Abstract
Genetic differences in acute behavioral responses to ethanol contribute to the susceptibility to alcohol use disorder and the reduction of anxiety is a commonly reported motive underlying ethanol consumption among alcoholics. Therefore, we studied the genetic variance in anxiolytic-like responses to ethanol across the BXD recombinant inbred mouse panel using the light-dark transition model of anxiety. Strain-mean genetic mapping and a mixed-model quantitative trait loci (QTL) analysis replicated several previously published QTL for locomotor activity and identified several novel anxiety-related loci. Significant loci included a chromosome 11 saline anxiety-like QTL (Salanq1) and a chromosome 12 locus (Etanq1) influencing the anxiolytic-like response to ethanol. Etanq1 was successfully validated by studies with BXD advanced intercross strains and fine-mapped to a region comprising less than 3.5 Mb. Through integration of genome-wide mRNA expression profiles of the mesocorticolimbic reward circuit (prefrontal cortex, nucleus accumbens, and ventral midbrain) across the BXD RI panel, we identified high priority candidate genes within Etanq1, the strongest of which was Ninein (Nin), a Gsk3β-interacting protein that is highly expressed in the brain.
Keywords: Ethanol, Anxiety, QTL, BXD Recombinant Inbred, Microarray, Ninein
Introduction
Individual differences in acute behavioral responses to ethanol are reported to contribute to the susceptibility of ethanol abuse and alcoholism (Schuckit & Smith, 1997), now collectively referred to as alcohol use disorder (AUD). Among the various acute ethanol behavioral responses, ethanol is a well-documented anxiolytic. Ethanol-induced anxiolysis has been observed using multiple rodent models of anxiety-like behavior, including the light-dark box (Bilkei-Gorzo et al., 1998, Boehm et al., 2002, Costall et al., 1988), elevated plus-maze (Boehm et al., 2002, Labuda & Fuchs, 2000), social interaction (Varlinskaya & Spear, 2002), and mirrored chamber (Cao et al., 1993, Kliethermes et al., 2003). Human studies have shown that in addition to a high comorbidity between ethanol and anxiety (Bibb & Chambless, 1986, Cornelius et al., 2003), the reduction of anxiety is a commonly reported motive among alcoholics (Conger, 1956, Newlin & Thomson, 1990, Pohorecky, 1981). Therefore, anxiety is hypothesized as a risk factor in the initiation of ethanol abuse and recidivism. While much is known about neural pathways and signaling mechanisms involved in anxiety, the molecular mechanisms underlying the anxiolytic-like response to ethanol are still poorly understood.
Acute behavioral responses to ethanol are modified by environmental and genetic factors. Although the genetic component of numerous acute ethanol behaviors has been extensively studied, minimal genetic research has been conducted on ethanol-induced anxiolysis. A common approach to explore genetic contributions to complex polygenic traits in animal models has been through mapping quantitative trait loci (QTL) in recombinant inbred (RI) mouse strains (Crabbe et al., 1994, Peirce et al., 2004). This method has been widely used to identify anxiety-related QTL (Henderson et al., 2004) and loci involved in various acute behavioral responses to ethanol (Browman & Crabbe, 2000, Buck et al., 1997, Cook et al., 2015, Crabbe, 1998, Demarest et al., 2001, Demarest et al., 1999, Dubose et al., 2013, Phillips et al., 1998). The BXD RI strains are of particular interest due to contrasting behavioral responses to ethanol between the C57BL/6J (B6) and DBA/2J (D2) inbred progenitor mouse strains. For example, while D2 mice show larger locomotor responses to acute ethanol, B6 mice consume more ethanol (Belknap et al., 1993, Lessov et al., 2001, Phillips et al., 1995). Additionally, since the genome of the B6 and D2 progenitor strains have been fully sequenced, the utilization of a full genome polymorphism database increases the precision with which one can identify candidate genes and polymorphisms within QTL.
Here, we initiated a genetic analysis of mechanisms underlying the anxiolytic effects of ethanol using the light-dark transition model to measure anxiety- and anxiolytic-like behavior in response to saline or ethanol, respectively. The light-dark transition model was chosen because it shows predictive validity, particularly in regard to clinically-used benzodiazepines, allows the simultaneous recording of locomotor activity, and is potentially less influenced by activity differences and investigator bias (Crawley, 1985). Furthermore, since the light-dark transition model of anxiety is based on an unconditioned fear paradigm, animal training was not required, thereby enabling high-throughput behavioral testing. Mounting evidence suggests that even similar behavioral models of anxiety identify different genetic aspects of anxiety-like behavior (Belzung & Le Pape, 1994, Brigman et al., 2009, File, 1995, Flint, 2003, Milner & Crabbe, 2008, Ramos et al., 1997, Turri et al., 2001). Thus, while employing only the light-dark transition model does not fully sample the genetic variation in anxiolytic-like responses to ethanol, the advantages of the model make it useful for an initial study on ethanol-induced anxiolysis.
Strain-mean genetic mapping and a mixed-model QTL analysis to identify genotype-by-treatment interactions enabled the detection of novel provisional QTL influencing anxiety-related behavioral responses, distinct from loci influencing locomotor activity. In particular, a significant QTL (Etanq1) modifying the anxiolytic-like response to ethanol was identified on mouse chromosome 12 and verified using BXD advanced recombinant inbred (ARI) strains (Peirce et al., 2004). Through integration of the B6 and D2 progenitor genome sequences and genome-wide transcription profiles for the prefrontal cortex (PFC), nucleus accumbens (NAc) and ventral midbrain (VMB), we identified candidate genes underlying Etanq1, the strongest of which was Ninein (Nin), a Gsk3β-interacting protein that is highly expressed in the brain.
Materials and Methods
Experimental Subjects
A total of 597 male mice across 38 strains (see Supplementary Table 2) were studied in three phases: provisional mapping, confirmation, and fine-mapping. While sex-specific QTL are of significant interest to anxiety and behavioral responses to ethanol, only male mice were used for this initial assessment to reduce phenotypic variation of anxiety-related behaviors and to make the number of animals tractable in terms of manpower and expense (see Discussion). For the provisional phase, 448 mice from 27 BXD RI strains and their B6 and D2 progenitor strains were purchased at 8–9 weeks of age from the Jackson Laboratory (Bar Harbor, ME, USA). All animals were habituated to the animal facility for a minimum of 2 weeks prior to testing and housed 4 per cage with ad libitum access to standard rodent chow (catalog #7912, Harlan Teklad, Madison, WI) and water in a 12h dark/light cycle with lights on at 0600 hours. Each BXD RI strain consisted of 6–8 mice per treatment and each progenitor strain had 13–16 mice per treatment. For the replication and fine-mapping analyses, we acquired, respectively, six and three novel BXD ARI strains (N = 5–10 per strain/ treatment) and additional B6 and D2 progenitor strains (N = 8 per strain/treatment) from Oak Ridge National Laboratory (Oak Ridge, TN, USA) at 6–9 weeks of age. These novel BXD ARI strains were derived from two independent advanced intercrosses between the B6 and D2 progenitor strains (Peirce et al., 2004). All Oak Ridge animals were housed 2–5 mice per cage and kept in a colony room until their age range corresponded with previously tested Jackson Laboratory animals. All experimental procedures were approved by Virginia Commonwealth University Institutional Animal Care and Use Committees in accordance with National Institutes of Health guidelines.
Light-Dark Box Behavioral Protocol and Apparatus
Genetic variation in anxiety- or anxiolytic-like behavior following saline or ethanol was measured across the BXD RI strains using the light-dark transition model of anxiety. Behavioral testing occurred between 10:00 A.M. and 1:00 P.M in multiple cohorts over a 12 month period. All behavioral testing was done by a male investigator. Each cohort contained a different combination of BXD strains with each strain balanced for saline or ethanol treatment. Following a 1 hour acclimation period to the behavioral testing room, animals were restrained in a 50 ml conical tube for 15 minutes followed by intraperitoneal (IP) injections with either physiological saline (0.9%) or 1.8 g/kg ethanol (12.8% w/v) in 0.9% saline. A mild restraint stress was employed to create an artificial baseline level of anxiety-like behavior. Fifteen minutes of immobilization has been shown to activate the sympathoadrenal system, as indicated by marked increases in plasma adrenocorticotropic hormone (ACTH), epinephrine, norepinephrine, and corticosterone (Tjurmina et al., 2002). Interestingly, ethanol antagonizes these neurochemical responses, including immobilization-induced plasma levels of epinephrine and norepinephrine (Deturck & Vogel, 1982, Patel & Pohorecky, 1988, Popper et al., 1977). Given this biochemical evidence for ethanol-induced anxiolysis, a mild restraint stress was incorporated into our behavioral paradigm to increase anxiety-like behavior and the robustness of anxiolytic-like responses. This method provided a common environmental stressor across all mice just prior to studying anxiolytic-like effects from a control (saline) or ethanol injection. We found this mild stress produced similar slight increases in anxiety-like behavior in the progenitor B6 and D2 strains compared to no restraint and showed a trend for reducing within-strain reproducibility of anxiety-like measures (data not shown). An activating dose of ethanol was chosen for this study based on its ability to elicit a significant anxiolytic-like response in the progenitor strains and allow for considerable phenotypic variation across the BXD RI strains. Following a 5 minute delay from the time of injection, each animal was placed in the light chamber facing the entrance to the dark chamber of the light-dark box. Once the animal entered the dark compartment, anxiety-like scores were collected in 5 minute intervals for a total of 10 minutes. Behavioral measures were recorded in both chambers of the light-dark box and included distance traveled (cm) and time spent in each compartment and total locomotor activity (TLA). Anxiety-like measures were reported as percent time spent in the light (TSL) and percent distance traveled in light (DTL) to control for locomotor activity. An increase in either measure was interpreted as decreased anxiety-like behavior.
Light-dark box studies were done with a commercial product adapted from the originally described apparatus (Crawley & Goodwin, 1980). An open field arena (27.3 cm x 27.3 cm x 20.3 cm) was divided into two equally-sized compartments using an opaque black plastic box insert with an opening in the middle to allow for light-dark transitions (catalog #s ENV-510 and ENV-511, Med Associates Inc , St. Albans, VT, USA). The arena was enclosed in a sound-attenuating box (catalog # MED-OFA-022) equipped with overhead lighting (two 100 mA bulbs; 22 lux in light compartment) and fan ventilation. The system was interfaced with Med Associates software enabling automatic measurement of activity using a set of 16 infrared beam sensors along the X-Y plane. The arena was cleaned with 5% ammonia solution between animals.
