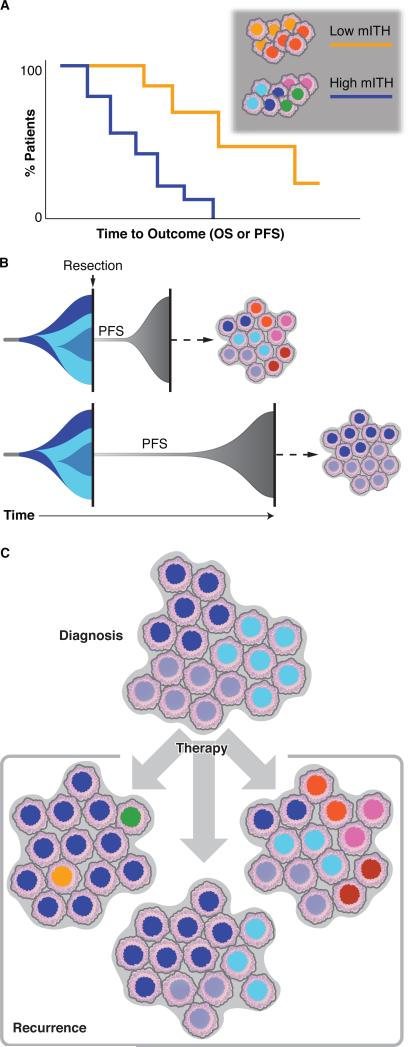
Figure 2. Clinical implications of intratumoral heterogeneity.
(A) Low levels of intratumoral heterogeneity in DNA methylation (mITH) have been associated with improved progression free survival (PFS) and overall survival (OS).
(B) Two different timeline models of the relationship between PFS and mITH. A tumor initiates (far left) and expands while acquiring a series of epigenetic alterations leading to three distinct subclones (shades of blue) at the time of surgical resection (vertical black line). In both scenarios, identical subclones are present at initial resection. Surgical resection removes the majority of tumor cells although a small number remain which continue to evolve and eventually develop into the recurrence after a short (top) or long (bottom) period of progression-free survival (PFS). Here we question whether the duration of PFS may correlate with the levels of mITH in the recurrent tumor (cell populations on right).
(C) The impact of therapy on mITH is understudied. For a given number of subclones at diagnosis (top), therapy may selectively kill some subclones leading to decreased mITH at recurrence (left), or it may not alter the diversity of subclones, leading to a recurrence with similar subclones as the diagnostic tumor (center), or it may promote increased mITH and expansion of novel subclones leading to a more heterogeneous recurrence (right).
It is unclear how mITH and patient outcome will relate in solid tumors. Increased gITH in solid tumors does correlate with poorer outcomes with some contribution of aberrant DNA methylation (Maley et al., 2006; Maley et al., 2004; Merlo et al., 2010). Single-cell analysis of gene expression in glioblastoma showed that detection of multiple expression-based subtypes (Verhaak et al., 2010) in an individual tumor is associated with worse overall survival (Patel et al., 2014). However, analysis of mITH at the single gene level suggests that this may not be universal. DNA methylation at the promoter/enhancer of MGMT is a prognostic marker in glioblastoma (Hegi et al., 2005). Several studies found that the MGMT methylation level is relatively consistent throughout individual gliomas (Dunn et al., 2009; Grasbon-Frodl et al., 2007; Hamilton et al., 2011; van Thuijl et al., 2015). This homogeneity may be a unique feature of MGMT or it may be indicative of a wider epigenetic pattern. Further analysis will be required to address the potential correlation between mITH and outcome in solid tumors.
Application of mITH in a clinical setting will require the development of clinically tractable assays. Methylation at MGMT provides a case study for bringing DNA methylation analyses into a clinical setting. The current standard is non-quantitative MSP, but several groups have investigated methods for quantifying methylation at the MGMT promoter, including pyrosequencing (Christians et al., 2012; Mikeska et al., 2007) and real-time MSP (Vlassenbroeck et al., 2008), the latter of which has been used in clinical trials (Gilbert et al., 2013). Such clinically tractable and quantitative methods could be applied to additional loci, as determined by genome-wide assays, for quantitative measures of mITH and their relationship to response and outcome in patients.