Microarray Data Generation
Mice used for behavioral studies above were sacrificed by cervical dislocation 4 hours following IP injection. Immediately thereafter, brains were extracted and chilled for 1 minute in iced phosphate buffer before being microdissected into 8 constituent regions as previously described (Kerns et al., 2005). These included PFC, NAc, and VMB, which contains ventral tegmental area and substantia nigra. Excised regions were placed in individual tubes, flash-frozen in liquid nitrogen, and stored at 80 C. Total RNA was extracted from 4–5 pooled brain region samples, processed, and hybridized to Mouse Genome 430 2.0 microarrays (Affymetrix, Santa Clara, CA, USA, part #900497) according to the manufacturer’s protocol as previously described (Kerns et al., 2005, Wolen & Miles, 2012). In total, this study incorporated PFC tissue from 27 BXD strains, NAc and VMB tissue from 35 BXD strains, and B6 and D2 tissue from all three regions in triplicate. A detailed description and network analysis of these microarray data were previously reported (Wolen et al., 2012).
Statistical Analysis
BXD strain genotypes used for all analyses consisted of approximately 4,000 genetic markers obtained from GeneNetwork (www.genenetwork.org/genotypes/BXD.geno). All behavioral measures collected across the BXD strains and their inbred progenitors were evaluated for strain, treatment, and strain-by-treatment differences using analysis of variance (ANOVA) with Tukey HSD post-hoc analysis. Narrow-sense heritability (h2) estimates for all behavioral measures were calculated as previously described by Belknap and colleagues (Belknap et al., 1996). Behavioral QTL were identified by strain-mean single locus interval mapping performed with the R/qtl package for R (Broman et al., 2003, Chesler et al., 2004). QTL genome-wide adjusted p-values were calculated empirically by permutation analysis with 1,000 iterations (Churchill & Doerge, 1994). Significant and suggestive thresholds corresponded to LOD scores with genome-wide p-values < 0.05, and 0.63, respectively (Lander & Kruglyak, 1995). QTL support intervals were estimated using the Bayes credible interval approach implemented in R/qtl with a 97% probability of coverage (Manichaikul et al., 2006).
To examine genotype-by-treatment interactions, a single marker mixed-model QTL analysis was conducted using the lme4 package for R (Bates et al., 2015). With genotype treated as a fixed effect, three models were fit using maximum likelihood to estimate the parameters. The reduced model approximated the previous strain-mean mapping, testing genotype effect only, on either the combined data (saline and ethanol), or separate data sets for each group. The additive model tested for genotype effects in addition to treatment effects from the reduced model. The full model included all parameters from the additive model and an interaction term that incorporated genotype-by-treatment effects. To reveal treatment-specific effects, the interaction model was analyzed by comparison of additive model parameters to those from the full model. QTL were identified as those markers with a Benjamini-Hochberg false discovery rate (FDR) < 10% in at least one measure of anxiety-related behavior across any model. To reveal the most parsimonious model, nested models were compared using the likelihood ratio test (χ2 = 10.8, p = 0.001) with degrees of freedom equal to differences in the number of parameters tested between models. Statistical significance for molecular studies was examined for each gene or protein using a Student’s t-test between the two strains or genotypes tested, with Bonferroni correction for multiple testing as necessary.
Bioinformatic Approaches to Identify Candidate Genes
To identify candidate genes underlying QTL, multiple standard bioinformatic approaches were utilized. First, PFC, NAc, and VMB gene expression datasets from BXD RI strains were correlated to light-dark box phenotypes following saline or ethanol treatment. Genes or predicted open-reading frames located within the QTL support interval and found to be significantly correlated (Spearman’s rank) with an anxiety-related behavioral measure were considered genes of interest. The significance of a correlation was determined by converting correlation coefficients to Z-scores with the Fisher transformation and using the false discovery rate (FDR) to adjust raw ρ-values in order to account for the multiple tests conducted within each brain region (Benjamini & Hochberg, 1995).
Next, microarray expression data were utilized to identify genetic loci regulating each gene’s level of expression in the three profiled brain regions by mapping expression QTL (eQTL) using GeneNetwork (Chesler et al., 2004). Any eQTL located within 5 Mb of its associated gene was considered a cis regulatory locus, or cis eQTL. Genes with cis eQTL overlapping a behavioral QTL were considered high priority candidates, especially if their expression levels strongly correlated with the behavior of interest. Genes with potentially spurious cis eQTL caused by D2 single nucleotide polymorphisms (SNPs) overlapping probe binding sites (Alberts et al., 2007) were evaluated and removed from further consideration.
Each gene of interest was examined for polymorphisms using the B6 and D2 genome sequences. Genes carrying non-synonymous, or putative functional polymorphisms, were considered high priority candidates. In order to assess the likelihood that a coding SNP would affect protein function, we employed the web-based bioinformatics tool, Variant Effect Predictor (VEP), which predicts whether an amino acid change might carry functional consequence for the protein (Mclaren et al., 2010). Finally, genes satisfying all criteria were examined for biological significance using evidence from literature associations with anxiety- and/or ethanol-related phenotypes.
Molecular Characterization of Candidate Genes Containing cis eQTL
To further investigate candidate genes containing cis eQTL on microarray analysis, B6 and D2 mRNA expression levels of each gene were compared by quantitative real-time PCR in the brain region of interest, NAc (n = 8 NAc per strain). If significant expression differences between B6 and D2 mice were verified, allele-specific sequencing of B6D2F1/J NAc tissue (n = 3 biological replicates of 2 pooled NAc) was performed in order to confirm cis regulation. Functional differences between B6 and D2 mice for the top priority candidate gene (Ninein), evidenced as differences in protein expression levels, were examined by immunoblotting from NAc samples (n = 5 NAc per strain). Additional details are provided in Supplementary Methods.
Results
Anxiolytic-like response to ethanol in B6 and D2 progenitors
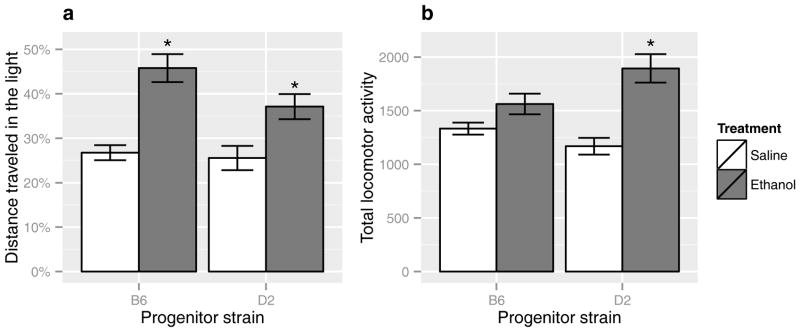
Acute ethanol elicited a significant anxiolytic-like response in B6 and D2 progenitor mouse strains, as measured using percent distance traveled in the light (DTL) in the light-dark transition model of anxiety (Figure 1a; p < 0.05 vs. within-strain saline; see Supplementary Table 1 for full statistical results). There also was a significant main effect of treatment across both DTL and percent time spent in the light (TSL; see Supplementary Table 1). Although there was no significant strain x treatment effect on ANOVA for DTL or TSL, following post-hoc testing the B6 strain showed significant ethanol anxiolytic-like responses with both DTL and TSL while D2 were only significant with the DTL measure. Consistent with earlier reports (Phillips et al., 1995, Tritto & Dudek, 1994), D2 and B6 strains showed contrasting total locomotor activity (TLA) in response to ethanol (Figure 1b). While ethanol induced locomotor activation in D2 mice, B6 mice lacked this response. Considering the B6 strain showed a significant anxiolytic-like response to ethanol with the absence of locomotor activation, ethanol-induced anxiolysis, as measured by DTL or TSL, did not appear to strictly dependent on ethanol induction of locomotor activity. Factor analysis of BXD strain phenotypes supports this observation, as detailed in the following section.
Figure 1. Ethanol-induced phenotypes in the light-dark transition model of anxiety.
Percent distance traveled in the light (a) following saline or 1.8 g/kg ethanol in B6 and D2 progenitors. An increase in the distance traveled in the light represents an anxiolytic-like response to ethanol. Total locomotor activity (b) following saline or 1.8g/kg ethanol was simultaneously recorded. Significance was determined by ANOVA with Tukey post-hoc analysis (*p < 0.05 vs. within-strain saline).
Anxiety-related behaviors across BXD RI strains
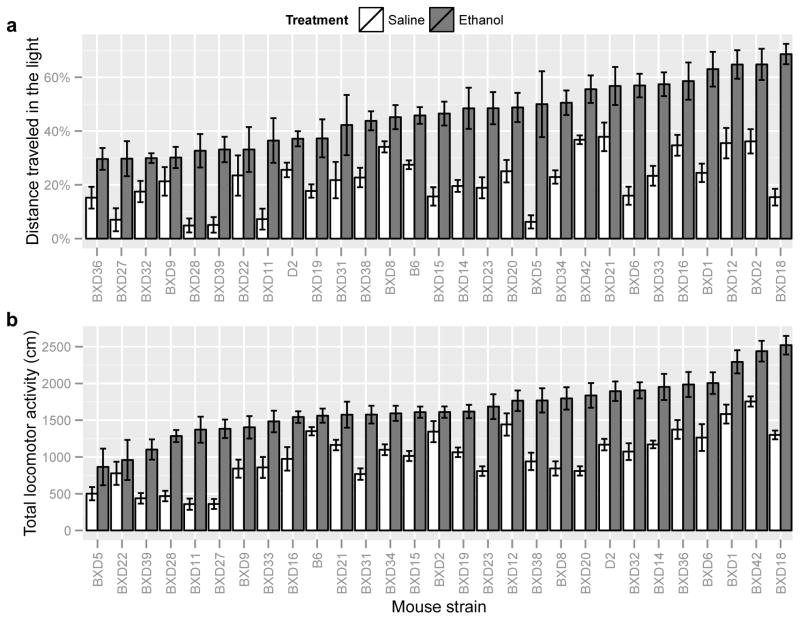
To identify provisional quantitative trait loci (QTL) contributing to genetic variance in ethanol-induced anxiolysis, anxiety-related behaviors in response to saline or ethanol were measured across 27 BXD RI strains and the progenitor B6 and D2 strains, as described in Methods. Behavioral phenotypes used for QTL mapping from the 0–5 minute and 0–10 minute intervals are summarized in Supplementary Table 2. As expected, results for DTL, TSL, and TLA were tightly correlated between the 0–5 and 0–10 minute data points (Pearson’s r > 0.96 for DTL, TSL, TLA, p < 0.0001). We therefore focus discussion of QTL analysis on the 0–5 minute interval due to greater anxiolytic-like responses to ethanol across all strains at this time interval. However, qualitatively identical results were obtained for QTL analysis of the 0–5 and 0–10 minute intervals. Distributions of the 0–5 minute saline and ethanol phenotypic (DTL, TLA) strain-means across the BXD RI strains are displayed graphically in Figure 2. Strikingly, nearly all BXD RI strains exhibited ethanol-induced anxiolytic-like responses. As mentioned above, the anxiolytic-like response to ethanol was not strictly dependent upon ethanol-induced locomotor activation, as demonstrated in B6 mice as well as other BXD RI strains (Fig. 2a vs. 2b; e.g. BXD12 and BXD2). We examined this more formally by factor analysis of post-ethanol behavioral phenotypes, which revealed that DTL and TLA load on different factors (Supplementary Figure 1).
Figure 2. Distribution of behavioral strain means.
Percent distance traveled in the light (a) and total locomotor activity (b) strain means + S.E. obtained in the light-dark box across 27 BXD RI strains and their progenitor strains following saline or 1.8 g/kg ethanol. Strains are sorted by ethanol strain means.
Analysis of variance across DTL and TLA revealed significant (p < 0.0001) strain (F(26, 364) = 6.91; F(26, 364) = 16.11, respectively) and treatment effects (F(1, 364) = 309.3; F(1, 364) = 357.2, respectively) across the BXD strains. Significant strain-by-treatment effects were also identified in DTL and TLA (F(26, 364) = 2.31, p< 0.0001; F(26, 364) = 1.98, p < 0.01), respectively). Within-strain statistics for DTL, TSL, and TLA are provided in Supplementary Table 3. As shown in Table 1, narrow-sense heritability estimates for DTL, TSL, and TLA following saline or ethanol ranged from 0.30 to 0.58. Table 1 also displays a phenotypic correlation matrix for DTL, TSL, and TLA recorded following saline or ethanol treatment. According to these data, both anxiety-related measures (DTL and TSL) were tightly correlated (Pearson’s r > 0.93, p < 0.001) within each treatment group, suggesting these behaviors measure similar anxiety-related phenotypes. We therefore focus our further discussion on DTL in additional studies on anxiety-like traits but results from TSL are displayed in Table 2, Supplementary Tables 1–2 and Supplementary Figures 1 and 5.
Table 1. Phenotype correlations.
Values in the lower diagonal represent Pearson's correlation coefficients between indicated phenotypes; p-values (upper diagonal) were obtained using Fisher's z-transformation.
| Saline TLA | Saline DTL | Saline TSL | Ethanol TLA | Ethanol DTL | Ethanol TSL | |
|---|---|---|---|---|---|---|
| Saline TLA | 1 | 7.84e-04 | 4.06e-04 | 1.99e-06 | 4.20e-03 | 5.39e-03 |
| Saline DTL | 0.589 | 1 | 2.70e-13 | 5.29e-02 | 5.91e-03 | 2.82e-03 |
| Saline TSL | 0.613 | 0.931 | 1 | 8.00e-02 | 2.71e-02 | 1.69e-02 |
| Ethanol TLA | 0.757 | 0.363 | 0.33 | 1 | 4.83e-03 | 1.16e-02 |
| Ethanol DTL | 0.516 | 0.499 | 0.41 | 0.509 | 1 | 2.22e-22 |
| Ethanol TSL | 0.503 | 0.534 | 0.44 | 0.462 | 0.986 | 1 |
|
| ||||||
| Heritability (h2) | 0.58 | 0.35 | 0.3 | 0.44 | 0.28 | 0.23 |
TLA: total locomotor activity; DTL: percent distance traveled in light; TSL: percent time spent in light
Table 2. Strain-mean QTL mapping results of light-dark box phenotypes.
Suggestive (p-value < 0.63) and significant (p-value < 0.05; bolded) QTL identified by single locus genome scans of strain-mean phenotypes. Support intervals define regions with a 97% probability of harboring the true QTL.
| Phenotype | Peak marker | Chr | Location | LOD | p-value | Support interval |
|---|---|---|---|---|---|---|
| Saline TLA | rs3696645 | 1 | 159.9 | 3.99 | 3.0e-02 | 147.39 to 184.42 |
| rs3689326 | 9 | 53.77 | 2.39 | 5.2e-01 | 29.75 to 106.32 | |
| Ethanol TLA | UT_1_113.684537 | 1 | 157.9 | 3.71 | 1.5e-02 | 147.39 to 184.42 |
| rs3675564 | 2 | 164.2 | 2.48 | 3.7e-01 | 9.75 to 172.85 | |
| rs13481014 | 11 | 47.93 | 2.16 | 6.2e-01 | 4.41 to 120.82 | |
| rs4231742 | 18 | 11.08 | 2.68 | 2.5e-01 | 3.52 to 86.75 | |
|
| ||||||
| Saline DTL | D1Mit139 | 1 | 128.4 | 2.41 | 5.6e-01 | 29.23 to 184.42 |
| gnf06.140.234 | 6 | 140.1 | 2.74 | 3.7e-01 | 16.01 to 149.21 | |
| rs13481251 | 11 | 116.9 | 2.7 | 4.0e-01 | 8.74 to 121.41 | |
| Ethanol DTL | rs13474399 | 1 | 182.5 | 2.59 | 4.5e-01 | 13.98 to 197.13 |
| rs3716547 | 12 | 68.67 | 4.32 | 2.7e-02 | 53.91 to 71.66 | |
|
| ||||||
| Saline TSL | rs13476098 | 1 | 128.5 | 3.35 | 9.8e-02 | 96.87 to 169.91 |
| rs13481251 | 11 | 116.9 | 3.75 | 4.1e-02 | 114.53 to 120.82 | |
| Ethanol TSL | rs13474399 | 1 | 182.5 | 2.9 | 2.8e-01 | 14.79 to 190.74 |
| rs3716547 | 12 | 68.67 | 4.37 | 2.3e-02 | 53.91 to 71.66 | |
Strain-mean QTL mapping
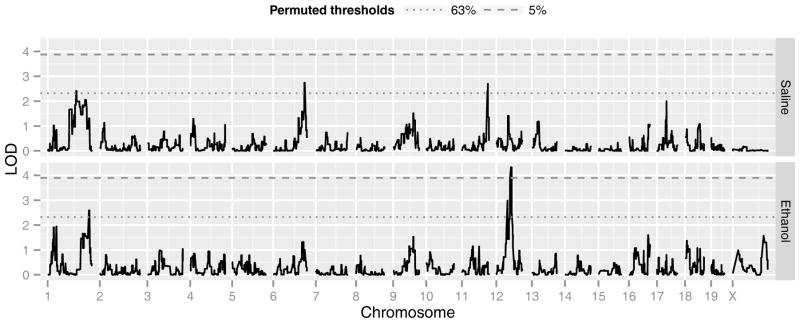
Strain-mean single factor interval mapping with permutation-corrected significance levels revealed multiple QTL influencing phenotypic variation in anxiety-related behaviors and TLA in response to saline or ethanol injections following mild restraint stress (Table 2). Three suggestive (p < 0.63) QTL altering DTL following saline were detected on chromosome (Chr) 1, 6 and 11 (Figure 3, Table 2). The Chr 1 and 11 loci also modified TSL following saline; the Chr 11 QTL (Saline anxiety-like QTL 1, Salanq1), exceeded the threshold for statistical significance (p < 0.05; Table 2). In addition, the post-saline TLA phenotype produced a significant locus (p < 0.05) on Chr 1 and a suggestive QTL (p < 0.63) on Chr 9 (Supplementary Figure 2). Although significant QTL were observed following saline injection for anxiety-like phenotypes, most notably the QTL on Chr 11 (Salanq1) that modified both DTL and TSL, the remaining analyses were focused on ethanol-induced phenotypes.
Figure 3. Anxiety-like behavior interval maps.
BXD strain-mean interval maps for percent distance traveled in the light (DTL) following saline (upper panel) or 1.8 g/kg ethanol (lower panel). Permuted genome-wide significance thresholds for falsely rejecting the null hypothesis (no linkage in the genome) at probabilities of 5% and 63% (corresponding to p < 0.05 and p < 0.63) are denoted by the dashed and dotted lines, respectively.
In contrast to results seen following saline injection, QTL modifying DTL following ethanol mapped to a suggestive locus (p < 0.63) on Chr 1 and a significant (p < 0.05) QTL on Chr 12 (Figure 3, Table 2). Interestingly, the Chr 11 locus that altered saline DTL and TSL did not appear to significantly influence ethanol-induced anxiolysis. For ethanol-induced TLA, a significant QTL (p < 0.05) was identified on Chr 1 and two suggestive loci were found on Chr 2 and Chr 18 (Table 2). The Chr 1 QTL that modified ethanol-induced TLA mapped to the same locus that alters TLA following saline. Since support intervals for all identified Chr 1 QTL influencing TLA following saline (159.8 Mb) and ethanol (157.9 Mb), saline anxiety-like behavior (128.4 Mb), and ethanol-induced anxiolysis (182.5 Mb) overlap, further analyses are required to determine whether these QTL represent distinct or overlapping loci. To focus on QTL more specific to ethanol-induced anxiolysis, anxiety-related QTL that overlapped TLA modifying QTL were omitted from additional analyses. Thus, the best provisional QTL altering the anxiolytic-like response to ethanol was the statistically significant (p < 0.05) QTL on Chr 12 (ethanol-induced anxiolysis QTL 1 (Etanq1) that modified both DTL and TSL (Table 2, Supplementary Figure 5).
Mixed-model QTL mapping
Although QTL studies measuring the effect of drug treatment have used difference values by calculating strain mean post-drug minus pre-drug scores, this method entails a variety of complications including increased error and spurious correlations (Nagoshi et al., 1986). An alternative approach to identify QTL influencing drug treatment-specific responses is through a nested model. Therefore, as described in Methods, we employed a single marker mixed-model QTL mapping procedure to examine genotype-by-treatment interactions across individual animals, thereby identifying saline and ethanol-specific effects on anxiety-related behaviors and TLA (Table 3; Supplementary Table 5).
Table 3. Mixed-model QTL mapping results of light-dark box DTL and TLA phenotypes.