These analyses examined mITH at an early time point and calculated how it relates to PFS. A related question is how heterogeneity of a tumor might change over time: given a particular state of heterogeneity of an initial tumor, is there a positive or negative correlation between the duration of PFS and the level of mITH at recurrence? Given that more heterogeneous initial tumors recur more quickly, it could be argued that continued high levels of mITH would be associated with shorter time to recurrence (Figure 2B). An alternate hypothesis is that the high level of mITH in the initial tumor could provide a larger population from which a single, most-fit subclone can emerge and rapidly regrow into a less heterogeneous recurrence in a shorter time. Future mITH studies with large cohorts of initial and recurrent tumors will be required to address this question across a range of malignancies.
Epigenetic heterogeneity as a driver of cell state and treatment response
Powerful in vitro and in vivo models have shown that epigenetic heterogeneity can drive variable responses to therapy and differences in tumor-propagating potential. Gupta et al. (2011) showed that upon separation of a breast cancer cell line into its basal, luminal, and stem-like cell populations, each cell type expanded into a heterogeneous culture that fully recapitulated the initial cell type heterogeneity through cell state interconversions. Kreso et al. (2013) further showed that after isolating individual cells from the same genetic background and transplanting them into mice, the separate transplants displayed differences in growth dynamics and treatment-response. Similarly, Sharma et al. (2010) found that while the majority of cells in a single cell derived non-small cell lung cancer subline were drug-sensitive, a small subpopulation of cells were drug-tolerant. Following removal of drug, these drug-tolerant persister cells expanded and reacquired drug-sensitivity. Persister cells displayed an altered chromatin landscape, suggesting that concurrent epigenetic therapies could reduce or eliminate them. Indeed, treatment of cell lines with HDAC inhibitors or knockdown of the histone demethylase KDM5A reduced the emergence of persister cells.
These persister cells may model observations that some patients responded initially to therapy, developed resistance, and then responded again to the same chemotherapy after a drug holiday (Cara and Tannock, 2001). Thus, one hypothesis is that eITH at the single-cell level may play a role in therapy responses in patients, and concurrent treatment with epigenetic therapies may improve drug responses (Fang et al., 2014; Matei and Nephew, 2010; Strauss and Figg, 2016). This also raises the question of how eITH is altered in tumors as a result of therapy. As in the in vitro cell models, a tumor may return to a state that is similar to the pre-treatment tumor (Figure 2C, center). Alternatively, a tumor may display increased eITH (Figure 2C, right) (Landau et al., 2014), or decreased eITH due to outgrowth of a small number of resistant cells that dominate the recurrence (Figure 2C, left) (Pan et al., 2015). Additional in vitro experiments with genomic and epigenomic profiling, along with increased analysis of patient tumors before and after therapy will help determine how often each of these models applies.
These models also highlight one of the problems with profiling bulk tumor samples. Measurements from bulk samples are a population average which will underestimate heterogeneity by masking rare subpopulations, even though rare populations can have a significant impact on treatment response. To fully understand eITH and to profile and study common and rare populations, emerging technologies for single-cell sequencing will be useful.
Coming soon: ITH at the single cell level
While numerous ITH-related discoveries have been made, current methods for determining the eITH profile of cancer are limited. Rare cell populations such as persister cells will be undetectable because profiling measures the dominant tumor cell population predominantly, while the rare populations contribute relatively little signal. Additionally, the presence of non-tumor cells can confound and mask signal from the tumor. The advancement of single-cell genome-wide profiling techniques has the potential to overcome the drawbacks associated with the standard population profiling methods.