Bold values indicate a statistically significant QTL ( FDR ≤ 10%).
| Phenotype | Chr | Peak marker | Location | Saline | Ethanol | Additive | Interaction |
|---|---|---|---|---|---|---|---|
| DTL | 1 | rs13476098 | 126.5 | 1.3e-03 | 7.6e-01 | 8.4e-02 | 7.4e-03 |
| 1 | rs13474399 | 180.6 | 1.9e-02 | 3.1e-04 | 2.3e-04 | 1.0e-02 | |
| 6 | gnf06.140.234 | 140.2 | 6.2e-04 | 2.5e-02 | 9.4e-04 | 7.6e-01 | |
| 11 | rs13481251 | 117 | 9.9e-04 | 2.0e-01 | 1.5e-02 | 1.5e-01 | |
| 12 | rs3716547 | 68.67 | 1.3e-01 | 4.3e-06 | 2.3e-04 | 1.0e-05 | |
| TLA | 1 | UT_1_113.684537 | 156.1 | 6.0e-05 | 2.3e-04 | 2.4e-05 | 9.1e-01 |
| 1 | rs3696645 | 157.9 | 6.0e-05 | 2.3e-04 | 2.4e-05 | 9.1e-01 | |
| 2 | rs8275857 | 164.4 | 1.3e-02 | 4.2e-04 | 1.0e-03 | 6.4e-03 | |
| 9 | rs13480205 | 54.53 | 3.6e-03 | 4.5e-03 | 2.1e-03 | 2.3e-01 | |
| 11 | rs13481014 | 48.12 | 6.4e-03 | 9.0e-04 | 1.1e-03 | 3.3e-02 | |
| 18 | rs4231742 | 11.08 | 4.0e-03 | 1.3e-03 | 9.3e-04 | 4.3e-01 |
Strain-mean QTL indicated that, with the exception of the Chr 1 QTL influencing TLA, most loci were treatment-specific, altering anxiety-like behavior in response to either ethanol or saline injections. In contrast, the mixed model analysis revealed these loci influenced anxiety-like behavior following both saline and ethanol, although to different degrees. For example, as shown in Table 3, isolating saline and ethanol responses by examining reduced model p-values for the Chr 6 (140.2 Mb) QTL showed a significant QTL for anxiety-related behavior following saline injection and a suggestive QTL following ethanol. Therefore, similarly to the TLA-related Chr 1 (156–158 Mb) QTL, it appears ethanol may partially elicit locomotor activation and anxiolytic-like effects through mechanisms involved in the basal (restraint stress + saline injection) regulation of these phenotypes. Regarding ethanol-specific anxiolytic-like effects, the interaction model validated the previously identified and significant Chr 12 QTL (68.67 Mb, p < 0.001). Although the Chr 12 QTL appeared unique to ethanol-induced anxiolysis following strain-mean genetic mapping, mixed-model QTL analysis showed a trend following saline injection also at this locus (p=0.13).
Replication of ethanol anxiolytic-like QTL
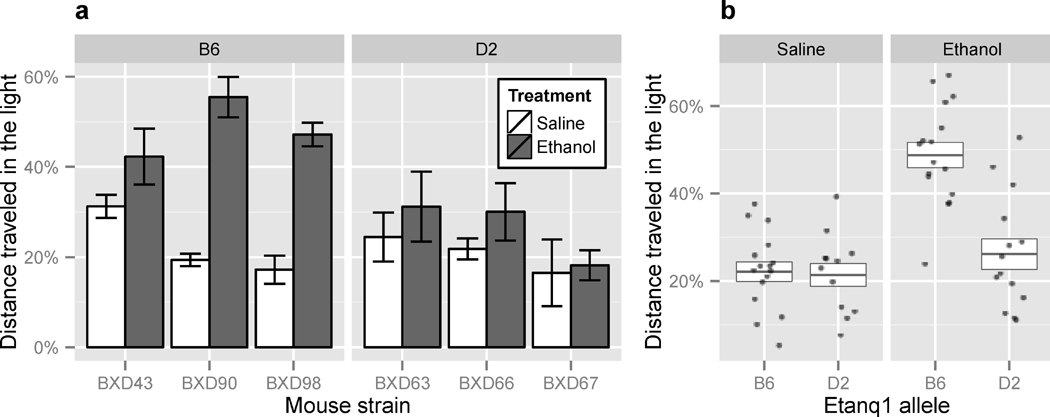
We utilized novel BXD RI strains (BXD ARI) derived from two independent advanced intercrosses between the B6 and D2 progenitor strains (Peirce et al., 2004) for initial confirmation of the significant ethanol-induced anxiolysis QTL (Etanq1) on Chr 12 (68.67 Mb). Using the peak genetic marker within Etanq1, 3 BXD ARI strains (BXD 43, 90, 98) containing the B6 allele were chosen, together with 3 strains (BXD 63, 66, 67) with the D2 allele. All 6 BXD ARI strains were balanced for genotype at the two significant post-saline anxiety-like QTL on Chr 1 and Chr 11, to minimize the influence of these loci on ethanol-induced anxiolysis. Anxiety-related behaviors in response to saline or ethanol were measured across each BXD ARI strain, as described in Methods. ANOVA revealed two of the three BXD ARI strains with the B6 allele showed a significant anxiolytic-like response to ethanol, whereas the BXD ARI strains with the D2 allele did not show significant anxiolytic-like responses to ethanol (Figure 4a). Measuring TSL in response to saline or ethanol resulted in similar findings (data not shown). Collapsing each of the 3 BXD ARI strains into genotypic groups identified a statistically significant difference in DTL following ethanol between strains with the B6 allele versus the D2 allele at the peak genetic marker underlying Etanq1 (Figure 4b). As expected, this response was ethanol-specific with no significant differences in anxiety-like behaviors between the two genotypic groups following saline injections.
Figure 4. Replication of chromosome 12 ethanol-related anxiety QTL.
a) Differences in percent distance traveled in the light (DTL) in response to ethanol across 6 BXD ARI strains containing either the D2 (BXD 63, 66, 67) or B6 (BXD 43, 90, 98) allele at the peak genetic marker (rs3716547, 68.67 Mb) within the chromosome 12 QTL, Etanq1. ANOVA with Tukey post-hoc test revealed a significant anxiolytic-like response to ethanol in most BXD strains containing the B6 allele (BXD43: p = 0.84, BXD90: p = 7.1e-05, BXD98: p = 3.4e-04), whereas the lowest p-value among strains containing the D2 allele was 0.97. b) Collapsing post-ethanol DTL scores across BXD ARI strains containing the D2 or B6 allele revealed a significant effect of the B6 allele on ethanol-induced anxiolysis (p = 2.9E-06 versus D2 allele ethanol, ANOVA with Tukey post-hoc analysis).
Fine-mapping of ethanol anxiolytic-like QTL
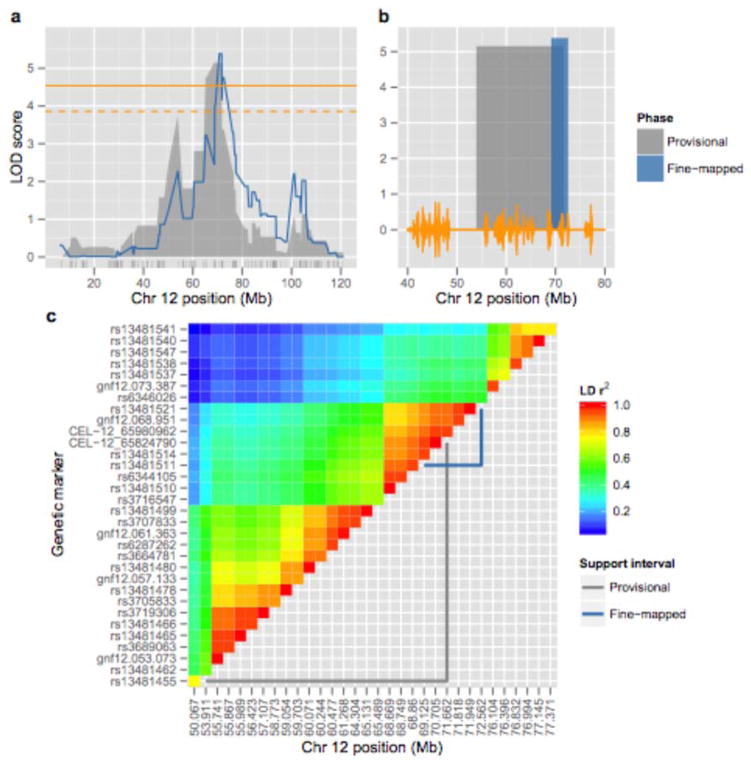
We examined the haplotype structure of Etanq1’s support interval using genotype data for all 93 BXD strains to calculate the r2 measure of linkage disequilibrium (LD) for all pairwise combinations of Chr 12 genetic markers. Results from this analysis revealed Etanq1’s provisional support interval was largely comprised of 3 haplotype blocks (Figure 5c). The independent effects of these haplotype blocks on ethanol-induced anxiolysis could not be discerned using only the BXD strains assayed for the provisional mapping of the QTL.
Figure 5. Etanq1’s fine-mapped support interval.
The original (blue) and fine- mapped (red) 97% support intervals for Etanq1 (DTL) across a subsection of Chr 12. Grey seismograph across the x-axis indicates the number of polymorphisms per kilobase between B6 and D2.
We therefore refined the Etanq1 support interval by conducting anxiety-related behavioral measurements for additional ARI BXD strains. These strains were chosen to contain informative recombination events within Etanq1’s support interval determined from the provisional and replication mapping studies (Supplementary Figure 3). The addition of these strains (BXD 50, 70, and 75) increased the peak LOD score of Etanq1 (DTL following ethanol) on interval mapping and proximally shifted the location of the linkage peak by 2 Mb (Figure 5a). Inclusion of the additional strains also narrowed Etanq1’s support interval by nearly 80%, to a region that spans from 69.13–72.56 Mb across Chr 12 (Figure 5b). To a lesser extent, the additional strains improved the Chr 1 QTL, increasing its peak LOD score from 2.59 to 3.03, but revealed a significant interaction with Etanq1 (Supplementary Figure 4).
The fine-mapped Etanq1 support interval centered on a very polymorphic region (70.9–71.5 Mb) flanked by areas comprising relatively few SNPs. The proximal region of the refined support interval was particularly SNP poor and may be identical by descent (IBD) between B6 and D2. As shown in Figure 5c, the refined support interval comprises a large portion of a single haplotype in which the r2 for all genetic markers is ≥ 0.87. As such, continuing to assay novel strains from the currently available BXD panel is unlikely to further resolve Etanq1 without additional genetic manipulation.
Bioinformatic Analysis of Etanq1 Candidate Genes
According to the Mouse Genome Informatics database (www.informatics.jax.org), Etanq1’s fine-mapped support interval harbors 41 protein-coding genes or non-coding RNAs. Given the limited potential for further refinement of the Etanq1 support interval by genetic mapping, we prioritized these positional candidates through a series of integrative analyses that combined the anxiety-like phenotype data with genomic sequence data from the B6 and D2 progenitor strains and genome-wide mRNA expression profiles of the mesocorticolimbic reward circuit: prefrontal cortex (PFC), nucleus accumbens (NAc) and ventral midbrain (VMB).
SNP analysis of Etanq1 candidate genes
Sequence variation analysis was performed between the B6 and D2 genomes for all Etanq1 positional candidates (Supplementary Table 4). There were a total of 3,484 SNPs within the Etanq1 support interval. While 62.3% of these SNPs fell within a transcript, only 130 occurred within an exon. Trim9 was the most polymorphic gene in the region, harboring over 400 B6/D2 SNPs, but only a few of these SNPs occur within a coding region. When considering only coding and untranslated region SNPs (Supplementary Table 4), Nin and Abhd12b were the most polymorphic genes within the Etanq1 support interval.