Studies using genome-wide single-cell measurements to study the genomic landscape in tumors have provided valuable insights that highlight their importance to cancer biology. Single-cell measures of CNA (Navin et al., 2011; Voet et al., 2013) have been used to perform phylogenetic analysis and elucidate the pattern of CNA evolution within tumors. Moreover, single-cell methods for identifying somatic mutations from exome or whole genome sequencing have also been developed and applied to a variety of malignancies (Hou et al., 2012; Li et al., 2012; Wang et al., 2014; Xu et al., 2012). Application of both techniques to primary breast cancer tissue has enhanced our understanding of genetic evolution. Navin et al. (2011) identified a population of pseudo-diploid cells, each with unique CNAs, in each of two primary breast tumors. However no pseudo-diploid cells were found in a patient-matched metastasis, suggesting that these cells may be the product of processes that generate genetic diversity, yet have failed to clonally expand. Wang et al. (2014) identified mutations and CNAs from single cells from breast cancers and inferred chromosome rearrangements that occurred early in tumor evolution, relative to the mutations that gradually accumulated over longer periods of time.
While valuable knowledge has been gained from single-cell genomic technologies, few studies have explored the single-cell epigenomic landscape of cancer tissues. Two recent studies measured chromatin accessibility by applying assay for transposase-accessible chromatin using sequencing (ATAC-seq) to single cells (Buenrostro et al., 2015; Cusanovich et al., 2015) and identified classes of transcription factors with high cell-to-cell variability in the accessibility of their binding motifs within cancer cell lines. Buenrostro et al. (2015) further found that transcription factor binding sites had the most variable accessibility in cell types for which those transcription factors drive cell state, like GATA transcription factors in K562 cells or Nanog in mouse embryonic stem cells, suggesting an important functional role for this variability. Single-cell ChIP-seq has also been developed and used to explore the variable chromatin landscape within populations of embryonic stem cells (Rotem et al., 2015). However the low complexity of reads produced by the approach – on the order of 800 peaks per cell – hampers its application to study ITH in cancer samples. The current state of the technology does not provide sufficient statistical power to reliably differentiate rare cell types in tumors from the technique-specific artifacts that can arise. Whole genome bisulfite sequencing (WGBS) has been performed on the single cell level (Farlik et al., 2015; Smallwood et al., 2014), but exact DNA methylation differences between cells cannot be made at base-pair resolution and therefore regulatory region sets were used to differentiate cell populations. Thus, while single-cell WGBS is available, the resolution of the technique is currently too sparse to reliably identify differentially methylated CpG sites. Higher coverage sequencing of DNA methylation can be obtained with single-cell RRBS (Guo et al., 2015), although in far fewer CpG sites compared to WGBS. While to date these methods have only been applied to cell lines, the next step will be to profile single cells dissociated from primary tumor tissue.
A complementary approach to measuring ITH of individual epigenetic marks is to examine RNA expression, in part as a readout of epigenetic states. Indeed, a recent study simultaneously assessed DNA methylation and RNA expression from the same single cells and were able to associate methylation changes in regulatory regions with same-cell transcriptional changes in mouse embryonic stem cells (Angermueller et al., 2016). In cancer, single-cell RNA sequencing has been applied to study drug tolerance in a metastatic breast cancer cell line (Lee et al., 2014), to detect two distinct subpopulations in a human lung adenocarcinoma patient-derived xenograft that could be separated by a module of cell cycle genes (Min et al., 2015), to understand the influence of promoter methylation on gene expression (Landau et al., 2014), and to investigate ITH in primary glioblastoma (Patel et al., 2014). From the single-cell expression data, Patel et al. (2014) showed that within a primary glioblastoma previously classified into one of the four expression-based tumor subtypes (Verhaak et al., 2010) from bulk expression measurements, individual cells were present that could be classified into the other three expression subtypes. The number of different expression subtypes present in each glioblastoma negatively correlated with patient outcome, suggesting increased ITH might lead to shorter overall survival. Moreover, expression of surface receptors including EGFR, EGFRvIII, PDGFRA and others were highly variably expressed between cells within a single tumor, raising concerns about the efficacy of targeted therapies against proteins with heterogeneous expression (Furnari et al., 2015; Nathanson et al., 2014; Patel et al., 2014; Snuderl et al., 2011). These studies highlight the power of single-cell analysis to better understand the biological and clinical relevance of ITH.