We used the Ensembl variant effect predictor (VEP; Version 73) script to determine whether any of these identified coding SNPs are likely to affect protein function (Mclaren et al., 2010). SNPs positions were re-mapped to the UCSC mm10 build of the genome using liftOver (https://genome.ucsc.edu/cgi-bin/hgLiftOver), in order to match the GRCm38.p1-based positions used by VEP. A total of 10 unique missense mutations were identified across 6 genes; Nin contained 4, Dact1 contained 2 and Atp5s, Abhd12b, Pygl and 2700049A03Rik each contained 1 (Supplementary Table 4b). Of these 10, the SIFT algorithm (Ng & Henikoff, 2003) identified only one SNP (rs29159683) within Nin as being potentially deleterious, located within the 16th exon (ENSMUST00000085314) of the Nin-001 transcript (ENSMUST00000085314).
Expression QTLs
In addition to functional changes in protein structure, genetic variation can affect gene expression levels and thus modify behavioral phenotypes. Thus, expression QTL (eQTL) analysis with brain expression data was utilized to identify genes with expression levels regulated by local (cis eQTL) B6/D2 polymorphisms. Only 5 positional candidate genes within the Etanq1 support interval were associated with significant cis eQTL in the PFC, NAc, or VMB: Sos2, Nin, Map4k5, Atp5s, and Trim9. All are located within the previously discussed polymorphic region near the center of the Etanq1 support interval (Figure 5c). However, polymorphisms overlapping microarray probe binding sites can produce false-positive cis eQTL-like effects by altering transcript hybridization in an allele-specific manner. We therefore used the previously published list of B6/D2 SNPs (Wolen et al., 2012) coincident with Affymetrix Mouse Genome 430 2.0 probe binding sites to identify such spurious cis eQTLs. Three of the probes within a Map4k5 probe-set (1440059_at) and one probe within the Sos2 probe-set (1443057_at) overlapped D2 SNPs. Removing the affected probes and repeating the QTL analysis nullified the cis eQTL for Map4k5, but not those for Sos2, narrowing the high priority candidate genes to four: Sos2, Nin, Atp5s, and Trim9 (Table 4).
Table 4. Genes with significant cis-eQTL within Etanq1.
Genes, location, brain region, peak marker, LOD scores, and p-values are shown (saline-treated only). Genes (Map4k5) with likely false-positive cis-eQTL were excluded.
| Gene | Mb | Region | Marker | LOD | p-value |
|---|---|---|---|---|---|
| Sos2 | 70.72 | NAc | rs6344105 | 9.11 | 1.0e-04 |
| Sos2 | 70.72 | VMB | rs6344105 | 10.24 | 6.0e-05 |
| Atp5s | 70.83 | NAc | rs3716547 | 5.15 | 2.9e-03 |
| Nin | 71.11 | NAc | rs6344105 | 4.09 | 1.3e-02 |
| Trim9 | 71.35 | NAc | rs6344105 | 4.71 | 1.4e-02 |
Correlation analysis with Etanq1 candidate genes
Further prioritization of positional candidate genes was completed by examining the association between DTL in response to ethanol and transcript abundance following saline or ethanol in the PFC, NAc or VMB. The strongest associations were observed in NAc (Figure 6). After correcting for multiple testing, significant phenotype/mRNA correlations (FDR ≤ 0.1) were detected for only 3 genes: Sos2, Trim9, and Nin (Table 5). The strongest was a negative correlation between DTL post ethanol and basal (saline) Ninein (Nin) expression in the NAc (p = −0.67).
Figure 6. Correlation of ethanol-induced anxiolysis and candidate gene expression in the mesocorticolimbic reward circuit.
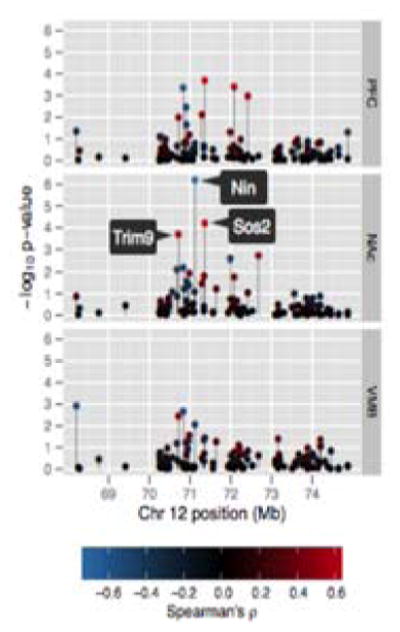
The y-axis indicates significance (Spearman ρ) of correlation between distance traveled in light (DTL) following ethanol and microarray expression values in the PFC, NAc, or VMB for candidate genes within the Etanq1 interval. Gene positions on Chr 12 are plotted on the x-axis. Correlation direction/significance is represented by point color. Gene symbols are provided for phenotype correlations significant on FDR analysis (q-value < .05).
Table 5. Gene expression and phenotype correlations.
Significant (FDR < 10%) Spearman's rank correlations between expression of Etanq1-region genes and distance traveled in the light following saline or ethanol. Within experimental group, only the most highly correlated probe-set is listed for each gene.
| Region | Treatment | Symbol | Probe-set | Position | n | ρ | p-value |
|---|---|---|---|---|---|---|---|
| NAc | Saline | Sos2 | 1443057_at | 70.72 | 35 | 0.56 | 3.4e-04 |
| Ethanol | 35 | 0.56 | 4.0e-04 | ||||
| Saline | Nin | 1419078_at | 71.12 | 35 | -0.67 | 3.6e-06 | |
| Saline | Trim9 | 1434595_at | 71.37 | 35 | 0.63 | 3.4e-05 | |
| Ethanol | 35 | 0.64 | 2.0e-05 | ||||
| PFC | Ethanol | Trim9 | 1434595_at | 71.37 | 29 | 0.68 | 1.9e-05 |
| VMB | Saline | Sos2 | 1443057_at | 70.72 | 35 | 0.63 | 3.1e-05 |
| Ethanol | 35 | 0.63 | 2.8e-05 | ||||
| Ethanol | Trim9 | 1454886_x_at | 71.35 | 35 | -0.57 | 2.5e-04 |
Biological relevance of Etanq1 candidate genes
To summarize the candidate gene list derived from bioinformatic analyses, six genes contained missense SNPs: Nin, Dact1, Atp5s, Abhd12b, Pygl, and 2700049A03Rik; four genes contained significant cis eQTL: Sos2, Nin, Atp5s, and Trim9; and only three genes had mRNA expression significantly correlated to the post-ethanol DTL phenotype: Sos2, Trim9, and Nin. Sos2 (Drosophila Son of sevenless homolog 2) encodes a RAS guanine nucleotide exchange factor affecting RAS/MAPK signal transduction (Bowtell et al., 1992). Trim9 (tripartite motif containing 9) is a brain-specific E3 ligase that self-ubiquitinates to target proteasomal degradation (Tanji et al., 2010). Nin (Ninein) anchors microtubules in the centrosome (Bouckson-Castaing et al., 1996, Mogensen et al., 2000) and influences neocortical axonal branching and growth (Srivatsa et al., 2015). Although the biological functions of these genes are still being discovered, their expression in the central nervous system (CNS), along with roles in signal transduction and neuronal plasticity make them all plausible candidate genes. Interestingly, of the three, only Nin satisfies all bioinformatic criteria and has been shown to interact with Gsk3b (Hong et al., 2000, Howng et al., 2004) a known ethanol-responsive gene (French & Heberlein, 2009, Luo, 2009, Wolen et al., 2012)
Expression of Ninein in the NAc
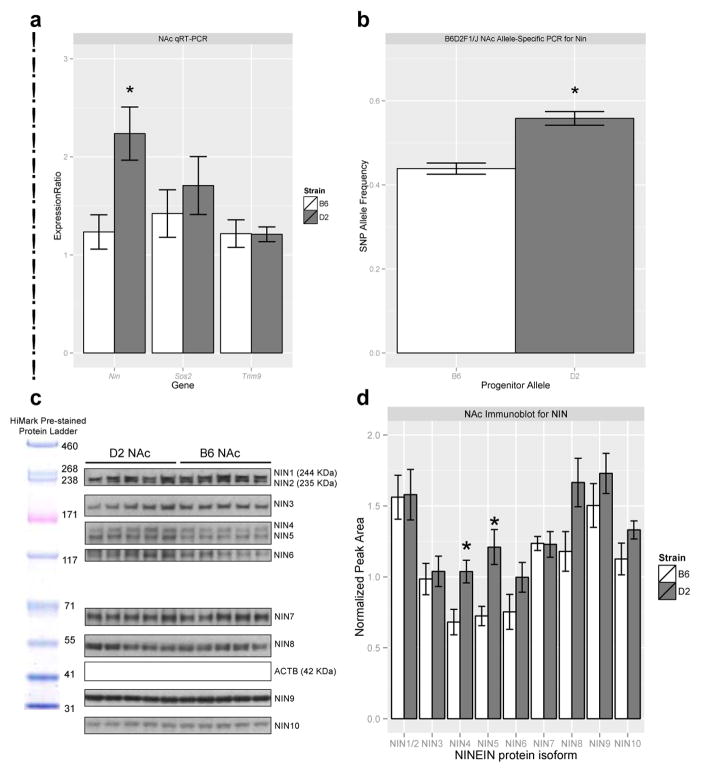
To further prioritize our list of candidate genes, differential expression of Nin, Sos2, and Trim9 was examined between B6 and D2 mice using qRT-PCR analysis. RNA from NAc samples were used since transcript abundance from the NAc had the strongest correlation with ethanol-induced anxiolysis, as described above. Only expression of Nin (Figure 7a) was significantly different between D2 and B6 progenitor strains (p < 0.01, n = 8/strain, Student’s t-test between strain). Allele-specific rtPCR pyrosequencing was performed to confirm cis regulation for Nin. Within the NAc of untreated B6D2F1/J mice, pyrosequencing of two missense SNPs within the Nin gene (rs29192398 and rs29159683 at 71.144373 and 71.144376 Mb of mouse build mm9, respectively) revealed a significantly greater frequency of the D2 alleles compared to B6 alleles (p<0.01, n= 2 NAc pooled per biological replicate, n= 3 biological replicates, n >4000 reads per group), confirming cis regulation of Nin (Figure 7b). Finally, we performed Western blot analysis of basal Ninein expression and confirmed significantly higher basal levels of two NIN protein isoforms in the NAc of D2 mice compared to B6 mice (p<0.05, Student’s t-test, n=5/group, Figure 7c–d), suggesting functional differences of NIN in the NAc between the B6 and D2 genotypes.
Figure 7. Confirmation of differential regulation of Ninein in the NAc.
a) Basal mRNA levels of Nin were significantly greater in D2 mice than B6 mice (*p<0.01, n=8 per group, Student’s t-test between strain) as confirmed by qRT-PCR. Nin expression was normalized to Ppp2r2a. b) Pyrosequencing of NAc samples of B6D2F1/J hybrid mice for an amplicon of Nin containing two progenitor SNPs confirmed cis regulation of Nin (*p<0.01, n=2 pooled per sample, n= 3 per group, n >4000 reads per group, Student’s t-test). c-d) Immunoblotting of NAc samples from untreated B6 and D2 mice revealed significantly higher levels of two isoforms (NIN4 and NIN5, ~120 KDa) in D2 mice compared to B6 (*p<0.05, Student’s t-test, n=5/strain). D2 mice also show a trend for higher levels of NIN6, NIN8, and NIN10 isoforms (p=0.1703, p=0.0589, p=0.1516, respectively, Student’s t-test). NIN levels were normalized to ACTB protein levels.