While there have been important discoveries from the application of single-cell technologies to study cancer biology, it is also important to consider their current limitations. Single-cell approaches require novel computational frameworks to take into account their relatively low signal resolution when compared to traditional bulk tumor sample sequencing. However, a main limitation is the potential for artifact introduced by the amplification of low amounts of nucleic acid input, at times making it difficult to distinguish a mutation or methylation call identified in a single cell from an error during amplification or sequencing. Additionally, it is currently not possible to determine if an event (e.g. expression of a given gene) that is not observed in a particular cell reflects true lack of expression or lack of sampling of that particular gene. As the field continues to improve the experimental techniques and analysis methods, the power of these methods will transform our understanding of ITH of the genome, epigenome and transcriptome, and will impact how tumors are classified and which therapies are indicated.
Future Perspectives
eITH can teach us about the evolutionary history of tumors, it may serve as a predictor of patient outcome, and may underlie variable responses to treatment and the re-sensitization of tumors following a drug holiday, and thus may guide new therapeutic strategies. However, many questions remain (Alizadeh et al., 2015). eITH derives from many sources, suggesting multiple future research directions. eITH may reflect tumor cells responding to unique microenvironments or various stages of differentiation from cancer stem cells (Mack et al., 2015; Schonberg et al., 2014). Alternatively, eITH may reflect a mix of subclones with distinct genomic and epigenomic features. Beyond the eITH within tumor cells, additional epigenome variability comes from the variety of other cells present in tumor tissue, including non-tumor stromal and immune cells. These cells can themselves have altered epigenomes, as in the case of tumor-adjacent stroma with altered DNA methylation (Fiegl et al., 2006; Hanson et al., 2006; Rodriguez-Canales et al., 2007). Given the epigenetic heterogeneity present in normal tissues, it will be interesting for future research to compare the level of eITH observed in cancer to the level of epigenetic heterogeneity in normal tissues.
As ITH research progresses, it will become important to standardize measures of mITH, to facilitate comparisons across studies and tumor types. Given the known relationships between genetics and epigenetics, it will be interesting to disentangle the two by investigating the effect of mITH, for example in the context of predicting patient outcomes, while accounting for the extent of gITH. Current measures of eITH significantly underestimate the levels of ITH for several reasons. Signal from bulk tumor samples is dominated by major subclones, rendering rare subpopulations undetectable, although emerging single-cell technologies will facilitate profiling rare cells. Additionally, in nearly every study to date the proportion of a tumor that is assayed is quite small relative to the full tumor mass in the patient. Finally, eITH analyses have been dominated by DNA methylation and including histone modifications and open chromatin in the future would be beneficial. Many questions remain and new research directions are emerging, making eITH an exciting and clinically important topic for investigation, with the potential to revolutionize our understanding of tumors.
Acknowledgements
Illustrations by Kenneth Xavier Probst. This project was generously supported by Accelerate Brain Cancer Cure and a gift from the Dabbiere family. Additional support by the National Institute Of General Medical Sciences T32GM067547 (A.P.); the National Science Foundation 1144247 (A.P.); the National Institutes of Health R01CA163336 (J.S.S.), R01CA169316 (J.F.C.), and P50CA097257 (J.F.C.); the Sontag Foundation (J.S.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
T.M., A.P. and J.F.C. contributed to the concept, writing and editing of the manuscript and figures. J.S.S. provided critical insights as well as oversight of the writing.
The authors declare no competing financial interests.