Discussion
Since ethanol’s effects on anxiety are thought to play an important role in the genesis of AUD and recidivism in alcoholics, our work sought to identify candidate genes underlying the acute anxiolytic-like response to ethanol in mice. Based on the overall design of this QTL mapping study, we not only gained insight into the genetic mechanisms underlying ethanol anxiolysis but multiple behaviors including saline- and ethanol-induced locomotor activity, and basal (saline) anxiety. The result is a fine-mapped ethanol anxiolysis QTL, Etanq1, and a narrowed list of underlying candidate genes, one of which, Ninein, is a high priority target based on comprehensive SNP, phenotype-expression correlation, eQTL, and molecular analyses.
Most rodent models of anxiety-like behavior are at least partially activity-based, complicating the identification of QTL specifically related to anxiety. This issue was demonstrated in a recent behavioral genetics correlation analysis of multiple anxiety-like behavior assays across a panel of inbred mouse strains (Milner & Crabbe, 2008). However, a detailed F2 mouse genetic analysis of multiple behavioral models of anxiety-like behavior suggested both separate and shared genetic loci contributing to each measure of anxiety and locomotor activity (Turri et al., 2001). A similar concept regarding anxiety-related responses to ethanol has also been previously proposed. Studies in selectively-bred mouse lines suggest that ethanol-induced locomotor activity and anxiolytic-like responses to ethanol are controlled by only partially overlapping sets of genes (Boehm et al., 2002). Furthermore, in a detailed bioinformatic of loci on Chr 1 contributing to the anxiolytic-like response to ethanol in the elevated zero-maze model of anxiety, showed at least one locus unique to ethanol anxiolytic-like activity (Cook et al., 2015).
In our studies described here, although anxiolytic-like responses to ethanol in the light-dark box were not dependent on ethanol-induced locomotor activation, as demonstrated in B6 progenitors (Figure 1) and various BXD RI mouse strains (Figure 2), we found significant phenotypic correlations between some anxiety-like measures and TLA in response to saline and ethanol (Table 1), and some overlap between anxiety-related and TLA QTL (see Table 2 and below). While the dissection of anxiety-related QTL in such regions may require extensive analysis (Cook et al., 2015, Willis-Owen & Flint, 2006), our results also identified anxiety-like QTL that appear distinct from locomotor influences. The most significant of these potentially unique anxiety-related loci consisted of a Chr 11 post-saline anxiety-like QTL (Salanq1) and a Chr 12 locus (Etanq1) that influenced anxiety-like behavior following ethanol (Figure 3 and Tables 2–3). Although the statistical power of mixed-model QTL analysis enabled the detection of additional loci involved in anxiety-related behavior, QTL identified by strain-mean mapping and confirmed using the mixed-model were given emphasis to minimize false positives. Of note, a recent report using the diversity outcross genetic mouse panel mapped an anxiety-like QTL in the light-dark transition model at a Chr 11 location (95.01–96.05 Mb) very close to the support interval identified here for Salanq1 (Logan et al., 2013). It should be recalled, however, that our protocol employed here used a mild restraint stress prior to either saline or ethanol injection and LD box testing. Thus, the anxiety-like QTL seen following saline actually represent the sum of basal anxiety, restraint-stress and saline injection stress effects, rather than merely “basal” anxiety. Regardless, further genetic dissection of Salanq1 may produce novel insight into the molecular neurobiology of anxiety.
Following ethanol treatment, we observed anxiolytic-like responses across nearly all BXD RI strains and their progenitors with a continuous pattern of phenotypic variation, suggesting a multi-gene quantitative trait (Figure 2). Loci influencing the anxiolytic-like response to ethanol consisted of a suggestive Chr 1 locus and a significant Chr 12 QTL, referred to as Etanq1 (Figure 3 and Table 2). Particularly apparent in the strain-mean mapping, Salanq1 was absent from our genetic mapping results in the ethanol cohort. This degree of genetic segregation of major loci influencing anxiety-like behavior following saline or ethanol suggests either that ethanol treatment abrogates the role of the influence of Salanq1, or that genetic and environmental variance following ethanol does not allow detection of Salanq1. Although a variety of ethanol-related behaviors, including acute and chronic ethanol withdrawal (Buck et al., 2000, Buck et al., 1997), and acute ethanol sensitivity phenotypes (Cook et al., 2015, Radcliffe et al., 2000), were previously mapped to similar loci on Chr 1, this complex region appeared to alter a variety of non anxiety-related phenotypes, as mentioned above. Therefore, these reports, along with the existence of large overlapping support intervals, suggest that speculation of candidate genes underlying this locus would be of limited value prior to further analysis.
In contrast, the Etanq1 Chr 12 locus was identified as a potentially exclusive ethanol anxiolysis-related QTL, distinct from TLA loci. An initial confirmation study completed using BXD RI strains derived from two independent advanced intercrosses between the B6 and D2 progenitor strains (Peirce et al., 2004) verified the existence of Etanq1 and the importance of the B6 allele in increasing ethanol-induced anxiolysis at this locus (Figure 4). Interestingly, mixed-model QTL analysis showed that although Etanq1 appeared unique to ethanol-induced anxiolysis, a suggestive effect of saline exists at this locus (Table 3). This result is supported by a previous report that identified a similar Chr 12 locus modifying basal anxiety-like behavior in the light-dark box (Turri et al., 2001) and suggests that identifying genes underlying Etanq1 may provide insight into basic mechanisms of anxiety, as well as ethanol-induced anxiolysis.
Since our provisional mapping and subsequent confirmation studies were performed in BXD RI strains, loci identified here may only represent a portion of natural genetic variation affecting the anxiolytic properties of ethanol. Similarly, our use of only the light-dark box for measurement of anxiety-like phenotypes clearly only captures some of the genetic variance linked to ethanol-induced anxiolysis, as seen with basal anxiety measures (Belzung & Le Pape, 1994, File, 1995, Flint, 2003, Ramos et al., 1997, Turri et al., 2001). Thus, ongoing studies in our laboratory are assessing the role of ethanol-induced anxiolysis QTL mapped here with other models of anxiety-like behavior. An additional factor possibly limiting the QTL identified here is the absence of female mice, thereby eliminating our ability to map sex-specific QTL as observed with other behavioral responses to ethanol (Downing et al., 2006, Radcliffe et al., 2000). Fluctuations in sex hormones during the ovarian cycle have been shown to influence anxiety-related behavior (Galeeva et al., 2003, Maguire et al., 2005). Using our behavioral paradigm in the light-dark box, we confirmed significant within-strain variation of anxiety-like behavior between male and female BXD mice (data not shown). Thus, to reduce total phenotypic variation and possibly increase our ability to successfully identify QTL, only male mice were used for this initial genetic analysis of ethanol-induced anxiolysis. Given the importance of sex-specific QTL to anxiety and behavioral responses to ethanol, sex-specific QTL will be elucidated in future studies but conducting those studies simultaneous, and on the same scale, with the current analysis would have been infeasible in terms of manpower and resources. Regardless of these limitations, this study is the first to report a confirmed QTL for ethanol-induced anxiolysis.
We performed extensive bioinformatic analyses to narrow down candidate genes for Etanq1. For molecular studies, we focused on the three genes with putative cis eQTL whose NAc expression significantly correlated to the DTL phenotype: Sos2, Trim9, and Nin (Table 4 and 5). Of these candidate genes, missense mutations within Sos2 have been associated with Noonan syndrome (Cordeddu et al., 2015, Yamamoto et al., 2015), an autosomal dominant disorder caused by functional dysregulation of RAS/MAPK signal transduction pathways; however, since Sos2 lacked missense mutations between the B6 and D2 progenitor sequences it was considered a less likely candidate gene. Trim9 is expressed in both developing and adult neurons (Winkle et al., 2014), with its highest expression found in the cortex and hippocampus. Reported associations between Trim9 expression or variants and disease are limited to severe repression of Trim9 in the brain of patients with Parkinson’s and Lewy body dementia (Tanji et al., 2010).
Of the top three candidates, Nin was the only gene to show PCR confirmed differential expression between the B6 and D2 progenitor strains in the NAc (Figure 7a). Additional analyses confirmed cis regulation of Nin using allele-specific PCR/sequencing in B6D2F1/J mice (Figure 7b) and showed differential expression of protein isoforms between progenitors in the NAc (Figure 7 c–d). Nin was also the only gene to possess missense mutation SNPs in a protein coding region predicted to alter protein function. In addition to these molecular data, biomedical literature further supports Nin as the strongest candidate gene underlying Etanq1. In the brain, Nin is known to function as a microtubule binding protein and an important regulator of neocortical axonal outgrowth and branching (Bouckson-Castaing et al., 1996, Mogensen et al., 2000, Srivatsa et al., 2015). Recent studies have reported an association between genetic variation within the Nin gene or altered expression patterns of Nin with a number of diseases and genetic disorders, including breast cancer (Olson et al., 2011), a type of skeletal dysplasia (Grosch et al., 2013), microcephalic primordial dwarfism (Dauber et al., 2012), polycystic ovary syndrome and non-alcoholic fatty liver disease (Baranova et al., 2013). This suggests that certain genetic variants of Nin might be a risk factor for multiple disease phenotypes.
As previously mentioned, Nin has been shown to interact with Gsk3β (Hong et al., 2000, Howng et al., 2004), a known ethanol-responsive gene (French & Heberlein, 2009, Luo, 2009, Wolen & Miles, 2012). Gsk3b is also a hub (highly inter-correlated) gene in a major ethanol-responsive gene network identified from the same microarray studies used in this report (Wolen et al., 2012). The two mouse Nin major protein isoforms share an overall ~78% sequence similarity to the human NIN protein, but strikingly, 94% similarity to the human phosphorylation binding site for Gsk3β suggesting conserved regulation of Nin by Gsk3β in mice (Howng et al., 2004). Interestingly, Gsk3b has also been shown to regulate anxiety-related behaviors in animals (Latapy et al., 2012, O'brien et al., 2004) and Gsk3b is associated with mood and psychiatric disorders including schizophrenia and bipolar disorder (Jope & Roh, 2006) and a known target of neuromodulators and psychotropic drugs (Li & Jope, 2010). In total, our findings suggest Nin may contribute strongly to genetic variance in ethanol-anxiolytic like activity in the BXD panel through genotype-specific differential transcriptional regulation, protein function, or both.