References
- Aggerholm A, Guldberg P, Hokland M, Hokland P. Extensive intra- and interindividual heterogeneity of p15INK4B methylation in acute myeloid leukemia. Cancer Res. 1999;59:436–441. [PubMed] [Google Scholar]
- Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, Caldas C, Califano A, Doherty M, Elsner M, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpar D, Barber LJ, Gerlinger M. Chapter 24. Genetic Intratumor Heterogeneity. In: Gray SG, editor. Epigenetic Cancer Therapy. Elsevier Inc.; 2015. pp. 569–594. [Google Scholar]
- Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood SA, Ponting CP, Voet T, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods. 2016;13:229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson WG, et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med. 2013;5:169ra110. doi: 10.1126/scitranslmed.3005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumann S, Abdel-Wahab O. Somatic alterations and dysregulation of epigenetic modifiers in cancers. Biochem Biophys Res Commun. 2014;455:24–34. doi: 10.1016/j.bbrc.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler T, Stölzel F, Heilig CE, Kunz C, Mohr B, Jauch A, Janssen JWG, Kramer M, Benner A, Bornhäuser M, et al. Clonal heterogeneity as detected by metaphase karyotyping is an indicator of poor prognosis in acute myeloid leukemia. J Clin Oncol. 2013;31:3898–3905. doi: 10.1200/JCO.2013.50.7921. [DOI] [PubMed] [Google Scholar]
- Brena RM, Costello JF. Genome-epigenome interactions in cancer. Hum Mol Genet 16 Spec No. 2007;1:R96–105. doi: 10.1093/hmg/ddm073. [DOI] [PubMed] [Google Scholar]
- Brocks D, Assenov Y, Minner S, Bogatyrova O, Simon R, Koop C, Oakes C, Zucknick M, Lipka DB, Weischenfeldt J, et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep. 2014;8:798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill N, Bergh A-C, Kanduri M, Göransson-Kultima H, Mansouri L, Isaksson A, Ryan F, Smedby KE, Juliusson G, Sundström C, et al. 450K-array analysis of chronic lymphocytic leukemia cells reveals global DNA methylation to be relatively stable over time and similar in resting and proliferative compartments. Leukemia. 2013;27:150–158. doi: 10.1038/leu.2012.245. [DOI] [PubMed] [Google Scholar]
- Cara S, Tannock IF. Retreatment of patients with the same chemotherapy: implications for clinical mechanisms of drug resistance. Ann Oncol. 2001;12:23–27. doi: 10.1023/a:1008389706725. [DOI] [PubMed] [Google Scholar]
- Christians A, Hartmann C, Benner A, Meyer J, von Deimling A, Weller M, Wick W, Weiler M. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS ONE. 2012;7:e33449. doi: 10.1371/journal.pone.0033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JF, Berger MS, Huang HS, Cavenee WK. Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res. 1996;56:2405–2410. [PubMed] [Google Scholar]
- Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994;14:6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Shaknovich R, Riester M, Elemento O, Geng H, Kormaksson M, Jiang Y, Woolcock B, Johnson N, Polo JM, et al. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet. 2013;9:e1003137. doi: 10.1371/journal.pgen.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Wendl MC, McMichael JF, Raphael BJ. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet. 2014;15:556–570. doi: 10.1038/nrg3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, Crooks D, Husband D, Shenoy A, Brodbelt A, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101:124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000a;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, et al. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000b;60:5021–5026. [PubMed] [Google Scholar]