In conclusion, we used QTL mapping across the BXD RI panel to identify loci that influence anxiety-like responses to ethanol in the light-dark transition model of anxiety. The Etanq1 QTL represents the first confirmed QTL influencing ethanol anxiolysis. We suggest Nin as a top priority candidate gene underlying Etanq1 and future studies confirming Nin’s role could provide valuable insight into the mechanisms of ethanol anxiolysis. Given the contribution of anxiety in drinking initiation and recidivism in alcoholics, Nin or other candidate genes elucidated here may ultimately aid the identification of new pharmacotherapies for alcohol use disorders.
Supplementary Material
Supplementary Figure 1: Factor analysis of anxiety-related phenotypes
Supplementary Figure 2: Total locomotor activity interval maps
Supplementary Figure 3: Genotype analysis of Etanq1 locus
Supplementary Figure 4: Chr1/Chr12 QTL interaction
Supplementary Figure 5: Comparison of DTL and TSL measures for ethanol anxiolytic-like QTL and BXD strain rank order
Supplementary Table 1: Progenitor strains light-dark box behavioral phenotype ANOVA results
Supplementary Table 2: Light-dark box behavioral phenotype strain means
Supplementary Table 3: Provisional BXD strains light-dark box behavioral phenotypes ANOVA results
Supplementary Table 4: Etanq1 SNP analysis
DTL and TLA Mixed model QTL analysis
Acknowledgments
This work was supported by grants R01AA014717, P20AA017828 and U01AA016667 to MFM and F31AA016052 (AHP) from the National Institute on Alcohol Abuse and Alcoholism. JLH was supported by T32DA007027 (William L. Dewey, PI) from the National Institute on Drug Abuse and ARW was supported by T32MH20030 (Michael Neale, PI) from the National Institute on Mental Health and UL1TR000058 from the NIH’s National Center for Advancing Translational Research. The authors thank Drs. John Crabbe (Oregon Health Sciences University) and Robert Williams (Univ. Tennessee Health Sciences University) for many helpful discussions during initiation of these studies and Dr. Tim York (Virginia Commonwealth University) for assistance with the mixed-model analysis. Additionally, the authors thank members of the Miles laboratory for helpful discussions and Dr. Jennifer Wolstenholme for reviewing the manuscript.
References
- Alberts R, Terpstra P, Li Y, Breitling R, Nap JP, Jansen RC. Sequence polymorphisms cause many false cis eQTLs. PloS one. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova A, Tran TP, Afendy A, Wang L, Shamsaddini A, Mehta R, Chandhoke V, Birerdinc A, Younossi ZM. Molecular signature of adipose tissue in patients with both Non-Alcoholic Fatty Liver Disease (NAFLD) and Polycystic Ovarian Syndrome (PCOS) Journal of Translational Medicine. 2013;11:1–1. doi: 10.1186/1479-5876-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Statistical Software. 2015;67:1–48. [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Mitchell SR, O'Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav Genet. 1996;26:149–160. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- Belzung C, Le Pape G. Comparison of different behavioral test situations used in psychopharmacology for measurement of anxiety. Physiol Behav. 1994;56:623–628. doi: 10.1016/0031-9384(94)90311-5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Bibb JL, Chambless DL. Alcohol use and abuse among diagnosed agoraphobics. Behav Res Ther. 1986;24:49–58. doi: 10.1016/0005-7967(86)90149-x. [DOI] [PubMed] [Google Scholar]
- Bilkei-Gorzo A, Gyertyan I, Levay G. mCPP-induced anxiety in the light-dark box in rats--a new method for screening anxiolytic activity. Psychopharmacology (Berl) 1998;136:291–298. doi: 10.1007/s002130050568. [DOI] [PubMed] [Google Scholar]
- Boehm SL, 2nd, Reed CL, McKinnon CS, Phillips TJ. Shared genes influence sensitivity to the effects of ethanol on locomotor and anxiety-like behaviors, and the stress axis. Psychopharmacology (Berl) 2002;161:54–63. doi: 10.1007/s00213-002-1000-y. [DOI] [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J Cell Sci. 1996;109(Pt 1):179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- Bowtell D, Fu P, Simon M, Senior P. Identification of murine homologues of the Drosophila son of sevenless gene: potential activators of ras. Proc Natl Acad Sci U S A. 1992;89:6511–6515. doi: 10.1073/pnas.89.14.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigman JL, Mathur P, Lu L, Williams RW, Holmes A. Genetic relationship between anxiety-related and fear-related behaviors in BXD recombinant inbred mice. Behavioural Pharmacology. 2009;20:204–209. doi: 10.1097/FBP.0b013e32830c368c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Browman KE, Crabbe JC. Quantitative trait loci affecting ethanol sensitivity in BXD recombinant inbred mice. Alcohol Clin Exp Res. 2000;24:17–23. [PubMed] [Google Scholar]
- Buck K, Lischka T, Dorow J, Crabbe J. Mapping quantitative trait loci that regulate sensitivity and tolerance to quinpirole, a dopamine mimetic selective for D(2)/D(3) receptors. Am J Med Genet. 2000;96:696–705. doi: 10.1002/1096-8628(20001009)96:5<696::aid-ajmg17>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Burkholder T, Wilkins L, Collins AC. A genetic comparison of behavioral actions of ethanol and nicotine in the mirrored chamber. Pharmacol Biochem Behav. 1993;45:803–809. doi: 10.1016/0091-3057(93)90124-c. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Wang J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7:485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conger JJ. Alcoholism: theory, problem and challenge. II. Reinforcement theory and the dynamics of alcoholism. Q J Stud Alcohol. 1956;17:296–305. [PubMed] [Google Scholar]
- Cook MN, Baker JA, Heldt SA, Williams RW, Hamre KM, Lu L. Identification of candidate genes that underlie the QTL on chromosome 1 that mediates genetic differences in stress-ethanol interactions. Physiological genomics. 2015;47:308–317. doi: 10.1152/physiolgenomics.00114.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeddu V, Yin JC, Gunnarsson C, Virtanen C, Drunat S, Lepri F, De Luca A, Rossi C, Ciolfi A, Pugh TJ, Bruselles A, Priest JR, Pennacchio LA, Lu Z, Danesh A, Quevedo R, Hamid A, Martinelli S, Pantaleoni F, Gnazzo M, Daniele P, Lissewski C, Bocchinfuso G, Stella L, Odent S, Philip N, Faivre L, Vlckova M, Seemanova E, Digilio C, Zenker M, Zampino G, Verloes A, Dallapiccola B, Roberts AE, Cave H, Gelb BD, Neel BG, Tartaglia M. Activating Mutations Affecting the Dbl Homology Domain of SOS2 Cause Noonan Syndrome. Hum Mutat. 2015;36:1080–1087. doi: 10.1002/humu.22834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius JR, Bukstein O, Salloum I, Clark D. Alcohol and psychiatric comorbidity. Recent Dev Alcohol. 2003;16:361–374. doi: 10.1007/0-306-47939-7_24. [DOI] [PubMed] [Google Scholar]
- Costall B, Kelly ME, Naylor RJ. The anxiolytic and anxiogenic actions of ethanol in a mouse model. J Pharm Pharmacol. 1988;40:197–202. doi: 10.1111/j.2042-7158.1988.tb05218.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC. Provisional mapping of quantitative trait loci for chronic ethanol withdrawal severity in BXD recombinant inbred mice. The Journal of pharmacology and experimental therapeutics. 1998;286:263–271. [PubMed] [Google Scholar]
- Crabbe JC, Belknap JK, Buck KJ. Genetic animal models of alcohol and drug abuse. Science. 1994;264:1715–1723. doi: 10.1126/science.8209252. [DOI] [PubMed] [Google Scholar]
- Crawley J, Goodwin FK. Preliminary report of a simple animal behavior model for the anxiolytic effects of benzodiazepines. Pharmacol Biochem Behav. 1980;13:167–170. doi: 10.1016/0091-3057(80)90067-2. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Exploratory behavior models of anxiety in mice. Neurosci Biobehav Rev. 1985;9:37–44. doi: 10.1016/0149-7634(85)90030-2. [DOI] [PubMed] [Google Scholar]
- Dauber A, Lafranchi SH, Maliga Z, Lui JC, Moon JE, McDeed C, Henke K, Zonana J, Kingman GA, Pers TH, Baron J, Rosenfeld RG, Hirschhorn JN, Harris MP, Hwa V. Novel microcephalic primordial dwarfism disorder associated with variants in the centrosomal protein ninein. J Clin Endocrinol Metab. 2012;97:E2140–2151. doi: 10.1210/jc.2012-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarest K, Koyner J, McCaughran J, Jr, Cipp L, Hitzemann R. Further characterization and high-resolution mapping of quantitative trait loci for ethanol-induced locomotor activity. Behav Genet. 2001;31:79–91. doi: 10.1023/a:1010261909853. [DOI] [PubMed] [Google Scholar]
- Demarest K, McCaughran J, Jr, Mahjubi E, Cipp L, Hitzemann R. Identification of an acute ethanol response quantitative trait locus on mouse chromosome 2. J Neurosci. 1999;19:549–561. doi: 10.1523/JNEUROSCI.19-02-00549.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeTurck KH, Vogel WH. Effects of acute ethanol on plasma and brain catecholamine levels in stressed and unstressed rats: evidence for an ethanol-stress interaction. The Journal of pharmacology and experimental therapeutics. 1982;223:348–354. [PubMed] [Google Scholar]
- Downing C, Carosone-Link P, Bennett B, Johnson T. QTL mapping for low-dose ethanol activation in the LXS recombinant inbred strains. Alcohol Clin Exp Res. 2006;30:1111–1120. doi: 10.1111/j.1530-0277.2006.00137.x. [DOI] [PubMed] [Google Scholar]
- DuBose CS, Chesler EJ, Goldowitz D, Hamre KM. Use of the Expanded Panel of BXD Mice Narrow QTL Regions in Ethanol-Induced Locomotor Activation and Motor Incoordination. Alcoholism-Clinical and Experimental Research. 2013;37:170–183. doi: 10.1111/j.1530-0277.2012.01865.x. [DOI] [PubMed] [Google Scholar]
- File SE. Animal models of different anxiety states. Adv Biochem Psychopharmacol. 1995;48:93–113. [PubMed] [Google Scholar]
- Flint J. Analysis of quantitative trait loci that influence animal behavior. J Neurobiol. 2003;54:46–77. doi: 10.1002/neu.10161. [DOI] [PubMed] [Google Scholar]
- French RL, Heberlein U. Glycogen synthase kinase-3/Shaggy mediates ethanol-induced excitotoxic cell death of Drosophila olfactory neurons. Proc Natl Acad Sci U S A. 2009;106:20924–20929. doi: 10.1073/pnas.0910813106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeeva AY, Tuohimaa P, Shalyapina VG. The role of sex steroids in forming anxiety states in female mice. Neuroscience and behavioral physiology. 2003;33:415–420. doi: 10.1023/a:1022864011385. [DOI] [PubMed] [Google Scholar]