- Elliott G, Hong C, Xing X, Zhou X, Li D, Coarfa C, Bell RJ, Maire CL, Ligon KL, Sigaroudinia M, et al. Intermediate DNA methylation is a conserved signature of genome regulation. Nat Commun. 2015;6:6363. doi: 10.1038/ncomms7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Munck J, Tang J, Taverna P, Wang Y, Miller DF, Pilrose J, Choy G, Azab M, Pawelczak KS, et al. The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer. Clin Cancer Res. 2014;20:6504–6516. doi: 10.1158/1078-0432.CCR-14-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schönegger A, Klughammer J, Bock C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Fiegl H, Millinger S, Goebel G, Muller-Holzner E, Marth C, Laird PW, Widschwendter M. Breast cancer DNA methylation profiles in cancer cells and tumor stroma: association with HER-2/neu status in primary breast cancer. Cancer Res66, 29-33. 2006 doi: 10.1158/0008-5472.CAN-05-2508. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–310. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MR, Wang M, Aldape KD, Stupp R, Hegi ME, Jaeckle KA, Armstrong TS, Wefel JS, Won M, Blumenthal DT, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol. 2013;31:4085–4091. doi: 10.1200/JCO.2013.49.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the E-cadherin 5' CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem. 2000;275:2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- Grasbon-Frodl EM, Kreth FW, Ruiter M, Schnell O, Bise K, Felsberg J, Reifenberger G, Tonn J-C, Kretzschmar HA. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J Cancer. 2007;121:2458–2464. doi: 10.1002/ijc.23020. [DOI] [PubMed] [Google Scholar]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger V, Debus N, Lohmann D, Höpping W, Passarge E, Horsthemke B. Frequency and parental origin of hypermethylated RB1 alleles in retinoblastoma. Hum Genet. 1994;94:491–496. doi: 10.1007/BF00211013. [DOI] [PubMed] [Google Scholar]
- Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Hognas G, Annala M, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zhu P, Guo F, Li X, Wu X, Fan X, Wen L, Tang F. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat Protoc. 2015;10:645–659. doi: 10.1038/nprot.2015.039. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Hamilton MG, Roldan G, Magliocco A, McIntyre JB, Parney I, Easaw JC. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol. 2011;102:255–260. doi: 10.1007/s11060-010-0307-5. [DOI] [PubMed] [Google Scholar]
- Hanson JA, Gillespie JW, Grover A, Tangrea MA, Chuaqui RF, Emmert-Buck MR, Tangrea JA, Libutti SK, Linehan WM, Woodson KG. Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Cancer Inst. 2006;98:255–261. doi: 10.1093/jnci/djj051. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, Whitehall VL, Leggett BA, Laird PW. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS ONE. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan LE, Meyer JA, Yang J, Wang J, Wong N, Yang W, Condos G, Hunger SP, Raetz E, Saffery R, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118:5218–5226. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju HX, An B, Okamoto Y, Shinjo K, Kanemitsu Y, Komori K, Hirai T, Shimizu Y, Sano T, Sawaki A, et al. Distinct profiles of epigenetic evolution between colorectal cancers with and without metastasis. Am J Pathol. 2011;178:1835–1846. doi: 10.1016/j.ajpath.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, Tan JL, Fogley RD, van Rooijen E, Hagedorn EJ, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016;351:aad2197. doi: 10.1126/science.aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klump B, Hsieh CJ, Holzmann K, Gregor M. Hypermethylation of the CDKN2/p16 promoter during neoplastic progression in Barrett's esophagus. Gastroenterology. 1998;115:1381–1386. doi: 10.1016/s0016-5085(98)70016-2. [DOI] [PubMed] [Google Scholar]
- Koestler DC, Christensen B, Karagas MR, Marsit CJ, Langevin SM, Kelsey KT, Wiencke JK, Houseman EA. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8:816–826. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova Y, Maloney RK, Lakey N, Citek RW, Bacher B, Budiman A, Ordway JM, McCombie WR, Leon J, Jeddeloh JA, McPherson JD. Massively parallel bisulphite pyrosequencing reveals the molecular complexity of breast cancer-associated cytosine-methylation patterns obtained from tissue and serum DNA. Genome Res. 2008;18:19–29. doi: 10.1101/gr.6883307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, O'Brien CA, van Galen P, Gan OI, Notta F, Brown AM, Ng K, Ma J, Wienholds E, Dunant C, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- Kulis M, Merkel A, Heath S, Queiros AC, Schuyler RP, Castellano G, Beekman R, Raineri E, Esteve A, Clot G, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landan G, Cohen NM, Mukamel Z, Bar A, Molchadsky A, Brosh R, Horn-Saban S, Zalcenstein DA, Goldfinger N, Zundelevich A, et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 2012;44:1207–1214. doi: 10.1038/ng.2442. [DOI] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, Stevenson K, Sougnez C, Wang L, Li S, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Lopez-Diaz FJ, Khan SY, Tariq MA, Dayn Y, Vaske CJ, Radenbaugh AJ, Kim HJ, Emerson BM, Pourmand N. Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc Natl Acad Sci U S A. 2014;111:E4726–4735. doi: 10.1073/pnas.1404656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu X, Song L, Hou Y, Li Z, Tsang S, Li F, Im KM, Wu K, Wu H, et al. Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. Gigascience. 2012;1:12. doi: 10.1186/2047-217X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Giannopoulou EG, Park K, Mosquera JM, Sboner A, Tewari AK, Garraway LA, Beltran H, Rubin MA, Elemento O. Epigenomic alterations in localized and advanced prostate cancer. Neoplasia. 2013;15:373–383. doi: 10.1593/neo.122146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowdon RF, Zhang B, Bilenky M, Mauro T, Li D, Gascard P, Sigaroudinia M, Farnham PJ, Bastian BC, Tlsty TD, et al. Regulatory network decoded from epigenomes of surface ectoderm-derived cell types. Nat Commun. 2014;5:5442. doi: 10.1038/ncomms6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, Kriegstein AR, Oldham MC. Radial glia require PDGFD-PDGFRbeta signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack SC, Hubert CG, Miller TE, Taylor MD, Rich JN. An epigenetic gateway to brain tumor cell identity. Nat Neurosci. 2015;19:10–19. doi: 10.1038/nn.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, Paulson TG, Blount PL, Risques RA, Rabinovitch PS, Reid BJ. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Li X, Sanchez CA, Paulson TG, Reid BJ. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett's esophagus. Cancer Res. 2004;64:3414–3427. doi: 10.1158/0008-5472.CAN-03-3249. [DOI] [PubMed] [Google Scholar]
- Matei DE, Nephew KP. Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecol Oncol. 2010;116:195–201. doi: 10.1016/j.ygyno.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor T, Pankov A, Johnson BE, Hong C, Hamilton EG, Bell RJA, Smirnov IV, Reis GF, Phillips JJ, Barnes MJ, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28:307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor K, Bernatsky S, Colmegna I, Hudson M, Pastinen T, Labbe A, Greenwood C. An evaluation of methods correcting for cell type heterogeneity in DNA methylation studies. bioRxiv. 2015 doi: 10.1186/s13059-016-0935-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5' CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- Merlo LMF, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, Maley CC. A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2010;3:1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeska T, Bock C, El-Maarri O, Hubner A, Ehrentraut D, Schramm J, Felsberg J, Kahl P, Buttner R, Pietsch T, Waha A. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JW, Kim WJ, Han JA, Jung YJ, Kim KT, Park WY, Lee HO, Choi SS. Identification of distinct tumor subpopulations in lung adenocarcinoma via single-cell RNA-seq. PLoS ONE. 2015;10:e0135817. doi: 10.1371/journal.pone.0135817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelans CB, de Groot JS, Pan X, van der Wall E, van Diest PJ. Clonal intratumor heterogeneity of promoter hypermethylation in breast cancer by MSMLPA. Mod Pathol. 2014;27:869–874. doi: 10.1038/modpathol.2013.207. [DOI] [PubMed] [Google Scholar]
- Myöhänen SK, Baylin SB, Herman JG. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res. 1998;58:591–593. [PubMed] [Google Scholar]
- Nakamura M, Watanabe T, Klangby U, Asker C, Wiman K, Yonekawa Y, Kleihues P, Ohgaki H. p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol. 2001a;11:159–168. doi: 10.1111/j.1750-3639.2001.tb00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest. 2001b;81:77–82. doi: 10.1038/labinvest.3780213. [DOI] [PubMed] [Google Scholar]
- Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Oakes CC, Claus R, Gu L, Assenov Y, Hullein J, Zucknick M, Bieg M, Brocks D, Bogatyrova O, Schmidt CR, et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2013;4:348–361. doi: 10.1158/2159-8290.CD-13-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS, Wang Q, Imbusch CD, Serva A, Koser SD, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016;48:253–264. doi: 10.1038/ng.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Jiang Y, Boi M, Tabbo F, Redmond D, Nie K, Ladetto M, Chiappella A, Cerchietti L, Shaknovich R, et al. Epigenomic evolution in diffuse large B-cell lymphomas. Nat Commun. 2015;6:6921. doi: 10.1038/ncomms7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper RO, Costello JF, Kroes RA, Futscher BW, Marathi U, Erickson LC. Direct correlation between methylation status and expression of the human O-6-methylguanine DNA methyltransferase gene. Cancer Commun. 1991;3:241–253. doi: 10.3727/095535491820873092. [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Canales J, Hanson JC, Tangrea MA, Erickson HS, Albert PS, Wallis BS, Richardson AM, Pinto PA, Linehan WM, Gillespie JW, et al. Identification of a unique epigenetic sub-microenvironment in prostate cancer. J Pathol. 2007;211:410–419. doi: 10.1002/path.2133. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Toguchida J, Ohtani N, Yandell DW, Rapaport JM, Dryja TP. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am J Hum Genet. 1991;48:880–888. [PMC free article] [PubMed] [Google Scholar]
- Schonberg DL, Lubelski D, Miller TE, Rich JN. Brain tumor stem cells: Molecular characteristics and their impact on therapy. Mol Aspects Med. 2014;39:82–101. doi: 10.1016/j.mam.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm A, Köster J, Assenov Y, Althoff K, Peifer M, Mahlow E, Odersky A, Beisser D, Ernst C, Henssen AG, et al. Mutational dynamics between primary and relapse neuroblastomas. Nature Genetics. 2015;47:872–877. doi: 10.1038/ng.3349. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Marjoram P, Tavare S, Shibata D. High DNA methylation pattern intratumoral diversity implies weak selection in many human colorectal cancers. PLoS ONE. 2011;6:e21657. doi: 10.1371/journal.pone.0021657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Marjoram P, Woo YJ, Tavare S, Shibata D. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci U S A. 2009;106:4828–4833. doi: 10.1073/pnas.0810276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013a;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Shibata D, Curtis C, Tavare S. Single-molecule genomic data delineate patient-specific tumor profiles and cancer stem cell organization. Cancer Res. 2013b;73:41–49. doi: 10.1158/0008-5472.CAN-12-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirzaker C, Millar DS, Paul CL, Warnecke PM, Harrison J, Vincent PC, Frommer M, Clark SJ. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229–2237. [PubMed] [Google Scholar]
- Strauss J, Figg WD. Using epigenetic therapy to overcome chemotherapy resistance. Anticancer Res. 2016;36:1–4. [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Thuijl HF, Mazor T, Johnson BE, Fouse SD, Aihara K, Hong C, Malmstrom A, Hallbeck M, Heimans JJ, Kloezeman JJ, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129:597–607. doi: 10.1007/s00401-015-1403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Mutch DG, Edmonston TB, Goodfellow PJ, Mitra RD. Intra-tumor heterogeneity of MLH1 promoter methylation revealed by deep single molecule bisulfite sequencing. Nucleic Acids Res. 2009;37:4603–4612. doi: 10.1093/nar/gkp457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassenbroeck I, Califice S, Diserens AC, Migliavacca E, Straub J, Di Stefano I, Moreau F, Hamou MF, Renard I, Delorenzi M, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10:332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet T, Kumar P, Van Loo P, Cooke SL, Marshall J, Lin ML, Zamani Esteki M, Van der Aa N, Mateiu L, McBride DJ, et al. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res. 2013;41:6119–6138. doi: 10.1093/nar/gkt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hypermethylation and homozygous deletion of the p14ARF and p16INK4a genes in oligodendrogliomas. Acta Neuropathol. 2001;101:185–189. doi: 10.1007/s004010000343. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VK, De S. An assessment of computational methods for estimating purity and clonality using genomic data derived from heterogeneous tumor tissue samples. Brief Bioinform. 2015;16:232–241. doi: 10.1093/bib/bbu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]