- Grosch M, Gruner B, Spranger S, Stutz AM, Rausch T, Korbel JO, Seelow D, Nurnberg P, Sticht H, Lausch E, Zabel B, Winterpacht A, Tagariello A. Identification of a Ninein (NIN) mutation in a family with spondyloepimetaphyseal dysplasia with joint laxity (leptodactylic type)-like phenotype. Matrix Biol. 2013;32:387–392. doi: 10.1016/j.matbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Henderson ND, Turri MG, DeFries JC, Flint J. QTL analysis of multiple behavioral measures of anxiety in mice. Behav Genet. 2004;34:267–293. doi: 10.1023/B:BEGE.0000017872.25069.44. [DOI] [PubMed] [Google Scholar]
- Hong YR, Chen CH, Chang JH, Wang S, Sy WD, Chou CK, Howng SL. Cloning and characterization of a novel human ninein protein that interacts with the glycogen synthase kinase 3beta. Biochimica et biophysica acta. 2000;1492:513–516. doi: 10.1016/s0167-4781(00)00127-5. [DOI] [PubMed] [Google Scholar]
- Howng SL, Hsu HC, Cheng TS, Lee YL, Chang LK, Lu PJ, Hong YR. A novel ninein-interaction protein, CGI-99, blocks ninein phosphorylation by GSK3β and is highly expressed in brain tumors. FEBS Letters. 2004;566:162–168. doi: 10.1016/j.febslet.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Jope RS, Roh MS. Glycogen synthase kinase-3 (GSK3) in psychiatric diseases and therapeutic interventions. Current drug targets. 2006;7:1421–1434. doi: 10.2174/1389450110607011421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliethermes CL, Finn DA, Crabbe JC. Validation of a modified mirrored chamber sensitive to anxiolytics and anxiogenics in mice. Psychopharmacology (Berl) 2003;169:190–197. doi: 10.1007/s00213-003-1493-z. [DOI] [PubMed] [Google Scholar]
- LaBuda CJ, Fuchs PN. Aspirin attenuates the anxiolytic actions of ethanol. Alcohol. 2000;21:287–290. doi: 10.1016/s0741-8329(00)00097-5. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Latapy C, Rioux V, Guitton MJ, Beaulieu JM. Selective deletion of forebrain glycogen synthase kinase 3beta reveals a central role in serotonin-sensitive anxiety and social behaviour. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2012;367:2460–2474. doi: 10.1098/rstb.2012.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessov CN, Palmer AA, Quick EA, Phillips TJ. Voluntary ethanol drinking in C57BL/6J and DBA/2J mice before and after sensitization to the locomotor stimulant effects of ethanol. Psychopharmacology (Berl) 2001;155:91–99. doi: 10.1007/s002130100699. [DOI] [PubMed] [Google Scholar]
- Li X, Jope RS. Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2010;35:2143–2154. doi: 10.1038/npp.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, Robledo RF, Recla JM, Philip VM, Bubier JA, Jay JJ, Harwood C, Wilcox T, Gatti DM, Bult CJ, Churchill GA, Chesler EJ. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes, brain, and behavior. 2013;12:424–437. doi: 10.1111/gbb.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. GSK3beta in ethanol neurotoxicity. Mol Neurobiol. 2009;40:108–121. doi: 10.1007/s12035-009-8075-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire JL, Stell BM, Rafizadeh M, Mody I. Ovarian cycle-linked changes in GABA(A) receptors mediating tonic inhibition alter seizure susceptibility and anxiety. Nat Neurosci. 2005;8:797–804. doi: 10.1038/nn1469. [DOI] [PubMed] [Google Scholar]
- Manichaikul A, Dupuis J, Sen S, Broman KW. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics. 2006;174:481–489. doi: 10.1534/genetics.106.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner LC, Crabbe JC. Three murine anxiety models: results from multiple inbred strain comparisons. Genes, brain, and behavior. 2008;7:496–505. doi: 10.1111/j.1601-183X.2007.00385.x. [DOI] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Nagoshi CT, Wilson JR, Plomin R. Use of regression residuals to quantify individual differences in acute sensitivity and tolerance to alcohol. Alcohol Clin Exp Res. 1986;10:343–349. doi: 10.1111/j.1530-0277.1986.tb05101.x. [DOI] [PubMed] [Google Scholar]
- Newlin DB, Thomson JB. Alcohol challenge with sons of alcoholics: a critical review and analysis. Psychol Bull. 1990;108:383–402. doi: 10.1037/0033-2909.108.3.383. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien WT, Harper AD, Jove F, Woodgett JR, Maretto S, Piccolo S, Klein PS. Glycogen synthase kinase-3beta haploinsufficiency mimics the behavioral and molecular effects of lithium. J Neurosci. 2004;24:6791–6798. doi: 10.1523/JNEUROSCI.4753-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JE, Wang X, Pankratz VS, Fredericksen ZS, Vachon CM, Vierkant RA, Cerhan JR, Couch FJ. Centrosome-related genes, genetic variation, and risk of breast cancer. Breast Cancer Res Treat. 2011;125:221–228. doi: 10.1007/s10549-010-0950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VA, Pohorecky LA. Interaction of stress and ethanol: effect on beta-endorphin and catecholamines. Alcohol Clin Exp Res. 1988;12:785–788. doi: 10.1111/j.1530-0277.1988.tb01346.x. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Belknap JK, Buck KJ, Cunningham CL. Genes on mouse chromosomes 2 and 9 determine variation in ethanol consumption. Mamm Genome. 1998;9:936–941. doi: 10.1007/s003359900903. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Huson M, Gwiazdon C, Burkhart-Kasch S, Shen EH. Effects of acute and repeated ethanol exposures on the locomotor activity of BXD recombinant inbred mice. Alcohol Clin Exp Res. 1995;19:269–278. doi: 10.1111/j.1530-0277.1995.tb01502.x. [DOI] [PubMed] [Google Scholar]
- Pohorecky LA. The interaction of alcohol and stress. A review. Neurosci Biobehav Rev. 1981;5:209–229. doi: 10.1016/0149-7634(81)90003-8. [DOI] [PubMed] [Google Scholar]
- Popper CW, Chiueh CC, Kopin IJ. Plasma catecholamine concentrations in unanesthetized rats during sleep, wakefulness, immobilization and after decapitation. The Journal of pharmacology and experimental therapeutics. 1977;202:144–148. [PubMed] [Google Scholar]
- Radcliffe RA, Bohl ML, Lowe MV, Cycowski CS, Wehner JM. Mapping of quantitative trait loci for hypnotic sensitivity to ethanol in crosses derived from the C57BL/6 and DBA/2 mouse strains. Alcohol Clin Exp Res. 2000;24:1335–1342. [PubMed] [Google Scholar]
- Ramos A, Berton O, Mormede P, Chaouloff F. A multiple-test study of anxiety-related behaviours in six inbred rat strains. Behav Brain Res. 1997;85:57–69. doi: 10.1016/s0166-4328(96)00164-7. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. Assessing the risk for alcoholism among sons of alcoholics. Journal of studies on alcohol. 1997;58:141–145. doi: 10.15288/jsa.1997.58.141. [DOI] [PubMed] [Google Scholar]
- Srivatsa S, Parthasarathy S, Molnár Z, Tarabykin V. Neuron. Elsevier Inc; 2015. Sip1 Downstream Effector ninein Controls Neocortical Axonal Growth, Ipsilateral Branching, and Microtubule Growth and Stability; pp. 998–1012. [DOI] [PubMed] [Google Scholar]
- Tanji K, Kamitani T, Mori F, Kakita A, Takahashi H, Wakabayashi K. TRIM9, a novel brain-specific E3 ubiquitin ligase, is repressed in the brain of Parkinson's disease and dementia with Lewy bodies. Neurobiology of Disease. 2010;38:210–218. doi: 10.1016/j.nbd.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjurmina OA, Armando I, Saavedra JM, Goldstein DS, Murphy DL. Exaggerated adrenomedullary response to immobilization in mice with targeted disruption of the serotonin transporter gene. Endocrinology. 2002;143:4520–4526. doi: 10.1210/en.2002-220416. [DOI] [PubMed] [Google Scholar]
- Tritto T, Dudek BC. Differential activating effects of ethanol in C57BL/6Abg and DBA/2Abg mice. Alcohol. 1994;11:133–139. doi: 10.1016/0741-8329(94)90054-x. [DOI] [PubMed] [Google Scholar]
- Turri MG, Datta SR, DeFries J, Henderson ND, Flint J. QTL analysis identifies multiple behavioral dimensions in ethological tests of anxiety in laboratory mice. Curr Biol. 2001;11:725–734. doi: 10.1016/s0960-9822(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Varlinskaya EI, Spear LP. Acute effects of ethanol on social behavior of adolescent and adult rats: role of familiarity of the test situation. Alcohol Clin Exp Res. 2002;26:1502–1511. doi: 10.1097/01.ALC.0000034033.95701.E3. [DOI] [PubMed] [Google Scholar]
- Willis-Owen SA, Flint J. The genetic basis of emotional behaviour in mice. Eur J Hum Genet. 2006;14:721–728. doi: 10.1038/sj.ejhg.5201569. [DOI] [PubMed] [Google Scholar]
- Winkle CC, McClain LM, Valtschanoff JG, Park CS, Maglione C, Gupton SL. A novel Netrin-1-sensitive mechanism promotes local SNARE-mediated exocytosis during axon branching. J Cell Biol. 2014;205:217–232. doi: 10.1083/jcb.201311003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolen AR, Miles MF. Identifying gene networks underlying the neurobiology of ethanol and alcoholism. Alcohol Res. 2012;34:306–317. [PMC free article] [PubMed] [Google Scholar]
- Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, York TP, Williams RW, Miles MF. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PloS one. 2012;7:e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto GL, Aguena M, Gos M, Hung C, Pilch J, Fahiminiya S, Abramowicz A, Cristian I, Buscarilli M, Naslavsky MS, Malaquias AC, Zatz M, Bodamer O, Majewski J, Jorge AA, Pereira AC, Kim CA, Passos-Bueno MR, Bertola DR. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J Med Genet. 2015;52:413–421. doi: 10.1136/jmedgenet-2015-103018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Factor analysis of anxiety-related phenotypes
Supplementary Figure 2: Total locomotor activity interval maps
Supplementary Figure 3: Genotype analysis of Etanq1 locus
Supplementary Figure 4: Chr1/Chr12 QTL interaction
Supplementary Figure 5: Comparison of DTL and TSL measures for ethanol anxiolytic-like QTL and BXD strain rank order
Supplementary Table 1: Progenitor strains light-dark box behavioral phenotype ANOVA results
Supplementary Table 2: Light-dark box behavioral phenotype strain means
Supplementary Table 3: Provisional BXD strains light-dark box behavioral phenotypes ANOVA results
Supplementary Table 4: Etanq1 SNP analysis
DTL and TLA Mixed model QTL analysis